Abstract
The modification of RNA with fluorophores, affinity tags and reactive moieties is of enormous utility for studying RNA localization, structure and dynamics as well as diverse biological phenomena involving RNA as an interacting partner. Here we report a labeling approach in which the RNA of interest—of either synthetic or biological origin—is modified at its 3′-end by a poly(A) polymerase with an azido-derivatized nucleotide. The azide is later on conjugated via copper-catalyzed or strain-promoted azide–alkyne click reaction. Under optimized conditions, a single modified nucleotide of choice (A, C, G, U) containing an azide at the 2′-position can be incorporated site-specifically. We have identified ligases that tolerate the presence of a 2′-azido group at the ligation site. This azide is subsequently reacted with a fluorophore alkyne. With this stepwise approach, we are able to achieve site-specific, internal backbone-labeling of de novo synthesized RNA molecules.
INTRODUCTION
In the post-genomic era, RNA has become one of the most interesting objects of investigation in diverse fields of research. Chemical RNA modification is often indispensable, not only for visualization (1), studies of structure and dynamics (2,3), purification (4), immobilization (5), or cross-linking of RNA to its biomolecular interaction partners (6,7), but also for modulation of its properties (8).
Certain modifications can be incorporated during chemical oligonucleotide synthesis (9,10), limited to shorter sequences that can be connected by enzymatic ligation to obtain longer ones. During in vitro RNA synthesis (e.g. by T7 RNA polymerase—T7 RNAP), it is possible to incorporate 5′-terminal modifications (11,12) and internal modifications, which are site-specific only when non-natural base pairs are used (13). Evidently, co-synthetic methods are unusable for RNA isolated from living organisms, where post-synthetic modification can be a valuable alternative. For natural and in vitro synthesized RNA, the 5′-terminus is often blocked by mono- and triphosphates, or cap structures. Thus, the 3′-terminus can be labeled more easily. Besides the chemical modification of the 3′-terminal vicinal diol (14,15), various enzymatic methods employing ligases (16–18) or nucleotidyl transferases, like terminal deoxynucleotidyl transferase (TdT) (19) or poly(A) polymerase (PAP) (20) have been reported. For post-synthetic, internal labeling of RNA, DNAzymes (21), twin ribozymes (22) or hybridization-based functionality transfer reactions (23) can be used, all of which rely on chemically synthesized, modified oligonucleotides. Although the conversion of terminal modifications to internal ones by ligases or nucleotidyl transferases has been reported (7), this approach is limited to the introduction of small modifications. Bulky modifications, such as fluorophores have not been introduced using these methods, presumably due to the severe sterical hindrance imposed by the modification. On the one hand, this sterical hindrance hampers the enzymatic introduction of the modified nucleotide, e.g. PAP is efficient only with certain modified NTPs (20). On the other hand, it also inhibits further enzymatic manipulations involving the modified end. One solution to circumvent both problems is to employ a modified nucleotide that does not contain the label itself, but rather provides a small reactive handle for a subsequent chemical reaction, in which the desired label can be attached to the RNA of interest. Here, the copper-catalyzed click reaction, or copper-catalyzed 1,3-dipolar azide–alkyne cycloaddition (CuAAC) (24,25) is of particular interest. In this reaction, which has been used to label DNA since 2003 (26–28), and more recently also to label RNA (6,29–33), the reactive groups (terminal alkynes and azides) are relatively small. Thus, they cause only minor structural changes in nucleic acids and their building blocks, so that enzymes are more likely to accept them as substrates. Indeed, for CuAAC labeling, enzymatic incorporation of alkyne-modified nucleotides into DNA in vitro (34–37), and of pyrimidines bearing an ethynyl modification at position C-5 into nucleic acids in vivo (29,38,39) are possible. Post-synthetic approaches, using methyltransferases to transfer alkyne-modified adenosine to DNA (40), or an extended alkynyl moiety to tRNA (30) have equally been reported. However, it can be advantageous to incorporate the azide instead of its alkyne counterpart, since azides have also been shown to participate in other, metal-free, click reactions, such as the strain promoted azide–alkyne cycloaddition (SPAAC) (41). While the introduction of alkyne groups into DNA (42–45) and RNA (31) has been achieved by standard phosphoramidite chemistry, the azido-group is chemically unstable under phosphoramidite coupling conditions (46). This has so far prevented the extensive exploitation of azides for both, DNA and RNA labeling. In contrast, enzymes have been successfully applied for providing nucleic acids with azides for further chemical conversion (32,47,48). For RNA, the incorporation of N3-modified nucleotides by a mutated T7 RNAP has been reported before the advent of CuAAC (48). In a recent study, an N3-modified initiator nucleotide has been incorporated at the 5′-terminus of RNA by T7 RNAP, for further conversion by CuAAC (32). In the same study, 3′-N3-2′,3′-ddATP was added to the 3′-terminus of RNA by Escherichia coli PAP, followed by CuAAC, which does, however, not permit further enzymatic 3′-modifications due to the missing 3′-hydroxyl group.
