Abstract
Flagella contribute to the virulence of bacteria through chemotaxis, adhesion to and invasion of host surfaces. Flagellar phase variation is believed to facilitate bacterial evasion of the host immune response. In this study, the flnA gene that encodes Escherichia coli H17 flagellin was examined by whole genome sequencing and genetic deletion analysis. Unilateral flagellar phase variation has been reported in E. coli H3, H47 and H17 strains, although the mechanism for phase variation in the H17 strain has not been previously understood. Analysis of phase variants indicated that the flagellar phase variation in the H17 strain was caused by the deletion of an ∼35 kb DNA region containing the flnA gene from diverse excision sites. The presence of covalently closed extrachromosomal circular forms of this excised 35 kb region was confirmed by the two-step polymerase chain reaction. The deletion and complementation test revealed that the Int1157 integrase, a tyrosine recombinase, mediates the excision of this region. Unlike most tyrosine recombinases, Int1157 is suggested to recognize diverse sites and mediate recombination between non-homologous DNA sequences. This is the first report of non-homologous recombination mediating flagellar phase variation.
INTRODUCTION
Bacterial flagella facilitate mobility, allowing the cell to move toward attractants and away from repellants. Flagella are critical to different stages of the bacterial life cycle, including pathogenesis and biofilm formation (1,2). Flagellar antigen, also known as H antigen, is one of the major antigens in Gram-negative bacteria. Flagellin is the protein subunit of the flagellar filament and determines the specificity of the flagellar antigen.
Phase variation of antigenic surface structures, such as flagella, fimbria, capsular polysaccharide and lipopolysaccharide, helps bacteria to evade the host immune system and to adapt to particular environments (3). Most studies investigating the molecular mechanisms of flagellar phase variation have been conducted in Salmonella enteria (4–13). Flagellar phase variation in the majority of S. enterica serovars is bilateral, alternating between the expression of two different flagellins, FljB and FliC. The fljBA operon encodes a negative regulator for fliC expression, fljA. The promoter for the fljBA operon is flanked by recombination sites that allow the promoter to be inverted. In one orientation, FljA and FljB are produced, and the expression of fliC is inhibited. In the alternate orientation, fljBA is not transcribed and fliC is expressed.
Salmonella enterica serovar Typhi does not normally undergo flagellar phase variation, as these isolates harbor only the fliC gene and lack fljB. Salmonella enterica serovar Typhi isolates from Indonesia, however, contain fljA and fljB, which encodes the novel flagellin Z66 (14). Upon deletion of the fljAB operon in these strains, the flagellar antigen is irreversibly switched from FljB to FliC (15). This is the only irreversible flagellar phase variation reported for S. enterica.
Although Escherichia coli is generally considered monophasic (16), unilateral flagellar phase variation has been reported in H3, H47 and H17 strains (17–20). Recently, we reported the molecular mechanism for unilateral flagellar phase variation in H3 and H47 strains (20). In H3 and H47 strains, the flagellin-specifying gene flkA and a repressor gene flkB are located in the genomic islet flk GI. When flk GI is present on the chromosome, the flkAB operon is expressed, which results in the production of FlkA and repression of fliC. When the flk GI is excised from the chromosome, flkAB is irreversibly deleted and fliC is derepressed.
The H17 strain can spontaneously and irreversibly switch flagellar antigens from H17 to H4 (21). Different from the other 52 of 53 E. coli flagellin genes that have been identified (20,22–24), the putative flnA gene encoding the H17 flagellin has only been partially sequenced using primers based on the conserved N- and C-terminal regions of flagellin genes (25). The role of FlnA in H17 flagella synthesis has not been confirmed, and the molecular mechanism for unilateral flagellar phase variation in the H17 strain has not been previously understood.
In this study, the flnA gene encoding the E. coli H17 flagellin was identified by whole genome sequencing and genetic deletion analysis. Experiments conducted with fliC-expressing variants indicated that unilateral flagellar phase variation in the H17 strain was caused by the deletion of an ∼35 kb DNA region containing the flnA gene from diverse excision sites. This region has been named the flnA region. A two-step polymerase chain reaction (PCR) assay confirmed the presence of covalently closed extrachromosomal circular forms of the excised flnA region. We determined that int1157, which is located upstream of the flnA region, encodes an integrase required for the excision of the flnA region. Int1157 belongs to the major tyrosine recombinase family, most members of which mediate recombination between direct repeat (DR) sequences. However, Int1157 was found to recognize a variety of sites and mediate recombination between non-homologous DNA. Here, the origin of flnA is discussed and a general model for flagellar phase variation in the H17 strain is proposed.
MATERIALS AND METHODS
Bacterial strains, media and antisera
Strains used in this study are listed in Table 1. Bacteria were grown in Luria-Bertani (LB) broth or agar supplemented with ampicillin (100 μg/ml) or chloramphenicol (20 μg/ml) when necessary. A semisolid medium (motility agar plates) containing 0.2% agar in LB broth was used to enhance the bacterial motility and to screen for motile strains. Antisera obtained from Tianjin Biochip Corporation, Tianjin, China, were used to probe for H antigen. Antisera were produced by immunizing rabbits with the H17 or H4 type strain, using the conventional method for preparing H antisera (26). There was no cross-reaction between H17 and H4 antisera after absorption using bacterial suspensions.
Table 1.
Escherichia coli strains and plasmids used in this study
| Strain noa | Strain | H antigen expressed | Flagellin genotype | Reference or source |
|---|---|---|---|---|
| P12b | H17 type strain | H17 | flnAonfliCH4off | (25) |
| G1780 | H4 type strain | H4 | fliCH4 | (22) |
| H2001 | flnA-deficient mutant of P12b | H4 | flnA− fliCH4 on | This study |
| H2002 | int1157-deficient mutant of P12b | H17 | flnAonfliCH4off | This study |
| H2003 | H2001 carrying pLW1547 | H17 | flnAonfliCH4off | This study |
| H2004 | H2002 carrying pLW1548 | H17 | flnAonfliCH4off | This study |
| H2097 | flnA-deficient mutant of H2002 | H4 | flnA− fliCH4 on | This study |
| H2017-H2049 | Phase variants of P12b | H4 | flnA− fliCH4 on | This study |
| Plasmid | Genotype or phenotype | Reference or source | ||
|---|---|---|---|---|
| pUC18 | Cloning vector; Apr | Takara | ||
| pKD3 | Chloramphenicol-resistant gene (cat) containing plasmid; Cmr Apr | (31) | ||
| pKD4 | Kanamycin-resistant gene (kan) containing plasmid; kmr Apr | (31) | ||
| pKD20 | Red recombinase expression plasmid; Apr | (31) | ||
| pLW1547 | pUC18 containing flnA | This study | ||
| pLW1548 | pUC18 containing int1157 | This study | ||
aEscherichia coli H antigen type strains were obtained from the Institute of Medical and Veterinary Science, Adelaide, Australia, and from the Mechnikov Research Institute for Vaccines and Sera, Russian Academy of Medical Sciences, Moscow, Russia.
