Abstract
FANCI and FANCD2 form a complex, and play essential roles in the repair of interstrand DNA crosslinks (ICLs) by the Fanconi anemia (FA) pathway. FANCD2 is monoubiquitylated by the FA core complex, composed of 10 FA proteins including FANCL as the catalytic E3 subunit. FANCD2 monoubiquitylation can be reconstituted with purified minimal components, such as FANCI, E1, UBE2T (E2) and FANCL (E3) in vitro; however, its efficiency is quite low as compared to the in vivo monoubiquitylation of FANCD2. In this study, we found that various forms of DNA, such as single-stranded, double-stranded and branched DNA, robustly stimulated the FANCD2 monoubiquitylation in vitro up to a level comparable to its in vivo monoubiquitylation. This stimulation of the FANCD2 monoubiquitylation strictly required FANCI, suggesting that FANCD2 monoubiquitylation may occur in the FANCI–FANCD2 complex. A FANCI mutant that was defective in DNA binding was also significantly defective in FANCD2 monoubiquitylation in vitro. In the presence of 5′ flapped DNA, a DNA substrate mimicking the arrested replication fork, about 70% of the input FANCD2 was monoubiquitylated, while less than 1% FANCD2 monoubiquitylation was observed in the absence of the DNA. Therefore, DNA may be the unidentified factor required for proper FANCD2 monoubiquitylation.
INTRODUCTION
Fanconi anemia (FA) is an autosomal recessive disorder, and 15 FA genes, FANCA, –B, –C, –D1 (BRCA2), –D2, –E, –F, –G, –I, –J (BRIP1), –L, –M, –N (PALB2), –O (RAD51C) and –P (SLX4), have been identified (1–3). In FA patients, mutations in these FA genes cause genomic instability with cancer susceptibility, progressive bone marrow failure and multiple developmental defects (1–4). These FA gene products are considered to function in a common DNA damage repair pathway, the ‘FA pathway’.
In the FA pathway, eight proteins, FANCA, FANCB, FANCC, FANCE, FANCF, FANCG, FANCL and FANCM, and two FANCA-associated polypeptides (FAAPs) form the FA core complex, and FANCI and FANCD2 form the ID complex (5–8). When cells are exposed to DNA crosslinking agents, the ID complex promptly becomes monoubiquitylated (6–9). This monoubiquitylation of the ID complex, especially for FANCD2, is a central process in the FA pathway, and the FA core complex is responsible for it, as the E3 ubiquitin ligase complex (5,9–11). FANCL was identified as the catalytic E3 subunit for the FANCD2 monoubiquitylation by the FA core complex (12). On the other hand, UBE2T, which is required for cellular resistance to interstrand DNA crosslinks (ICLs), has been shown to interact with FANCL, and functions as the E2 ubiquitin-conjugating enzyme required for the FANCL-dependent FANCD2 monoubiquitylation (13). An in vitro study showed that E1, UBE2T, and FANCL are sufficient for FANCD2 monoubiquitylation (14). However, the efficiency of FANCD2 monoubiquitylation is extremely low, as compared to that of FANCD2 monoubiquitylation during ICL repair in vivo (9). Therefore, an essential factor(s) required for mediating FANCD2 monoubiquitylation is missing in the in vitro FANCD2 monoubiquitylation assay.
In the present study, we prepared the ID complex with recombinant FANCI and FANCD2, and performed in vitro monoubiquitylation assays with recombinant E1, UBE2T (E2) and FANCL (E3). We found that FANCD2 monoubiquitylation in the ID complex was robustly stimulated by DNA, such as single-stranded, double-stranded and branched DNAs, up to the level comparable to the in vivo monoubiquitylation of FANCD2.
MATERIALS AND METHODS
Purification of chicken FANCD2, FANCI, FANCL and human UBE2T
The DNA fragments encoding chicken FANCD2 and FANCI were ligated into the NdeI-BamHI and NdeI-XhoI sites of the pET-15 b vector, respectively, and the proteins were overexpressed in the Escherichia coli BL21(DE3) strain, which also carried an expression vector for the minor tRNAs [Codon(+)RIL, Stratagene]. The cells were cultured in LB medium supplemented with ampicillin (100 µg/ml) and chloramphenicol (35 µg/ml) at 30°C to an OD600 = 0.6, and then protein production was induced by adding 0.5 mM Isopropyl β-D-1-thiogalactopyranoside and culturing the cells overnight at 18°C. The cells were collected, resuspended in buffer A containing 50 mM Tris–HCl (pH 8.0), 10% glycerol, 0.5 M NaCl, 1 mM PMSF, 12 mM imidazole and 5 mM 2-mercaptoethanol, and disrupted by sonication. After disruption, the supernatant was separated from the cell debris by centrifugation (27 200g) at 4°C for 20 min, and was mixed gently with nickel-nitrilotriacetic acid (Ni-NTA) agarose resin (3 ml; Qiagen) at 4°C for 1 h. Later, the Ni-NTA beads were packed into an Econo-Column (Bio-Rad), and were washed with 150 ml buffer A. His6-tagged FANCD2 or FANCI was eluted with a 60 ml linear gradient of 12 to 400 mM imidazole in buffer A. Since the His6-tag may affect the DNA-binding property of the proteins, the His6-tag was removed by digestion with thrombin protease (GE Healthcare; 3 U/mg protein for FANCD2, 20 U/mg protein for FANCI) during dialysis against 4 L of buffer B, containing 20 mM Tris–HCl (pH 8.0), 10% glycerol, 5 mM 2-mercaptoethanol and 0.2 M NaCl. After removal of the His6-tag, the protein solutions were then loaded onto a Heparin Sepharose CL-6B column (3 ml; GE Healthcare) equilibrated with buffer B. The column was washed with 150 ml of buffer B containing 280 mM NaCl, and the proteins were eluted with a 60 ml linear gradient of 280 to 1000 mM NaCl in buffer B. Peak fractions were collected, concentrated and then applied to a Superdex 200 gel filtration column (HiLoad 26/60 preparation grade; GE Healthcare) equilibrated with buffer B. Purified proteins were concentrated to 5 mg/ml, and were stored at −80°C. All of the FANCI and FANCD2 mutants, except for FANCI Ex6, were purified by the same method as that used for the wild-type FANCI or FANCD2 protein. For the FANCI Ex6 mutant purification, Q Sepharose Fast Flow column chromatography (3 ml; GE Healthcare) was employed, instead of Heparin Sepharose column chromatography. The column was washed with 100 ml of buffer B containing 215 mM NaCl, and the proteins were eluted with a 60 mL linear gradient of 215–450 mM NaCl in buffer B. The subsequent purification procedure for FANCI Ex6 was the same as that for the wild-type FANCI.
