Abstract
The yeast protein Ebp2 is required for early steps in production of 60S ribosomal subunits. To search for cofactors with which Ebp2 functions, or substrates on which it acts, we screened for mutants that were synthetically lethal (sl) with the ebp2-14 mutation. Four different mutant alleles of the 60S ribosomal subunit assembly factor Brx1 were found. To investigate defects of the double mutant, we constructed strains conditional for the ebp2-14 brx1- synthetic lethal phenotype. These ebp2-14 brx1 mutants were defective in processing of 27S pre-rRNA and production of 60S subunits, under conditions where each single mutant was not. Ebp2 and Brx1 exhibit a strong two-hybrid interaction, which is eliminated by some combinations of brx1 and ebp2 mutations. In one such mutant, Ebp2 and Brx1 can still associate with pre-ribosomes, but subunit maturation is perturbed. Depletion of either Ebp2 or Brx1 revealed that Brx1 requires Ebp2 for its stable association with pre-ribosomes, but Ebp2 does not depend on the presence of Brx1 to enter pre-ribosomes. These results suggest that assembly of 60S ribosomal subunits requires cooperation of Ebp2 with Brx1, together with other molecules present in pre-ribosomes, potentially including several found in assembly subcomplexes with Brx1 and Ebp2.
INTRODUCTION
Yeast ribosomes contain four RNAs and 79 ribosomal proteins (r proteins). The mature 25S, 18S and 5.8S rRNAs are derived from a single long precursor rRNA, the 35S pre-rRNA, transcribed by RNA polymerase I. The 5S rRNA is transcribed from separate genes by RNA polymerase III (1). As these rRNAs are transcribed, they must fold into secondary and tertiary structures that enable modification of the RNA, removal of spacer sequences and binding of the ribosomal proteins. Thus, constructing these complex ribonucleoprotein particles requires the establishment and remodeling of RNA–RNA, RNA–protein and protein–protein interactions. Genetic and proteomic studies have revealed that there are more than 180 proteins, in addition to r proteins, required for these dynamic processes occurring during ribosome assembly (2,3). The effects on ribosome production and pre-rRNA processing have been examined when each of these factors was depleted or inactivated. Most factors have been assigned to function in production of one or the other ribosomal subunit, and to participate in one or more steps of pre-rRNA processing.
The challenge before us now is to elucidate precisely how each assembly factor (and r protein) facilitates accurate and efficient production of functional ribosomes. To understand in better detail the mechanisms of ribosome assembly, it will be critical to answer the following questions: When does each protein associate with pre-ribosomes, and when does each assembly factor dissociate? Which molecules are necessary for the stable docking of each protein with pre-rRNPs, and for dissociation of each? Once bound to pre-ribosomes, with which proteins or RNAs does each factor and r protein interact? These pre-ribosomal ligands will include cofactors (both positive and negative regulators), as well as substrates upon which each factor might act. Where in pre-ribosomes is each factor located with respect to the others? How do these ligands and locations change as particles undergo maturation? The recent determination of the crystal structure of mature eukaryotic ribosomes (4,5) provides a valuable structural context to facilitate answering some of these questions.
One such assembly factor is Ebp2, which was previously shown to be essential for maturation of 25S rRNA and assembly of 60S ribosomal subunits (6–8). To investigate the function of Ebp2 in more detail, we carried out a genetic screen for mutations that are synthetically lethal (sl) with the ebp2-14 mutation. Such a screen should identify proteins that functionally or physically interact with Ebp2. We found that mutations in the gene encoding 60S ribosomal subunit assembly factor Brx1 exhibit synthetic lethality with ebp2-14. We constructed GAL-EBP2 ebp2-14 brx1 strains conditional for this synthetic lethality, and demonstrated that the ebp2 brx1 double mutant strains are unable to assemble 60S subunits under conditions where each single mutant is functional in subunit biogenesis. Wild-type Ebp2 and Brx1 associate with each other tightly in a two-hybrid assay (9). However, three out of four brx1 mutations (brx1-52, brx1-102 and brx1-227) in combination with ebp2-14 prevent this interaction. Interestingly, in the ebp2-14 brx1-172 double mutant, the two proteins can still interact. Therefore, we studied in more detail changes in pre-ribosomal particles in one of the double mutants where the interaction is disrupted and compared them to the double mutant where the interaction is not abolished. Surprisingly, in both cases, both mutant proteins were able to assemble into pre-ribosomes. This result suggests that other molecules in pre-ribosomes help anchor Ebp2 and Brx1 within pre-rRNPs. Consistent with these findings, we also established that in the absence of Brx1, Ebp2 can stably associate with pre-ribosomes, but Brx1 cannot enter pre-ribosomes without Ebp2. Thus, these ebp2 and brx1 mutations may provide insight into functions of Ebp2 and Brx1 in 60S subunit maturation, independent of their recruitment into nascent ribosomes.
MATERIALS AND METHODS
Construction and growth of yeast strains
The yeast strains used in this study are listed in Supplementary Table S1. The ebp2-14 conditional allele was generated by random-PCR mutagenesis (8). Strain 4795-408 (a gift from Lee Hartwell) was crossed twice with W303α, then subsequently crossed with the Ts− ebp2-14 strain containing Ycp50-EBP2-ADE3 (see below), to generate KM822, the parental strain used to screen for synthetic lethal mutants (Figure 1A). A yeast strain for conditional expression of EBP2 was constructed by transformation of a plasmid containing EBP2 expressed from the GAL1 promoter. A yeast strain for conditional expression of BRX1 was constructed as described in Longtine et al. (10). Briefly, sequences containing the KANMX6 gene expressing resistance to geneticin (G418), plus the GAL1 promoter sequence followed by an ATG and codons encoding 3HA were amplified by PCR. The PCR products were transformed into yeast strain JWY6147, and the strain was grown in selective media containing galactose. Transformants were screened for correct integration of the GAL1 promoter and the 3HA tag upstream and in-frame with BRX1, by western blotting with anti-HA antisera. Yeast strains expressing C-terminal tandem-affinity purification (TAP)-tagged Nop7, Rpf2 or Rrp5, C-terminal triple hemagglutinin (3HA) tagged proteins or C-terminal 13Myc-tagged proteins were created by PCR of the tag sequence and a selectable marker (URA3 for the TAP tag and HIS3 or TRP1 for other tags), transformation and selection, as described in Rigaut et al. (11) and Longtine et al. (10). Transformants were screened by western blotting for those expressing the tagged proteins.
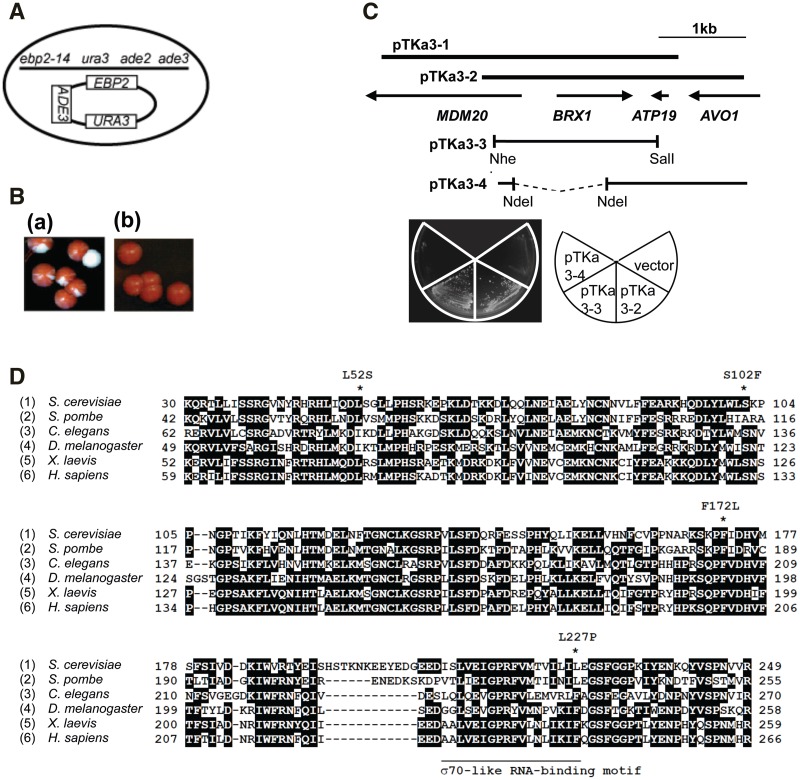
Figure 1.
Isolation of mutants exhibiting synthetic lethality with ebp2-14. (A) Schematic representation of the strain used for the sl screen. (B) Non-sectoring phenotype of a mutant. The parent strain (a) and isolated mutants (b) were streaked on YEPD media and incubated at 30°C for 7 days. (C) Cloning of alleles responsible for synthetic lethality with ebp2-14. pKTa3-1 and pKTa3-2 were cloned from a yeast genomic library (cloned in plasmid vector YCplac111), and subcloned into pKTa3-3 (in pRS315) and pKTa3-4 (in YCplac111). The m3 (KM635) strain was transformed with these plasmids or a vector plasmid (pRS315), and cultured on 5-FOA-containing medium at 25°C for 3 days. (D) Synthetic lethal brx1 mutations are in conserved amino acid residues. The amino acid sequences are from the GenBank database. Restricted regions with a homologous sequence are shown in single-letter code; 1, S. cerevisiae (accession Q08235, 30–249 amino acids aligned from full length of 291 amino acids); 2, Schizosaccharomyces pombe (Q9HGL6, 42–255 amino acids from full length of 295 amino acids); 3, Caenorhabditis elegans (P34524, 62–270 amino acids from 352 amino acids); 4, Drosophila melanogaster (Q9VZE6, 49–258 amino acids from full length of 359 amino acids); 5, Xenopus laevis (Q8UVY2, 52–258 amino acids from 339 amino acids); 6, Homo sapiens (Q8TDN6, 59–266 amino acids from 353 amino acids). A sequence alignment was generated by BLAST and ClustalW. Deletions needed for alignment are indicated by dashes. Residues boxed in black are identical in at least three of the sequences compared. The amino acids substituted in the brx1 alleles are indicated with asterisks above the sequence.