Genome sequencing
Genomic DNA isolated from E. coli H17 type strain P12b was sequenced using Roche 454 sequencing in combination with whole-genome random shotgun sequencing. The 454 sequencing was performed using a 454/Roche FLX machine according to the manufacturer's protocols and produced 280 242 reads with an average length of 238 bp, representing a theoretical 13.5-fold coverage of the genome. In shotgun sequencing, double-ended plasmid sequencing reactions were carried out at the Tianjin Biochip Corporation using an ABI BigDye Terminator V3.1 Cycle Sequencing Kit and an ABI 3730 Automated DNA Analyzer (Applied Biosystems). In total, 17 665 reads were generated, providing a 2.86-fold coverage. Sequence gaps were closed by primer walking on linked clones or by sequencing PCR products amplified from genomic DNA. All repeated DNA regions and low-quality regions were amplified and sequence verified.
Annotation and analysis
Open reading frames were predicted using Glimmer 3.0 (27). Artemis (28) was used to collate data and facilitate annotation. BLASTp was used to make predictions of protein function.
PCR and reverse transcription PCR
The primers used in this study are listed in Table 2. PCRs were carried out in 50 μl reactions containing 2 mM MgCl2, 0.2 mM dNTPs, 1 μM primers and 2 U of Taq DNA polymerase. PCR was performed under the following condition: 95°C for 5 min; 30 cycles of 95°C for 30 s, 55°C for 30 s and 72°C for 1 min; a final extension at 72°C for 5 min. RNA was purified from mid-log phase cells using the RNeasy RNA extraction kit (Qiagen) according to the manufacturer's guidelines. The quality and quantity of purified RNA was examined on an ND-1000 spectrophotometer (Nanodrop). RNA was treated with DNase and subsequently tested by PCR to ensure efficient hydrolysis of the DNA, then reverse transcribed using superscript II reverse transcriptase (Invitrogen). cDNA was amplified with primers specific for fliCH4 (wl-13727/wl-13728) and flnA (wl-13432/wl-13433), respectively. PCR was performed as described above. Amplified products were examined on a 2% agarose gel.
Table 2.
Primers used in this study
| Primer | Sequence (5′–3′) | Amplified gene or region |
|---|---|---|
| wl-13727 | ACGAACTCTGATTCCGACCT | Part of fliCH4 |
| wl-13728 | TTACCCCCAGCATCCTTGAC | |
| wl-13432 | GCTAACCGTTTCACCTCTAAC | Part of flnA |
| wl-13433 | AACCAGTAACACCAGCATTG | |
| wl-13518 | CGGGATCCTAAAAAACCCCGCCGGAGCG | flnA |
| wl-13519 | CCCAAGCTTTTGAGTCATTACCCAATCGC | |
| wl-13439 | CATGGAATTCGATGCAAAAATCCTTCCGATTC | int1157 |
| wl-13440 | CCCAAGCTTTCACTGTTCTTTACTGGTAGCTAC | |
| wl-14410a | TTTACCCGGTCAGACGTTATCCCGA | Part of flnA region |
| wl-16182a | GGTGTAACCGCAACAGCGACAAGAAT | |
| wl-22864b | GCGTGAGATGTGGCTGTCGG | Part of flnA region |
| wl-22865b | GAAACGACGCAATCCCAACT | |
| wl-14407 | GAAAACCAATATCAGAACCCCGGTGGGCCTATG | int1157-flhD region |
| wl-16181 | GAATCTTGCGTCAACTGAGTAATCGTCTGG | |
| wl-13501 | AATCACGTAAACCGTAGATTTGAACACAGGAAACTAAATCGTGTAGGCTGGAGCTGCTTC | cat gene in pKD3 (used in flnA deletion) |
| wl-13502 | AAAAACCCCGCCGGAGCGGGGTTTGAGCATCGTCAGGCGACATATGAATATCCTCCTTA | |
| wl-13436 | ATGATATCAATCAGATATATGAGGATAAATATGCAAAAATGTGTAGGCTGGAGCTGCTTC | cat gene in pKD3 (used in int1157 deletion) |
| wl-13437 | TCACTGTTCTTTACTGGTAGCTACGGGGGGTAAAAGAAGGCATATGAATATCCTCCTTA | |
| wl-47283 | TGTGCAGAAAAAGAGTACTATAAGCAAAGCTGCCGTAGATGATTGAACAAGATGGATTG | kan gene in pKD4 (used in flnA deletion) |
| wl-47284 | GAATTTATCGACAGAAGCGATAGCTGCATCCAGAGCCGTCAGAAGAACTCGTCAAGAAG |
Flagellin isolation and MALDI-TOF MS analysis
Flagellins were isolated using the method as described by Ibrahim et al. (29), and were analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE). The expected bands of flagellin were excised from gels, digested with trypsin and generated peptides were analyzed using a 4700 Proteomics Analyzer (Applied Biosystems). Proteins were identified by automated peptide mass fingerprinting using the Global Proteome Server Explorer software (Applied Biosystems) and an in-house built sequence database of P12b proteins.
Selection of spontaneous variants with altered H antigen
Bacteria were grown on semisolid medium containing antiserum raised against the expressed H antigen. This provided strict conditions for selecting variants expressing an alternative flagellar antigen phase (17,30). While the parental strain was immobilized, the variants remained motile.
Deletion of flnA and int1157
The flnA and int1157 genes of strain P12b were replaced by a chloramphenicol resistance gene (cat) using the RED recombination system of the phage lambda (31,32) to generate strains H2001 and H2002, respectively. The cat gene was amplified by PCR from the plasmid pKD3 by using the primer pairs wl-13501/wl-13502 and wl-13436/wl-13437, respectively, binding to the 5′- and 3′-ends of the genes. Each primer contained 40 bp based on the sequence of strain P12b, which flanks flnA and int1157, respectively. The PCR products were transformed into P12b carrying pKD20, and chloramphenicol-resistant transformants were selected after induction of the RED genes by the protocol described by Datsenko and Wanner (31). Deletions were verified by PCR. The flnA and int1157 genes were amplified using the primer pairs wl-13518/wl-13519 and wl-13439/wl-13440, respectively, and were cloned into pUC18 to obtain the complementing plasmids pLW1547 and pLW1548. These two plasmids were transformed into H2001 and H2002 to generate strains H2003 and H2004, respectively.