The DNA fragments encoding chicken FANCL and human UBE2T were each ligated separately into the EcoRI-XhoI sites of the pGEX6P-1 vector, and the proteins were expressed by the same methods as for chicken FANCD2 and FANCI. The cells producing FANCL or UBE2T were collected, resuspended in buffer C, containing 50 mM Tris–HCl (pH 8.0), 10% glycerol, 0.5 M NaCl, 1 mM PMSF, 1 mM EDTA, 0.1% Nonidet P-40 and 5 mM 2-mercaptoethanol, and disrupted by sonication. After disruption, the supernatant was separated from the cell debris by centrifugation for 30 min, and was then mixed gently with Glutathione Sepharose 4B beads (3 ml; GE Healthcare) at 4°C for 2 h. The Glutathione Sepharose 4B beads were packed into an Econo-Column, and were washed with 150 ml of buffer C containing 1 M NaCl. The GST-tagged FANCL or UBE2T was eluted with 50 ml of buffer C containing 20 mM reduced glutathione. Peak fractions were collected, concentrated and then applied to a Superdex 200 gel filtration column equilibrated with buffer B containing 150 mM NaCl. Purified GST-FANCL was concentrated to 3 mg/ml, and was stored at −80°C. The GST-tag of the GST-UBE2T was removed by an overnight treatment with PreScission protease (GE Healthcare; 1.5 U/mg protein) at 4°C. After the GST-tag removal, the protein solution was mixed gently with Glutathione Sepharose 4B at 4°C for 3 h. The flow-through fractions containing purified UBE2T were collected, concentrated to 3 mg/ml, and stored at −80°C. The protein concentration was determined by the Bradford method (15), using bovine serum albumin as the standard protein.
DNA substrates
49-mer single-stranded oligonucleotides, 1, 2, 3 and 4, (16–18) with the sequences 5′-ATCGA TGTCT CTAGA CAGCT GCTCA GGATT GATCT GTAAT GGCCT GGGA-3′, 5′-GTCCC AGGCC ATTAC AGATC AATCC TGAGC ATGTT TACCA AGCGC ATTG-3′, 5′-TGATC ACTTG CTAGC GTCGC AATCC TGAGC AGCTG TCTAG AGACA TCGA-3′ and 5′-CCAAT GCGCT TGGTA AACAT GCTCA GGATT GCGAC GCTAG CAAGT GATC-3′, respectively, were used to prepare the synthetic Holliday junction (HJ) (1, 2, 3 and 4) by annealing. The splayed arm DNA was prepared by annealing oligonucleotides 1 and 2. The dsDNA was prepared by annealing oligonucleotide 1 with its complementary oligonucleotide. The 5′ flapped DNA was prepared by annealing 5′-CAATGCGCTTGGTAAACA-3′ to the 3′ ssDNA region of the splayed arm DNA. The static fork DNA was prepared by annealing 5′-GCTGTCTAGAGACATCGAT-3′ to the 5′ ssDNA region of the 5′ flapped DNA. All of the oligonucleotides were purified by HPLC, and the DNA concentrations are expressed in moles of nucleotides.
DNA binding assay
The synthetic HJ (4.5 µM), splayed arm DNA (4.5 µM), and dsDNA (4.5 µM) were mixed with 0.05–0.20 µM of the ID complex in 10 µL of reaction buffer, containing 20 mM Tris–HCl (pH 8.0), 70 mM NaCl, 2% glycerol, 0.3 mM MgCl2, 5 mM dithiothreitol and 10 µg/ml bovine serum albumin. For the experiments with three-way branched DNAs, splayed arm DNA (4.5 µM), 5′ flapped DNA (4.5 µM) and static fork DNA (4.5 µM) were mixed with 0.05–0.20 µM of the ID complex in 10 µl of reaction buffer. The samples were incubated at 37°C for 15 min, and were then analyzed by 8.0% or 3.5% PAGE in TBE (18 mM Tris-borate and 0.4 mM EDTA) buffer. DNAs were visualized by SYBR Gold (Invitrogen) staining.
In vitro ubiquitylation assay
The purified FANCD2 protein (1 μM) or the ID complex (1 μM) was mixed with UBE2T (2–8 μM), GST–FANCL (2 μM), human recombinant E1 (75 nM) (Boston Biochem) and HA-tagged ubiquitin (10 μM) (Boston Biochem), in the presence or absence of DNA substrates, in 20 μL of reaction buffer, containing 0.5 mM DTT, 2 mM ATP, 2 mM MgCl2, 50 mM Tris–HCl (pH 7.5), 3.6% glycerol and 64 mM NaCl. The reactions were incubated for 90 min at 30°C, and were then stopped by the addition of 2% sodium dodecyl sulphate. The reaction products were separated by 7% sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE), and were transferred to a polyvinylidene fluoride membrane. Ubiquitylated proteins were detected by an anti-HA antibody (F-7; Santa Cruz Biotechnology, Inc.), and protein bands were visualized by Coomassie Brilliant Blue staining.
