Abstract
Nuclear architecture as well as gene nuclear positioning can modulate gene expression. In this work, we have analyzed the nuclear position of the interferon-β (IFN-β) locus, responsible for the establishment of the innate antiviral response, with respect to pericentromeric heterochromatin (PCH) in correlation with virus-induced IFN-β gene expression. Experiments were carried out in two different cell types either non-infected (NI) or during the time course of three different viral infections. In NI cells, we showed a monoallelic IFN-β promoter association with PCH that strongly decreased after viral infection. Dissociation of the IFN-β locus away from these repressive regions preceded strong promoter transcriptional activation and was reversible within 12 h after infection. No dissociation was observed after infection with a virus that abnormally maintained the IFN-β gene in a repressed state. Dissociation induced after virus infection specifically targeted the IFN-β locus without affecting the general structure and nuclear distribution of PCH clusters. Using cell lines stably transfected with wild-type or mutated IFN-β promoters, we identified the proximal region of the IFN-β promoter containing YY1 DNA-binding sites as the region regulating IFN-β promoter association with PCH before as well as during virus infection.
INTRODUCTION
Positioning of a gene at the vicinity of pericentromeric heterochromatin (PCH) clusters has been described as a means to induce heritable gene silencing. On the contrary, re-positioning of gene loci away from PCH has been correlated with its transcriptional activation (1–3). In mammals, such a phenomenon has been mainly observed until now during cell differentiation as in the case of the β-globin locus during erythroid differentiation (4), lymphoid-associated genes in cycling versus non-cycling primary B lymphocytes (5) and cytokine genes IL-4 and IFN-γ during activation of naive T cells (6). Nevertheless, there are some examples of gene silencing through PCH association that have been observed in differentiated cells (7,8).
In this work, we have investigated the nuclear positioning of the IFN-β gene with respect to PCH in correlation with the expression of the interferon-β (IFN-β) gene. Beyond the essential role of IFN-β on the establishment of the innate antiviral response (9,10), IFN-β has profound effects on many other aspects of cell physiology for most of them linked to immune and inflammatory responses (9,11,12). Small fold changes in IFN-β production can lead to the activation of important signaling cascades, and an abnormal accumulation of IFN-β is deleterious for the organism. Therefore, the repressive state of the IFN-β gene needs to be tightly regulated in order to avoid an inappropriate production of IFN-β in the absence of stimulus.
Contrary to the molecular mechanisms leading to activation of the IFN-β promoter that have been thoroughly documented and require promoter transcription factor recruitment coupled to chromatin remodeling events (13–19), little information is available concerning the mechanisms responsible for maintaining the promoter in its heritable silent state. The presence of deacetylated histones and repressor complex containing SAP30/Sin3A/NCoR/HDAC3 on the silent promoter (20–22) suggested that a dense non-permissive chromatin structure could play an important role during maintenance of IFN-β gene silencing.
Using two-color DNA fluorescent in situ hybridization (FISH), we measured the degree of association of the IFN-β promoter with PCH in non-stimulated cells as well as during the time course of different viral infections. Nuclear positioning of the endogenous IFN-βετα locus was analyzed in two different IFN-βετα producing cell types (fibroblasts and macrophages) and three viruses were used in this work: Newcastle Disease Virus (NDV) and Clone 13 of Rift Valley fever virus (RVFV) that are non-pathogenic for mice and induce interferon-β gene expression and the ZH 548 strain of RVFV, which is highly pathogenic for mice through mechanisms that suppress IFN-β induction and action (13, 23). Experiments were performed on the endogenous wild-type IFN-β promoter as well as on stably transfected IFN-β promoters either wild-type or mutated on their binding sites for transcription factor YY1. In the case of the endogenous promoter, a predominant monoallelic association of the silent promoter with PCH was observed in fibroblasts as well as macrophages, a situation that is reminiscent of what has been observed in the case of immunoglobulin genes (8). Infection with NDV and Clone 13 induced re-positioning of the endogenous and wild-type-integrated IFN-β promoters away from PCH clusters in correlation with the activation of the promoter transcriptional capacity. Dissociation occurred at the onset of strong promoter transcriptional activation and was reversible independently of cell cycle progression. On the contrary, no dissociation was observed after infection with a pathogenic virus that abnormally maintained the promoter under a repressed state. Using cells lines containing IFN-β promoters mutated on one or the other or the two YY1 DNA-binding sites present on the IFN-β promoter, we have demonstrated that binding of transcription factor YY1 to the IFN-β promoter was necessary for promoter pericentromeric recruitment in non-infected (NI) cells as well as for virus-induced re-positioning of the IFN-β promoter away from PCH.
MATERIALS AND METHODS
Viruses and cells
Experiments on murine macrophages were carried out using the ATCC cell line RAW264.7 (ref: TIB-71). Murine fibroblastic L929 cells and L929wt330, mut90, mut122, mut122/90 cell lines and NDV infections were described previously (24,18). RVFV ZH548 and Clone 13 infections were carried out as previously described (22).
Antibodies
Antibodies used for chromatin immunoprecipitations were: anti-CBP A-22 (sc-369), anti-YY1 H-10 (sc-7341), anti-IRF3 FL-425 (sc-9082) and anti-C23 H-250 (sc-13057) from Santa Cruz; anti-acetyl-Histone H4 (Lys8) (06–760), anti-acetyl-Histone H3 (Lys14) (06–911), anti-acetyl-Histone H3 (Lys9) (07–442) and anti-trimethyl-Histone H3 (Lys9) (06-942) from Upstate (Millipore). Primary antibody used for immunofluorescence was anti-YY1 H-10 (sc-7341) from Santa Cruz. Secondary antibody used for YY1 immunofluorescence was Alexa fluor-conjugated chicken anti-mouse from Invitrogen (A21200).
Immunofluorescence
For immunofluorescence, cells grown in 24-well plates on coverslips were fixed with 4% formaldehyde in PBS for 15 min and permeabilized with 0.5% Triton X-100 in PBS for 30 min. Then cells were incubated for 1 h at 37°C with mouse anti-YY1 monoclonal antibody diluted in PBS/BSA 1%. Cells were then washed with PBS and incubated for 45 min at room temperature with corresponding secondary antibodies.
FISH
FISH was carried as previously described (25). Plasmid pBluescript-γ-satellite (a generous gift of Dr Niall Dillon) which contains eight mouse γ-satellite repeats cloned into the EcoRI site of a low copy version of pBluescript (26) was used as pericentromeric γ-satellite probe. The murine IFN-β locus regions from −1153 to +1660 (27) cloned into plasmid pBR322 were used as a probe for the endogenous IFN-β promoter. Plasmid pBLCAT3-muIFN-β-wt330 which contains the wild-type murine IFN-β promoter fragment from −330 to +20 cloned in front of the CAT reporter gene of plasmid pBLCAT3 (24) was used as a probe for the integrated IFN-β promoters. Probes were labeled by nick translation (Amersham) with either rhodamine-5-dUTP (Enzo Life Sciences) or with biotin-11-dUTP (Enzo Life Sciences) as indicated in figure legends. For visualization of biotin, rabbit anti-biotin concentrate (43861) from Enzo was used as primary antibody and Alexa fluor conjugated donkey anti-rabbit from Invitrogen (A31572) was used as secondary antibody.
Chromatin immunoprecipitation
Chromatin immunoprecipitation experiments were carried out as previously described (22). PCR analysis of inputs or immunoprecipitated DNAs was performed using oligonucleotides F-40 (5′-GTT TTC CCA GTC ACG AC-3′) and CAT (5′-CCA TTT TAG CTT CCT TAG-3′) to reveal the integrated wt330 IFN-β promoter. Sense oligonucleotide 5′-TAT GGC GAG AAC CTG AAA-3′ and anti-sense oligonucleotide 5′-TTC ACG TCC TAA AGT GTG TAT-3′ were used as 5′ and 3′ primers, respectively, to reveal the γ-satellite sequence. For the F-40, CAT set of primers, PCR conditions were as follows: 1 cycle at 94°C for 5 min; 20 cycles at 94°C for 30 s, 53°C for 30 s and 72°C for 30 s; 1 cycle at 72°C for 10 min. For the γ-satellite set of primers, PCR conditions were as follows: 1 cycle at 94°C for 5 min; 9 cycles at 94°C for 30 s, 54°C for 30 s and 72°C for 30 s; 1 cycle at 72°C for 10 min. A first ‘cold’ PCR was carried out in the presence of 25 pmol of each primer; 1.5 µl of the product of the first PCR was subjected to a second ‘hot’ PCR carried out in the presence of 0.1 µl α-32PdATP (6000 Ci/mmol) and 25 pmol of each primer.
Image acquisition and manipulation
Samples were analyzed at room temperature by confocal laser scanning microscopy using an Axiovert 200 M (Zeiss LSM510 confocal system) at the Service Commun de Microscopie (SCM) of Université Paris Descartes. This system is equipped with a 63×, 1.4 O.N oil immersion lens (Plan Neofluor). For oil immersion microscopy, we used oil with refractive index of 1.518 (Zeiss). Images were collected in the direction of the z-axis corresponding to the optical axis of the microscope at 0.37 µm intervals with the z-axis going through the image planes. LSM 510 imaging software was used for image capture (512 × 512 pixels, 8 bit data). The images were analyzed by the LSM5 Image browser or Image J software. Double-labeled pixels were displayed in yellow on the merge images.
RT-qPCR
Total RNA was extracted using TriPure Isolation Reagent (Roche) according to the manufacturer's protocol. Of total RNA, 1 μg was digested with RQ1 RNase-Free DNase (Promega) to remove contaminated DNA. Reverse transcription was carried out with Random Primers (Promega) and M-MLV Reverse Transcriptase (Invitrogen) according to the manufacturers’ recommendations. RT-qPCR was performed using SYBR Green (Thermo scientific) reagents: 1 cycle at 95°C for 15 min, and then 40 cycles at 95°C for 15 s and 60°C for 1 min, followed by a dissociation step. GAPDH (forward: 5′-GAACATCATCCCTGCATCCA and reverse: 5′-CCAGTGAGCTTCCCGTTC) levels were used for normalization and relative IFN-β (forward: 5′-ATGAACAACAGGTGGATCCTCC and reverse: 5′-AGGAGCTCCTGACATTTCCGAA) mRNA levels were calculated using the ΔΔCt method. Each experiment was carried out twice in triplicate, and the results are expressed as mean (SD).