Yeast cells were grown in YEPD (yeast extract, polypeptone, glucose) rich medium, YEPGal (yeast extract, polypeptone, galactose) rich medium, or synthetic complete medium containing 2% glucose (SC) or SC dropout medium, depending on the plasmid markers. Standard techniques were used for manipulation of yeast (12).
Plasmid construction
To construct plasmid YCp50 (CEN URA3)-EBP2-ADE3, the SmaI–SalI fragment of pDK255 (13) containing ADE3 was cloned between the NruI and SalI sites of YCp50-EBP2 (6). pRS314-GAL1-myc-BS, a pRS314-based expression vector that contains the GAL1 promoter, sequences encoding the three myc epitopes just downstream of the initiation codon, a multicloning site and the TDH3 terminator, was kindly provided by Kazuma Tanaka. pRS314-GAL1-myc-BRX1 was constructed by inserting a BamHI–SmaI PCR fragment of the BRX1 ORF into pRS314-GAL1-myc-BS. A plasmid expressing myc-tagged Brx1 from its own promoter, pRS314-myc-BRX1, was constructed as follows. The NotI–EcoRI (blunt-ended) fragment containing the GAL1 promoter from pRS314-GAL1-myc-BRX1 was replaced with the NotI–SmaI PCR fragment of the BRX1 promoter, produced from yeast genomic DNA by PCR. pRS304-myc-BRX1 was generated by inserting the NotI–XhoI region from pRS314-myc-BRX1 into pRS304. pRS304-myc-BRX1 was digested with SpeI and used for integration into the genome to express myc-Brx1. Plasmids expressing mutant myc-brx1 were constructed similarly. PCR was performed with KOD plus (TOYOBO, Japan). The entire PCR products were sequenced and the structures of all plasmids were confirmed by restriction site analysis. Primers used for PCR are available upon request.
Isolation of sl mutants
To generate mutations that cause synthetic lethality with the ebp2-14 allele, 5.0 × 105 cells of strain KM822 containing the ebp2-14, ura3, ade2 and ade3 mutations in the genome, plus plasmid YCp50 (URA3)-EBP2-ADE3 (Figure 1A), were plated on YEPD, mutagenized with UV to 15–40% viability, and incubated at 30°C for 7–9 days. Colonies that could not grow without the plasmid, i.e. those showing a red non-sectoring phenotype, were isolated (Figure 1B). Of the 289 colonies that showed this phenotype, 121 colonies could not grow on 5-FOA medium at 30°C. These strains were subsequently transformed with plasmids pRS313-EBP2 or pRS313-ebp2-14, which also contained the HIS3 gene, to test whether pRS313-EBP2, and not pRS313-ebp2-14, could replace YCp50-EBP2-ADE3 in cells grown on medium containing 5-FOA. Nine mutants (m1–m9) were obtained. After crossing each mutant with an EBP2 or ebp2–14 strain, tetrad analysis revealed 8 alleles derived from single mutations of genomic DNA.
Cloning and sequencing of the BRX1 gene
In order to identify the gene defined by the single complementation group of sl mutants, we transformed the m3 sl mutant strain (KM635) with a library of partial Sau3A fragments of Saccharomyces cerevisiae genomic DNA constructed in the single copy yeast vector YCplac111 (a gift from Kouichi Funato). This plasmid contains LEU2 as a selectable marker, and our parental sl mutant strain is Leu+, because EBP2 was disrupted by insertion of the LEU2 gene. Therefore, we first replaced the LEU2 gene in the m3 sl mutant strain with the HIS3 gene. To do so, the strain was transformed with a DNA fragment prepared by digestion of plasmid pRS304 ebp2::HIS3 with EcoRI and XbaI. The replacement of ebp2::LEU2 by ebp2::HIS3 was checked by PCR and DNA sequencing, and the strain was named KM823. We then transformed the Leu− sl mutant strain KM823 with the plasmid library. Of 1.8 × 104 Leu+ transformants, 10 colonies grew at 25°C on medium containing 5-FOA. Four colonies grew well at 31°C but not at 37°C on SC–Leu medium, which is expected for complementation of the sl mutation but not of the Ts− phenotype of ebp2-14. Each of the four plasmids that were isolated from these colonies complemented the synthetic lethality of the mutant cells when retransformed. Partial sequencing of the genomic DNA inserts and subcloning revealed that the complementing activity contained the BRX1 gene (Figure 1C). In order to confirm that mutations(s) in BRX1 are responsible for the synergistic temperature sensitivity of the m3 mutant, DNA fragments containing the BRX1 gene were amplified by PCR using chromosomal DNA extracted from the wildtype and the m3 strains. The DNA fragment from wildtype cells, but not that from the m3 mutant strain, could complement the temperature sensitivity of the m3 cells (data not shown). This result confirms that BRX1 is mutated in the m3 strain and is responsible for the synthetic defect with ebp2-14. The wildtype BRX1 gene and the mutated allele of the chromosomal BRX1 gene from each of the five sl mutant strains (KM634, KM635, KM638, KM639, KM641) was isolated by PCR, using total chromosomal DNA isolated from wild type and each mutant. The PCR was done in duplicate and the products of each reaction were independently sequenced. PCR fragments were digested with BamHI and PstI and inserted into pRS315 (pRS315-brx1).
Gene disruption and construction of brx1 mutants
A deletion–insertion mutation of BRX1 was constructed in the diploid W303. The BRX1 gene was subcloned into pRS306 as a 1.9-kb NheI–SalI fragment. HpaI–XbaI fragments (amino acids 71–262) were deleted from the BRX1 gene and a 1.8-kb long SmaI–XbaI fragment containing the HIS3 gene was inserted at the site of the BRX1 gene. The resulting brx1Δ::HIS3 construct was excised with NotI and SalI as a 3.0-kb fragment and used to transform the diploid W303 strain to His+. Correct integration of the brx1Δ::HIS3 gene at the homologous locus in His+ transformants was confirmed by PCR. The diploid BRX1/brx1Δ::HIS3 was transformed with pRS316-BRX1 (URA3) and sporulated, and tetrads were dissected to obtain the brx1Δ pRS316-BRX1 (URA3) strain (KM642). To construct brx1 strains, pRS315-brx1-52, pRS315-brx1-102, pRS315-brx1-172 and pRS315-brx1-227 were digested with BamHI and PstI, and each 2.1-kb BamHI-SalI fragment was subcloned into pRS304. The brx1 genes were excised from the resulting plasmids with EcoRI and integrated in the genomic DNA of the KM642 strain. The brx1-52, brx1-102, brx1-172 and brx1-227 strains (KM1721–KM1724) were obtained by selection on medium containing 5-FOA. The wildtype control strain KM1720 was constructed by similar means.
Gradient assays of ribosomes and polyribosomes
Yeast whole cell extracts were overlaid on top of 11 ml 7–47% (wt/vol) sucrose gradients and centrifuged for 3.4 h at 35 000 rpm at 4°C in a Hitachi RPS40T rotor, as previously described (9). Gradients were collected by pumping up using a peristaltic pump, and fractions were monitored at 254 nm.
Two-hybrid assays
To carry out yeast two-hybrid assays, plasmids pBTM116 and pACT2 encoding lexA binding domain-fusion proteins and Gal4 activation domain–fusion proteins, respectively, were co-transformed into the yeast L40 strain cells containing a HIS3 reporter gene as previously described (6). Leu+ Trp+ transformants were selected. Five-fold serial dilutions of the cell cultures were stamped on SC–Leu, Trp, His plates containing various concentrations of 3-amino-1,2,4-triazole (3-AT), an inhibitor of the HIS3 gene product, and incubated. A plasmid expressing the lexA binding domain-Adh1 fusion protein (a gift from David Shore) was used as a negative control.
Primer extension analysis and northern blotting
Yeast strains expressing TAP-tagged assembly factors Ebp2, Brx1 or Nob1, were grown in rich medium containing galactose at 30°C to 3–5 × 107 cells/ml, harvested, and subjected to affinity purification, as described in Sahasranaman et al. (14). RNA enriched from these purified pre-ribosomes and ribosomes was extracted as described in Sahasranaman et al. (14). These pre-rRNAs and rRNAs were assayed by primer extension or northern blotting, as described in Horsey et al. (15). The sequences of primers used are available upon request.
Affinity purifications of pre-ribosomes
Ribosome assembly intermediates were affinity-purified from whole cell extracts with magnetic Dynabeads (Invitrogen), using TAP-tagged assembly factors Nop7, Rpf2 or Rrp5, as described in Sahasranaman et al. (14).
Western analysis
Western blotting was performed following standard techniques. Signals were visualized by Enhanced Chemiluminescence (Amersham), as instructed by the manufacturer, or otherwise using the colorimetric method (Promega). Mouse anti-myc monoclonal antibodies (9E10; BAbCO) and rabbit anti-Ebp2 polyclonal antibodies (8) were used. Horseradish peroxidase-conjugated anti-rabbit or anti-mouse IgG secondary antibodies (Amersham) were used in 1:1000 dilutions. TAP-tagged proteins were detected using alkaline phosphatase conjugated to IgG (Pierce). Otherwise, antibodies specific for r proteins or ribosomal assembly factors were used to detect proteins in purified pre-ribosomes. AP-conjugated anti-mouse or anti-rabbit secondary antibodies (Promega) were used in 1:7500 dilutions in these experiments.