The flnA gene of strain H2002 was replaced by a kanamycin resistance gene (kan) using the method described above to generate strain H2097. The kan gene was amplified by PCR from the plasmid pKD4 by using the primer pairs wl-47283/wl-47284.
Nucleotide sequence accession numbers
The genome sequence reported in this study has been deposited in DDBJ/EMBL/GenBank under the accession number CP002291.
RESULTS
Identification of the flnA gene encoding the H17 flagellin
The E. coli H17 strain can irreversibly switch its flagellar antigen from H17 to H4 (21). To identify the flnA gene that encodes the H17 antigen, and reveal the molecular mechanism of unilaterally flagellar phase variation in the H17 strain, we sequenced the genome of the H17 type strain P12b. The genome consists of a circular chromosome (4 935 294 bp) with 4394 predicted open reading frames.
As expected, two putative flagellin genes were found in the genome. One of them (from positions 1 238 617 to 1 239 666) is at the fliC locus, sharing 100% identity to the fliCH4 gene of the H4 type strain (22). Therefore, we suggest that this gene encodes the H4 flagellin. The other putative flagellin gene (from positions 1 259 891 to 1 261 414) is 1524 bp in length, and contains the typical N-terminal and C-terminal conserved regions of E. coli flagellin genes. It shares 100% identity to the partial putative flnA gene sequence (1445 bp in length) (25) in the overlap region. This gene is located 20.2 kb downstream from fliC, consistent with previous observations that the fliC and flnA genes in the H17 strain are closely linked to each other (17–19).
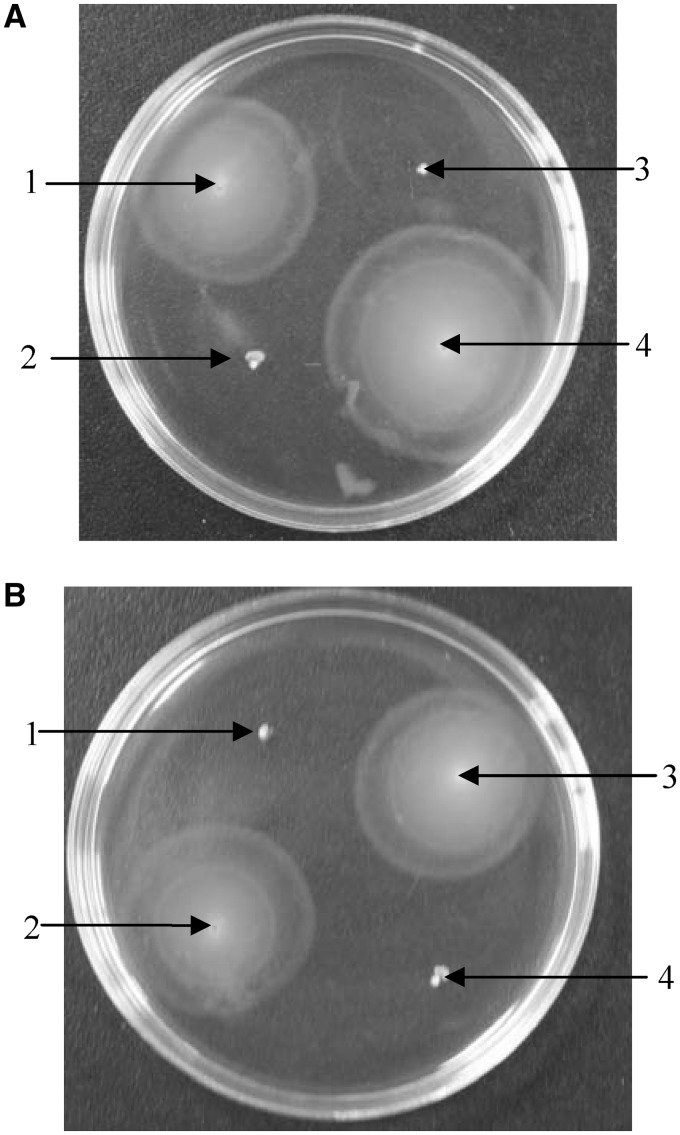
Motile ability of the strains P12b, H2001 (flnA-deficient mutant of P12b) and H2003 (H2001 carrying pLW1547) were tested on semisolid media containing antisera against H4 and H17, respectively. We observed that the strains P12b and H2003 could swim on the motility agar containing antiserum against H4, but remained fixed on the motility agar containing antiserum against H17 (Figure 1), thus indicating that P12b and H2003 produce agglutination with H17 antiserum but not with H4 antiserum. In contrast, H2001 was found to have motility ability on the motility agar containing antiserum against H17, but to be fixed on the motility agar containing antiserum against H4, thereby indicating that H2001 produces agglutination with H4 antiserum but not with H17 antiserum (Figure 1). These results suggested that P12b and H2003 express the H17 antigen, and H2001 expresses the H4 antigen.
Figure 1.
Motility test of strains P12b, H2001, H2003 and G1780. (A) The strains were tested on semisolid media containing antiserum against H4 (1, P12b; 2, G1780; 3, H2001; 4, H2003). (B) The strains were tested on semisolid media containing antiserum against H17 (1, P12b; 2, G1780; 3, H2001; 4, H2003).