RNA interference experiments (RNAi)
L929wt330 cells were transfected using Lipofectamine 2000 (Invitrogen Life Technologies) with a pool of siRNA oligos specific for yy1 (Thermo Scientific Dharmacon® L-050273-00-0005) or with a pool of control siRNA sequences (Thermo Scientific Dharmacon® D-001810-10-05) at a final concentration of 50 nM each. Cells were fixed for immunofluorescence or harvested for RNA isolation 48 h after transfection.
Statistical analyses
Statistical analysis was done applying the chi-square test. After determining the homogeneity of the corresponding cell populations from the various independent experiments, data were considered as statistically significant at P < 0.05, as very significant at P < 0.01 and highly statistically significant at P < 0.001.
RESULTS
Transcriptional activation of the interferon-β gene is correlated with a strong decrease of the association of the IFN-β locus with pericentromeric heterochromatin clusters
In murine cells, PCH is constituted of clusters of γ-satellite sequences (28,29). In order to analyze the degree of association of the endogenous IFN-β locus with clusters of γ-satellite sequences with respect to IFN-β expression, we carried out two-color DNA FISH experiments using a rhodamine-labeled γ-satellite probe (red) and a biotin-labeled IFN-β probe (green) containing the IFN-β locus region from −1153 to +1660 cloned into pBR322 plasmid. Experiments were carried out in murine fibroblastic L929 cells either NI or infected with the non-pathogenic Clone13 strain (C13) of the bunyavirus RVFV, which has been described as a good inducer of IFN-β expression (23). As a control, we used the virulent ZH548 strain (ZH) of RVFV known to be lethal for mice and to abnormally maintain the IFN-β promoter in a transcriptionally silent state (23). For each condition, the percentage of IFN-β signals colocalizing with γ-satellite FISH signals was quantified and correlated to the induction of IFN-β mRNA synthesis that was assessed by qRT-PCR.
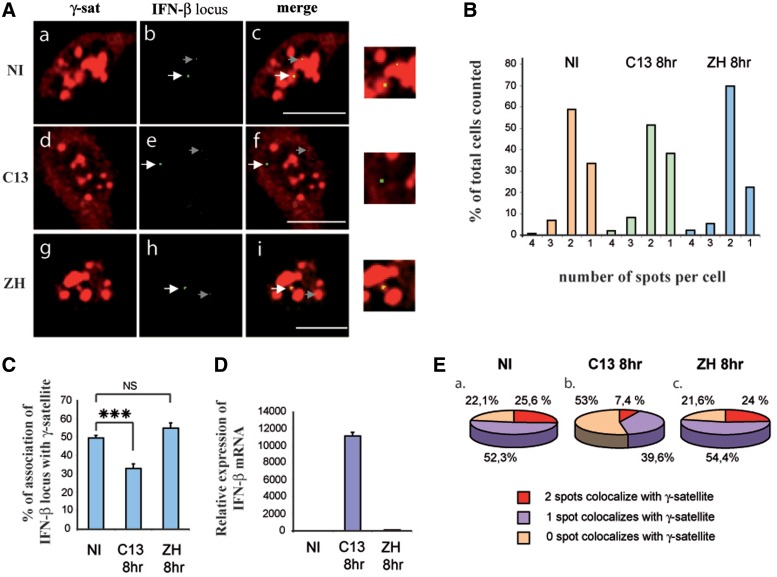
The images corresponding to single confocal sections of nuclei of L929 cells either NI (a–c) or 8 h after infection with the C13 (d–f) or the ZH (g–i) strain of RVFV are shown in Figure 1A. The number of IFN-β signals present per cell was counted in a total of at least 137 cells from two independent experiments under NI, C13 and ZH conditions. As expected for an endogenous locus in asynchronously growing cells, the majority of the cells displayed two IFN-β FISH signals per cell (Figure 1B) with only a small amount of cells displaying three or four IFN-β FISH signals corresponding to those cells that have duplicated their genome and where the corresponding sister chromatides could be individually visualized. However, in ∼30% of the cells, only one IFN-β FISH signal was visible per cell. This is a situation that could result from allelic pairing, similarly to what has been observed in the case of immunoglobulin loci Igh and Igk (30) or correspond to two alleles separated by a distance inferior to the resolution of the objective used for image acquisition (200 nm). The FISH signals obtained with the IFN-β probe were specific since no signals were obtained with the empty pBR322 plasmid under the same conditions (Supplementary Figure S1A).
Figure 1.
In murine fibroblasts, the silent endogenous IFN-β promoter is associated with clusters of γ-satellite sequences. Murine fibroblastic L929 cells either NI or virus-infected by non-pathogenic C13 (C13) or pathogenic ZH548 (ZH) RVFV strain were collected 8 h p.i. (A) Single confocal section displaying rhodamine-labeled γ-satellite FISH (γ-sat) in red (a, d, g) combined with biotin-labeled endogenous IFN-β FISH (IFN-β) in green (b, e, h) and the corresponding merge images (c, f, i) with enlargement of selected regions in the far right column. In middle and right panels, the white arrows indicate the IFN-β FISH signals visible in the shown confocal sections and gray arrows indicate the position of IFN-β signals present on other confocal sections of the same nuclei, weakly visible on the confocal sections shown here. (B) Percentage of cells displaying 1, 2, 3 or 4 IFN-β FISH spots per cell, and (C) the number of IFN-β FISH signals colocalizing with γ-satellite clusters was determined for each condition. (D) The corresponding IFN-β mRNA induction fold was measured by RT-qPCR, and (E) percent of cells displaying 0, 1 or 2 IFN-β spots colocalizing with clusters of γ-satellite sequences was determined among cells displaying two IFN-β spots per cell. ***P < 0.001, NS, non-significantly different. Scale Bars = 10 µm.
In NI cells, the endogenous IFN-β FISH signals appeared colocalizing with clusters of γ-satellite DNA (Figure 1A, a–c). Measurement of the overlapping distance of the respective fluorescence intensities through the region of colocalization translated a true colocalization event (Supplementary Figure S1B). From a total of 256 FISH signals counted from 153 cells from two independent experiments, 49.6 ± 1.5% of the endogenous IFN-β signals colocalized with γ-satellite clusters (Figure 1C). As expected, no IFN-β mRNA was detected under NI conditions (Figure 1D) confirming the silent state of the promoter in the absence of virus infection.
After infection with C13 strain (Figure 1A, d–f), that activated the transcriptional capacity of the IFN-β promoter (Figure 1D), we observed dissociation of the IFN-β promoter from clusters of γ-satellite sequences. From 276 FISH signals counted (from a total of 167 cells from two independent infections), only 32.97 ± 2.6% of the endogenous IFN-β signals still colocalized with γ-satellite clusters at 8 h after infection by C13 (Figure 1C). Contrary to the results obtained after infection with C13, no dissociation was observed after infection with pathogenic ZH strain (Figure 1A, g–i) that maintained the IFN-β promoter in a transcriptionally silent state (Figure 1D). From 242 FISH signals counted in 137 cells from two independent experiments, 54.95 ± 3% of IFN-β signals (Figure 1C) remained colocalizing with clusters of γ-satellite sequences after infection with the pathogenic ZH548 strain of RVFV.
Overall, these results showed a correlation between association with PCH clusters and the absence of transcriptional activation as well as vice versa a correlation between statistically significant virus-induced dissociation from PCH and virus-induced IFN-β transcriptional activation.
In NI cells, a maximum of ∼50% of the silent endogenous IFN-β loci appeared colocalizing with γ-satellite clusters suggesting the possibility of a monallelic pericentromeric recruitment. In order to determine if the association of IFN-β locus with γ-satellite clusters was indeed monoallelic, we counted, among the population of cells displaying two IFN-β signals per cell, the percentage of cells displaying 2, 1 or 0 FISH signals colocalizing with γ-satellite clusters. As shown in Figure 1E, under NI conditions, 52.3% of these cells displayed a monoallelic pericentromeric recruitment with only one out of the two FISH signal colocalizing with γ-satellite clusters, whereas under the same conditions, only 25.5% of the cells displayed biallelic pericentromeric recruitment.
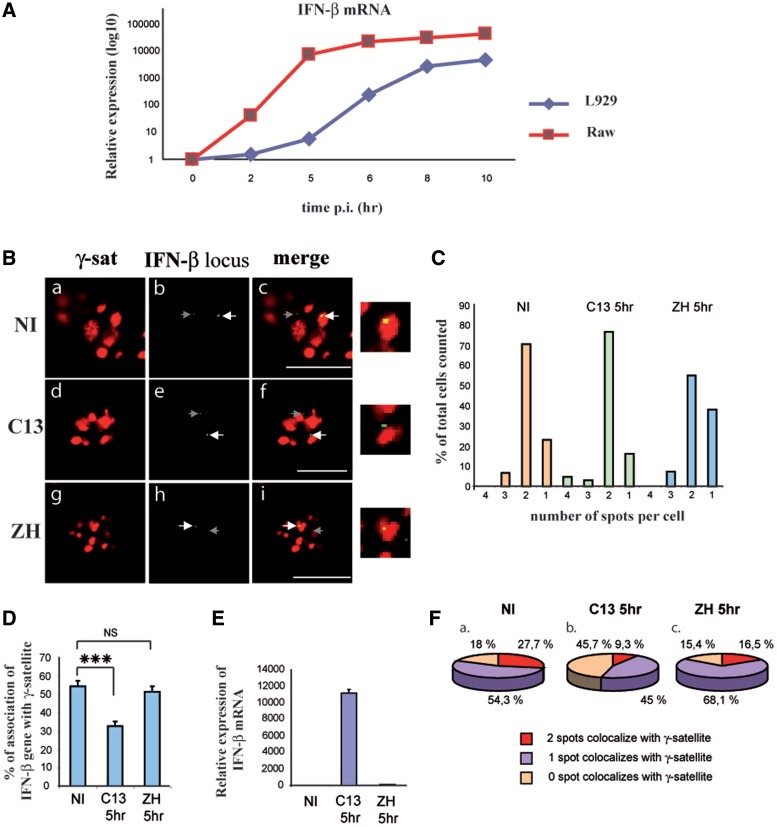
In order to further confirm and by doing so enhance the relevance of monoallelic PCH recruitment as a mechanism to regulate IFN-β expression before and after virus infection, FISH experiments as those described in Figure 1 were carried out on macrophages that constitute a major IFN-β producing cell type. Compared to fibroblasts, macrophages displayed a much faster kinetics of virus-induced IFN-β mRNA expression (Figure 2A) displaying 5 h p.i. a degree of IFN-β expression analogous to the one displayed, under the same conditions, by fibroblasts 8 h p.i. Therefore, in the case of macrophages, the percentage of association was measured 5 h p.i. instead of 8 h p.i. as in the case of fibroblasts. As it can be observed in Figure 2, results obtained on macrophages 5 h p.i. were very similar to those obtained on fibroblasts 8 h p.i. indicating that monoallelic PCH recruitment of the IFN-β locus was not specific of a single cell type.