Here, we set out to expand the scope of RNA click labeling. For 3′-terminal RNA labeling, we tested four different nucleotidyl transferases [yeast and E. coli PAP, Cid 1 poly(U) polymerase (PUP) (49) and TdT] for their ability to incorporate a set of modified NTPs bearing azides at different positions. We developed a toolbox for reliable labeling of RNA with fluorophores and other relevant functional tags via copper-catalyzed (CuAAC) or metal-free strain-promoted (SPAAC) click chemistry for all four possible nucleotides (A, C, G and U), at a wide range of concentrations. Moreover, after introduction of nucleotides modified at position C-8 or C-2′, the resulting 3′-termini contain free hydroxyl groups. Those RNAs could be subjected to enzymatic manipulations involving their modified 3′ termini, namely, addition of poly(A)-tails, 3′-adapter ligation and splinted ligation to other RNA sequences. Doing so, we converted the terminal azide modifications into internal ones, for which we optimized CuAAC reaction conditions, to achieve site-specific, internal RNA labeling of de novo synthesized RNA. This protocol does not demand either de novo synthesis of modified entities or characterization and manipulation of new enzymes, and involves only commercially available reagents and enzymes, making it more accessible to future users.
MATERIALS AND METHODS
Materials
RNA1 and all DNAs (Table 1) were purchased from IBA. RNA2 and RNA3 were kindly provided by Prof. Mark Helm (Institute of Pharmacy and Biochemistry, University of Mainz). RNA4 was purchased from Dharmacon. Modified NTPs were purchased either from BIOLOG, IBA or tebu-bio (Supplementary Table S1). Yeast PAP was purchased from USB. Escherichia coli PAP, Cid1 PUP and RNA ligase 2, truncated (RNL2, tr.) were purchased from New England Biolabs. T4 polynucleotide kinase (PNK), TdT, T4 DNA ligase (DNL), T4 RNA ligase (RNL1) and DNase I were purchased from Fermentas. T4 RNA ligase 2 (RNL2) was prepared in-house. Alexa Fluor 647 alkyne, Alexa Fluor 488 alkyne and biotin-alkyne were purchased from Invitrogen. Aza-dibenzocyclooctyne (DIBAC) Fluor 488 (sold as Dibenzylcyclooctyne-Fluor 488) was purchased from Jena Bioscience. Tris-(3-hydroxypropyltriazolylmethyl)-amine (THPTA) was synthesized according to the published procedure (50). Escherichia coli cells of strain JM 109 were purchased from Promega.
Table 1.
Sequences of oligonucleotides
| Name | Sequence | Features |
|---|---|---|
| RNA1 | 5′-GUGACCGCGGAUCGACUUCACCGCGCAGUG-3′ | 10 nt loop, 5 nt dangling ends |
| RNA2 | 5′-GCAAGCUGACCCUGAAGUUCAU-3′ | siRNA (sense) |
| RNA3 | 5′-GAACUUCAGGGUCAGCUUGCCG-3′ | siRNA (antisense) |
| RNA4 | 5′-pGCCCGACGAUGCUUCAGCAACCAGUGUAAUGGCG-3′ | Ligation fragmenta |
| DNA1 | 5′-CGCCATTACACTGGTTGCTGAAGCATCGTCGGGCCCACTGCGCGGTGAGT CGATCCGCGGTCAC-3′ | DNA splinta |
| DNA2 | 3′-Ap-5′-5′-pCTGTAGGCACCATCAAT-3′-block | Adapter: 3′-block, 5′ adenylated |
| DNA3 | 5′-CGCCATTACACTGGTTGCTGAAGCATCGTCGCGGTGAAGTCGATCCGCGGTCAC-3′ | Helper DNA, assisting CuAAC |
aFor splinted ligation of RNA1 (+G) to RNA4.