For SDS-PAGE, silver staining, and western blotting with samples from purified particles, proteins were recovered from affinity-purified pre-ribosomes by precipitation with 10% TCA, and suspended in SDS sample buffer. Proteins were resolved by SDS–PAGE on 4–20%Tris–Glycine Novex precast gels (Invitrogen), stained with silver by standard methods, and assayed by western blot analysis (16), with the following modification. To enable detection of multiple different proteins on one blot, and to conserve antiserum by using less volume of blotting buffer, nitrocellulose membranes were cut into smaller sections based on the previously established mobility of the different proteins.
RESULTS
brx1 mutations cause a synthetic growth defect with the ebp2-14 mutation
Previously we isolated temperature-sensitive ebp2 mutants by random PCR mutagenesis of EBP2, and showed that they were defective in growth and 60S subunit biogenesis at 37°C, but not at 25°C (8). In order to obtain more details about the function of Ebp2, we sought yeast mutants that exhibit a synthetic growth defect with ebp2-14 at temperatures permissive for its growth. We identified eight strains, each containing recessive mutations in a single gene (Figure 1A–C). Three strains were conditionally lethal (m1, m4 and m8), and were not studied here. Five mutants (m2, m3, m6, m7 and m9) were unconditionally synthetic lethal and formed one complementation group (data not shown), which upon cloning, proved to be the BRX1 gene. BRX1 encodes a ribosome assembly factor consisting of 291 amino acid residues. DNA sequence analysis revealed that each of the five sl brx1-mutants had a single point mutation in BRX1, leucine 52 to serine (m9; KM641), serine 102 to phenylalanine in two mutants (m2; KM634 and m6; KM638), phenylalanine 172 to leucine (m3; KM635), and leucine 227 to proline (m7; KM639). We named the alleles brx1-52, brx1-102, brx1-172 and brx1-227, respectively. Brx1 is highly conserved in eukaryotes as a member of a superfamily that possesses a σ70-like RNA-binding motif (17,18). Alignment of Brx1 homologs reveals that the mutated amino acids in brx1-52, brx1-102 and brx1-172 are highly conserved (Figure 1D). However, these amino acids are not conserved among the superfamily that consists of Brx1, Ssf1, Rpf1, Rpf2 and Imp4 (for alignment, see ref.19), suggesting that these residues are important for a specific function of Brx1. Leucine 227 is located at the C-terminal end of the σ70-like RNA-binding motif in Brx1.
Three of the four brx1 single mutant strains exhibit no growth defect at any temperature, whereas brx1-227 is temperature-sensitive
We constructed brx1 single mutant strains, by integrating each of the four cloned mutant alleles into the genome of a brx1 null strain, as described in ‘Materials and Methods’. The resulting brx1-52, brx1-102 and brx1-172 strains grew as well as wild-type BRX1 strains at 16°C, 25°C, 30°C, 32°C, 35°C and 37°C, whereas the brx1-227 strain exhibited weak temperature sensitivity at 37°C (Figure 2A). This result indicates that each brx1 allele has little impact on cell growth, but is lethal in combination with ebp2-14.
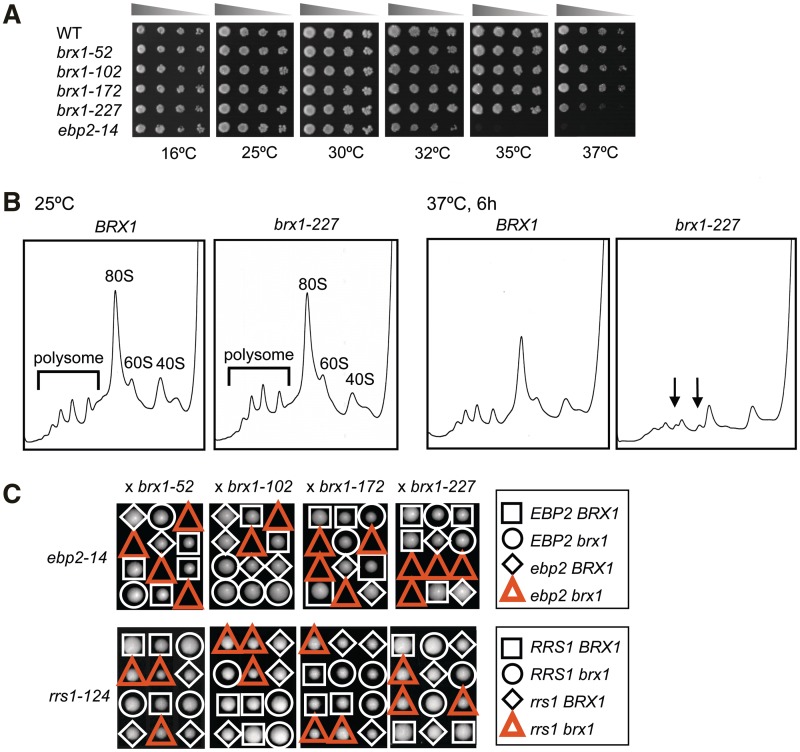
Figure 2.
The synthetic lethality of ebp2-14 with brx1. (A) brx1 and ebp2-14 single mutants grow under conditions that are lethal in the double mutants. The wild-type BRX1 strain and brx1-52, brx1-102, brx1-172, brx1-227 and ebp2-14 single mutants were cultured in SC medium at 25°C overnight. Five-fold serial dilutions of the culture (OD600 = ∼1) were spotted onto YEPD medium, and incubated at 16°C for 10 days, and at 25°C, 30°C, 32°C, 35°C and 37°C for 3 days. (B) The temperature-sensitive brx1-227 mutation leu227pro leads to a defect in 60S subunit biogenesis at 37°C. The KM1720 (BRX1) and KM1724 (brx1-227) strains were grown at 25°C overnight. The cultures were then shifted to 37°C and grown for 6 h. Cell extracts were used for polysome analysis by sucrose density gradient ultracentrifugation. Ribosomal profiles were determined by OD254 measurement of the gradient (7–47%) fractions. Arrows indicate halfmer polysomes. (C) Specificity of the synthetic lethality of ebp2-14 with brx1. Tetrad analysis of ebp2 brx1 and rrs1 brx1 diploids. Diploids constructed by crossing the ebp2-14 or rrs1-124 mutant with each brx1 mutant were sporulated and meiotic progeny were analyzed by tetrad dissection. Colony formation was scored on plates containing glucose-medium incubated at 25°C for 7–8 days for ebp2-14 (top) or 6 days for rrs1-124 (bottom). Spores corresponding to double mutants are indicated by red triangles.
To ascertain whether the brx1-227 temperature-sensitive mutant is defective in 60S ribosome biogenesis at the restrictive temperature, we assayed levels of ribosomal subunits and polyribosomes in the mutant. The brx1-227 strain was grown at 25°C and shifted to 37°C for 6 h, and extracts were prepared and subjected to centrifugation on 7–47% sucrose gradients. Amounts of free 60S subunits relative to 40S subunits decreased, and halfmer polysomes appeared in the extracts from the brx1-227 mutant grown at 37°C (Figure 2B). In contrast, the brx1-227 mutant maintained at 25°C or the wild-type BRX1 strain shifted from 25°C to 37°C exhibited wild-type polysome profiles (Figure 2B). These results indicate that 60S subunit production is compromised in the brx1-227 mutant at the restrictive temperature, 37°C, but not at temperatures permissive for growth.
The synthetic growth defect with brx1 is specific for the ebp2 mutant
To further verify that ebp2 brx1 double mutants are synthetically lethal, we constructed doubly heterozygous mutant diploids by crossing the ebp2-14 mutant with each brx1 mutant. These diploids were sporulated, and tetrads were dissected to obtain haploid spore clones. Each of the spores corresponding to ebp2-14 brx1 double mutants, indicated by red triangles in Figure 2C, was inviable.
Then, we checked whether the synthetic lethality was specific for brx1 mutants. To do so, we assayed whether a mutation in a different 60S ribosomal subunit assembly factor, Rrs1 (20), exhibits synthetic lethality with brx1. EBP2 was first identified in a two-hybrid screen for interactions with Rrs1 (see Figure 3A; ref.6). In temperature sensitive rrs1-124 yeast, 60S subunit biogenesis is compromised at the restrictive temperature (21). Tetrad analysis showed that rrs1-124 did not exhibit synthetic lethality or a synthetic growth defect with any of the brx1 alleles (Figure 2C). These results indicate that synthetic lethality is specific for ebp2 and brx1.
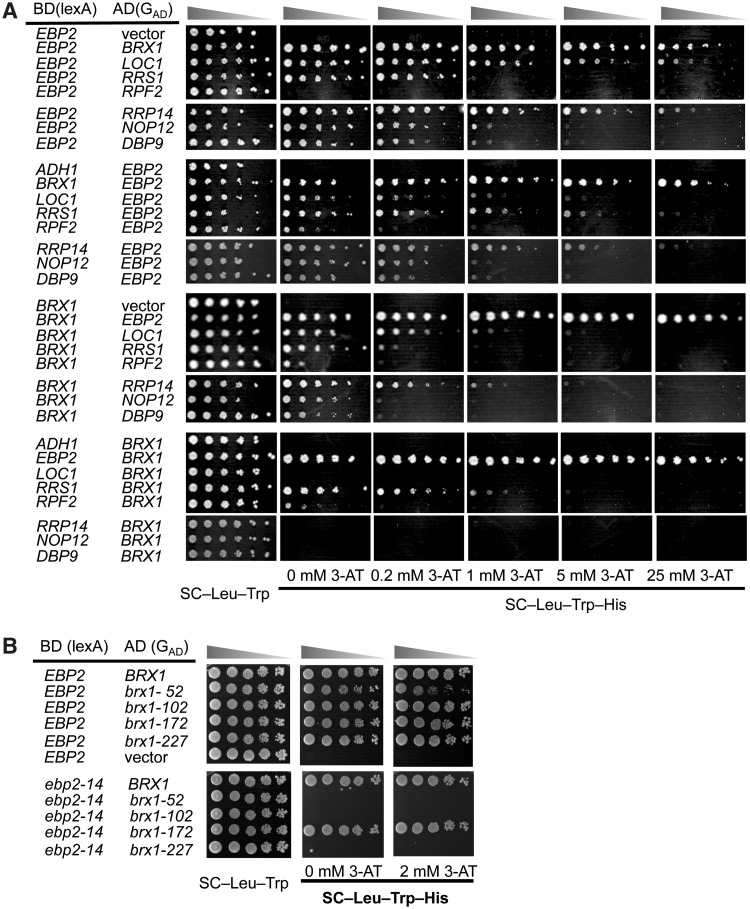
Figure 3.