Figure 2.
In murine macrophages, the silent endogenous IFN-β promoter is associated with clusters γ-satellite sequences. (A) Kinetics of induction (in log10) of IFN-β expression in murine L929 fibroblasts and RAW264.7 macrophages after infection with the C13 strain of RVFV. Relative expression of IFN-β mRNA was measured by RT-qPCR at indicated times. (B–F) Murine macrophages RAW264.7 either NI or virus-infected by non-pathogenic C13 (C13) or pathogenic ZH548 (ZH) RVFV strain were collected 5 h p.i. FISH experiments were carried out as in Figure 1 using a rhodamine-labeled γ-satellite probe and combined with biotin-labeled endogenous IFN-β probe. (B) Single confocal section displaying rhodamine-labeled γ-satellite FISH (γ-sat) in red (a, d and g) combined with biotin-labeled endogenous IFN-β FISH (IFN-β) in green (b, e and h) and the corresponding merge images (c, f, i) with enlargement of selected regions in the far right column. In middle and right panels, the white arrows indicate the IFN-β FISH signals visible in the shown confocal sections and gray arrows indicate the position of IFN-β signals present on other confocal sections of the same nuclei, weakly visible on the confocal sections shown here. (C) Percent of cells displaying 1, 2, 3 or 4 IFN-β FISH spots per cell, (D) the number of IFN-β FISH signals colocalizing with γ-satellite clusters was determined for each condition. (E) The corresponding IFN-β mRNA induction fold was measured by RT-qPCR. (F) Percent of cells displaying 0, 1 or 2 IFN-β spots colocalizing with clusters of γ-satellite sequences was determined among cells displaying two IFN-β spots per cell. ***P < 0.001, NS = non-significantly different.
The proximal upstream region of the IFN-β promoter-induced association of the silent IFN-β promoter with pericentromeric heterochromatin
In order to determine if the proximal region of the IFN-β promoter was sufficient to induce the association of the promoter with PCH, FISH experiments were carried out with the murine fibroblastic L929wt330 cell line. This cell line corresponds to a pool of 10 independent L929 clones each one containing the murine IFN-β wt330 promoter region, from −330 to +20 cloned into plasmid pBLCAT3 in front of the CAT reporter gene, randomly integrated into their genome (24). Until now, this pool of murine IFN-β wt330 promoters has been shown to display the same properties than the endogenous murine IFN-β promoter (24,19,22). For FISH experiments, we used here a biotin-labeled γ-satellite probe (green) and a rhodamine-labeled pBLCAT3IFN-βwt330 probe (red) specific of the integrated IFN-βwt330 promoter. During the establishment of the stably transfected L929wt330 clones, the entire plasmid was integrated into the genome of L929 cells so that during FISH experiments the entire probe hybridized with the integrated locus.
Experiments were carried out in NI cells, when the promoter is maintained in a transcriptionally silent state as well as in cells infected with two different viruses corresponding to the avian paramyxovirus NDV and the previously described Clone13 strain (C13) of RVFV, both being good inducers of IFN-β promoter activity (13,23). As before, we used as a control the virulent ZH548 strain (ZH) of RVFV that maintained the promoter in a transcriptionally silent state. For each condition, the percentage of colocalizing FISH signals was quantified and correlated to the transcriptional capacity of the promoter that was determined by measuring the corresponding CAT activities.
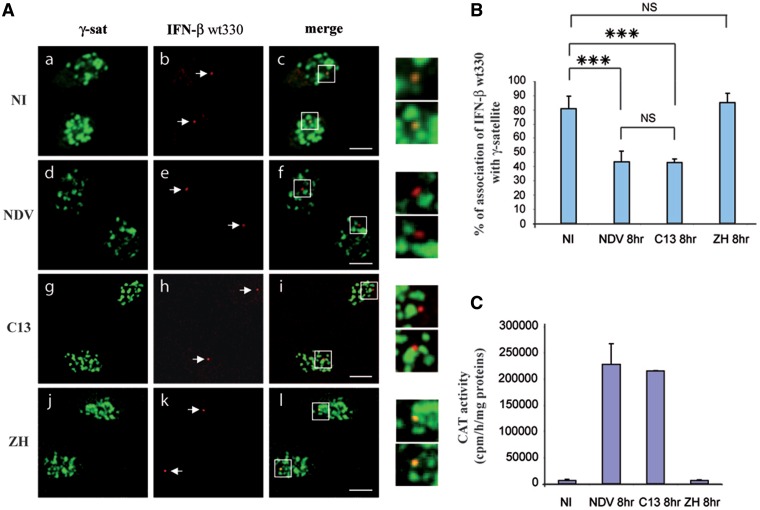
The images corresponding to single confocal sections of nuclei of L929wt330 cells either NI (a–c) or collected at 8 h after infection with NDV (d–f), C13 (g–i) or ZH (j–l) are shown in Figure 3A. The FISH signal obtained with the 5300-bp long pBLCAT3IFN-βwt330 probe was specific of the integrated promoter since no signal corresponding to the endogenous IFN-β promoter was detected with this probe in L929 cells (Supplementary Figure S2A). As a matter of fact, the only region that the pBLCAT3IFN-βwt330 probe shares with the endogenous IFN-β locus corresponds to the −330 to +20 IFN-β promoter region. This region of 350 bp is much too short to give rise to a visible fluorescent signal compared to the 5300 bp pBLCAT3IFNβwt330 probe that hybridizes with the integrated locus. Also, no signal was obtained in L929wt330 cells with plasmid pENTR/U6 used here as a rhodamine-labeled irrelevant non-specific probe (Supplementary Figure S2B) further confirming the specificity of the hybridization signal obtained with the pBLCAT3IFN-βwt330 probe on L929wt330 cells.
Figure 3.
The −330 to +20 region of the IFN-β promoter induces association of the silent promoter with clusters of γ-satellite sequences. Murine fibroblastic L929wt330 cells either NI or virus-infected by non-pathogenic NDV or RVFV C13 (C13) strain or by pathogenic RVFV ZH548 (ZH) strain were collected 8 h p.i. (A) Single confocal section displaying biotin-labeled γ-satellite FISH (γ-sat) in green (a, d, g and j) combined with rhodamine-labeled IFN-β wt330 promoter FISH (IFN-β) in red (b, e, h and k) and the corresponding merge images (c, f, i, l) with enlargement of selected regions in the far right column. (B) The number of IFN-β wt330 promoter FISH signals colocalizing with γ-satellite clusters was quantified with n > 90 for each condition and (C) the corresponding CAT activities were measured. Arrows indicate rhodamine-labeled IFN-β wt330 promoter FISH signals. ***P < 0.001, NS, non-significantly different. Scale Bars = 10 µm.
In NI cells, the transcriptionally silent integrated IFN-βwt330 promoter appeared in its great majority colocalizing with clusters of γ-satellite DNA (Figure 3A, a–c). As expected for an integrated locus, the large majority of the cells (90.4 ± 3.85%) displayed only one FISH signal per cell (data not shown). From a total of 247 FISH signals counted from 220 cells from six independent experiments, 80.97 ± 8.78% of IFN-βwt330 FISH signals appeared colocalizing with clusters of γ-satellite DNA (Figure 3B). As expected, no CAT activity was detected in NI cells (Figure 3C) confirming that in the absence of virus infection the integrated IFN-βwt330 promoter was, as the endogenous promoter, maintained in a constitutive silent state. As in the case of the endogenous promoter, measurement of the overlapping distance of the respective fluorescence intensities through the region of colocalization translated a true colocalization event (Supplementary Figure S2C).
Overall, these results suggested that the proximal upstream region from −330 to +20 of the IFN-β promoter could be sufficient to drive association of the silent IFN-β with PCH clusters in NI cells. The −330 to +20 region of the promoter appeared all the more sufficient by itself to drive association with clusters of γ-satellite sequences that association occurred independently of the genomic environment of the integration site. Indeed, in the great majority of NI L929wt330 cells, the IFN-βwt330 FISH signals appeared colocalizing with PCH even though the pool of L929wt330 cells derives from 10 independent clones that have, each one, randomly integrated IFN-β promoter in the genome of L929 cells. Contrary to the endogenous promoter that displayed a monoallelic PCH association (with a maximum of ∼50% of IFN-β loci associated with PCH), 80% of the integrated IFN-β loci were observed associated with PCH. This observation suggests that the presence of two alleles could be necessary to regulate monoallelic distribution with respect to PCH as described in the case of immunoglobulin loci (31).