Preparation of 5′-radioactively labeled oligonucleotides
For 5′-labeling, RNA and [γ-32P]-ATP (10 µCi/µl, 3000 Ci/mmol; Hartmann Analytic), (end concentration: both 1 µM) were incubated with PNK (1 U/µl) in 1× T4 PNK buffer A (50 mM Tris–HCl (pH 7.6 at 25°C), 10 mM MgCl2, 5 mM DTT, 100 µM spermidine) for 60 min at 37°C. Gel-purified oligonucleotides were recovered by precipitation with ethanol (RNA1) or isopropanol (RNA2, RNA3). To image radioactive bands, we exposed storage phosphor screens (GE Healthcare) to the gels and scanned them with a Typhoon 9400 scanner (GE Healthcare). If not stated otherwise, whenever quantifications are given, radioactive bands were background-corrected (to the object average of a region in the gel not containing any radioactive bands), and quantified using ImageQuant software (Molecular Dynamics; version 5.2).
Screening and optimization of enzymatic incorporation of azide-containing nucleotides to the 3′ terminus of RNA oligonucleotides
To enable visualization of RNA after analysis by denaturing sequencing PAGE (seqPAGE), non-labeled RNA was generally doped with the respective 5′-radiolabeled RNA (final concentrations: 1–20 nM). For initial tests, 5 pmol of RNA1 was incubated with the respective N3-NTP (500 µM) and the respective nucleotidyl transferase in its recommended buffer at 37°C for 60 min (both PAPs and Cid1 PUP) or 90 min (TdT). Amounts of nucleotidyl transferase and buffer used were: 600 U of yeast PAP in 1× USB yeast poly(A) polymerase buffer [20 mM Tris–HCl (pH 7.0 at 25°C), 0.6 mM MnCl2, 0.02 mM EDTA, 100 μg/ml acetylated BSA, 10% glycerol] in a final volume of 25 µl; 5 U of E. coli PAP in 1× NEB E. coli poly(A) polymerase buffer [50 mM Tris–HCl (pH 7.9 at 25°C), 250 mM NaCl, 10 mM MgCl2] in a final volume of 20 µl; 2 U of Cid1 PUP in 1× NEBuffer 2 [10 mM Tris–HCl (pH 7.9 at 25°C), 50 mM NaCl, 10 mM MgCl2, 1 mM DTT] in a final volume of 25 µl; 20 U of TdT in 1× Fermentas terminal deoxynucleotidyl transferase buffer [25 mM Tris (pH 7.2 at 25°C), 200 mM potassium cacodylate, 0.01% (v/v) Triton X-100, 1 mM CoCl2] in a final volume of 20 µl. Reaction mixtures were optionally stored at −20°C until analysis by 15 or 18% seqPAGE. For unit definitions, see Supplementary Methods.
For N3-NTPs modified at positions C-2′ and C-8, in combination with yeast and E. coli PAP, RNA concentrations and incubation times were varied, as indicated. Buffer conditions were maintained, if not stated otherwise. For the 2′-N3-NTPs (except for the very initial study), the PAP was heat-inactivated by incubation at 65°C for 10 min (yeast) or 20 min (E. coli). For 2′-N3-2′-dATP, in combination with yeast PAP, lower NTP concentrations (50 µM) were also employed, whereas with E. coli PAP, double enzyme concentration (0.5 U/µl) and addition of MnCl2 (2.5 mM) were tested at 500 µM NTP concentration.
Modification of RNA with single 2′-N3-nucleotides for CuAAC
The exact reaction conditions used for single nucleotide incorporation at 4 µM or 5 µM RNA concentration, employing yeast PAP or E. coli PAP, respectively, are summarized in Table 2.
Table 2.