RESULTS
In vitro monoubiquitylation of the FANCI–FANCD2 complex
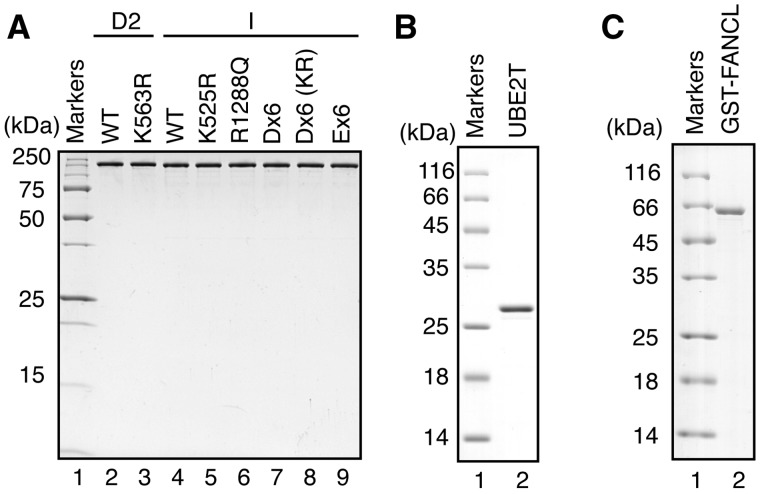
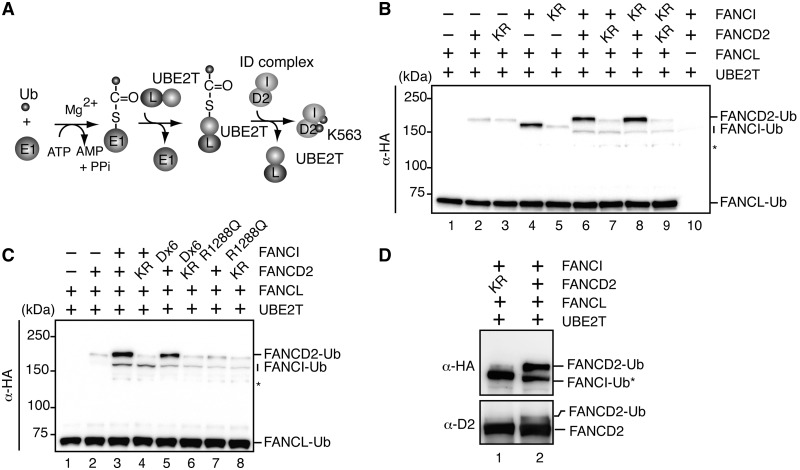
Chicken FANCI and FANCD2 were purified as recombinant proteins (Figure 1A, lanes 2 and 4), and the ID complex was prepared by mixing them in 1:1 stoichiometry. We also purified UBE2T and FANCL, which are known as the E2 and E3 proteins for FANCD2 monoubiquitylation, respectively, as recombinant proteins (Figure 1B and C). We then performed the in vitro monoubiquitylation assay (14). The ID complex was incubated with E1, UBE2T and FANCL in the presence of HA-tagged ubiquitin (Figure 2A), and the ubiquitylated proteins were detected by a western blotting analysis with an anti-HA antibody (Figure 2B).
Figure 1.
Purification of chicken FANCD2, FANCI, FANCL and human UBE2T. (A) Purified chicken FANCD2 and FANCI, used for in vitro assays, were analyzed by 15% SDS-PAGE with Coomassie Brilliant Blue staining. Lane 1 indicates the molecular mass markers. Lanes 2 and 3 indicate purified FANCD2 and FANCD2 K563R, respectively. Lanes 4–9 indicate purified FANCI, FANCI K525R, FANCI R1288Q, FANCI Dx6, FANCI Dx6 K525R and FANCI Ex6, respectively. (B) Purified human UBE2T, used for in vitro monoubiquitylation assays, was analyzed by 15% SDS-PAGE with Coomassie Brilliant Blue staining. Lane 1 indicates the molecular mass markers. Lane 2 indicates purified UBE2T. (C) Purified GST-tagged chicken FANCL, used for in vitro monoubiquitylation assays, was analyzed by 15% SDS-PAGE with Coomassie Brilliant Blue staining. Lane 1 indicates the molecular mass markers. Lane 2 indicates purified GST-tagged FANCL.
Figure 2.
In vitro monoubiquitylation of FANCD2. (A) Schematic diagram of the monoubiquitylation assay. The monoubiquitylation assay was performed by incubating FANCD2 (D2) or FANCI (I)-FANCD2 with E1, UBE2T and FANCL (L) in the presence of ATP and HA-tagged ubiquitin (Ub). (B) Ubiquitylated proteins were separated by 7% SDS-PAGE, and were detected by western blotting with an anti-HA monoclonal antibody (α-HA). Lanes 1 and 2 indicate control experiments without FANCI-FANCD2 and FANCI, respectively. Lane 3 indicates an experiment with FANCD2 K563R in the absence of FANCI. Lane 4 indicates an experiment with FANCI in the absence of FANCD2. Lane 5 indicates an experiment with FANCI K525R in the absence of FANCD2. Lane 6 indicates an experiment in the presence of the complete set of proteins. Lane 7 indicates an experiment with FANCD2 K563R, in the presence of FANCI. Lane 8 indicates an experiment with FANCI K525R, in the presence of FANCD2. Lane 9 indicates an experiment with FANCD2 K563R and FANCI K525R. Lane 10 indicates an experiment with FANCD2 and FANCI, without FANCL. Asterisk indicates the degradation product of monoubiquitylated FANCI. (C) Experiments were performed as in panel (B). Lanes 1 and 2 indicate control experiments without FANCI-FANCD2 and FANCI, respectively. Lane 3 indicates an experiment in the presence of the complete set of proteins. Lane 4 indicates an experiment with FANCD2 K563R, in the presence of FANCI. Lanes 5 and 6 indicate experiments with FANCI Dx6, in the presence of FANCD2 and FANCD2 K563R, respectively. Lanes 7 and 8 indicate experiments with FANCI R1288Q, in the presence of FANCD2 and FANCD2 K563R, respectively. Asterisk indicates the degradation product of monoubiquitylated FANCI. (D) Experiments were performed as in panel (B). Enlarged images of the monoubiquitylated FANCD2 band detected by α-HA (upper panel) and an anti-chicken FANCD2 polyclonal antibody (lower panel). FANCI-Ub* indicates non-specific monoubiquitylation of FANCI. Lane 1 indicates a negative control experiment in the presence of FANCD2 K563R. Lane 2 indicates an experiment in the presence of the complete set of proteins.