As previously observed in the case of the endogenous promoter, dissociation of the integrated IFN-βwt330 promoter from clusters of γ-satellite sequences was observed after infection with non-pathogenic viruses NDV and C13 (Figure 3A, d–i) that induced the activation of the transcriptional capacity of the IFN-β promoter (Figure 3C). From 91 (NDV) and 140 (C13) FISH signals counted (from a total of two NDV and three C13 independent infections), only 38.4 ± 2% and 42.85 ± 2.64% of the IFN-β signals still colocalized with γ-satellite clusters at 8 h after infection by NDV and C13, respectively (Figure 3B), as opposed to 80.97 ± 8.78% of colocalization observed in NI cells. Contrary to results obtained after infection with NDV and C13, no dissociation was observed after infection with pathogenic ZH strain (Figure 3A, j–l) that maintained the IFN-β promoter in a transcriptionally silent state (Figure 3C). From 216 FISH signals counted from 195 cells from four independent experiments, 85.18 ± 6.45% of IFN-β signals (Figure 3B) remained colocalizing with clusters of γ-satellite sequences after infection with pathogenic ZH548 strain of RVFV.
Given the stronger homogeneity of the population IFN-βwt330 FISH signals compared to endogenous IFN-β FISH signals, the subsequent experiments aiming at characterizing the mechanism regulating IFN-β promoter association with γ-satellite clusters before and after virus infection were carried out on the integrated IFN-β promoters.
Virus infection affected the IFN-β promoter region without affecting the structure or organization of γ-satellite sequences
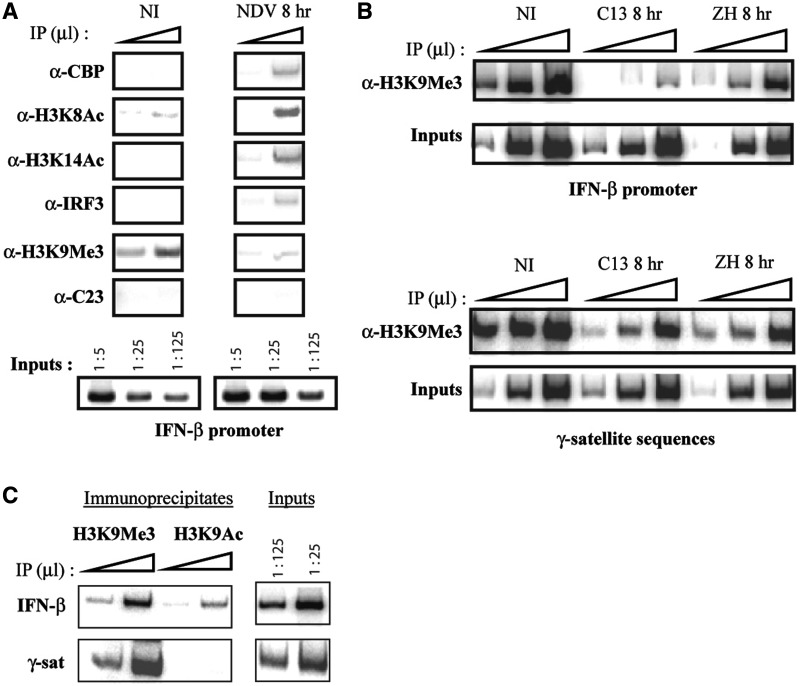
Some viruses, such as HERPES simplex virus type 1, severely damage the structure of cellular centromeres (32). In order to determine if the viruses used in this work affected the general structure of PCH, we analyzed here the effect of virus infection on the presence of the trimethylated form of lysine 9 of histone H3 (H3K9me3) on γ-satellite sequences as compared to IFN-β promoter as well as on the number and the radial distribution of γ-satellite clusters. Indeed, H3K9me3 is an epigenetic mark concentrated within clusters of γ-satellite sequences that plays an essential role in the general organization of PCH and that it is also commonly found associated with transcriptionally silent regions of the genome in relation with heterochromatization (33–35).
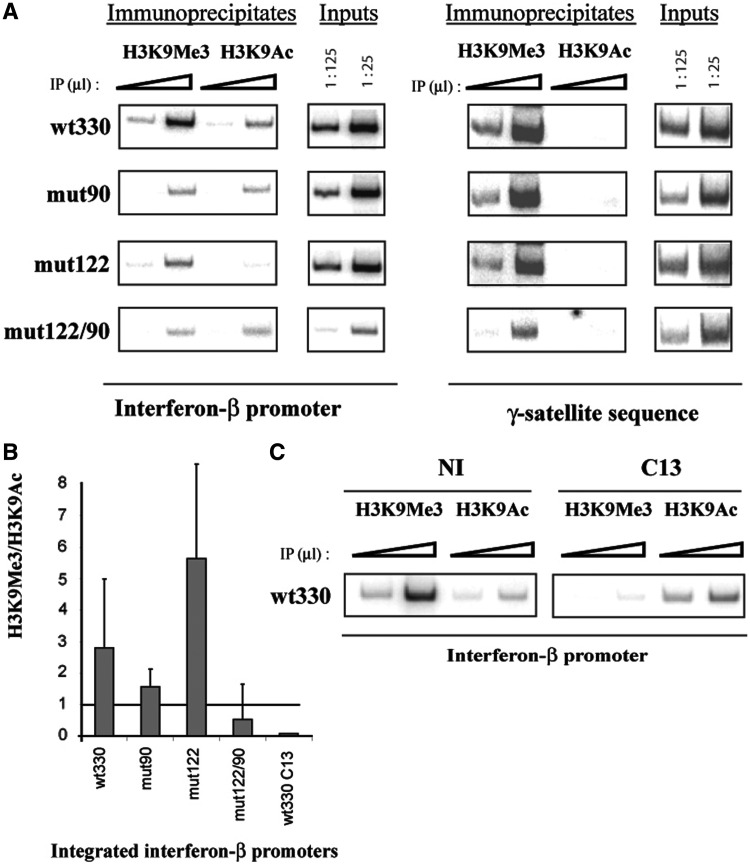
As shown in Figure 4A, H3K9me3 was found present over the IFN-βwt330 promoter under NI conditions, whereas, epigenetic marks predominantly associated with transcriptionally active regions such as the acetylated forms of lysine 8 of histone H4 (H4K8Ac) and of lysine 14 of histone H3 (H3K14Ac), as well as co-activator CBP and transcription factor IRF3, were absent from the silent promoter under NI conditions. The presence of H3K9me3 on the promoter region strongly diminished after NDV-induced promoter activation, contrary to the presence of H4K8Ac, H3K14Ac, CBP and IRF3 that was enhanced. The specificity of the interaction was assessed: no signal was obtained from immunoprecipitates carried out with antibody anti-C23 directed against nucleoline that was used here as an irrelevant antibody.
Figure 4.
Virus infection regulates the rate of H3K9me3 present over the IFN-β promoter region without affecting γ-satellite sequences. (A and B) DNA immunoprecipitated (IP) with the indicated antibodies and inputs corresponding to non-immunoprecipitated genomic DNA, collected from L929wt330 cells either NI or 8 h p.i. by NDV or by non-pathogenic C13 (C13) or pathogenic ZH548 (ZH) RVFV strain was amplified with primers specific for either the IFN-β wt330 promoter or γ-satellite DNA sequences. (C) DNA immunoprecipitated (IP) with anti-H3K9me3 (H3K9me3) and anti-H3K9Ac (H3K9Ac) antibodies and inputs corresponding to non-immunoprecipitated genomic DNA, collected from NI L929 wt330, was amplified with primers specific for either IFN-β promoters or γ-satellite sequences. In panels (A, B and C), the triangles indicate increasing amounts of IPs or inputs.
The effect of virus infection upon the presence of H3K9me3 on γ-satellite sequences as compared to the integrated IFN-βwt330 promoter was analyzed in NI and C13- with respect to ZH-infected cells. As shown in Figure 4B, the transcriptionally silent promoter from either NI or ZH-infected cells appeared associated with H3K9me3, whereas the presence of H3K9me3 on the transcriptionally active promoter strongly diminished in C13-infected cells as previously observed after NDV infection. This diminution was specific of the integrated IFN-βwt330 promoter with respect to γ-satellite sequences since no change was observed when the same immunoprecipitates were amplified with primers specific for γ-satellite sequences (Figure 4B). In order to further confirm the lack of effect of virus infection upon PCH structures, γ-satellite cluster formation and distribution were analyzed before and after virus infection. The total number of γ-satellite clusters present per nuclei as well as the radial distribution of these clusters with respect to the edge or the center of the nucleus remained unchanged during virus infection (data not shown). Overall, these results strongly suggested that virus infection affected the degree of association of the IFN-β promoter with clusters of γ-satellite sequences without affecting the organization and structure of PCH itself.
Interestingly, ChIP experiments carried out with antibodies directed against H3K9me3 as well as against the acetylated form of lysine 9 of histone H3 (H3K9Ac) demonstrated that in NI cells both these epigenetic marks were present on the integrated IFN-βwt330 promoter associated with γ-satellite clusters, whereas as expected only the H3K9me3 mark was present over γ-satellite sequences (Figure 4C). Therefore, even though the integrated IFN-βwt330 promoter colocalized with PCH clusters, it displayed a characteristic combination of epigenetic marks different from that of its proximal surrounding environment.