Wildtype Ebp2 and Brx1 exhibit a strong two-hybrid interaction, that is disrupted in some of the ebp2 brx1 double mutants. (A) Protein–protein interactions were analyzed by the yeast two-hybrid system. Five-fold serial dilutions of cultures of L40 yeast transformed with lexA binding domain-fused and Gal4p activation domain-fused genes were plated on SC–Trp, Leu, or SC–Trp, Leu, His containing the indicated concentrations of 3-AT, and cultured at 25°C for 4–5 days. (B) Interactions were assayed as described in (A) above.
The combination of ebp2-14 and brx1 mutations eliminates the interaction of Ebp2 with Brx1 in three out of four double mutant strains
Consistent with the genetic interactions that we observe between ebp2 and brx1 mutations, we previously found that Ebp2 and Brx1 interact physically. BRX1 was isolated in a two-hybrid screen using a yeast cDNA library, with EBP2 as bait (9). In the screen, 55 out of 94 clones were identified as BRX1. The remaining clones included four other 60S assembly factor genes, DBP9, LOC1, NOP12 and RRP14. EBP2 and RPF2 were isolated in the screen using RRS1 as bait (6,22). The two-hybrid interaction of Ebp2 with Brx1 generated the strongest response of reporter genes among all combinations (Figure 3A).
One possible explanation for the synthetic lethal phenotype of the ebp2 brx1 double mutants is that the combination of mutations disrupts interaction between Ebp2 and Brx1. Therefore, we tested the effect of the ebp2 and brx1 mutations on the Ebp2-Brx1 two-hybrid interactions. A strong interaction, similar to that for wild-type proteins, was observed between wild-type Ebp2 and each mutant brx1 protein; these two-hybrid strains grew on 2 mM 3-AT (Figure 3B). However, when ebp2-14 was combined with brx1-52, brx1-102 or brx1-227, the interaction was abolished; none of these two-hybrid strains grew on medium even without 3-AT. Interestingly, the combination of ebp2-14 and brx1-172 did not affect their interaction.
A combination of the ebp2-14 and brx1-102 or brx1-172 mutations leads to synergistic defects in 60S subunit biogenesis and pre-rRNA processing
The observation that three of the ebp2 brx1 sl mutations prevented physical interactions between Brx1 and Ebp2, but the ebp2 brx1-172 double mutation did not, led us to investigate the importance of the pairwise interaction between Brx1 and Ebp2 in ribosome biogenesis, downstream of docking of these proteins into pre-ribosomes. Because ebp2 brx1 double mutants are lethal at all growth temperatures, we needed to construct a conditional lethal strain to investigate phenotypes of the double mutants in more detail. To do so, we built strains KM1732 and KM1733 containing ebp2-14 brx1-102 or ebp2-14 brx1-172, respectively, plus a plasmid containing wild-type HA-tagged EBP2 under the control of the galactose-dependent GAL1 promoter (GAL1-HA-EBP2:ADE2). When these two strains were shifted from galactose-containing medium to glucose-containing medium, expression of EBP2 was turned off, and the phenotypes of the ebp2 brx1 double mutant could be revealed.
To examine effects on ribosome production, extracts were prepared from these strains grown at 25°C in galactose, and 16 and 24 h after shifting cultures from galactose to glucose medium. In the shifted strains, amounts of free 60S ribosomal subunits, 80S monoribosomes and polyribosomes decreased, and halfmer polyribosomes appeared (Figure 4A). Polysome profiles of strains grown in galactose where EBP2 was expressed but brx1 is mutant, looked wild-type (Figure 4A, 0 h time point). Similarly, the original ebp2-14 single mutant grown at 25°C had a wild-type polysome profile (8). Thus at 25°C, 60S subunit production is compromised in the ebp2 brx1 double mutants, but not in any of the ebp2 or brx1 single mutants.
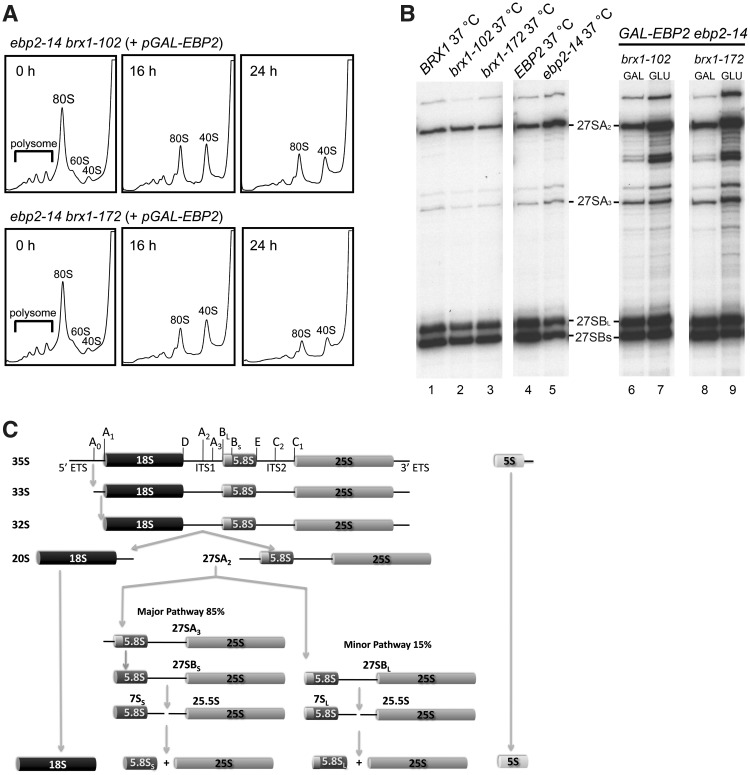
Figure 4.
The synergistic defect of ebp2 and brx1 in biogenesis of 60S ribosomal subunits. The single mutant strains KM414 (ebp2-14), KM1722 (brx1-102) and KM1723 (brx1-172), were grown at 25°C and shifted to 37°C for 4 h. The KM1732 (ebp2-14 brx1-102 pGAL1-HA-EBP2) and KM1733 (ebp2-14 brx1-172 pGAL1-HA-EBP2) strains were grown in galactose medium at 25°C and shifted to glucose medium and grown at 25°C for 16 h and 24 h. (A) Cell extracts were used for polysome analysis by sucrose density gradient ultracentrifugation. (B) Pre-rRNA processing was assayed by primer extension. (C) Pre-rRNA processing pathway.
We examined pre-rRNA processing in the brx1 and ebp2 single mutants grown at 25°C and shifted to 37°C, as well as the conditional double mutants grown at 25°C, using primer extension assays to assay 5′-ends of pre-rRNAs. As shown in Figure 4B, lanes 1–3, amounts of pre-rRNAs containing A2, A3, BS or BL 5′-ends in the brx1-102 or brx1-172 single mutants were similar to those in wild-type BRX1 yeast. The ebp2-14 mutant contained slightly higher levels of 27SA2 and 27SA3 pre-rRNAs relative to other pre-rRNAs, compared to the wild-type EBP2 strain (Figure 4B, lanes 4 and 5). Lastly, both double mutant strains KM1732 and KM1733 shifted to glucose medium contained increased amounts of 27SA2 and 27SA3 pre-rRNAs compared to the unshifted strains expressing EBP2 (Figure 4B, lanes 6–9).
Brx1 and Ebp2 assemble into pre-ribosomes early, with pre-rRNPs containing 35S pre-rRNA
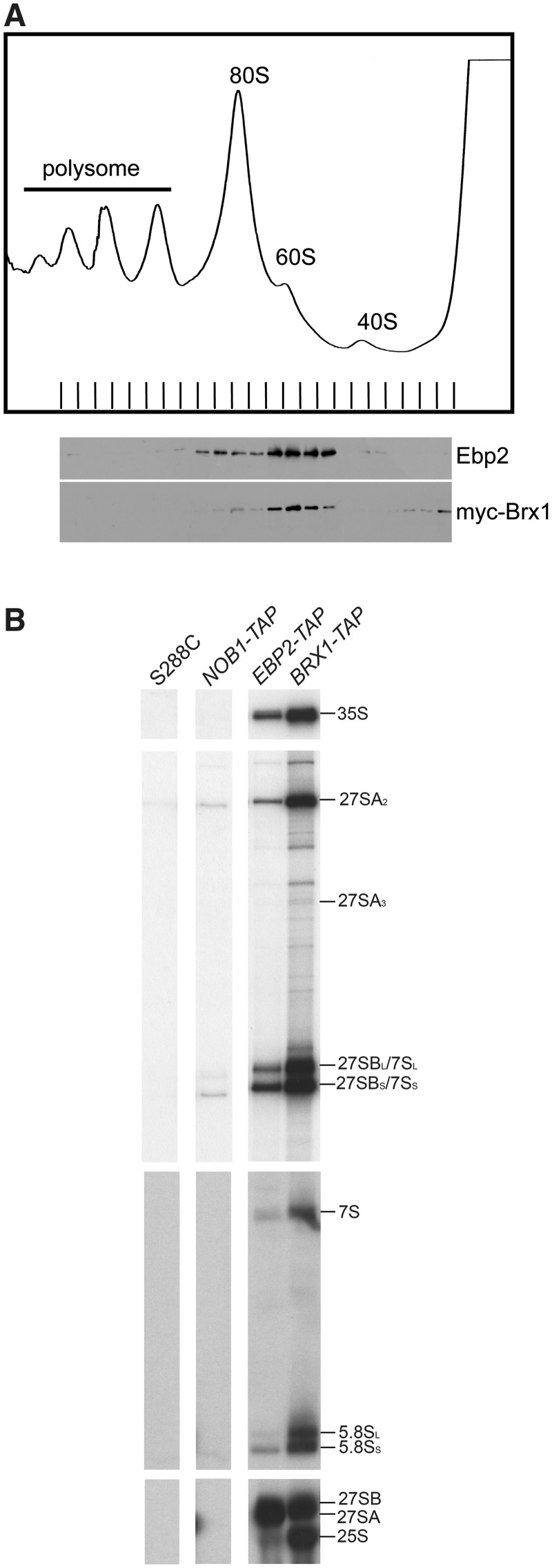
Before investigating the effect of the ebp2 and brx1 mutations on association of Ebp2 and Brx1 with pre-ribosomes, we wanted to determine with which pre-ribosomes they are associated. Brx1 and Ebp2 are routinely detected by mass spectrometry in pre-ribosomes affinity-purified using TAP-tagged early or middle assembly factors, but not with late assembly factors (reviewed in ref. 2). Sucrose gradients confirmed co-sedimentation of Brx1 and Ebp2 with 66S pre-ribosomes (Figure 5A). Myc-tagged Brx1 also sedimented in fractions at the top of the gradient.