As shown in Figure 2B, FANCD2 was monoubiquitylated in this assay (lane 6). The FANCD2 K563R mutant, which is defective in monoubiquitylation in vivo (9), was also defective in this monoubiquitylation assay (Figure 2B, lane 7), suggesting that the FANCD2 monoubiquitylation properly occurred in this in vitro system. The FANCD2 monoubiquitylation may predominantly occur in the ID complex, because only a background level of FANCD2 monoubiquitylation was observed in the absence of FANCI (Figure 2B, lanes 2 and 3). Consistently, FANCD2 monoubiquitylation was not stimulated when the chicken FANCI R1288Q mutant, corresponding to the human FANCI R1285Q mutant, which is reportedly defective in both FANCD2 and DNA binding (19), was added instead of FANCI (Figure 2C, lane 7). Interestingly, the FANCI K525R mutant, which is defective in FANCI monoubiquitylation in vivo, was proficient in stimulating FANCD2 monoubiquitylation in vitro (Figure 2B, lane 8). In our in vitro system, FANCI monoubiquitylation was weakly observed in the presence of FANCD2 (Figure 2B, lane 6). However, the weak FANCI monoubiquitylation observed in the ID complex may not properly occur on the Lys525 residue of FANCI, for the following reasons. First, this weak monoubiquitylation was also observed with the FANCI K525R mutant (Figure 2B, lanes 8 and 9). Second, the FANCI monoubiquitylation occurred on the Lys525 residue without FANCD2 (Figure 2B, lane 4), as previously reported (20), but the weakly monoubiquitylated FANCI in the ID complex migrated slightly faster than the Lys525-monoubiquitylated FANCI (Figure 2B, lane 4 versus lane 6).
DNAs robustly stimulate FANCD2 monoubiquitylation
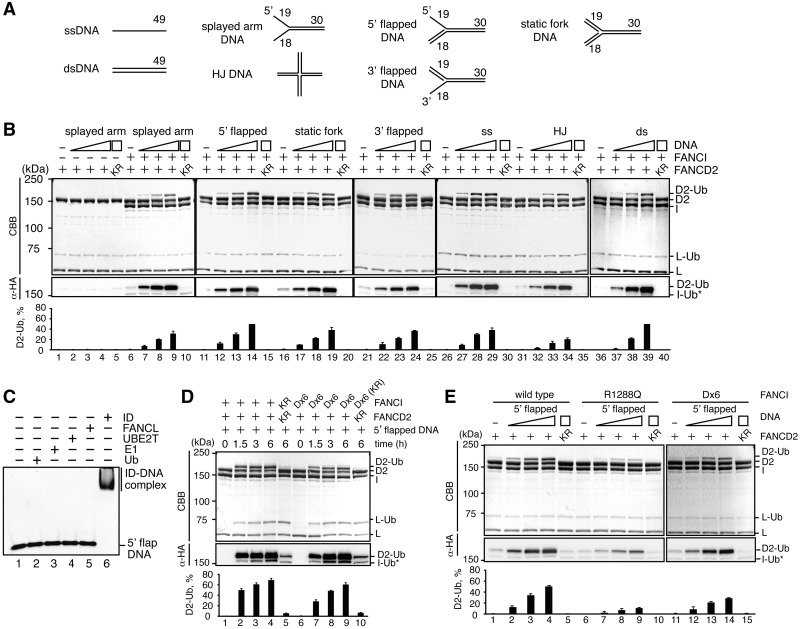
Upon the induction of DNA crosslinking damage, 40–70% of FANCD2 was reportedly monoubiquitylated in cells (11,21). However, we observed less than 1% monoubiquitylation of the input FANCD2 in vitro (Figure 2D). This indicated that an essential component required for the proper monoubiquitylation of FANCD2 was missing in this in vitro system. The FANCD2 monoubiquitylation site is reportedly buried in the binding interface with FANCI in the ID complex (22). Interestingly, the putative DNA-binding path in the ID complex is located near the FANCD2 monoubiquitylation site (22). This suggested that DNA binding by the ID complex may induce a conformational change of the ID complex, and may expose the FANCD2 monoubiquitylation site on an accessible surface.
We therefore tested whether the DNA affected the FANCD2 monoubiquitylation in the ID complex in vitro. We tested seven DNA structures, including splayed arm DNA, 5′ flapped DNA, 3′ flapped DNA, static fork DNA, HJ DNA, ssDNA and dsDNA, as substrates (Figure 3A). To our surprise, we found that the FANCD2 monoubiquitylation in the ID complex was robustly enhanced in the presence of these DNAs (Figure 3B). This FANCD2 monoubiquitylation stimulation by DNA was not observed in the absence of FANCI (Figure 3B, lanes 1–4) or in the presence of the FANCD2 K563R mutant, instead of FANCD2 (Figure 3B, lanes 5, 10, 15, 20, 25, 30, 35 and 40). The ID complex bound to the 5′ flapped DNA, but E1, UBE2T and FANCL did not (Figure 3C). Therefore, we conclude that DNA binding by the ID complex is required for the robust stimulation of the FANCD2 monoubiquitylation. Since about 70% of FANCD2 was monoubiquitylated after a 6 h reaction in the presence of FANCI and 5′ flapped DNA, the FANCD2 monoubiquitylation can be promoted to the level observed in cells with DNA and FANCI in vitro (Figure 3D).
Figure 3.