Virus-induced dissociation of the IFN-β promoter region from γ-satellite clusters preceded strong promoter transcriptional activation
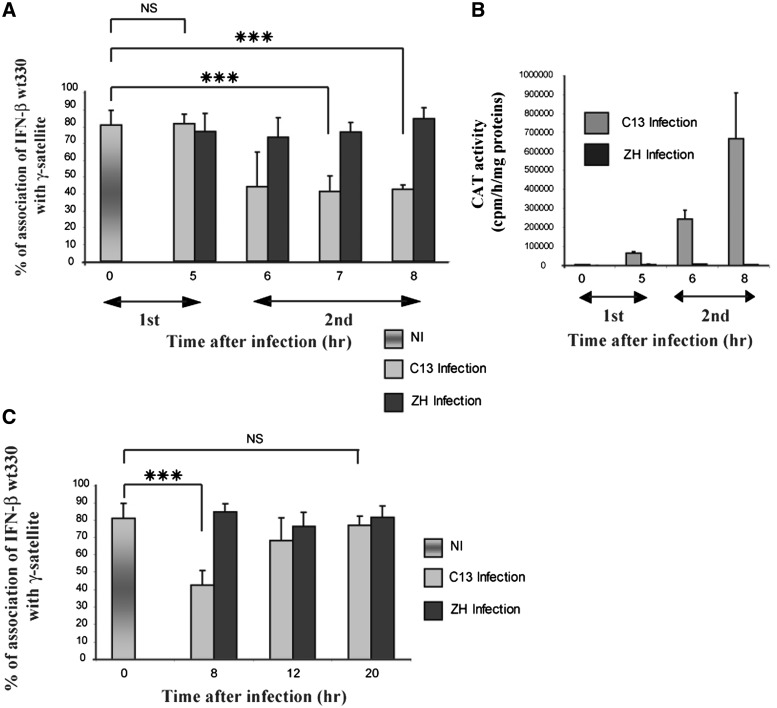
In order to further analyze the correlation between dissociation and transcriptional activation, the percentage of association of the integrated IFN-βwt330 promoter with γ-satellite clusters was analyzed during the time course of infection with either Clone 13 or ZH 548 strains of the RVFV virus. As shown in Figure 5A, the degree of association of the integrated IFN-βwt330 promoter with γ-satellite clusters observed in NI cells, corresponding to around 80% of the FISH signals, remained constant up to 5 h after C13 infection. In contrast, 6 h p.i., at the onset of strong transcriptional activity, the percentage of FISH signals showing association with γ-satellite clusters was strongly reduced to ∼40%. Dissociation and transcriptional activation were not linearly correlated since the highest percentage of dissociation that was reached 6 h p.i. remained constant between 6 and 8 h p.i. in spite of the strong increase of the promoter transcriptional capacity measured between 6 and 8 h p.i. (Figure 5B). Dissociation therefore appeared as a phenomenon occurring between 5 and 6 h p.i. preceding the onset of strong transcriptional activity. Results shown in Figure 5A correspond to the average obtained after counting cells from at least three independent experiments. Statistical analysis using the chi-square test showed that the respective population of cells counted was homogenous for all the time points except for 6 h p.i. The stronger variability observed at this time suggested that 6 h p.i. was probably the time of transition from the state of association to the state of dissociation induced after virus infection.
Figure 5.
Dissociation of the IFN-β promoter from γ-satellite clusters preceded strong virus-induced promoter transcriptional activation and was reversible. (A–C) L929 wt330 cells infected with either non-pathogenic RVFV C13 (C13) strain or pathogenic RVFV ZH548 (ZH) strain were hybridized at different times after infection with γ-satellite and IFN-β wt330 promoter DNA probes as in Figure 2. (A and C) The number of IFN-β wt330 promoter FISH signals colocalizing with γ-satellite sequences was quantified with n > 120 for each condition. (B) CAT activities of cells counted in (A) were measured. (A and B) Time course of the first and second phase of IFN-β transcriptional activation (4,25) are indicated. ***P < 0.001, NS, non-significantly different.
Contrary to the results obtained during infection with Clone 13, the transcriptionally silent IFN-β promoter remained associated with PCH during the complete time course of ZH infection (Figure 5A and B).
Virus-induced dissociation of the IFN-β promoter from clusters of γ-satellite sequences is a reversible phenomenon
Virus-induced IFN-β transcriptional activation is known as a transient, reversible phenomenon. Reversibility is regulated by a post-infection transcriptional turn-off mechanism that is set up 10–12 h after infection that is necessary to bring the IFN-β gene back to its pre-infection silent state (13,36). Therefore, if association with PCH was required for the establishment of IFN-β promoter silencing, the IFN-β promoter virus-induced dissociation from PCH was expected to be a reversible phenomenon. In order to analyze the reversibility of the dissociation of the integrated IFN-βwt330 promoter from γ-satellite clusters, we measured the percentage of IFN-βwt330 FISH signals colocalizing with γ-satellite sequences at 8, 12 and 20 h p.i. with C13 or ZH strains of RVFV with respect to NI cells. As shown in Figure 5C, between 8 and 12 h p.i., the percentage of IFN-βwt330 FISH signals colocalizing with γ-satellite clusters increased from 40% to 70% reaching at 12 h p.i., a percentage of association almost identical to the one observed for the silent integrated IFN-βwt330 promoter in NI cells. All infections were carried out on a population of asynchronously growing L929wt330 cells whose cycling time is of ∼20 h. As observed in Figure 5C, the percentage of association observed 12 h after infection remained constant from 12 to 20 h p.i. indicating that reversibility occurred independently of cell division. Results shown here correspond to the average obtained after counting cells from at least three independent experiments. The strongest variability was observed at 12 h p.i., suggesting that this time was probably the time of transition from the state of virus-induced dissociation to the state of post-infection reassociation.
YY1 binding to the IFN-β promoter regulated the promoter's association with γ-satellite clusters
The IFN-β promoter contains two functional YY1 DNA binding sites present at positions −90 and −122 (18,37) that regulate the promoter transcriptional capacity with a dual repressor/activator role (18). In NI cells, YY1 is predominantly bound to its −90 site. The binding of YY1 to its −122 site is enhanced after NDV or C13 infection but not after ZH infection (19,22). After virus infection, simultaneous binding of YY1 to both its −90 and −122 sites was shown to be essential for the recruitment of an activator complex containing histone acetyltransferase CBP.
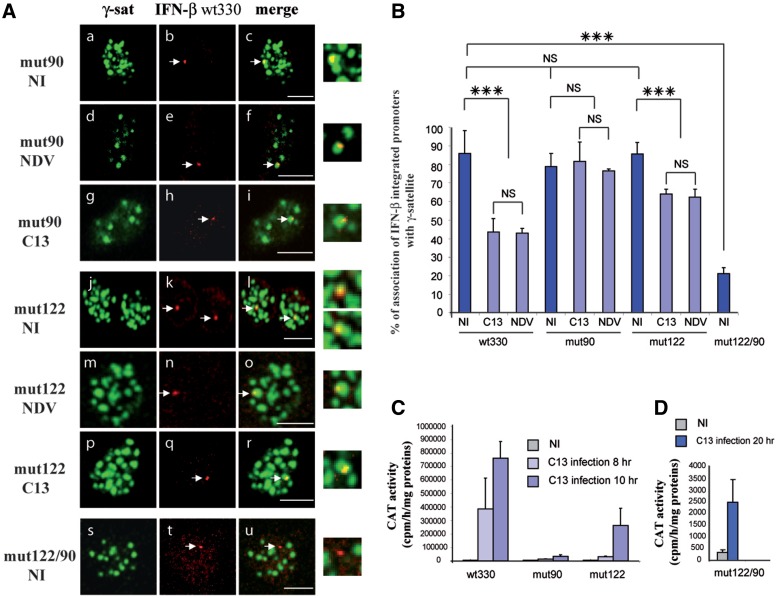
In order to analyze the eventual role of YY1 binding to the IFN-β promoter upon the regulation of the association/dissociation rate of the promoter with/from γ-satellite clusters, we carried out double FISH experiments on previously constructed L929mut90 and L929mut122 cell lines. These cell lines were constructed as the L929wt330 cell line except that the corresponding IFN-β promoters (from −330 to +20) carry a single point mutation either on the YY1 −90 (L929mut90) or −122 (L929mut122) sites (18).
The percentage of the corresponding IFN-β FISH signals associated with γ-satellite clusters was measured before and at 8 h after NDV and C13 infection. Results shown in Figure 6A and B indicated that virus-induced promoter dissociation from γ-satellite clusters was strongly impaired by the introduction of either mutation mut90 or mut122. Contrary to the results obtained with the wild-type wt330 IFN-β promoter, both the mut90 and the mut122 promoter remained predominantly associated with clusters of γ-satellite sequences after NDV as well as C13 infection (Figure 6A and B). The inability of promoter mut90 and mut122 to reposition away from γ-satellite clusters was correlated with the particularly weak transcriptional capacity characteristic of both these promoters (Figure 6C) and (18,19). Interestingly, the mut122 promoter that displayed a slightly, but statistically significant, higher degree of dissociation after infection than the mut90 promoter (Figure 6A and B) also corresponded, as previously shown (18,19), to the promoter displaying a slightly higher virus-induced transcriptional capacity (Figure 6C), indicating once more a correlation between repositioning away from PCH and activation of the promoter transcriptional capacity.
Figure 6.
Binding of transcription factor YY1 to the IFN-β promoter regulated the rate of association and dissociation of the promoter with and from γ-satellite sequences. (A) Confocal sections of murine fibroblastic cells containing the IFN-β promoter carrying a single point mutation in either the YY1 −90 site (mut90) (a–i) or YY1 –122 site (mut122) (j–r) or simultaneous –90 and −122 mutations (mut122/90) (s–u) either NI (a–c, j–l and s–u) or infected by NDV (d–f and m–o) or non-pathogenic RVFV C13 (C13) (g–i and p–r) strain displaying biotin-labeled γ-satellite FISH in green (a, d, g, j, m, p, s) combined with rhodamine-labeled IFN-β promoter (IFN-β) in red (b, e, h, k, n, q and t) with the corresponding merge images in (c, f, i, l, o, r and u). (B) The number of IFN-β promoter FISH signals colocalizing with γ-satellite cluster as displayed in (A) was quantified with n > 60 for each condition. (C and D) the corresponding CAT activities were measured. ***P < 0.001, NS = non-significantly different. Scale Bars = 10 µm.
In NI cells, both the mut90 and mut122 IFN-β promoters displayed a percentage of association with clusters of γ-satellite sequences identical to that determined for the wild-type wt330 promoter (Figure 6A and B). This indicated that contrary to the dissociation process, either one of the two YY1 binding sites was dispensable for the establishment of IFN-β association with γ-satellite clusters. This could be the result of two different situations, either association was totally independent of YY1 binding to the IFN-β promoter or the presence of only one site was sufficient to allow association.
In order to answer this question, we carried out double FISH experiments on the L929mut122/90 cell line that was generated as previous cell lines except that the corresponding IFN-β promoter (from −330 to +20) was simultaneously mutated on both the −90 and −122 YY1 binding sites (18). As shown in Figure 6A and B, only 20% of mut122/90 promoter FISH signals (247 FISH signals counted from 200 cells from three independent experiments) were found associated with γ-satellite clusters in NI cells. As opposed to ∼80% of association observed under the same conditions for the wild-type wt330 or the mut90 and mut122 promoters, indicating that binding of YY1 to the promoter was also necessary for the promoter to associate with γ-satellite clusters in NI cells.