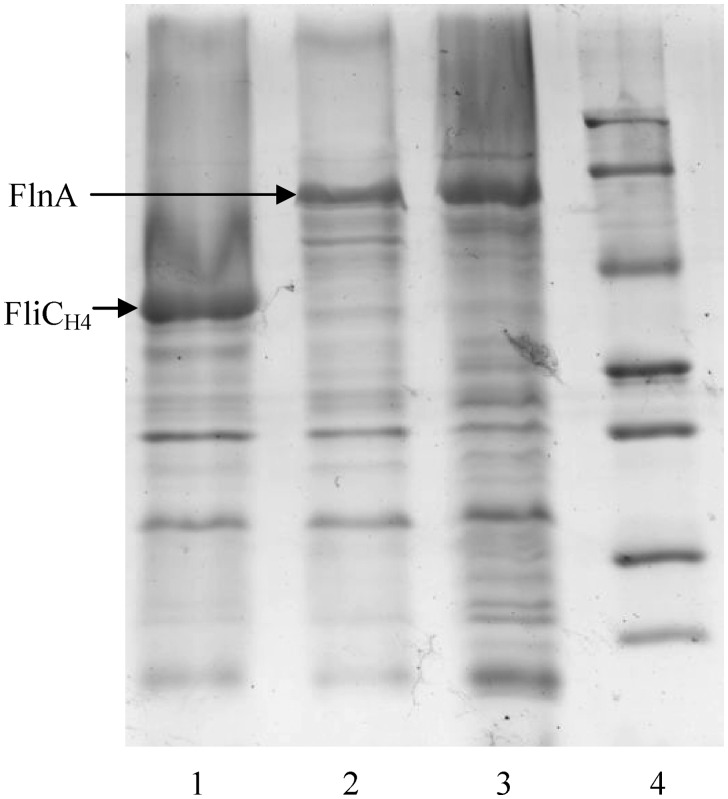
Flagellins isolated from the strains P12b, H2001 and H2003 were analyzed by SDS–PAGE. P12b and H2003 were found to produce a band with an estimated molecular mass (58.2 kDa) corresponding to the calculated mass of FlnA (52.1 kDa). H2001 was found to produce a band with an estimated molecular mass (40.3 kDa) corresponding to the calculated mass of FliCH4 (36.3 kDa) (Figure 2). The bands of P12b, H2003 and H2001 were identified by MALDI-TOF/MS analysis and were found to match the amino acid sequences of FlnA or FliCH4 well, as their protein scores were 310, 292 and 285, respectively (significance level P < 0.05). Thus, the gene from positions 1 259 891 to 1 261 414 encodes the H17 flagellin, and is the flnA gene.
Figure 2.
SDS–PAGE analysis of flagellins isolated from P12b, H2001 and H2003 (Lane 1, H2001; lane 2, P12b; lane 3, H2003; lane 4, molecular mass markers).
Deletion of the flnA region mediates flagellar phase variation from H17 to H4
To investigate the molecular mechanism for unilateral flagellar phase variation in the H17 strain, we generated a spontaneous phase variant (H2017) by growing the strain P12b on motility agar containing antiserum against H17 and screening for clone which exhibited spreading growth. H2017 produced agglutination with H4 antiserum but not with H17 antiserum, and was strongly immobilized on semisolid motility medium containing H4 antiserum. The strong immobilization of H2017 by the H4 antiserum indicated that unilateral phase change occurred, as previously reported (17,18).
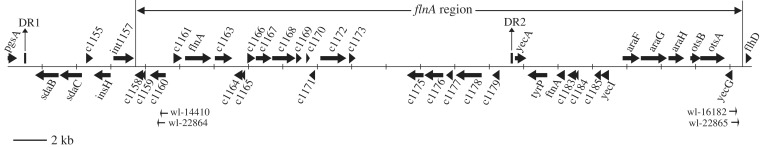
The flnA gene was not detected in H2017 by PCR when using the primer pairs wl-13432/wl-13433, thus indicating that it was absent from this strain. A series of PCRs (data not shown) were performed to screen for the presence of genes in close proximity to flnA. It was observed that all genes between int1157 and flhD were absent in H2017, excluding c1158 (from positions 1 256 950 to 1 257 453) that encodes a putative transposase, and c1159 (from positions 1 257 372 to 1 257 599) that encodes a putative IS1 repressor protein (Figure 3). Sequencing the int1157-flhD region of H2017 by using the primer pair wl-14407/wl-16181 confirmed that the DNA region in P12b from positions 1 257 702 to 1 292 151 had been deleted (see first line of Table 3).
Figure 3.
Map of the region between pgsA and flhD in the P12b genome. Arrows indicate the orientations of open reading frames (marked with gene names or locus tag). The imperfect 133 bp DRs are indicated by bars. The primer pairs wl-16182/wl4410 and wl-22864/wl-22865 were used for detection of the excised circular forms of the flnA region.
Table 3.
Excision sites of the flnA region in flagellar phase variants of the strain P12b
| Excised regiona | Excision site (upstream)b | Excision site (downstream)c | No. of variants with the same excision sites |
|---|---|---|---|
| cagcatcacctcacctgtca … aaaactttttaacagattga | 1257702 | 1292151 | 2 |
| ctgcagttgccatgttttac … gtgacgagtacagttgcgtc | 1257531 | 1292288 | 2 |
| cagcatcacctcacctgtca … caaaacaaaagtatgactta | 1257702 | 1292180 | 2 |
| cgtgcgcaacaaccgtcttc … tattttaactgtgcgcaaca | 1257243 | 1292084 | 3 |
| cagcatcacctcacctgtca … ttaattagtttgttgtgcgg | 1257702 | 1291986 | 1 |
| cagcatcacctcacctgtca … atttacggtgagttatttta | 1257702 | 1292071 | 1 |
| cagcatcacctcacctgtca … tttacggtgagttattttaa | 1257702 | 1292072 | 2 |
| cagcatcacctcacctgtca … attcctgtttcatttttgtc | 1257702 | 1292117 | 2 |
| cagcatcacctcacctgtca … ttcctgtttcatttttgtct | 1257702 | 1292118 | 2 |
| cagcatcacctcacctgtca … gctagcgtagcgaaaaactt | 1257702 | 1292139 | 1 |
| cagcatcacctcacctgtca … aaaactttttaacagattga | 1257702 | 1292152 | 2 |
| cagcatcacctcacctgtca … tacacccaaaacaaaagtat | 1257702 | 1292174 | 2 |
| cagcatcacctcacctgtca … tctgattattagtaaagaca | 1257702 | 1291825 | 3 |
| cagcatcacctcacctgtca … aaagtgattatttatagcag | 1257702 | 1292045 | 2 |
| cagcatcacctcacctgtca … ccgttgtatgtgcgtgtagt | 1257702 | 1292271 | 1 |
| cagcatcacctcacctgtca … ttaggaaaaatcttagataa | 1257702 | 1292312 | 3 |
| cagcatcacctcacctgtca … aagaaaataatgtactgatt | 1257702 | 1291891 | 2 |
aThe excised flnA region is underlined.
bThe first base upstream of the excised flnA region.
cThe first base downstream of the excised flnA region.