DNA stimulates FANCD2 monoubiquitylation in vitro. (A) Schematic representation of the DNA substrates. (B) FANCD2 (lanes 1–4) or FANCI–FANCD2 (lanes 6–9, 11–14, 16–19, 21–24, 26–29, 31–34 and 36–39) was incubated with E1, UBE2T and FANCL in the presence of ATP and HA-tagged ubiquitin. The reactions were performed in the presence of splayed arm DNA (lanes 2–5 and 6–10), 5′ flapped DNA (lanes 12–15), static fork DNA (lanes 17–20), 3′ flapped DNA (lanes 22–25), ssDNA (lanes 27–30), HJ DNA (lanes 32–35), and dsDNA (lanes 37–40). The DNA concentrations were 0 µM (lanes 1, 6, 11, 16, 21, 26, 31 and 36), 5 µM (lanes 2, 7, 12, 17, 22, 27, 32 and 37), 20 µM (lanes 3, 8, 13, 18, 23, 28, 33 and 38), and 50 µM (lanes 4, 5, 9, 10, 14, 15, 19, 20, 24, 25, 29, 30, 34, 35, 39 and 40). Lanes 5, 10, 15, 20, 25, 30, 35 and 40 indicate negative control experiments with FANCD2 K563R. Proteins were separated by 7% SDS-PAGE, blotted onto a membrane, and detected by Coomassie Brilliant Blue staining (upper panel) or α-HA antibody staining (middle panel). The amounts of monoubiquitylated FANCD2 were estimated, and the averages of three independent experiments are indicated as black bars with the standard deviation values (lower panel). (C) Gel shift assay. 5′ flapped DNA (5 µM) was incubated with ubiquitin (10 µM) (lane 2), E1 (75 nM) (lane 3), UBE2T (2 µM) (lane 4), GST-FANCL (2 µM) (lane 5), or FANCI-FANCD2 (1 µM) (lane 6) at 30°C for 15 min. Samples were analyzed by 3.5% PAGE in 0.2 × TBE buffer (18 mM Tris base, 18 mM boric acid, and 0.4 mM EDTA) with SYBR Gold staining. (D) Time course experiments of the FANCD2 monoubiquitylation. Experiments were performed as in panel (B). FANCI–FANCD2 (lanes 1–4) or FANCI Dx6–FANCD2 (lanes 6–9) was incubated with 50 µM 5′ flapped DNA. Reaction times were 0 h (lanes 1 and 6), 1.5 h (lanes 2 and 7), 3 h (lanes 3 and 8) and 6 h (lanes 4, 5, 9 and 10). Lanes 5 and 10 indicate negative control experiments with FANCD2 K563R and FANCI K525R or FANCI Dx6 K525R, respectively. The results are presented as in panel B. (E) The stimulation of FANCD2 monoubiquitylation by FANCI R1288Q and FANCI Dx6. Experiments were performed as in panel (B), with 5′ flapped DNA and FANCI R1288Q (lanes 6–10) or FANCI Dx6 (lanes 11–15), instead of FANCI (lanes 1–5). The DNA concentrations were 0 µM (lanes 1, 6 and 11), 5 µM (lanes 2, 7 and 12), 20 µM (lanes 3, 8 and 13), and 50 µM (lanes 4, 5, 9, 10, 14 and 15). Lanes 5, 10 and 15 indicate negative control experiments with FANCD2 K563R.
The FANCI R1288Q mutant is defective in stimulating FANCD2 monoubiquitylation
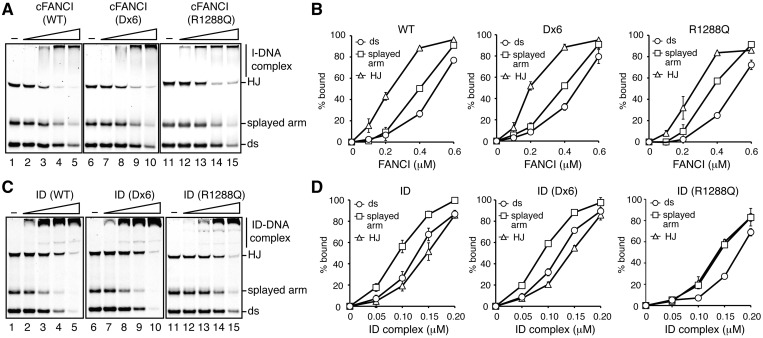
The human FANCI R1285Q mutant, corresponding to the chicken FANCI R1288Q mutant, is reportedly defective in both FANCD2 binding and DNA binding (19). We found that the FANCI R1288Q mutant was significantly defective in stimulating the FANCD2 monoubiquitylation in the presence of DNA in vitro (Figure 3E, lanes 6–10). The DNA-binding activity of FANCI alone was consistent with that reported previously (20), in which FANCI preferentially binds to the HJ DNA (Figure 4A and B). The FANCI R1288Q mutant alone was proficient in the DNA binding (Figure 4A and B), unlike the human FANCI R1285Q mutant (19); however, the DNA-binding activity of the ID complex containing the FANCI R1288Q mutant was clearly reduced (Figure 4C and D), as compared to the wild-type ID complex (Figure 4C and D). Therefore, both the FANCD2-binding and DNA-binding activities of FANCI may be required for stimulating the FANCD2 monoubiquitylation.
Figure 4.
DNA binding activities of the ID complex and FANCI. (A) Competitive DNA-binding assay with FANCI. The synthetic HJ DNA (4.5 µM), splayed arm DNA (4.5 µM), and dsDNA (4.5 µM) were incubated with increasing amounts (0, 0.1, 0.2, 0.4 and 0.6 µM) of FANCI (lanes 1–5), FANCI Dx6 (lanes 6–10), or FANCI R1288Q (lanes 11–15) at 37°C for 15 min. The samples were then separated by 8% PAGE in 0.2 × TBE buffer, and the bands were visualized by SYBR Gold staining. (B) Graphic representations of the experiments with FANCI (left), FANCI Dx6 (center), and FANCI R1288Q (right) shown in panel (A). The DNAs bound to FANCI were estimated, and the averages of three independent experiments are plotted against the protein concentrations with the standard deviation values. (C) Competitive DNA-binding assay with the ID complex. Experiments were performed as in panel (B). The FANCI–FANCD2 (lanes 1–5), FANCI Dx6–FANCD2 (lanes 6–10), and FANCI R1288Q–FANCD2 (lanes 11–15) concentrations were 0, 0.05, 0.10, 0.15 and 0.20 µM. (D) Graphic representations of the experiments with FANCI–FANCD2 (left), FANCI Dx6–FANCD2 (center), and FANCI R1288Q–FANCD2 (right) shown in panel (C). The DNAs bound to the ID complex were estimated, and the averages of three independent experiments are plotted against the protein concentrations with the standard deviation values.