The mut122/90 IFN-β promoter is only weakly activated after virus infection (Figure 6D) and (18). Nevertheless, in NI cells the mut122/90 promoter remained silent and did not reach the transcriptional activity displayed by this promoter after virus infection even though 80% of mut122/90 IFN-β promoter were dissociated from γ-satellite clusters (Figure 6D). Therefore, dissociation from γ-satellite clusters was not by itself sufficient to allow transcriptional activation.
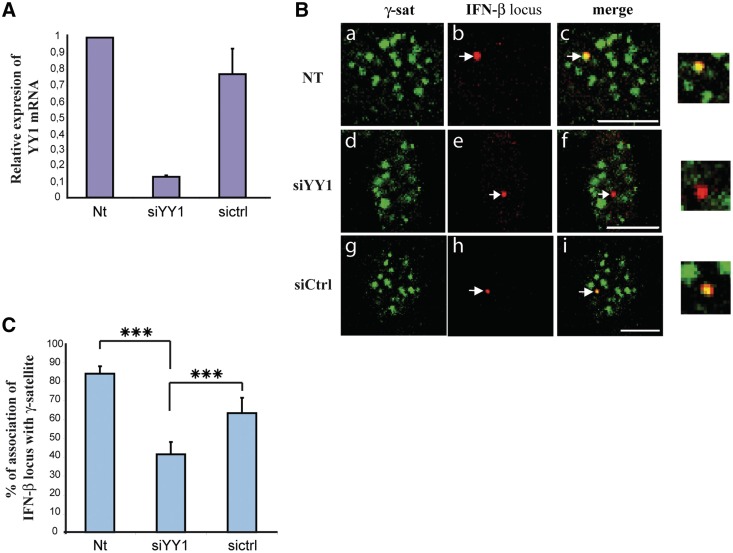
In order to further analyze the role of YY1 in the establishment of the association of the IFN-β locus to PCH, we used RNA interference (RNAi) to suppress YY1 in NI L929wt330 cells (Figure 7A). Diminution of YY1 expression was correlated with a clear diminution of the association of the IFN-β promoter with PCH (Figure 7B and C). From a minimum of 120 cells counted from two independent experiments, the percentage of association dropped from 84.7% in NI and non-treated cells to 42.7% in NI cells treated with siRNA directed against YY1 (siYY1). A slight diminution of YY1 expression was also observed under the same conditions after treatment of the cells with random control siRNAs (Figure 7A). Interestingly, this slight diminution of YY1 expression was also correlated with a slight diminution of the percentage of association of the IFN-β promoter with PCH (Figure 7C) but as expected, the diminution observed after transfection with control siRNAs was significantly less pronounced than the one observed after treatment with siYY1. Overall, these results confirmed the role of YY1 regulating the association of the IFN-β promoter to PCH.
Figure 7.
Transcription factor YY1 regulated the association of IFN-β promoter to clusters of γ-satellite sequences in NI cells. (A) RT-qPCR analysis of the expression of the yy1 gene after transfection of NI L929wt330 cells with 50 nM final concentration of siRNA directed against YY1 (siYY1) or scrambled control siRNAs (siCtrl) respect to non-transfected cells (NT). (B) Confocal sections of NI L929wt330 cells either not transfected (NT) (a to c) or transfected as in A with siRNA directed against yy1 (siYY1) (d–f) or with scrambled control siRNAs (siCtrl) (g–i) displaying biotin-labeled γ-satellite FISH in green (a, d and g) combined with rhodamine-labeled IFN-βwt330 promoter (IFN-βwt330) in red (b, e, h) with the corresponding merge images in (c, f and i). (C) The number of IFN-βwt330 promoter FISH signals colocalizing with γ-satellite cluster as displayed in (B) was quantified from two independent experiments with n > 120 for each condition. ***P < 0.001. Scale bars = 10 µm.
As previously shown in Figure 4D, a particular combination of opposite epigenetic marks, H3K9me3 and H3K9Ac, was detected over the integrated IFN-βwt330 promoter in NI cells. Since simultaneous binding of YY1 to both its sites present on the IFN-β promoter has been shown to regulate CBP promoter recruitment as well as the acetylation of histones positioned over the promoter region (19), we analyzed the effect of YY1 binding upon the presence over the promoter region of H3K9me3 and H3K9Ac (Figure 8). The presence of both these epigenetic marks appeared diminished on YY1-mutated promoters with mutation mut90 predominantly affecting the presence of H3K9me3 and mutation mut122 predominantly affecting the presence of H3K9Ac. Nevertheless, the ratio H3K9me3:H3K9Ac remained >1 in the case of promoters wt330, mut90 and mut122 under NI conditions when these promoters were observed associated with γ-satellite clusters, whereas the H3K9me3:H3K9Ac ratio became <1 in the case of promoters that were dissociated from γ-satellites clusters such as promoter mut122/90 or the wild-type wt330 promoter 8 h after infection with C13. It is therefore tempting to hypothesize that YY1-dependent regulation of the ratio of H3K9me3:H3K9Ac over the IFN-β promoter region could influence the degree of association of this promoter with clusters of PCH.
Figure 8.
YY1 binding and virus infection regulates the rate of H3K9me3/H3K9Ac present over the integrated IFN-β promoters. (A and C) DNA immunoprecipitated (IP) with the indicated antibodies and inputs corresponding to non-immunoprecipitated genomic DNA, collected from (A) NI L929wt330, mut90, mut122, and mut122/90 cells or from (C) L929wt330 cells either NI or 8 h p.i. by non-pathogenic RVFV C13 (C13) strain, was amplified with primers specific for either the integrated IFN-β promoters or γ-satellite DNA sequences. (B) The corresponding rates of H3K9me3/H3K9Ac were calculated after PhosphoImager quantification of PCR products from three independent ChIP experiments equivalent to those shown in (A) and of the ChIP experiment shown in (C).
DISCUSSION
Monoallelic pericentromeric recruitment of the silent IFN-β locus
In this work, we have demonstrated that the silent IFN-β promoter displayed PCH recruitment under non-stimulated conditions adding an additional epigenetic level to the transcriptional regulation of this inducible gene whose silent state needs to be tightly regulated. Reversible dissociation from PCH was observed after virus infection at the onset of strong promoter transcriptional activation in two different IFN-β producing cell lines (fibroblasts and macrophages) during the time course of infections carried out with two different viruses, a bunyavirus and a paramyxovirus.
With respect to previously published data that described considerable cell-to-cell variability of virus-induced IFN-β expression occurring independently of fluctuations in viral load (38), it is interesting to note that our results also show considerable cell-to-cell variability concerning the number of IFN-β spots per cell associated with PCH. Among cells displaying two IFN-β spots, we observed cells displaying either 0/2, 1/2 or 2/2 IFN-β spots associated with PCH (Figures 1E and 2F). The strong promoter transcriptional activation (observed 8 h p.i. in the case of fibroblasts or 5 h p.i. in the case of macrophages) could be the result of transcriptional activation taking place on only a certain population of these cells. Such as cells displaying 0/2 spots colocalizing with PCH for which the percentage was enhanced from 22.1% under NI conditions to 53% 8 h p.i. with C13 in the case of fibroblasts and from 18% to 45.7% 5 h p.i. with C13 in the case of macrophages. This phenomenon was correlated with a strong diminution of cells displaying 2/2 spots colocalizing with PCH that after infection dropped from 25.6% to 7.4% in fibroblasts and from 27.7% to 9.2% in macrophages, whereas a less strong diminution was observed in the case of cells displaying 1/2 IFN-β spots associated with PCH whose percentage only dropped after infection from 52.3% to 39.6% in the case of fibroblasts and from 54.3% to 45% in the case of macrophages.
The association of the endogenous IFN-β locus with PCH appeared as predominantly monoallelic. An observation that is in agreement with previously published results describing allelic IFN-β expression heterogeneity in human as well as in murine cells (38,39). Apostolou and Thanos (39) have described a monoallelic expression of the human IFN-β gene taking place during the first phase of weak transcriptional activation occurring at early times (0–5 h) after infection with monoallelic expression occurring on the allele present within euchromatin associated with Alu sequences rich on NF-kB binding sites. Biallelic IFN-β promoter activation was observed during the second phase of strong expression starting 6 h after infection that coincides with the moment when we observe promoter dissociation from clusters of PCH. Therefore, according to previously published data (38,39) and data obtained in this work, we propose that, in NI cells, one allele would be placed within euchromatin associated with human Alu or murine Alu-like (40,41) sequences rich in NF-kB sites whereas the other allele would be associated with PCH. The allele placed within euchromatin would be transcribed during the first phase of weak transcription that takes place at early times after infection, whereas the allele associated with PCH would dissociate from these repressive regions at later times after infection allowing the onset of the second phase of strong biallelic transcription starting 6 h after infection.
Monoallelic pericentromeric recruitment is a situation that has been previously observed in the case of other genes, as in the case of immunoglobulin loci Igh and Igk (8,30). Even though the mechanism regulating monoallelic PCH recruitment of Igh and Igk genes has not been deciphered yet, it has been described to require allelic pairing (31) similarly to what has been observed during female X chromosome inactivation (42,43). Interestingly, in the case of the endogenous IFN-β gene, 30% of NI cells displayed only one IFN-β spot per cell, a situation that could be translating IFN-β allelic pairing. A requirement for allelic pairing to induce monoallelic PCH recruitment would explain why, contrary to the endogenous IFN-β locus, 80% of the integrated IFN-β loci displayed PCH recruitment.
In olfactory sensory neurons, olfactory receptor (OR) gene expression is also monoallelic and OR allelic repression is also associated with the deposition of an epigenetic signature corresponding to the molecular hallmarks of constitutive heterochromatin that sets up the conditions to allow stochastic selection of the OR gene and allele to be transcribed (44). In the case of immunoglobulin alleles as well as in the case of OR genes, silencing of one allele serves a qualitative function necessary to ensure, respectively, a unique recombination product and the expression of a unique OR per cell, whereas, in the case of the IFN-β gene, monoallelic PCH recruitment would have a quantitative function, necessary to control gene dosage as in the case of imprinted genes or X-linked genes in female cells.