Further analysis of additional 32 spontaneous phase variants (H2018-H2049) indicated that an ∼35 kb region between int1157 and flhD had been lost in all variants (Figure 3, Table 3). This region, which we named the flnA region, contains 34 genes (Supplementary Table S1). The excision sites flanking the flnA region in the 33 phase variants are diverse. At the 3′-end of the flnA region, 17 different excision sites were identified within a 487-bp region (from positions 1 291 825 to 1 292 312) upstream of flhD (Table 3). At the 5′-end, most excision sites were found at position 1 257 702 (Table 3), and some at positions 1 257 531 and 1 257 243.
Many of the genes in the flnA region encode transposases or hypothetical proteins (Supplementary Table S1). Other genes in this region include araFGH (arabinose transporter) (33), otsB (trehalose-6-phosphate phosphatase), otsA (trehalose-6-phosphate synthase) and ftnA (ferritin) (Supplementary Table S1).
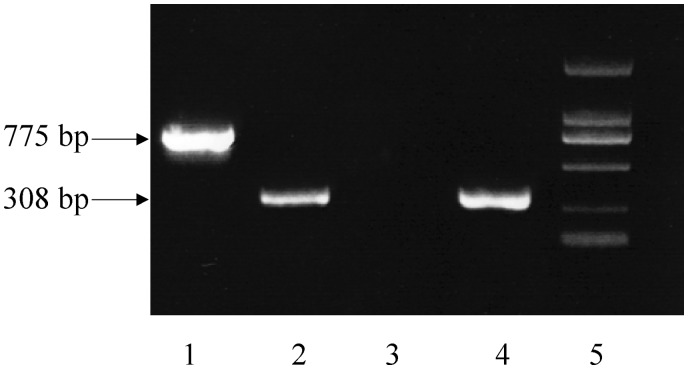
Detection of the excised extrachromosomal circular form of the flnA region
A two-step PCR was carried out with the strain P12b grown in LB broth. The first-round PCR was performed using the primers wl-16182/wl-14410 oriented toward the left and right ends of the flnA region, respectively (Figure 3). The second-round PCR was carried out using an aliquot of the product from the first round PCR as template and the primers wl-22864/ wl-22865, which were designed based on the sequence of the expected product from the first-round PCR (Figure 3). The PCR amplicons were cloned into the pGEM-T Easy vector and sequenced to screen PCR products connecting the two ends of the flnA region. Six types of PCR products (ranging from 581 to 1276 bp) connecting the two ends of the flnA region were detected in 15 independent experiments (50 clones were sequenced in each experiment). This indicated that six different recombination events had occurred between the two ends of the flnA region, thus generating six different excised extrachromosomal circular forms of the flnA region (Table 4). The 11 of 12 junction sites (Table 4) in the excised circular forms of the flnA region are within the range for excision sites of the flnA region in the chromosome (from positions 1 257 243 to 1 257 702 at the 5′-end; from positions 1 291 825 to 1 292 312 at the 3′-end) (Table 3). However, the sequence comparison showed that no junction site in the extrachromosomal circular forms was consistent with the excision sites of the flnA region in H2017-H2049, which indicates that there are more potential excision sites of the flnA region than those that we found.
Table 4.
Junction sites in excised extrachromosomal circular forms of the flnA regions
| Junction region (a/b) | Positions of the junction sitesc |
|---|---|
| … ccactacgac/aaagtacgtt … | 1257693/1292037 |
| … gcaaccggag/aagaatatta … | 1257417/1291885 |
| … cgagagtgac/aagtacgtta … | 1257544/1292036 |
| … ggcctgtagt/tgagttaatg … | 1257565/1292213 |
| … gaggacaagt/ttattttaca … | 1257612/1291943 |
| … gcccttgtcc/gatttagctg … | 1258352/1292305 |
aJunction region corresponding to the 5′-end of the flnA region.
bJunction region corresponding to the 3′-end of the flnA region.
cCorresponding positions in P12b genome for the junction sites.
Int1157 mediates excision of the flnA region
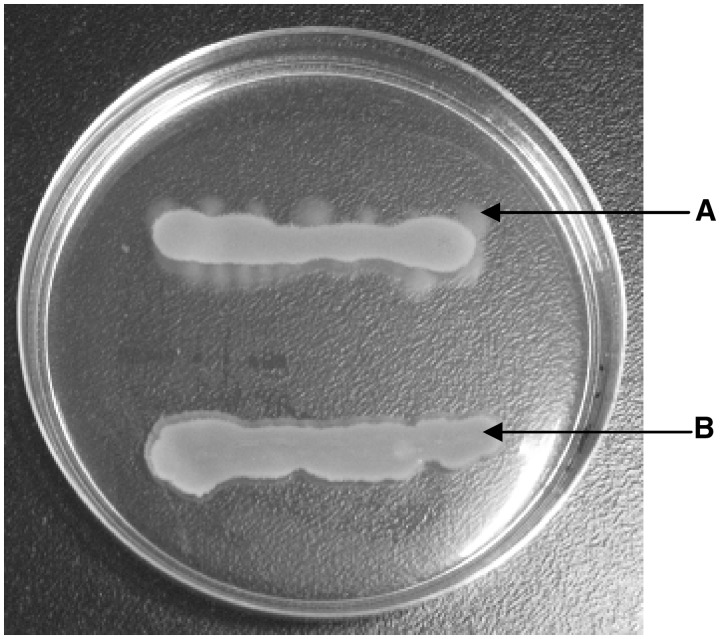
The int1157 gene (from positions 1 255 726 to 1 256 928) located upstream of the flnA region encodes a putative integrase. To test if Int1157 is the integrase responsible for excision of the flnA region, we tested for the presence of extrachromosomal circular forms of the flnA region in the strains H2002 (int1157-deficient mutant of P12b) and H2004 (H2002 carrying pLW1548) using the two-step PCR assay described above. Cloning and sequencing of the amplicons revealed that PCR products indicative of the circular forms of the flnA region were generated from H2004. A total of 300 clones generated from H2002 were sequenced. However, no product connecting the two ends of the flnA region was detected, except for non-specific PCR products. We subsequently grew both strains on motility agar containing H17 antiserum and were able to isolate spontaneous phase variants with the H4 phenotype for H2004, but not for H2002 (Figure 4). Our results strongly indicated that Int1157 is a functional integrase required for the excision of the flnA region.
Figure 4.