The FANCI Dx6 mutant stimulates FANCD2 monoubiquitylation at a reduced rate
The FANCI Dx6 mutant, in which Ser558, Ser561, Thr567, Ser598, Ser619 and Ser631 are replaced by Asp, reportedly induces constitutive FANCD2 monoubiquitylation in vivo (11). We found that the FANCI Dx6 mutant also supported the stimulation of the FANCD2 monoubiquitylation in the presence or absence of DNA, at a reduced rate as compared to FANCI (Figure 2C, lane 5, and Figure 3E, lanes 11–15). The FANCD2 monoubiquitylation stimulation by the FANCI Dx6 mutant was still significant, because about 55% of the FANCD2 was monoubiquitylated in the ID complex containing the FANCI Dx6 mutant during the 6 h reaction in the presence of 5′ flapped DNA (Figure 3D). These results are consistent with the previous in vivo results, in which the FANCD2 monoubiquitylation in the DT40 cells harboring the FANCI Dx6 mutant occurs at a slightly reduced rate, as compared to the wild-type cells, in the presence of mitomycin C (11). The FANCI Dx6 mutant alone and the ID complex containing the FANCI Dx6 mutant were completely proficient in DNA binding (Figure 4). Therefore, the FANCI phosphorylation mimicked by the FANCI Dx6 mutation may not significantly affect the DNA-binding and FANCI–FANCD2-binding activities of FANCI, although the mechanism of the constitutive FANCD2 monoubiquitylation by the FANCI Dx6 mutant in the absence of DNA damage remains unknown (11).
The DNA-binding activity of FANCI is required to stimulate FANCD2 monoubiquitylation
Finally, we tested whether the DNA-binding activity of FANCI is required to stimulate FANCD2 monoubiquitylation in vitro. To do so, we designed the FANCI Ex6 mutant, in which Lys293, Lys296, Lys305, Lys333, Arg338 and Lys341 were replaced by Glu. These amino acid residues are located near the DNA-binding surface of FANCI (22). As anticipated, the FANCI Ex6 mutant alone and the ID complex containing the FANCI Ex6 mutant were significantly defective in DNA binding (Figure 5A–D). Interestingly, the FANCI Ex6 mutant was also quite defective in the DNA-stimulated FANCD2 monoubiquitylation in vitro (Figure 5F). In the absence of DNA, the FANCI Ex6 mutant induced the FANCD2 monoubiquitylation as well as FANCI (Figure 5E), indicating the proficiency of FANCD2 binding by the FANCI Ex6 mutant. These results strongly suggested that the DNA-binding activity of FANCI plays an essential role in the DNA-stimulated FANCD2 monoubiquitylation.
Figure 5.
The DNA-binding activity of FANCI is required for robust stimulation of the FANCD2 monoubiquitylation. (A) Competitive DNA-binding assay with FANCI Ex6. Experiments were performed as in Figure 4A. (B) Graphic representation of the experiments shown in panel (A). The DNAs bound to FANCI Ex6 were estimated, and the averages of three independent experiments are plotted against the protein concentrations with the standard deviation values. (C) Competitive DNA-binding assay with FANCI Ex6–FANCD2. Experiments were performed as in Figure 4C. (D) Graphic representation of the experiments shown in panel (C). The DNAs bound to the ID (Ex6) complex were estimated, and the averages of three independent experiments are plotted against the protein concentrations with the standard deviation values. (E) In vitro monoubiquitylation assay with FANCI Ex6 in the absence of DNA. Proteins were separated by 7% SDS-PAGE, blotted onto a membrane, and detected by α-HA antibody staining. (F) In vitro monoubiquitylation assay with FANCI Ex6 in the presence of 5′ flapped DNA. Experiments were performed as in Figure 3E. Proteins were separated by 7% SDS-PAGE, blotted onto a membrane and detected by Coomassie Brilliant Blue staining (upper panel) or α-HA antibody staining (middle panel). The amounts of monoubiquitylated FANCD2 were estimated, and the averages of three independent experiments are indicated as black bars with the standard deviation values (lower panel).
DISCUSSION
Monoubiquitylation of FANCD2 is the central process for DNA crosslink repair by the FA pathway. FANCL has been identified as a catalytic component of the E3 ligase FA core complex, containing FANCA, –B, –C, –E, –F, –G, –L, –M and two FANCA-associated polypeptides (FAAPs). UBE2T has been identified as an E2 ubiquitin-conjugating enzyme for the FANCD2 monoubiquitylation (13). Alpi et al. (14) successfully reconstituted the FANCD2 monoubiquitylation in vitro with purified UBE2T and FANCL, and in the present study, we also observed the in vitro FANCD2 monoubiquitylation in the ID complex with purified E1, UBE2T and FANCL. However, only slight FANCD2 monoubiquitylation (less than 1% of the input FANCD2) was detected in vitro, although robust FANCD2 monoubiquitylation (40–70%) has been detected in vivo (11,21). These results, therefore, indicated that the factor(s) required for proper FANCD2 monoubiquitylation may be missing in the in vitro FANCD2 monoubiquitylation system.
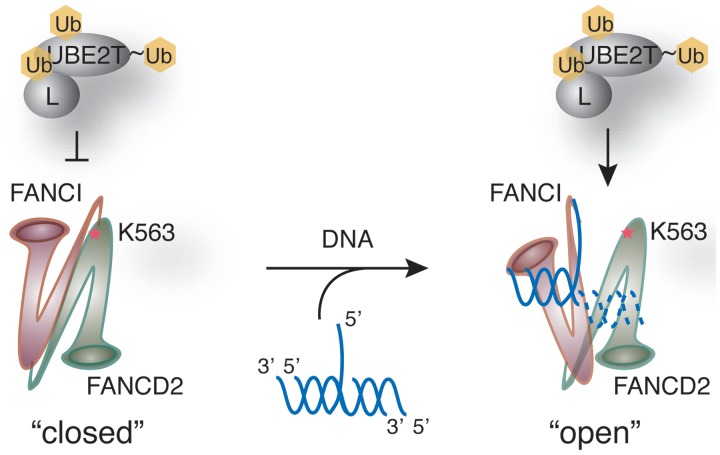
The crystal structure of the ID complex was recently reported (22). In the ID complex structure, however, the monoubiquitylation site of FANCD2 is buried in the FANCI–FANCD2 interface. This raises a new question: How does the ubiquitin ligase gain access to its target sites on the ID complex? This may be explained if a conformational change of the ID complex occurs upon DNA binding, thus relocating the FANCD2 monoubiquitylation site on the accessible surface of the complex. Since the three-way branched DNA is predicted to bind near the FANCD2 monoubiquitylation site in the ID complex (22), it may induce such a conformational change of the ID complex when it binds (Figure 6).