Figure 5.
Both Brx1 and Ebp2 assemble early into pre-ribosomes. (A) KM1725 (myc-BRX1) cells were grown at 25°C to mid-log phase, then cell extracts were prepared and subjected to sucrose density gradient ultracentrifugation to resolve ribosomal subunits, monoribosomes and polyribosomes. A ribosomal profile was determined by measurement of OD254 of gradient (7–47%) fractions. Each fraction was collected and subjected to SDS–PAGE and western analysis using antibodies against Ebp2 and the myc epitope. (B) Both Ebp2 and Brx1 associate with 90S pre-ribosomes containing 35S pre-rRNA, as well as 66S pre-ribosomes containing 27S pre-rRNAs. TAP-tagged Ebp2 and Brx1 were used to affinity-purify pre-ribosomes. Pre-RNAs contained in these pre-rRNPs were identified by primer extension and gel electrophoresis. The untagged parent strain S288c and Nob1-Tap were used as controls.
To determine more precisely when Brx1 and Ebp2 enter the pathway of ribosome assembly, we assayed with which of the consecutive pre-rRNA processing intermediates TAP-tagged Brx1 and Ebp2 co-purify. Primer extension and northern blotting indicated that 35S, 27S and 7S pre-rRNAs co-purified with Brx1-TAP and Ebp2-TAP, whereas significantly smaller amounts of pre-rRNAs were detected with Nob1-TAP, a component of 43S precursors to 40S subunits, or with untagged control samples (Figure 5B). The 5.8S and 25S rRNAs also copurified with TAP-tagged Ebp2 or Brx1 above control levels, although the amount that copurified with Ebp2-TAP was much less than with Brx1-TAP. This suggests that Ebp2 dissociates from pre-ribosomes before Brx1 or else that Ebp2 is less stably associated with late pre-ribosomes and fails to copurify with large amounts of mature rRNAs. Taken together, these results suggest that both Brx1 and Ebp2 assemble into 90S pre-ribosomal particles containing 35S pre-rRNA and are also present in all 66S pre-ribosomal particles (Figure 5B), consistent with their role in early steps of pre-rRNA processing (Figure 4B).
Interaction between Ebp2 and Brx1 is not necessary for their assembly into pre-ribosomes
To determine the effect of sl mutations of ebp2 and brx1 on association of these proteins with pre-ribosomes, we began by assaying the ability of the ebp2-14 mutant protein to compete with wild-type Ebp2 for association with pre-ribosomes. When myc-tagged ebp2-14 was co-expressed with wildtype Ebp2 in a wild-type BRX1 background, only wild-type Ebp2 protein cosedimented with pre-ribosomes on sucrose gradients, even though both forms of the Ebp2 protein were present in cells (Figure 6A). Upon depletion of Ebp2 only ebp2-14 was expressed, and it cosedimented with pre-ribosomes, although an equal amount sedimented at the top of gradients (Figure 6A). This result demonstrates that the ebp2 mutant protein does not stably associate with pre-rRNPs as strongly as the wild-type Ebp2 even at 25°C. However, when the wild-type Ebp2 protein is absent, the mutant ebp2 protein can enter pre-ribosomes.
Figure 6.
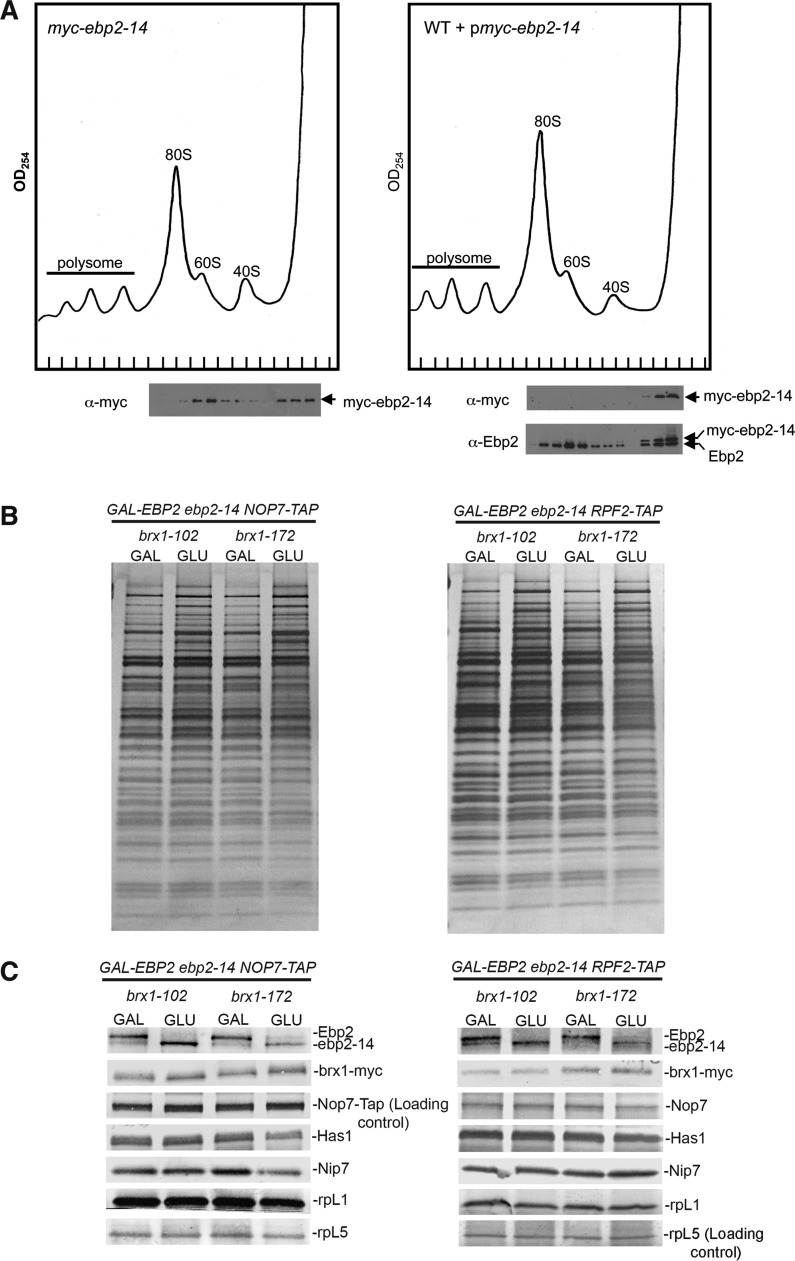
The synthetic lethal mutant ebp2-14 and brx1-102 proteins can assemble into pre-ribosomes, even though they have lost ability to interact with each other in the two-hybrid system. (A) A defect of the mutant ebp2-14 protein at 25°C is revealed by a competition assay. The ebp2–14 mutant protein can assemble into pre-ribosomes, but not when wild-type Ebp2 protein is present. KM1514 (myc-ebp2; left) and wild-type (EBP2) transformed with a plasmid expressing myc-ebp2-14 under the EBP2 promoter (right) were cultured at 25°C. Extracts were prepared from the unshifted and shifted strains and subjected to centrifugation on 7–47% sucrose gradients. Proteins were precipitated from each fraction with 10% TCA. Amounts of wildtype Ebp2 or mutant ebp2 in pre-ribosomes were assayed by western blot analysis using anti-Ebp2 antisera. Anti-myc antibody was used to detect only myc-tagged ebp2-14 protein. (B) The GAL-EBP2 ebp2-14 brx1-102 and GAL-EBP2 ebp2-14 brx1-172 strains containing NOP7-TAP (left), and the GAL-EBP2 ebp2-14 brx1-102 and GAL-EBP2 ebp2-14 brx1-172 strains containing RPF2-TAP (right) were grown in galactose medium at 25°C and shifted to glucose-containing medium for 16 h. Pre-ribosomes were affinity-purified using TAP-tagged assembly factor Nop7 (left) or Rpf2 (right). Proteins present in purified pre-ribosomes were resolved by SDS–PAGE and stained with silver. (C) Amounts of HA-tagged wild-type Ebp2 protein, ebp2-14 mutant protein and myc-tagged brx1 mutant protein were assayed by western blotting using antibodies against Ebp2 and myc. Antibodies against other assembly factors and rp were used as controls. Loading controls are marked with an asterisk. Wild-type HA-tagged Ebp2 could be distinguished from mutant ebp2-14 due to the altered electrophoretic moblity caused by the HA tag.