Selection of spontaneous flagellar phase variants from H2002 and H2004 on motility agar containing antiserum against H17 antigen. Arrow A indicates spreading growth produced by the phase variants from H2004. Arrow B indicates immobilized growth of the H2002 in the presence of H17 antiserum.
fliCH4 is transcribed in P12b
We further tested the presence of fliCH4 and flnA mRNA in P12b and H2001 by reverse transcription PCR (RT–PCR). flnA mRNA was detected only in P12b, whereas fliCH4 mRNA was detected in both strains (Figure 5), indicating that fliCH4 is transcribed in P12b. The fliC repressor, such as fljA or flkB (4,6,9,20), was not detected in the P12b genome, indicating that this strain might have a different mechanism for inhibiting fliCH4. It is likely that there is competition between FlnA and FliCH4. Such a mechanism has been reported in S. enterica, where competition between the FljB and FliC proteins for translation and assembly of the growing flagellum has been observed (4). The mechanism for the repression of FliCH4 will be the subject of future studies.
Figure 5.
Agarose gel electrophoresis of RT–PCR products. Lane 1, P12b (using primer pair wl-13432/wl-13433 for flnA); lane 2, P12b (using primer pair wl-13727/wl-13728 for fliCH4); lane 3, H2001 (using primer pair wl-13432/wl-13433 for flnA); lane 4, H2001 (using primer pair wl-13727/wl-13728 for fliCH4); lane 5, DL2000 size marker.
Strain H2097 (flnA-deficient mutant of H2002) was generated and was found to express the H4 antigen. It implies that strain H2002 possibly switches its flagellar antigen from H17 to H4 when point mutations or deletions that affect the expression of FlnA occur (the repression of FliCH4 is released), although such spontaneous phase variant was not isolated in our experiment.
DISCUSSION
Flagellar phase variation has been found to contribute to bacterial survival and virulence in animal infection models (34). Two flagellin types appear to elicit different responses from eukaryotic cells (35). In E. coli, flagellar phase variation has been reported in the H3, H47 and H17 strains. The mechanisms for flagellar variation in the H3 and H47 strains are well understood. In this study, we found that the flagellar phase variation in the H17 strain is caused by excision of the flnA region, which is mediated by non-homologous recombination. The possible principal mechanism for phase variation between FlnA and FliCH4 is that, when the flnA region is present in the chromosome, FlnA is produced. The translation and/or assembly for FliCH4 is inhibited by FlnA, which may prevent the assembly of flagella with a mixture of subunit types. When the flnA region is irreversibly excised from the chromosome, the repression of the FliCH4 is released.
Flagellar phase variation in E. coli is irreversible due to the loss of the genetic elements that contain flagellin genes (flk GI in the H3 and H47 strains, and the flnA region in the H17 strain). Bilateral flagellar phase variation is found in most S. enterica serovars, although some Typhi isolates are unidirectional (15). It is likely that bilateral flagellar phase variation is more advantageous than unidirectional phase variation, as the expression of two different flagellar antigens would allow the bacteria to adapt more readily to different environments. Salmonella enteria and E. coli diverged ∼140 million years ago (36), and it appears possible that S. enteria strains have obtained a suitable flagellar phase variation mechanism since then, which is beneficial for their survival and virulence. The fact that unidirectional flagellar phase variation is only found in 3 of 53 E. coli H type strains may be related with the different living environments and pathogenic mechanisms between E. coli and S. enteria.
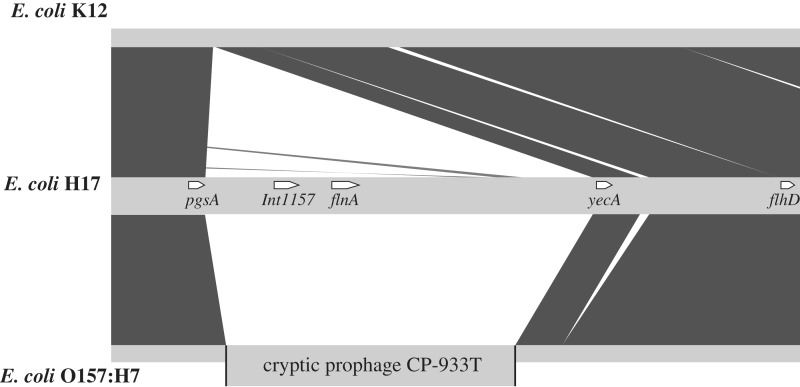
Origin of flnA
Comparison of the P12b genome to the other 30 available E. coli full genomes revealed that the region between pgsA and yecA in P12b, which overlaps with the flnA region, is unique to P12b (Figure 6). DRs are always present at the two ends of mobile elements, such as genomic islands and islets (GI) (37). Two imperfect 133 bp DRs (DR1 and DR2) were found at the two ends of the pgsA-yecA region (Figure 3). tRNA sequences were also found upstream of the pgsA-yecA region, a typical feature of GIs (37). It is likely that the pgsA-yecA region is a GI that has integrated into the P12b chromosome, and that flnA was introduced into the chromosome along with the pgsA-yecA GI.
Figure 6.
Comparison of the regions between pgsA and flhD from the genomes of the E. coli H17 (P12b), K12, and O157:H7 strains. Black blocks indicate regions of high level sequence similarity between genomes.
In most E. coli genomes, such as E. coli K12, there is no GI or prophage present between pgsA and yecA (Figure 6). Exceptions are the E. coli O157:H7 (NC_013008, NC_002695, NC_002655 and NC_011353) and 55989 (NC_011748) strains, which possess a cryptic prophage between pgsA and yecA, respectively (Figure 6). The same 122 bp DRs were found at the two ends of the prophages in the O157:H7 and 55989 strains, which correspond to base pairs 1–122 of the 133 bp DRs in P12b. The same integrase gene was found upstream of yecA in the O157:H7 and 55989 strains and probably mediates prophage integration. However, no homolog of that integrase gene could be found in the pgsA-yecA GI. We propose that the integrase gene which mediates the integration of the pgsA-yecA GI may have degraded after it integrated into the chromosome.
It is apparent that Int1157 is not the integrase which mediates the integration of the pgsA-yecA GI, as the site that it recognizes is different from the DRs of the GI. A possible explanation could be that int1157 was introduced into the chromosome after integration of the pgsA-yecA GI.