Figure 6.
Model for the role of branched DNA in FANCD2 monoubiquitylation. According to the crystal structure of the ID complex (22), the monoubiquitylation site (K563) of FANCD2 is located near the interface with FANCI. Therefore, UBE2T and FANCL may not allow access to the FANCD2 monoubiquitylation site in the ID complex (left panel). Branched DNA may bind to FANCI, as revealed by the crystal structure of the FANCI–DNA complex (22), and thus induce a conformational change of the ID complex to expose the FANCD2 monoubiquitylation site for UBE2T and FANCL (right panel).
Consistent with the idea described above, in the present study, we found that DNA robustly stimulates the FANCD2 monoubiquitylation, up to a level comparable to the in vivo FANCD2 monoubiquitylation. This is consistent with the previous observation that DNA fragments trigger FANCD2 monoubiquitylation in crude Xenopus laevis egg extracts (23). It has been proposed that the FA core complex monoubiquitylates FANCD2, and that its monoubiquitylation is required to recruit the ID complex to chromatin (10,24). However, our findings suggested that FANCD2 monoubiquitylation may occur after it binds to the damaged DNA. This discrepancy may be explained, if the FA core complex is considered to function in the chromatin targeting of the ID complex, and then the FANCD2 monoubiquitylation occurs after its chromatin targeting. The FA core complex has been proposed to have multiple functions within the FA pathway (10), and thus the ID complex recruitment to chromatin may be one of them. Alternatively, monoubiquitylation of the ID complex by the FA core complex in chromatin may prevent the dissociation of the ID complex from the damaged chromatin. This idea is consistent with the previous observation that the FANCD2 K563R mutant, which was defective in its monoubiquitylation, did not stably associate with chromatin (10).
Monoubiquitylated FANCD2 in chromatin is proposed to function in recruiting ubiquitin-binding proteins, such as FAN1 and SLX4, to chromatin (1,25). FAN1 is the Fanconi anemia associated nuclease, which is considered to promote nucleolytic incisions of the crosslink and/or the processing of HR intermediates (26–29). SLX4 is considered to function as a scaffold that interacts with the other nucleases, SLX1, XPF and MUS81 (30–32). The DNA-stimulated monoubiquitylation of FANCD2 reported here may occur just before the recruitment of these nucleases required for DNA crosslink repair.
In the present in vitro assay, unlike FANCD2, FANCI was not properly monoubiquitylated in the ID complex, although the FANCD2-free FANCI was properly monoubiquitylated. This may suggest that the essential factor(s) for FANCI monoubiquitylation in the ID complex is still missing in this system. The subunits of the FA core complex may be potential candidates for the factor required for proper monoubiquitylation of FANCI in the ID complex. The development of an in vitro system for FANCI monoubiquitylation in the ID complex will be needed to evaluate the significance of the monoubiquitylations of both FANCI and FANCD2.
FUNDING
This work was supported in part by Grants-in-Aid from the Japanese Society for the Promotion of Science (JSPS), and the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan. Waseda Research Institute for Science and Engineering (to H.K.). Ichiro Kanehara Foundation and the Mochida Memorial Foundation for Medical and Pharmaceutical Research (to M.I.). Funding for open access charge: Waseda University.
Conflict of interest statement. None declared.
REFERENCES
- 1.Garner E, Smogorzewska A. Ubiquitylation and the Fanconi anemia pathway. FEBS Lett. 2011;585:2853–2860. doi: 10.1016/j.febslet.2011.04.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kee Y, D'Andrea AD. Expanded roles of the Fanconi anemia pathway in preserving genomic stability. Genes Dev. 2010;24:1680–1694. doi: 10.1101/gad.1955310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kitao H, Takata M. Fanconi anemia: a disorder defective in the DNA damage response. Int. J. Hematol. 2011;93:417–424. doi: 10.1007/s12185-011-0777-z. [DOI] [PubMed] [Google Scholar]
- 4.Auerbach AD. Fanconi anemia and its diagnosis. Mutat. Res. 2009;668:4–10. doi: 10.1016/j.mrfmmm.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat. Rev. Genet. 2007;8:735–748. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- 6.Smogorzewska A, Matsuoka S, Vinciguerra P, McDonald ER, 3rd, Hurov KE, Luo J, Ballif BA, Gygi SP, Hofmann K, D'Andrea AD, et al. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sims AE, Spiteri E, Sims RJ, 3rd, Arita AG, Lach FP, Landers T, Wurm M, Freund M, Neveling K, Hanenberg H, et al. FANCI is a second monoubiquitinated member of the Fanconi anemia pathway. Nat. Struct. Mol. Biol. 2007;14:564–567. doi: 10.1038/nsmb1252. [DOI] [PubMed] [Google Scholar]
- 8.Dorsman JC, Levitus M, Rockx D, Rooimans MA, Oostra AB, Haitjema A, Bakker ST, Steltenpool J, Schuler D, Mohan S, et al. Identification of the Fanconi anemia complementation group I gene, FANCI. Cell Oncol. 2007;29:211–218. doi: 10.1155/2007/151968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia-Higuera I, Taniguchi T, Ganesan S, Meyn MS, Timmers C, Hejna J, Grompe M, D'Andrea AD. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol. Cell. 2001;7:249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- 10.Matsushita N, Kitao H, Ishiai M, Nagashima N, Hirano S, Okawa K, Ohta T, Yu DS, McHugh PJ, Hickson ID, et al. A FancD2-monoubiquitin fusion reveals hidden functions of Fanconi anemia core complex in DNA repair. Mol. Cell. 2005;19:841–847. doi: 10.1016/j.molcel.2005.08.018. [DOI] [PubMed] [Google Scholar]