We then investigated the effect of the synthetic lethal ebp2 brx1 mutations on association of Ebp2 and Brx1 with pre-ribosomes. In order to purify pre-ribosomes, we TAP-tagged assembly factors Nop7 or Rpf2 in KM1732 (GAL-HA-EBP2 ebp2-14 myc-brx1-102) and KM1733 (GAL-HA-EBP2 ebp2-14 myc-brx1-172). Both Nop7 and Rpf2 co-purify with 90S and 66S pre-ribosomes (23,24), and thus can be used to purify the precursors to 60S subunits and survey changes in them. These strains were grown at 25°C in galactose-containing medium, and shifted to glucose-containing medium to deplete wild-type Ebp2 protein and uncover the sl interaction. Pre-ribosomes were affinity-purified from each strain and proteins in them were resolved by SDS–PAGE (Figure 6B). Western blotting was used to assay amounts of Ebp2 and Brx1 in each sample of pre-ribosomes (Figure 6C). The wild-type HA-tagged Ebp2 and the mutant ebp2-14 protein could be distinguished using anti-Ebp2 antisera, which cross-reacted with both proteins. The HA tag on wild-type Ebp2 causes it to migrate more slowly on gels than the mutant ebp2-14 protein. The mutant Brx1 proteins were identified using a myc tag. In both ebp2-14 brx1-102 and the ebp2-14 brx1-172 double mutant strains, amounts of brx1-102 and brx1-172 mutant proteins in pre-ribosomes were unchanged when wild-type Ebp2 was depleted and ebp2-14 could enter pre-ribosomes (Figure 6C). Since the interaction at permissive temperature between ebp2-14 and brx1-102 was shown to be disrupted (Figure 3), but was still intact between ebp2-14 and brx1-172, we conclude that the strong interaction between Ebp2 and Brx1 is not necessary for their assembly into pre-ribosomes. Alternatively, it is possible that the interaction seen between the mutant ebp2 and brx1 proteins in the context of the two-hybrid assay is disrupted, but within pre-ribosomal particles these two proteins are held in close proximity by the surrounding proteins or RNA structure.
Brx1 depends on Ebp2, but Ebp2 does not require Brx1 to assemble into pre-ribosomes
To further investigate the interdependence of Brx1 and Ebp2 for assembly into pre-ribosomes, we assayed the effects of depleting either protein, using GAL promoter fusions. As previously reported (6,25), depletion of either protein is lethal, causing a block in processing of 27SA2 and 27SA3 pre-rRNA to 27SB pre-rRNA (Figure 7A), and consequently decreased production of 60S ribosomal subunits. To purify pre-ribosomes, we TAP-tagged Rpf2, an assembly factor required for pre-rRNA processing steps after those involving Ebp2 or Brx1 (24). To assay association of Ebp2 and Brx1 with pre-ribosomes when either protein was depleted, we epitope-tagged Brx1 in the GAL-EBP2 strain, and used Ebp2 antibody in the GAL-BRX1 strain. SDS-PAGE of proteins in pre-ribosomes purified from either depletion strain revealed significant changes compared to undepleted strains (Figure 7B, left, Figure 7C, left). Although many bands decreased upon depletion of Ebp2 and Brx1, we noticed an enrichment of early assembly factors relative to late factors and r proteins. Thus these effects, including those on Ebp2 and Brx1, may simply reflect the absence of middle and late assembly intermediates in these two depleted strains blocked in the production of the later pre-ribosomes. Therefore, in order to be able to better compare pre-ribosomes from undepleted and depleted samples, we TAP-tagged Rrp5, an assembly factor that enters pre-ribosomes early, associating with 90S pre-ribosomes, and that leaves the pathway before the 27SA3 pre-rRNA processing step (J. Talkish, unpublished). Thus using Rrp5-TAP, we could more specifically assay changes in early particles. When Rrp5-TAP was used to purify pre-ribosomal particles in the presence and the absence of Ebp2 or Brx1, many fewer changes in pre-ribosomal proteins in depleted strains were visible by silver staining (Figure 7B, right). Western blotting of proteins in these pre-ribosomes demonstrated that HA-tagged Brx1 was absent upon depletion of Ebp2. However, when Brx1 was depleted, amounts of Ebp2 in pre-ribosomes were not changed (Figure 7C, right). Thus, Brx1 depends on Ebp2 to stably associate with early pre-ribosomes, but not vice versa. This result suggests that there may be molecules in pre-ribosomes other than Brx1 that are important for association of Ebp2 with pre-rRNPs, but which do not strictly depend on the presence of Brx1 to associate with pre-rRNPs or to maintain Ebp2 in pre-rRNPs.
Figure 7.
Brx1 depends on Ebp2 to assemble into pre-ribosomes, but Ebp2 does not require Brx1. GAL-EBP2 and GAL-BRX1 yeast cells were grown in galactose-containing medium, or shifted to glucose-containing medium for 16 h. (A) The pre-rRNA processing defect in GAL-EBP2 and GAL-BRX1. RNA was extracted from unshifted and shifted strains and assayed by primer extension. (B) SDS–PAGE of proteins in pre-ribosomes purified from strains containing or lacking Ebp2 and Brx1 are shown. Pre-ribosomes were affinity-purified from each culture, using TAP-tagged assembly factor Rpf2 (left) or Rrp5 (right), and proteins in them were resolved by SDS-PAGE and stained with silver. (C) Western blot analysis of association of HA-tagged Ebp2 and HA-tagged Brx1 with pre-ribosomes, upon depletion of Brx1 or Ebp2 respectively. Loading controls are marked with an asterisk. (D) Schematic representation of pre-ribosomal particles co-purifying with Rpf2-Tap and with Rrp5-Tap.
DISCUSSION
Previously Ebp2 and Brx1 were identified as phylogenetically conserved nucleolar proteins involved in early steps of 60S ribosomal subunit assembly in yeast. In the ebp2-1 temperature-sensitive mutant, processing of 27SA2 pre-rRNA is perturbed (7). Upon depletion of Brx1, 27SA2 and 27SA3 pre-rRNAs accumulate and 27SBS pre-rRNA is diminished (25). Here, we have identified brx1 mutations that exhibit synthetic lethal interactions with the ebp2-14 mutation. Three of these four brx1 mutations also eliminate the strong two-hybrid interaction between Ebp2 and Brx1. Despite this inability to interact with each other, both mutant ebp2 and brx1 proteins can associate stably with pre-ribosomes. Nevertheless, these sl interactions result in defects in pre-rRNA processing. Thus, these interactions may enable us to investigate functions of Ebp2 and Brx1 after their initial docking into pre-rRNPs. We discuss a broader network of physical and functional interactions in which Ebp2 and Brx1 might participate to enable early steps of 60S subunit biogenesis.
We previously suggested that Ebp2 might enter 90S pre-ribosomal particles based on two-hybrid data demonstrating that Ebp2 associates with the 40S subunit assembly factors Utp11 and Faf1, and r protein S16 (9). Consistent with Ebp2 and Brx1 functioning early in assembly, both proteins copurify with ribosome assembly factors that function at early or middle steps in the pathway (2). Using the consecutive pre-rRNA processing intermediates as landmarks, we also found that Ebp2 and Brx1 are associated with the 35S pre-rRNA present in 90S pre-ribosomes, as well as each of the 27S pre-rRNAs found in 66S pre-rRNPs (Figure 5B). Neither of these experimental approaches distinguishes the order in which Ebp2 and Brx1 dock with nascent particles. However, two observations suggest that Ebp2 might assemble before Brx1: (i) Brx1 depends on Ebp2 to associate with pre-ribosomes, but not vice versa (Figure 7C, right). (ii) Depletion of Brx1 blocks 27SA3 pre-rRNA processing downstream of the step blocked by depletion of Ebp2 (Figure 7A). Nevertheless, a higher resolution view of the assembly hierarchy awaits better tools to investigate ribosome biogenesis.
The genetic interactions between ebp2-14 and brx1 could provide additional insight into the function of Ebp2 and Brx1. At 25°C, each of the single ebp2 or brx1 missense mutants has no apparent defect in growth or in production of 60S ribosomal subunits (Figure 2A, Figure 4A, see ref.8). Even the brx1-227 mutant, which is temperature sensitive for growth, appears not to have a defect in production of 60S ribosomal subunits at 25°C (Figure 2B). In contrast, the ebp2-14 brx1 double mutants are unable to grow, and are defective in production of 60S subunits and processing of 27SA2 and 27SA3 pre-rRNAs (Figure 2C, 4A). The ebp2-14 mutation together with brx1-52, brx1-102 or brx1-227 mutations prevents stable interaction of ebp2 and brx1 mutant proteins with each other in a two-hybrid assay (Figure 3B). The combination of ebp2-14 and brx1-172 has little effect on the interaction. As the combination of ebp2-12 (other allele, see ref. 8) and brx1-172 also has no effect on the interaction (data not shown), we speculate that the brx1-172 mutant protein, similar to Brx1, might interact with ebp2 mutant proteins as well as with Ebp2. Despite the loss of interaction, both ebp2-14 and brx1-102 mutant proteins can efficiently assemble into nascent ribosomes in the double mutants, as well as the ebp2-14 brx1-172 mutant proteins where the two-hybrid interaction is not abolished (Figure 6B). This result indicates that other protein–protein or protein–RNA interactions within assembling ribosomes must enable Ebp2 and Brx1 to stably associate with pre-ribosomes. A similar result for assembly factors Imp4 and Mpp10 was found previously by Gallagher and Baserga (26). Mutations in Imp4 that disrupt two-hybrid interactions with Mpp10, also cause a slow growth phenotype and perturb pre-rRNA processing, but nevertheless do not prevent either protein from associating with the SSU processome.