Conditions that lead to single nucleotide addition
| Modified NTP | PAP from | Reaction time |
|---|---|---|
| 2'-N3-2'-dATP | Escherichia coli (0.5 U/µl) | 16 h (−MnCl2) |
| Escherichia coli (0.25 U/µl) | 20 min (+MnCl2)a | |
| 2'-N3-2'-dCTP | Yeast | 20 min |
| 2'-N3-2'-dGTP | Yeast | 2 h |
| 2'-N3-2'-dUTP | Yeast | 5 min |
| 3'-N3-2',3'-ddATP | Yeast | Not optimized |
| 3'-N3-2',3'-ddTTP | Yeast | Not optimized |
| 8-N3-ATP | Not possible | – |
General conditions: 5 µM RNA, 0.25 or 0.5 U/µl PAP (E. coli) or 4 µM RNA, 24 U/µl PAP (yeast), commercial buffers (additives as indicated), 500 µM NTP, 37°C. Samples were heat inactivated by incubation at 65°C for 10 min (yeast PAP) or 20 min (E. coli PAP).
aPreferred conditions (2.5 mM MnCl2).
CuAAC with RNA after PAP reaction with N3-modified NTPs
PAP reaction mixtures were purified either by ethanol precipitation (RNA1), isopropanol precipitation (RNA2, RNA3), by size exclusion chromatography employing MicroSpin™ G-25 columns (GE Healthcare) or by QIAquick Nucleotide Removal Kit (Qiagen).
The CuAAC was generally performed with an RNA concentration of 50 nM, 1 µM or 10 µM in 50 mM phosphate buffer (pH 7 at 25°C), 100 µM CuSO4, 500 µM THPTA, 1 mM sodium ascorbate [for in situ generation of Cu(I)] in a final volume of 10–100 µl, and varying concentrations of either Alexa Fluor 488 alkyne, Alexa Fluor 647 alkyne or biotin alkyne. Reactions were performed for 2 h at 37°C. At RNA concentrations above 10 µM (e.g. 50 µM), the concentrations of CuSO4, THPTA and sodium ascorbate were increased 5-fold.
If not stated otherwise, reaction mixtures were purified by phenol extraction (only for Alexa Fluor alkynes) in presence of EDTA (10 mM) and Na-acetate (0.3–0.6 M, pH 5.5), followed by ethanol or isopropanol precipitation, and then analyzed by 12 or 15% seqPAGE. Bands were visualized by Storage Phosphor Screen scan, as well as fluorescence scan on the Typhoon 9400 (Alexa Fluor 488: ex: 488 nm, em: 520 nm BP 40; Alexa Fluor 647: ex: 633 nm, em: 670 nm BP 30). To compare the different scans, gels were registered using the ImageJ plugin Turboreg (51). For monitoring biotinylation by streptavidin gel shift, the oligonucleotides were incubated at room temperature with a streptavidin solution (Thermo Scientific) of at least 5-fold molar excess for 5 min prior to gel loading.
Fluorescent labeling of N3-modified RNA by SPAAC
RNA 1 (doped with radioactively labeled RNA 1) was labeled with a single N3-A, -C, -G and -U, using the reaction conditions described in Table 2 and was purified by ethanol precipitation. N3-modified RNA (1 µM) was reacted with DIBAC Fluor 488 (50 µM) in 50 mM phosphate buffer (pH 7 at 25°C) for 2 h at 37°C. The reacted RNA was purified by ethanol precipitation in the presence of 0.3 M Na-acetate and analyzed by 15% seqPAGE. Bands were visualized by Storage Phosphor Screen scan, as well as fluorescence scan on the Typhoon 9400 (fluorescence scan: ex: 488 nm, em: 520 BP 40). To compare the different scans, gels were registered, as described above.
Liquid chromatography–mass spectrometry analysis of PAP and CuAAC reaction products
Liquid chromatography–mass spectrometry (LC–MS) experiments were carried out using a microTOFQII ESI mass spectrometer (Bruker) connected to a 1200 Series HPLC system (Agilent), equipped with a Phenomenex kinetex C18 2.6 µm column (2.1 × 100 mm). The solvent system used for elution consisted of 100 mM hexafluoroisopropanol/8.6 mM triethylamine (pH 8.3) as solvent A and methanol (LC–MS grade) as solvent B. The column was pre-equilibrated with 95% solvent A and 5% solvent B for 10 min. Precipitated oligonucleotides were re-dissolved in water, injected onto the column and were eluted with a gradient starting from 5% solvent B to 35% over 30 min. Absorption signals at 254 nm (oligonucleotide absorption) and at 492 nm (Alexa Fluor 488 maximal absorption) were recorded with a multi wavelength detector. For internal calibration (enhanced quadratic mode) of high-resolution mass spectra, we used ESI Tune Mix (Fluka). Data analysis was carried out using the DataAnalysis software (version 4.0 SP 4) for analysis of mass spectra that were deconvoluted using the maximum entropy deconvolution, and Hyphenation Star PP software (version 3.2.44.0) for analysis of LC chromatograms (both Bruker Daltonik). Molecular masses refer to the m/z values given by the DataAnalysis software.