- 11.Ishiai M, Kitao H, Smogorzewska A, Tomida J, Kinomura A, Uchida E, Saberi A, Kinoshita E, Kinoshita-Kikuta E, Koike T, et al. FANCI phosphorylation functions as a molecular switch to turn on the Fanconi anemia pathway. Nat. Struct. Mol. Biol. 2008;15:1138–1146. doi: 10.1038/nsmb.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meetei AR, de Winter JP, Medhurst AL, Wallisch M, Waisfisz Q, van de Vrugt HJ, Oostra AB, Yan Z, Ling C, Bishop CE, et al. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat. Genet. 2003;35:165–170. doi: 10.1038/ng1241. [DOI] [PubMed] [Google Scholar]
- 13.Machida YJ, Machida Y, Chen Y, Gurtan AM, Kupfer GM, D'Andrea AD, Dutta A. UBE2T is the E2 in the Fanconi anemia pathway and undergoes negative autoregulation. Mol. Cell. 2006;23:589–596. doi: 10.1016/j.molcel.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 14.Alpi AF, Pace PE, Babu MM, Patel KJ. Mechanistic insight into site-restricted monoubiquitination of FANCD2 by Ube2t, FANCL, and FANCI. Mol. Cell. 2008;32:767–777. doi: 10.1016/j.molcel.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 15.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 16.Iwasaki H, Takahagi M, Nakata A, Shinagawa H. Escherichia coli RuvA and RuvB proteins specifically interact with Holliday junctions and promote branch migration. Genes Dev. 1992;6:2214–2220. doi: 10.1101/gad.6.11.2214. [DOI] [PubMed] [Google Scholar]
- 17.Yokoyama H, Kurumizaka H, Ikawa S, Yokoyama S, Shibata T. Holliday junction binding activity of the human Rad51B protein. J. Biol. Chem. 2003;278:2767–2772. doi: 10.1074/jbc.M210899200. [DOI] [PubMed] [Google Scholar]
- 18.Horikoshi N, Morozumi Y, Takaku M, Takizawa Y, Kurumizaka H. Holliday junction-binding activity of human SPF45. Genes Cells. 2010;5:373–383. doi: 10.1111/j.1365-2443.2010.01383.x. [DOI] [PubMed] [Google Scholar]
- 19.Yuan F, Hokayem EI, J, Zhou W, Zhang Y. FANCI protein binds to DNA and interacts with FANCD2 to recognize branched structures. J. Biol. Chem. 2009;284:24442–24452. doi: 10.1074/jbc.M109.016006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Longerich S, San Filippo J, Liu D, Sung P. FANCI binds branched DNA and is monoubiquitinated by UBE2T-FANCL. J. Biol. Chem. 2009;284:23182–23186. doi: 10.1074/jbc.C109.038075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andreassen PR, D'Andrea AD, Taniguchi T. ATR couples FANCD2 monoubiquitination to the DNA-damage response. Genes Dev. 2004;18:1958–1963. doi: 10.1101/gad.1196104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joo W, Xu G, Persky NS, Smogorzewska A, Rudge DG, Buzovetsky O, Elledge SJ, Pavletich NP. Structure of the FANCI-FANCD2 complex: insights into the Fanconi anemia DNA repair pathway. Science. 2011;333:312–316. doi: 10.1126/science.1205805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sobeck A, Stone S, Hoatlin ME. DNA structure-induced recruitment and activation of the Fanconi anemia pathway protein FANCD2. Mol. Cell. Biol. 2007;27:4283–4292. doi: 10.1128/MCB.02196-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cohn MA, D'Andrea AD. Chromatin recruitment of DNA repair proteins: lessons from the fanconi anemia and double-strand break repair pathways. Mol. Cell. 2008;32:306–312. doi: 10.1016/j.molcel.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang M, D'Andrea AD. A new nuclease member of the FAN club. Nat. Struct. Mol. Biol. 2010;17:926–928. doi: 10.1038/nsmb0810-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kratz K, Schöpf B, Kaden S, Sendoel A, Eberhard R, Lademann C, Cannavó E, Sartori AA, Hengartner MO, Jiricny J. Deficiency of FANCD2-associated nuclease KIAA1018/FAN1 sensitizes cells to interstrand crosslinking agents. Cell. 2010;142:77–88. doi: 10.1016/j.cell.2010.06.022. [DOI] [PubMed] [Google Scholar]
- 27.MacKay C, Déclais AC, Lundin C, Agostinho A, Deans AJ, MacArtney TJ, Hofmann K, Gartner A, West SC, Helleday T, et al. Identification of KIAA1018/FAN1, a DNA repair nuclease recruited to DNA damage by monoubiquitinated FANCD2. Cell. 2010;142:65–76. doi: 10.1016/j.cell.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu T, Ghosal G, Yuan J, Chen J, Huang J. FAN1 acts with FANCI-FANCD2 to promote DNA interstrand cross-link repair. Science. 2010;329:693–696. doi: 10.1126/science.1192656. [DOI] [PubMed] [Google Scholar]
- 29.Yoshikiyo K, Kratz K, Hirota K, Nishihara K, Takata M, Kurumizaka H, Horimoto S, Takeda S, Jiricny J. KIAA1018/FAN1 nuclease protects cells against genomic instability induced by interstrand cross-linking agents. Proc. Natl Acad. Sci. USA. 2010;107:21553–21557. doi: 10.1073/pnas.1011081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fekairi S, Scaglione S, Chahwan C, Taylor ER, Tissier A, Coulon S, Dong MQ, Ruse C, Yates JR, 3rd, Russell P, et al. Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell. 2009;138:78–89. doi: 10.1016/j.cell.2009.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Svendsen JM, Smogorzewska A, Sowa ME, O'Connell BC, Gygi SP, Elledge SJ, Harper JW. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell. 2009;138:63–77. doi: 10.1016/j.cell.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamamoto KN, Kobayashi S, Tsuda M, Kurumizaka H, Takata M, Kono K, Jiricny J, Takeda S, Hirota K. Involvement of SLX4 in interstrand cross-link repair is regulated by the Fanconi anemia pathway. Proc. Natl Acad. Sci. USA. 2011;108:6492–6496. doi: 10.1073/pnas.1018487108. [DOI] [PMC free article] [PubMed] [Google Scholar]