Int1157 mediates non-homologous recombination
There are two major families of site-specific recombinases: the tyrosine recombinases family (also known as λ integrase family), such as XerC in E. coli (38), and the serine recombinase family (also known as resolvase family), such as Hin in S. enterica (5). Int1157 belongs to the phage integrase family (PF00589, E value= 1.7e−24) and to the site-specific recombinase XerC family (COG4973, E value= 4.5e−10), and contains a conserved catalytic tyrosine residue at the C-terminus of the protein, a feature of tyrosine recombinases (39).
Recombinations mediated by most tyrosine recombinases occur between DRs (38), resulting in excision of GIs from the chromosome. For instance, in E. coli flk-positive strains, the tyrosine recombinase Orf486 mediates recombination of the DRs at the two ends of the flk GI, resulting in excision of the flk GI (20). However, DRs were not found flanking the flnA region in P12b, and the sequence near the excision sites at the two ends of the flnA region shows no homology, thus indicating that recombination mediated by Int1157 occurs between non-homologous DNA regions. Non-homologous recombination mediated by tyrosine recombinases has previously been observed for the conjugative transposons Tn916 and Tn4555. The excision of Tn916 and Tn4555 occurs by recombination of non-homologous flanking DNA sequences, resulting in a covalently closed circular molecule (40–44).
A novel feature of Int1157 is that it recognizes diverse sites spread over ∼500 bp at each end of the flnA region. The integrases for Tn916 and Tn4555 have also been reported to have diverse recognition sites, most of which have intrinsic curvature attributed to stretches of polyA and polyT within the sequence (40). Further studies should therefore aim to investigate the mechanisms by which Int1157 utilizes a variety of target sites.
ACCESSION NUMBER
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1.
FUNDING
Chinese National Science Fund for Distinguished Young Scholars (30788001); National Natural Science Foundation of China (NSFC) Key Program Grant (31030002); NSFC General Program Grant (30771175, 30900041); Tianjin Research Program of Application Foundation and Advanced Technology (10JCYBJC10000); Research Fund for the Doctoral Program of Higher Education of China (20090031120023); Fundamental Research Funds for the Central Universities (65020121 and 65020061); National 973 Program of China Grant (2009CB522603, 2011CB504900, 2012CB721101 and 2012CB721001); National Key Programs for Infectious Diseases of China (2009ZX10004-108). Funding for open access charge: NSFC Key Program Grant (31030002).
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Ramos HC, Rumbo M, Sirard JC. Bacterial flagellins: mediators of pathogenicity and host immune responses in mucosa. Trends Microbiol. 2004;12:509–517. doi: 10.1016/j.tim.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 2.Van Houdt R, Michiels CW. Role of bacterial cell surface structures in Escherichia coli biofilm formation. Res. Microbiol. 2005;156:626–633. doi: 10.1016/j.resmic.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Saunders JR. Population genetics of phase variable antigens. In: Baumberg S, Young JPW, Wellington EMH, Saunders JR, editors. Population genetics of Bacteria. Cambridge, UK: Cambridge University Press; 1994. pp. 247–268. [Google Scholar]
- 4.Aldridge PD, Wu C, Gnerer J, Karlinsey JE, Hughes KT, Sachs MS. Regulatory protein that inhibits both synthesis and use of the target protein controls flagellar phase variation in Salmonella enterica. Proc. Natl Acad. Sci. USA. 2006;103:11340–11345. doi: 10.1073/pnas.0602127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson RC, Simon MI. Hin-mediated site-specific recombination requires two 26 bp recombination sites and a 60 bp recombinational enhancer. Cell. 1985;41:781–791. doi: 10.1016/s0092-8674(85)80059-3. [DOI] [PubMed] [Google Scholar]
- 6.Yamamoto S, Kutsukake K. FljA-mediated posttranscriptional control of phase 1 flagellin expression in flagellar phase variation of Salmonella enterica serovar Typhimurium. J. Bacteriol. 2006;188:958–967. doi: 10.1128/JB.188.3.958-967.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kutsukake K, Iino T. A trans-acting factor mediates inversion of a specific DNA segment in flagellar phase variation of Salmonella. Nature. 1980;284:479–481. doi: 10.1038/284479a0. [DOI] [PubMed] [Google Scholar]
- 8.Silverman M, Zieg J, Hilmen M, Simon M. Phase variation in Salmonella: genetic analysis of a recombination switch. Proc. Natl Acad. Sci. USA. 1979;76:391–395. doi: 10.1073/pnas.76.1.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silverman M, Simon M. Phase variation: genetic analysis of switching mutants. Cell. 1980;19:845–854. doi: 10.1016/0092-8674(80)90075-6. [DOI] [PubMed] [Google Scholar]
- 10.Zieg J, Silverman M, Hilmen M, Simon M. Recombinational switch for gene expression. Science. 1977;196:170–172. doi: 10.1126/science.322276. [DOI] [PubMed] [Google Scholar]
- 11.Zieg J, Simon M. Analysis of the nucleotide sequence of an invertible controlling element. Proc. Natl Acad. Sci. USA. 1980;77:4196–4200. doi: 10.1073/pnas.77.7.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neidhardt FC, Ingraham JL, Magasanik B, Schaechter M, Umbarger HE. Washington, D.C: American Society for Microbiology; 1987. Escherichia and Salmonella typhimurium: cellular and Molecular Biology. [Google Scholar]
- 13.Heichman KA, Johnson RC. The Hin invertasome: protein-mediated joining of distant recombination sites at the enhancer. Science. 1990;249:511–517. doi: 10.1126/science.2166334. [DOI] [PubMed] [Google Scholar]
- 14.Baker S, Hardy J, Sanderson KE, Quail M, Goodhead I, Kingsley RA, Parkhill J, Stocker B, Dougan G. A novel linear plasmid mediates flagellar variation in Salmonella Typhi. PLoS Pathog. 2007;3:e59. doi: 10.1371/journal.ppat.0030059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baker S, Holt K, Whitehead S, Goodhead I, Perkins T, Stocker B, Hardy J, Dougan G. A linear plasmid truncation induces unidirectional flagellar phase change in H:z66 positive Salmonella Typhi. Mol. Microbiol. 2007;66:1207–1218. doi: 10.1111/j.1365-2958.2007.05995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Macnab RM. Genetics and biogenesis of bacterial flagella. Annu. Rev. Genet. 1992;26:131–158. doi: 10.1146/annurev.ge.26.120192.001023. [DOI] [PubMed] [Google Scholar]