Binding of transcription factor YY1 to the IFN-β promoter regulated promoter PCH recruitment before and after virus infection
Transcription factor YY1 is a highly conserved, ubiquitously expressed transcription factor that regulates the expression of many cellular genes (45); the IFN-β gene is one among them (18,19,37). We show here that YY1 binding to the IFN-β promoter regulated the association/dissociation rate of the IFN-β locus to and from PCH before as well as after virus infection. On the one hand, our results showed that IFN-β virus-induced dissociation from PCH was strongly diminished in the case of IFN-β promoters mutated in one or the other of the two YY1 binding sites. The virus-induced simultaneous occupancy of both YY1 binding sites present over the IFN-β promoter was demonstrated as necessary for the promoter to recruit CBP (19). Creation of a potentially active chromatin environment through YY1-dependent recruitment of CBP could be a prerequisite for repositioning of the IFN-β promoter away from PCH. Similarly to what has been described by Huang et al. (46) in the case of the chicken β-globin locus.
On the other hand, our results showed that under NI conditions, promoters mutated on both YY1 binding sites were unable to associate with γ-satellite clusters in NI cells, indicating that YY1 binding to the IFN-β promoter also played a role establishing promoter association with PCH. Treatment of NI cells with siRNAs directed against YY1 induced significant dissociation of the IFN-β promoter from PCH clusters further confirming a role for YY1 on the establishment of IFN-β promoter association with PCH. Even though transcription factor YY1 has been shown to bind γ-satellite sequences that are rich in YY1 DNA-binding sites (47), recruitment of the IFN-β promoter to γ-satellite clusters could not occur only through YY1 since YY1 does not self-associate. Several members of the repressor SAP30/Sin3A/NCoR/HDAC3 complex that is recruited over the silent IFN-β promoter through YY1 (22) have also been shown to interact with PCH either directly or indirectly (48–50) and could therefore also intervene in anchoring the IFN-β promoter within the environment of γ-satellite clusters.
Results obtained in this work showed that virus-induced dissociation of the IFN-β promoter from PCH was reversible with reversibility occurring between 8 and 12 h p.i. that corresponds to the time period during which IFN-β promoter transcriptional turnoff is established. Even though not much is known concerning the molecular mechanisms governing IFN-β transcriptional turnoff, some previously published data suggest that association with PCH could play a role in this process. Indeed, Gyory et al. (51) have shown that methylation of lysine 9 of histone H3 could be induced on the IFN-β promoter region by histone methyltransferase (HMT) G9a through PRDI-BF1, which is a factor that was shown to participate on the regulation of IFN-β transcriptional turnoff (52). Also, we have shown that YY1 binding to the IFN-β promoter was necessary for the IFN-β transcriptional turnoff to be correctly established (18), a result that has been recently confirmed by Siednienko et al. (53) and in this work we have demonstrated that YY1 regulates the association of the IFN-β promoter to PCH. Nevertheless, even though these results suggest that association with PCH could play a role in establishing IFN-β transcriptional turnoff, more experiments are required to decipher this issue.
Alongside with regulating IFN-β promoter recruitment to PCH, YY1 also appeared regulating the presence of H3K9me3 over the promoter region under NI conditions. Therefore, the role of YY1 establishing PCH recruitment could be related to its capacity to regulate trimethylation of H3K9. Transcription factor YY1 belongs to the polycomb group of proteins that contain HMTs such as Enhancer of zeste 2 (Ezh2) and Suppressor of zeste 12 (Suz12) and even though these HMTs are mainly responsible of inducing trimethylation of H3K27, Suz12 has also been characterized as capable of inducing trimethylation of H3K9 (54).
Altogether, we propose that under NI conditions, YY1 (predominantly bound to its −90 site) and a corepressor complex containing SAP30/NCoR/HDAC3/Sin3a would participate in establishing IFN-β promoter PCH recruitment through their capacity to induce H3K9me3 over the promoter region and interact with PCH, whereas, after virus infection, release of the corepressor complex occurring concomitantly with enhanced YY1 binding to its −122 as well as virus-induced IFN-β enhanceosome formation, CBP recruitment and enhanced acetylation of K9H3 would induce dissociation of the IFN-β promoter from PCH.
As mentioned before, immunoglobulin Igh and Igk genes as well as OR genes are other examples of genes displaying monoallelic expression regulation in somatic differentiated cells. In both these examples, monoallelic repression was linked with the establishment of PCH epigenetic hallmarks over the repressed gene. Interestingly, OR genes have an unusual cluster of YY1 binding sites (55) and YY1 binding as well as YY1-dependent locus rearrangement regulation has been described in the case of the IgH locus (56). We could therefore speculate on a more general role for YY1 in monoallelic gene expression regulation linked with PCH recruitment.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1 and 2.
FUNDING
The Centre National de la Recherche Scientifique, Institut Pasteur (Paris, France) and by grants from Agence Nationale de la Recherche (ANR-08-MIE-022 to E.B. and M.B.). Funding for open access charge: Agence National de la Recherche.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank S. Ronsseray for help with statistical analysis.
REFERENCES
- 1.Fraser P, Bickmore W. Nuclear organization of the genome and the potential for gene regulation. Nature. 2007;447:413–417. doi: 10.1038/nature05916. [DOI] [PubMed] [Google Scholar]
- 2.Schneider R, Grosschedl R. Dynamics and interplay of nuclear architecture, genome organization, and gene expression. Genes Dev. 2007;21:3027–3043. doi: 10.1101/gad.1604607. [DOI] [PubMed] [Google Scholar]
- 3.Zhao R, Bodnar MS, Spector DL. Nuclear neighborhoods and gene expression. Curr. Opin. Genet. Dev. 2009;19:172–179. doi: 10.1016/j.gde.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Francastel C, Magis W, Groudine M. Nuclear relocation of a transactivator subunit precedes target gene activation. Proc. Natl Acad. Sci. USA. 2001;98:12120–12125. doi: 10.1073/pnas.211444898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown KE, Guest SS, Smale ST, Hahm K, Merkenschlager M, Fisher AG. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 1997;91:845–854. doi: 10.1016/s0092-8674(00)80472-9. [DOI] [PubMed] [Google Scholar]
- 6.Grogan JL, Mohrs M, Harmon B, Lacy DA, Sedat JW, Locksley MR. Early transcription and silencing of cytokine genes underlie polarization of T helper cell subsets. Immunity. 2001;14:205–215. doi: 10.1016/s1074-7613(01)00103-0. [DOI] [PubMed] [Google Scholar]
- 7.Guasconi V, Pritchard LL, Fritsch L, Mesner LD, Francastel C, Harel-Bellan A, Ait-Si-Ali S. Preferential association of irreversibly silenced E2F-target genes with pericentromeric heterochromatin in differentiated muscle cells. Epigenetics. 2010;5:704–709. doi: 10.4161/epi.5.8.13025. [DOI] [PubMed] [Google Scholar]
- 8.Fitzsimmons SP, Bernstein RM, Max EE, Skok JA, Shapiro MA. Dynamic changes in accessibility, nuclear positioning, recombination, and transcription at the Igk locus. J. Immunol. 2007;179:5264–5273. doi: 10.4049/jimmunol.179.8.5264. [DOI] [PubMed] [Google Scholar]
- 9.Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu. Rev. Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 10.Stetson DB, Medzhitov R. Type I interferons in host defense. Immunity. 2006;25:373–381. doi: 10.1016/j.immuni.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 11.Dafny N, Yang PB. Interferon and the central nervous system. Eur. J. Pharrmacol. 2005;523:1–15. doi: 10.1016/j.ejphar.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 12.Axtell RC, Steinman L. Type 1 interferons cool the inflamed brain. Immunity. 2008;28:600–602. doi: 10.1016/j.immuni.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 13.Higashi Y. Changes of chromatin conformation around mouse interferon-β gene associated with induction of interferon synthesis. Nucleic Acids Res. 1985;13:5157–5172. doi: 10.1093/nar/13.14.5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maniatis T, Falvo JV, Kim TH, Kim TK, Lin CH, Parekh BS, Wathelet MG. Structure and function of the Interferon-β enhanceosome. Cold Spring Harb. Symp. Quant. Biol. 1998;63:609–620. doi: 10.1101/sqb.1998.63.609. [DOI] [PubMed] [Google Scholar]
- 15.Agalioti T, Lomvardas S, Parekh B, Yie J, Maniatis T, Thanos D. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-β promoter. Cell. 2000;103:667–678. doi: 10.1016/s0092-8674(00)00169-0. [DOI] [PubMed] [Google Scholar]
- 16.Sato M, Taniguchi T, Tanaka T. The interferon system and interferon regulatory factor transcription factors-studies from gene knockout mice. Cytokine Growth Factor Rev. 2001;12:133–142. doi: 10.1016/s1359-6101(00)00032-0. [DOI] [PubMed] [Google Scholar]
- 17.Agalioti T, Chen G, Thanos D. Deciphering the transcriptional histone acetylation code for a human gene. Cell. 2002;111:381–392. doi: 10.1016/s0092-8674(02)01077-2. [DOI] [PubMed] [Google Scholar]