Although the ebp2-14 and the brx1-102 mutant proteins can assemble into pre-ribosomes, the ebp2-14 brx1-102 double mutant is defective in processing of 27SA2 and 27SA3 pre-rRNAs, more so than either single mutant (Figure 4B). Furthermore, the ebp2-14 brx1-172 double mutant in which the Ebp2–Brx1 interaction is not diminished, nevertheless produces fewer 60S subunits than either single mutant, and is defective in processing of 27SA3 as well as 27SA2 pre-rRNAs (Figure 4B).
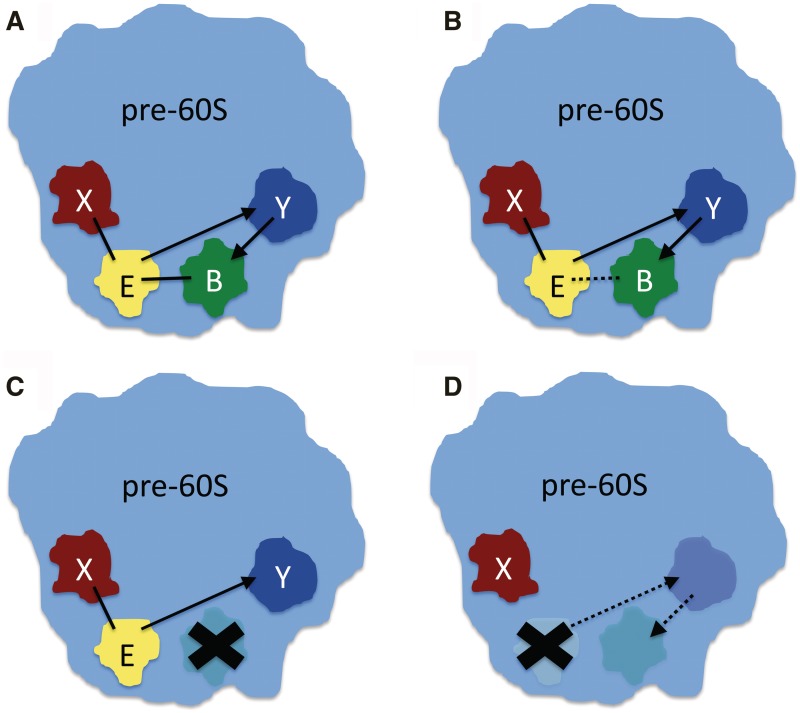
The effects on ribosome assembly upon depleting either Ebp2 or Brx1 support the existence of a broader network of pre-ribosomal interactions with Brx1 and Ebp2 (Figure 8). Brx1 depends on Ebp2 to associate with early pre-ribosomes, but Ebp2 does not require Brx1 to do so (Figure 7C, right). Thus, there must be other molecules in pre-rRNPs not affected by the absence of Brx1 that help anchor Ebp2 in pre-rRNPs. On the other hand, Brx1 may not depend on direct interactions with Ebp2 to assemble, but on a third molecule in pre-ribosomes, whose presence or anchoring function is diminished by the absence of Ebp2 (Figure 8).
Figure 8.
Model of dependence of Brx1 on Ebp2 for assembly into pre-ribosomes. Ebp2 (E) and Brx1 (B) are represented with yellow and green, respectively. X (red) and Y (blue) represent other proteins present in pre-ribosomes. Each X and each Y can represent one or more proteins. Black lines mark interactions. Loss of interaction is shown as dashed line. Arrows point FROM the protein required to recruit another protein into pre-ribosomes TO the protein dependent on the first protein to be in pre-ribosomes. (A) In the wild-type, Ebp2 and Brx1 interact with each other and each interacts with other proteins within pre-ribosomes (pre-60S). (B) Loss of interaction between Ebp2 and Brx1 does not cause either protein to be absent from pre-ribosomal particles, presumably due their interactions with other proteins. (C) In the absence of Brx1, Ebp2 is still present in pre-ribosomes. (D) The absence of Ebp2 affects one or more proteins that stabilize the association of Brx1 with pre-ribosomes. In their absence, Brx1 is absent from pre-ribosomes.
The best candidates for factors that help Ebp2 or Brx1 enter pre-ribosomes and function in these particles are four other ribosome assembly factors and r proteins found in subcomplexes together with Ebp2 or Brx1. Krogan et al. (27) found that Brx1 copurified with Nop12 and Pwp1, when TAP-tagged Pwp1 was used for affinity purification from high speed supernatants of whole cell extracts from which intact pre-ribosomes and ribosomes were removed. By a similar approach, Zhang et al. (28) found that Ebp2, Brx1, Nop12 and r proteins L8 and L15 copurified with TAP-tagged Pwp1. Consistent with these observations, Ebp2 interacts not only with Brx1 in two-hybrid assays, but also with Nop12 (Figure 3A).
The association of Brx1 and Ebp2 with these four assembly factors and r proteins provides some hints about their possible location within pre-ribosomes. The r proteins L8 and L15 lie adjacent to each other in mature 60S ribosomal subunits, near the proximal stem formed by base-pairing of the 3′-end of 5.8S rRNA with the 5′-end of 25S rRNA (4,5). During biogenesis of the 60S subunit, these 5′-and 3′-ends of rRNA are formed by processing of the internal transcribed spacer 2 (ITS2) that lies between them. Nop12 cross-links to nucleotides immediately adjacent to the proximal stem, but distal from ITS2 (29). This physical neighborhood of six pre-ribosomal proteins, including Brx1 and Ebp2, is also functionally interconnected. The phenotype of nop12, pwp1 and rpl8 mutants is similar to that of brx1 and ebp2 mutants: 27SA2 and 27SA3 processing are perturbed (JJ and JT, unpublished).
The ‘A3 cluster’ of assembly factors required for processing of 27SA3 pre-rRNA (14) may also be part of a larger local neighborhood of proteins that interact with Ebp2 and Brx1 and function together with them. Erb1 and Nop7 crosslink to rRNA in domains I and III near the proximal stem (29), and Ytm1 binds directly to Erb1 (30). Nop15 and Cic1 cross-link to sequences in ITS2, which lies adjacent to the proximal stem (29). Thus, it seems likely that Ebp2 and Brx1 are located in pre-ribosomes near the proximal stem and function together with Nop12, Pwp1, r proteins L8 and L15, and perhaps other nearby A3 factors, to create RNP structures necessary for early steps of 27S pre-rRNA processing.
How might the genetic and physical interactions between Ebp2 and Brx1, and between these two proteins and the other pre-ribosomal proteins described above, relate to their function in ribosome assembly and pre-rRNA processing? Their overlapping mutant phenotypes indicate that these proteins participate either directly or indirectly in processing of 27SA2 pre-RNA and subsequently 27SA3 pre-rRNA. This first processing step involves endonucleolytic cleavage at the A3 site in the internal transcribed spacer 1 (ITS1) by the RNase MRP endonuclease (31–33). Subsequently, 77 nt at the 5′-end of the resulting 27SA3 pre-rRNA are removed by the 5′–3′ exonucleases Rat1, Xrn1 and Rrp17, stopping precisely at the B1S site, to produce 27SBS pre-rRNA (34,35). In both steps, the proper timing and efficiency of cleavage and processing of these sequences in ITS1 must depend upon the pre-rRNA substrates presenting the appropriate conformation to be recognized and acted upon by the nucleases. Curiously, five A3 factors, as well as r proteins L8 and L15, are associated with pre-rRNA near the 3′-end of 5.8S rRNA, perhaps some distance from ITS1 located upstream of the 5′-end of 5.8S rRNA (29). It seems likely that these seven proteins, which bind RNA but lack any apparent enzymatic activities, may function in these processing steps by establishing conformations of pre-rRNA to create long-range interactions between the 5′-and 3′-ends of 5.8S rRNA, analogous to allosteric interactions that occur within the ribosome during protein synthesis (36). As pointed out by Granneman et al. (29), the A3 factors may coordinate folding and processing at the 5′-and 3′-ends of 5.8S rRNA to ensure that ITS1 processing occurs after proper folding of ITS2, and before removal of ITS2.
We recently found that Ebp2 and Rrs1 are localized at the nuclear periphery as well as the nucleolus and play roles in telomere organization and silencing (37). Although it remains to be elucidated if Brx1 has similar functions, the synthetic lethality of brx1 with ebp2 reflects cooperative functions of the two proteins in ribosome biogenesis, not in other functions, because suppression of a defect in telomere organization of ebp2-14 did not suppress either a defect in temperature sensitivity for growth or a defect in ribosome biogenesis (37).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table S1.
FUNDING
Japan-US Cooperative Science Program and Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (to K.M.); Grant from the National Institutes of Health (grant GM28301 to J.W.); The Richard King Mellon Foundation Presidential Graduate Fellowship and the Semon H. Stupakoff Scholarship (to J.T.). Funding for open access charge: Management Expenses Grant from Japanese government.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Leland Hartwell for a yeast strain, David Shore and Kazuma Tanaka for plasmids, and Kouichi Funato for the yeast genomic DNA library. We thank the following people for their generous gifts of antibodies: Jesús de la Cruz and Patrick Linder (Has1), David Goldfarb (Nip7), Michael McAlear (Ebp2), Cosmin Saveanu and Micheline Fromont-Racine (Tif6), Arlen Johnson (rpL8), Sabine Rospert (rpL17), Elizabeth Tosta (Cic1/Nsa3) and Francois Lacroute (rpL1). We are grateful to members of our laboratories for critical discussions and reading the article.