- 17.Ratiner A. Two genetic arrangement determining flagellar antigen specificities in two diphasic Escherichia coli strains. FEMS Microbiol. Lett. 1985;29:317–323. [Google Scholar]
- 18.Ratiner A. Different alleles of the flagellin gene hagB in Escherichia coli standard H test strains. FEMS Microbiol. Lett. 1987;48:97–104. [Google Scholar]
- 19.Ratiner YA. New flagellin-specifying genes in some Escherichia coli strains. J. Bacteriol. 1998;180:979–984. doi: 10.1128/jb.180.4.979-984.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feng L, Liu B, Liu Y, Ratiner YA, Hu B, Li D, Zong X, Xiong W, Wang L. A genomic islet determines flagellar antigen phase variation in Escherichia coli H3, H35, H36, H47 and H53. J. Bacteriol. 2008;190:4470–4471. doi: 10.1128/JB.01937-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ratiner A. Mutation of E. coli with regard to the H-antigen. I. Isolation of H-antigen mutants from test H-strains of Escherichia culture. Zh. Mikrobiol. Epidemiol. Immunobiol. 1967;44:23–29. [PubMed] [Google Scholar]
- 22.Wang L, Rothemund D, Curd H, Reeves PR. Species-wide variation in the Escherichia coli flagellin (H-antigen) gene. J. Bacteriol. 2003;185:2936–2943. doi: 10.1128/JB.185.9.2936-2943.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tominaga A. Characterization of six flagellin genes in the H3, H53 and H54 standard strains of Escherichia coli. Genes Genet. Syst. 2004;79:1–8. doi: 10.1266/ggs.79.1. [DOI] [PubMed] [Google Scholar]
- 24.Tominaga A, Kutsukake K. Expressed and cryptic flagellin genes in the H44 and H55 type strains of Escherichia coli. Genes Genet. Syst. 2007;82:1–8. doi: 10.1266/ggs.82.1. [DOI] [PubMed] [Google Scholar]
- 25.Ratiner YA, Sihvonen LM, Liu Y, Wang L, Siitonen A. Alteration of flagellar phenotype of Escherichia coli strain P12b, the standard type strain for flagellar antigen H17, possessing a new non-fliC flagellin gene flnA, and possible loss of original flagellar phenotype and genotype in the course of subculturing through semisolid media. Arch. Microbiol. 2010;192:267–278. doi: 10.1007/s00203-010-0556-x. [DOI] [PubMed] [Google Scholar]
- 26.Ewing WH. Edwards and Ewing's Identification of Enterbacteriaceae. 4th edn. New York: Elsevier Science Publishing, Inc.; 1986. [Google Scholar]
- 27.Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. Artemis: sequence visualisation and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 29.Ibrahim GF, Fleet GH, Lyons MJ, Walker RA. Method for the isolation of highly purified Salmonella flagellins. J. Clin. Microbiol. 1985;22:1040–1044. doi: 10.1128/jcm.22.6.1040-1044.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ratiner A. Phase variation of the H antigen in Escherichia coli strain Bi7327-41, the standard strain for Escherichia coli flagellar antigen H3. FEMS Microbiol. Lett. 1982;15:33–36. [Google Scholar]
- 31.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl Acad. Sci. USA. 2000;91:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horazdovsky BF, Hogg RW. Genetic reconstitution of the high-affinity L-arabinose transport system. J. Bacteriol. 1989;171:3053–3059. doi: 10.1128/jb.171.6.3053-3059.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ikeda JS, Schmitt CK, Darnell SC, Watson PR, Bispham J, Wallis TS, Weinstein DL, Metcalf ES, Adams P, O'Connor CD, et al. Flagellar phase variation of Salmonella enterica serovar Typhimurium contributes to virulence in the murine typhoid infection model but does not influence Salmonella-induced enteropathogenesis. Infect. Immun. 2001;69:3021–3030. doi: 10.1128/IAI.69.5.3021-3030.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ciacci-Woolwine F, Blomfield IC, Richardson SH, Mizel SB. Salmonella flagellin induces tumor necrosis factor alpha in a human promonocytic cell line. Infect. Immun. 1998;66:1127–1134. doi: 10.1128/iai.66.3.1127-1134.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ochman H, Wilson AC. Evolutionary history of enteric bacteria. In: Neidhardt FC, editor. Escherichia coli and Salmonella typhimurium Cellular and Molecular Biology. Vol. 2. Washington, DC: American Society for Microbiology; 1987. pp. 1649–1654. [Google Scholar]
- 37.Dobrindt U, Hochhut B, Hentschel U, Hacker J. Genomic islands in pathogenic and environmental microorganisms. Nat. Rev. Microbiol. 2004;2:414–424. doi: 10.1038/nrmicro884. [DOI] [PubMed] [Google Scholar]
- 38.Grindley ND, Whiteson KL, Rice PA. Mechanisms of site-specific recombination. Annu. Rev. Biochem. 2006;75:567–605. doi: 10.1146/annurev.biochem.73.011303.073908. [DOI] [PubMed] [Google Scholar]
- 39.Esposito D, Scocca JJ. The integrase family of tyrosine recombinases: evolution of a conserved active site domain. Nucleic Acids Res. 1997;25:3605–3614. doi: 10.1093/nar/25.18.3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tribble GD, Parker AC, Smith CJ. The Bacteroides mobilizable transposon Tn4555 integrates by a site-specific recombination mechanism similar to that of the gram-positive bacterial element Tn916. J. Bacteriol. 1997;179:2731–2739. doi: 10.1128/jb.179.8.2731-2739.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tribble GD, Parker AC, Smith CJ. Transposition genes of the Bacteroides mobilizable transposon Tn4555: role of a novel targeting gene. Mol. Microbiol. 1999;34:385–394. doi: 10.1046/j.1365-2958.1999.01616.x. [DOI] [PubMed] [Google Scholar]
- 42.Marra D, Scott JR. Regulation of excision of the conjugative transposon Tn916. Mol. Microbiol. 1999;31:609–621. doi: 10.1046/j.1365-2958.1999.01201.x. [DOI] [PubMed] [Google Scholar]
- 43.Salyers AA, Shoemaker NB, Stevens AM, Li LY. Conjugative transposons: an unusual and diverse set of integrated gene transfer elements. Microbiol. Rev. 1995;59:579–590. doi: 10.1128/mr.59.4.579-590.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Caparon MG, Scott JR. Excision and insertion of the conjugative transposon Tn916 involves a novel recombination mechanism. Cell. 1989;59:1027–1034. doi: 10.1016/0092-8674(89)90759-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.