- 18.Weill L, Shestakova E, Bonnefoy E. Transcription factor YY1 binds to the murine interferon-beta promoter and regulates its transcriptional capacity with a dual activator/repressor role. J. Virol. 2003;77:2903–2914. doi: 10.1128/JVI.77.5.2903-2914.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mokrani H, Sharaf el Dein O, Mansuroglu Z, Bonnefoy E. Binding of YY1 to the proximal region of the murine beta interferon promoter is essential to allow CBP recruitment and K8H4/K14H3 acetylation on the promoter region after virus infection. Mol. Cell. Biol. 2006;26:8551–8561. doi: 10.1128/MCB.00420-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parekh BS, Maniatis T. Virus infection leads to localized hyperacetylation of histone H3 and H4 at the IFN-β promoter. Mol. Cell. 1999;3:125–129. doi: 10.1016/s1097-2765(00)80181-1. [DOI] [PubMed] [Google Scholar]
- 21.Shestakova E, Bandu MT, Doly J, Bonnefoy E. Inhibition of histone deacetylation induces constitutive derepression of the beta interferon promoter and confers antiviral activity. J. Virol. 2001;75:3444–3452. doi: 10.1128/JVI.75.7.3444-3452.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Le May N, Mansuroglu Z, Léger P, Josse T, Blot G, Billecocq A, Flick R, Jacob Y, Bonnefoy E, Bouloy M. A SAP30 complex inhibits IFN-beta expression in Rift Valley Fever Virus infected cells. PloS Pathog. 2008;4:e13. doi: 10.1371/journal.ppat.0040013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Billecocq A, Spiegel M, Vialat P, Kohl A, Weber F, Bouloy M, Haller O. NSs protein of Rift Valley fever virus blocks interferon production by inhibiting host gene transcription. J. Virol. 2004;78:9798–9806. doi: 10.1128/JVI.78.18.9798-9806.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bonnefoy E, Bandu MT, Doly J. Specific binding of High-Mobility-Group I (HMGI) protein and histone H1 to the upstream AT-rich region of the murine beta interferon promoter: HMGI protein acts as a potential antirepressor of the promoter. Mol. Cell. Biol. 1999;19:2803–2816. doi: 10.1128/mcb.19.4.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mansuroglu Z, Josse T, Gilleron J, Billecocq A, Leger P, Bouloy M, Bonnefoy E. Nonstructural NSs protein of Rift Valley Fever Virus interacts with pericentromeric DNA sequences of the host cell, inducing chromosme cohesion and segregation defects. J. Virol. 2010;84:928–939. doi: 10.1128/JVI.01165-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lundgren M, Chow CM, Sabbattini P, Georgiou A, Minaee S, Dillon N. Transcription factor dosage affects changes in higher order chromatin structure associated with activation of a heterochromatic gene. Cell. 2000;103:733–743. doi: 10.1016/s0092-8674(00)00177-x. [DOI] [PubMed] [Google Scholar]
- 27.Vodjdani G, Coulombel C, Doly J. Structure and characterization of a murine chromosomal fragment containing the interferon-β gene. J. Mol. Biol. 1988;204:221–231. doi: 10.1016/0022-2836(88)90571-2. [DOI] [PubMed] [Google Scholar]
- 28.Guenatri M, Bailly D, Maison C, Almouzni G. Mouse centric and pericentric satellite repeats form distinct functional heterochromatin. J. Cell. Biol. 2004;166:493–505. doi: 10.1083/jcb.200403109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eymery A, Callanan M, Vourc'h C. The secret message of heterochromatin: new insights into the mechanisms and function of centromeric and pericentric repeat sequence transcription. Int. J. Dev. Biol. 2009;53:259–268. doi: 10.1387/ijdb.082673ae. [DOI] [PubMed] [Google Scholar]
- 30.Hewitt SL, Yin B, Ji Y, Chaumeil J, Marszalek K, Tenthorey J, Salvagiotto G, Steinel N, Ramsey LB, Ghysdael J, et al. RAG-1 and ATM coordinate monoallelic recombination and nuclear positioning of immunoglobulin loci. Nat. Immunol. 2009;10:655–664. doi: 10.1038/ni.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brandt VL, Hewitt SL, Skok JA. It takes two: communication between homologous alleles preserves genomic stability during V(D)J recombination. Nucleus. 2010;1:23–29. doi: 10.4161/nucl.1.1.10595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lomonte P, Merency E. Centromeric protein CENP-B proteosomal degradation induced by the viral protein ICP0. FEBS Lett. 2007;581:658–662. doi: 10.1016/j.febslet.2007.01.027. [DOI] [PubMed] [Google Scholar]
- 33.Richards EJ, Elgin SCR. Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell. 2002;108:489–500. doi: 10.1016/s0092-8674(02)00644-x. [DOI] [PubMed] [Google Scholar]
- 34.Peters AH, Kubicek S, Mechtler K, O'Sullivan RJ, Derijck AA, Perez-Burgos L, Kohlmaier A, Opravil S, Tachibana M, Shinkai Y, et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol. Cell. 2003;12:1577–1589. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- 35.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 36.Whittemore LA, Maniatis T. Postinduction turnoff of beta-interferon gene expression. Mol. Cell. Biol. 1990;10:1329–1337. doi: 10.1128/mcb.10.4.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klar M, Bode J. Enhanceosome formation over the beta interferon promoter underlies a remote-control mechanism mediated by YY1 and YY2. Mol. Cell. Biol. 2005;25:10159–10170. doi: 10.1128/MCB.25.22.10159-10170.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu J, Sealfon SC, Hayot F, Jayaprakash C, Kumar M, Pendleton AC, Ganee A, Fernandez-Sesma A, Moran TM, Wetmur JG. Chromosome-specific and noisy IFNB1 transcription in individual virus-infected human primary dendritic cells. Nucleic Acids Res. 2007;35:5232–5241. doi: 10.1093/nar/gkm557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Apostolou E, Thanos D. Virus infection induces NF-κB dependent interchromosomal associations mediating monoallelic IFN-β gene expression. Cell. 2008;134:85–96. doi: 10.1016/j.cell.2008.05.052. [DOI] [PubMed] [Google Scholar]
- 40.Mighell AJ, Markham AF, Robinson PA. Alu sequences. FEBS Lett. 1997;417:1–5. doi: 10.1016/s0014-5793(97)01259-3. [DOI] [PubMed] [Google Scholar]
- 41.Liang R, Igarashi H, Tsuzuki T, Nakabeppu Y, Sekiguchi M, Kasprzak KS, Shiao YH. Presence of potential nickel-responsive element(s) in the mouse MTH1 promoter. Ann. Clin. Lab. Sci. 2001;31:91–98. [PubMed] [Google Scholar]
- 42.Xu N, Tsai CL, Lee JT. Transient homologous chromosome pairing marks the onset of X inactivation. Science. 2006;311:1149–1152. doi: 10.1126/science.1122984. [DOI] [PubMed] [Google Scholar]
- 43.Bacher CP, Guggiari M, Brors B, Augui S, Clerc P, Avner P, Eils R, Heard E. Transient colocalization of X-inactivation centres accompanies the initiation of X inactivation. Nat. Cell Biol. 2006;8:293–299. doi: 10.1038/ncb1365. [DOI] [PubMed] [Google Scholar]
- 44.Magklara A, Yen A, Colquitt BM, Clowney EJ, Allen W, Markenscoff-Papadimitriou E, Evans ZA, Kheradpour P, Mountoufaris G, Carey C, et al. An epigenetic signature for monoallelic olfactory receptor expression. Cell. 2011;145:555–570. doi: 10.1016/j.cell.2011.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Affar elB, Gay F, Shi Y, Liu H, Huarte M, Wu S, Collins T, Li E, Shi Y. Essential dosage-dependent functions of the transcription factor Yin Yang 1 in late embryonic development and cell cycle progression. Mol. Cell. Biol. 2006;26:3565–3581. doi: 10.1128/MCB.26.9.3565-3581.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang S, Li X, Yusufzai TM, Qiu Y, Felsenfeld G. USF1 recruits histone modification complexes and is critical for maintenance of a chromatin barrier. Mol. Cell. Biol. 2007;27:7991–8002. doi: 10.1128/MCB.01326-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shestakova E, Mansuroglu Z, Mokrani H, Ghinea N, Bonnefoy E. Transcription factor YY1 associates with pericentromeric γ-satellite DNA in cycling but not in quiescent (G0) cells. Nucleic Acids Res. 2004;32:4390–4399. doi: 10.1093/nar/gkh737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Underhill C, Qutob MS, Yee SP, Torchia J. A novel nuclear receptor corepressor complex, N-CoR, contains components of the mammalian SWI/SNF complex and the corepressor KAP-1. J. Biol. Chem. 2000;275:40463–40470. doi: 10.1074/jbc.M007864200. [DOI] [PubMed] [Google Scholar]
- 49.David G, Turner GM, Yao Y, Protopopov A, DePinho RA. mSin3-associated protein, mSds3, is essential for pericentric heterochromatin formation and chromosome segregation in mammalian cells. Genes Dev. 2003;17:2396–2405. doi: 10.1101/gad.1109403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vaute O, Nicolas E, Vandel L, Trouche D. Functional and physical interaction between the histone methyl transferase Suv39H1 and histone deacetylases. Nucleic Acids Res. 2002;30:475–481. doi: 10.1093/nar/30.2.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gyory I, Wu J, Fejér G, Seto E, Wright KI. PRDI-BF1 recruits the histone H3 methyltransferase G9a in transcriptional silencing. Nat. Immunol. 2004;5:299–308. doi: 10.1038/ni1046. [DOI] [PubMed] [Google Scholar]
- 52.Keller AD, Maniatis T. Identification and characterization of a novel repressor of β-interferon gene expression. Genes Dev. 1991;5:868–879. doi: 10.1101/gad.5.5.868. [DOI] [PubMed] [Google Scholar]
- 53.Siednienko J, Maratha A, Yang S, Mitkiewicz M, Miggin SM, Moynagh PN. The NF-κB subunits RelB and cRel negatively regulate TLR3-mediated IFN-β production via induction of the transcriptional repressor protein YY1. J. Biol. Chem. 2011;286:44750–44763. doi: 10.1074/jbc.M111.250894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de la Cruz C, Kirmizis A, Simon MD, Isono K, Koseki H, Panning B. The Polycomb Group Protein SUZ12 regulates histone H3 lysine 9 methylation and HP1a distribution. Chromosome Res. 2007;15:299–314. doi: 10.1007/s10577-007-1126-1. [DOI] [PubMed] [Google Scholar]
- 55.Faulk CD, Kim J. YY1's DNA-binding motifs in mammalian olfactory receptor genes. BMC Genomics. 2009;10:576. doi: 10.1186/1471-2164-10-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu H, Schmidt-Supprian M, Shi Y, Hobeika E, Barteneva N, Jumaa H, Pelanda R, Reth M, Skok J, Rajewsky K, et al. Yin Yang 1 is a critical regulator of B-cell development. Genes Dev. 2007;21:1179–1189. doi: 10.1101/gad.1529307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bolte S, Cordelières FP. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.