REFERENCES
- 1.Raue HA. Pre-ribosomal RNA processing and assembly in Saccharomyces cerevisiae: the machine that makes the machine. In: Olson MOJ, editor. The Nucleolus. NY: Kulwer Academic/Plenum Publishers; 2003. pp. 1–24. [Google Scholar]
- 2.Henras AK, Soudet J, Gérus M, Lebaron S, Caizergues-Ferrer M, Mougin A, Henry Y. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol. Life Sci. 2008;65:2334–2359. doi: 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kressler D, Hurt E, Bassler J. Driving ribosome assembly. Biochim. Biophys. Acta. 2010;1803:673–683. doi: 10.1016/j.bbamcr.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 4.Ben-Shem A, Jenner L, Yusupova G, Yusupov M. Crystal structure of the eukaryotic ribosome. Science. 2010;330:1203–1209. doi: 10.1126/science.1194294. [DOI] [PubMed] [Google Scholar]
- 5.Klinge S, Voigts-Hoffmann F, Leibundgut M, Arpagaus S, Ban N. Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science. 2011;334:941–948. doi: 10.1126/science.1211204. [DOI] [PubMed] [Google Scholar]
- 6.Tsujii R, Miyoshi K, Tsuno A, Matsui Y, Toh-e A, Miyakawa T, Mizuta K. Ebp2p, yeast homologue of a human protein that interacts with Epstein-Barr virus Nuclear Antigen 1, is required for pre-rRNA processing and ribosomal subunit assembly. Genes Cells. 2000;5:543–553. doi: 10.1046/j.1365-2443.2000.00346.x. [DOI] [PubMed] [Google Scholar]
- 7.Huber MD, Dworet JH, Shire K, Frappier L, McAlear MA. The budding yeast homolog of the human EBNA1-binding protein 2 (Ebp2p) is an essential nucleolar protein required for pre-rRNA processing. J. Biol. Chem. 2000;275:28764–28773. doi: 10.1074/jbc.M000594200. [DOI] [PubMed] [Google Scholar]
- 8.Horigome C, Okada T, Matsuki K, Mizuta K. A ribosome assembly factor Ebp2p, the yeast homolog of EBNA1-binding protein 2, is involved in the secretory response. Biosci. Biotechnol. Biochem. 2008;72:1080–1086. doi: 10.1271/bbb.70817. [DOI] [PubMed] [Google Scholar]
- 9.Shirai C, Takai T, Nariai M, Horigome C, Mizuta K. Ebp2p, the yeast homolog of Epstein-Barr virus nuclear antigen 1-binding protein 2, interacts with factors of both the 60S and 40S ribosomal subunit assembly. J. Biol. Chem. 2004;279:25353–25358. doi: 10.1074/jbc.M403338200. [DOI] [PubMed] [Google Scholar]
- 10.Longtine MS, McKenzie A, III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 11.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteomic exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 12.Kaiser C, Michaelis S, Mitchell A. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 13.Koshland D, Kent JC, Hartwell LH. Genetic analysis of the mitotic transmission of minichromosomes. Cell. 1985;40:393–403. doi: 10.1016/0092-8674(85)90153-9. [DOI] [PubMed] [Google Scholar]
- 14.Sahasranaman A, Dembowski J, Strahler J, Andrews P, Maddock J, Woolford JL., Jr Assembly of Saccharomyces cerevisiae 60S ribosomal subunits: role of factors required for 27S pre-rRNA processing. EMBO J. 2011;30:4020–4032. doi: 10.1038/emboj.2011.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horsey EW, Jakovljevic J, Miles TD, Harnpicharnchai P, Woolford JL., Jr Role of the yeast Rrp1 protein in the dynamics of pre-ribosome maturation. RNA. 2004;5:813–827. doi: 10.1261/rna.5255804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. NY: John Wiley & Sons Inc.; 1994. [Google Scholar]
- 17.Eisenhaber F, Wechselberger C, Kreil G. The Brix domain protein family, a key to the ribosomal biogenesis pathway? Trends Biochem. Sci. 2001;26:345–347. doi: 10.1016/s0968-0004(01)01851-5. [DOI] [PubMed] [Google Scholar]
- 18.Wehner KA, Baserga SJ. The sigma(70)-like motif: a eukaryotic RNA binding domain unique to a superfamily of proteins rerquired for ribosome biogenesis. Mol. Cell. 2002;9:329–339. doi: 10.1016/s1097-2765(02)00438-0. [DOI] [PubMed] [Google Scholar]
- 19.Fatica A, Cronshaw AD, Dlakic M, Tollervey D. Ssf1p prevents premature processing of an early pre-60S ribosomal particle. Mol. Cell. 2002;9:341–351. doi: 10.1016/s1097-2765(02)00458-6. [DOI] [PubMed] [Google Scholar]
- 20.Tsuno A, Miyoshi K, Tsujii R, Miyakawa T, Mizuta K. RRS1, a conserved essential gene, encodes a novel regulatory protein required for ribosome biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 2000;20:2066–2074. doi: 10.1128/mcb.20.6.2066-2074.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyoshi K, Shirai C, Horigome C, Takenami K, Kawasaki J, Mizuta K. Rrs1p, a ribosomal protein L11-binding protein, is required for nuclear export of the 60 S pre-ribosomal subunit in Saccharomyces cerevisiae. FEBS Lett. 2004;565:106–110. doi: 10.1016/j.febslet.2004.03.087. [DOI] [PubMed] [Google Scholar]
- 22.Morita D, Miyoshi K, Matsui Y, Toh-e A, Shinkawa H, Miyakawa T, Mizuta K. Rpf2p, an evolutionarily conserved protein, interacts with ribosomal protein L11 and essential for the processing of 27SB pre-rRNA to 25S rRNA and is the 60S ribosomal subunit assembly in Saccharomyces cerevisiae. J. Biol. Chem. 2002;277:28780–28786. doi: 10.1074/jbc.M203399200. [DOI] [PubMed] [Google Scholar]
- 23.Harnpicharnchai P, Jakovljevic J, Horsey E, Miles T, Roman J, Rout M, Meagher D, Imai B, Guo Y, Brame CJ, et al. Composition and functional characterization of yeast 66S ribosome assembly intermediates. Mol. Cell. 2001;8:505–515. doi: 10.1016/s1097-2765(01)00344-6. [DOI] [PubMed] [Google Scholar]
- 24.Zhang J, Harnpicharnchai P, Jakovljevic J, Tang L, Guo Y, Oeffinger M, Rout MP, Hiley SL, Hughes T, Woolford JL., Jr Assembly factors Rpf2 and Rrs1 recruit 5S rRNA and ribosomal proteins rpL5 and rpL11 into nascent ribosomes. Genes Dev. 2007;21:2580–2592. doi: 10.1101/gad.1569307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaser A, Bogengruber E, Hallegger M, Doppler E, Lepperdinger G, Jantsch M, Breitenbach M, Kreil G. Brix from Xenopus laevis and Brx1p from yeast define a new family of proteins involved in the biogenesis of large ribosomal subunits. Biol. Chem. 2001;382:1637–1647. doi: 10.1515/BC.2001.199. [DOI] [PubMed] [Google Scholar]
- 26.Gallagher JE, Baserga SJ. Two-hybrid Mpp10 interaction-defective Imp4 proteins are not interaction defective in vivo but do confer specific pre-rRNA processing defects in Saccahromyces cerevisiae. Nucleic Acids Res. 2003;32:1404–1413. doi: 10.1093/nar/gkh318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krogan NJ, Peng WT, Cagney G, Robinson MD, Haw R, Zhong G, Guo X, Zhang X, Canadien V, Richards DP, et al. High definition macromolecular composition of yeast RNA-processing complexes. Mol. Cell. 2004;13:225–239. doi: 10.1016/s1097-2765(04)00003-6. [DOI] [PubMed] [Google Scholar]
- 28.Zhang W, Morris QD, Chang R, Shai O, Bakowski MA, Mitsakakis N, Mohammad N, Robinson MD, Zirngibl R, Somogyi E, et al. The functional landscape of mouse gene expression. J. Biol. 2004;3:e21. doi: 10.1186/jbiol16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Granneman S, Petfalski E, Tollervey D. A cluster of ribosome synthesis factors regulate pre-rRNA folding and 5.8S rRNA maturation by the Rat1 exonuclease. EMBO J. 2011;30:4006–4019. doi: 10.1038/emboj.2011.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miles TD, Jakovljevic J, Horsey EW, Harnpicharnchai P, Tang L, Woolford JL., Jr Ytm1, Nop7, and Erb1 form a complex necessary for maturation of yeast 66S pre-ribosomes. Mol. Cell Biol. 2005;25:10419–10432. doi: 10.1128/MCB.25.23.10419-10432.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmitt ME, Clayton DA. Nuclear RNase MRP is required for correct processing of pre-5.8S rRNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 1993;13:7935–7941. doi: 10.1128/mcb.13.12.7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chu S, Archer RH, Zengel JM, Lindahl L. The RNA of RNase MRP is required for normal processing of ribosomal RNA. Proc. Natl Acad. Sci. USA. 1994;91:659–663. doi: 10.1073/pnas.91.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lygerou Z, Allmang C, Tollervey D, Seraphin B. Accurate processing of a eukaryotic precursor ribosomal RNA by ribonuclease MRP in vitro. Science. 1996;272:268–270. doi: 10.1126/science.272.5259.268. [DOI] [PubMed] [Google Scholar]
- 34.Henry Y, Wood H, Morrissey JP, Petfalski E, Kearsey S, Tollervey D. The 5' end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. EMBO J. 1994;13:2452–2463. doi: 10.1002/j.1460-2075.1994.tb06530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oeffinger M, Zenklusen D, Ferguson A, Wei KE, El Hage A, Tollervey D, Chait BT, Singer RH, Rout MP. Rrp17p is a eukaryotic exonuclease required for 5' end processing of Pre-60S ribosomal RNA. Mol. Cell. 2009;36:768–781. doi: 10.1016/j.molcel.2009.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmeing TM, Ramakrishnan V. What recent ribosome structures have revealed about the mechanism of translation. Nature. 2009;461:1234–1242. doi: 10.1038/nature08403. [DOI] [PubMed] [Google Scholar]
- 37.Horigome C, Okada T, Shimazu K, Gasser SM, Mizuta K. Ribosome biogenesis factors bind a nuclear envelope SUN domain protein to cluster yeast telomeres. EMBO J. 2011;30:3799–3811. doi: 10.1038/emboj.2011.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.