Fluorescent labeling of a natural RNA isolate
Total RNA was isolated from E. coli JM 109 using the GenElute Total RNA Purification Kit (Sigma Aldrich). In all, 28 µg of total RNA (∼50 pmol) were modified in a reaction containing 2′-N3-2′-dUTP (500 µM), yeast PAP (600 U) in 1× USB yeast poly(A) polymerase buffer (25 µl) for 30 min at 37°C. The enzyme was removed by phenol extraction and the RNA recovered by ethanol precipitation in the presence of Na-acetate.
To perform CuAAC, the modified total RNA (∼1 µM) was incubated with Alexa Fluor 647 alkyne (500 µM), CuSO4 (100 µM), THPTA (500 µM), Na-ascorbate (1 mM) in phosphate buffer (50 mM, pH 7) at 25°C for 30 min. RNA was purified immediately after the reaction by phenol extraction in the presence of EDTA (10 mM) and Na-acetate (0.6 M, pH 5.5) followed by ethanol precipitation. About 5 pmol of non-reacted, PAP-reacted and clicked RNA were analyzed by 8% denaturing PAGE and first scanned for Alexa Fluor 647 signals. Then, RNA was stained unspecifically with SYBR Gold (Invitrogen) and scanned again (ex: 532 nm, em: 526 nm SP). Gel images were registered as described above.
Polyadenylation of N3-modified RNA
Polyadenylation of N3-nucleotide-containing, purified RNA by yeast PAP or E. coli PAP was performed by adding 2.5 pmol of ethanol-precipitated, 2′-N3-2′-dNTP-reacted RNA, 2 µl of unpurified N3-NTP containing yeast PAP mix or 1.6 µl of N3-NTP containing E. coli PAP mix into a PAP reaction as described above, containing ATP (500 µM) instead of the modified NTP and fresh PAP. All reaction mixtures were analyzed by 12 or 15% seqPAGE.
Shifted bands or smears, representing polyadenylated RNA were quantified against the different bands representing non-polyadenylated sequences, differentiating between non-modified and N3-nucleotide-bearing RNA, whenever different bands were distinguishable.
Adapter ligation of N3-modified RNA
The 3′-adapter ligation was performed using an adenylated adapter (DNA2) prepared according to a published protocol (52). In all, 2 µl of unpurified N3-NTP containing yeast PAP or 1.6 µl of N3-NTP containing E. coli PAP mix were added to the ligation mixture, consisting of adapter (7.5–10 µM), RNL1 (1 U/µl) and RNL2, tr. (20 U/µl) in ligation buffer (50 mM Tris–HCl, pH 7.4, 10 mM MgCl2, final volume: 10 µl). Ligation mixtures were incubated at 4°C overnight and were analyzed by 12 or 15% seqPAGE. Bands containing non-ligated RNA or ligated RNA (shifted bands) were quantified. The ligation yield (consumption of non-ligated material) was determined ratiometrically, differentiating between non-N3-modified and N3-nucleotide-bearing RNA, whenever different bands were distinguishable.
Splinted ligation of N3-modified RNA
To convert the 3′-terminal modification into an internal one, splinted ligation was carried out, employing non-purified, 2′-N3-2′-dGTP-reacted RNA1 (0.5 µM), RNA4 (2.5 µM), DNA1 splint DNA (2.25 µM), and DNL (1.5 U/µl) or RNL2 (500 nmol/µl) in 1× ligation buffer [50 mM Tris–HCl (pH 7.4 at 25°C), 10 mM MgCl2, 200 µM ATP and 5 mM DTT; final volume: 20 µl]. For those samples subjected to CuAAC, the DTT concentration in the ligase buffer was decreased to 0.5 mM. The mixture was denatured for 30 s at 90°C and then cooled to room temperature for 15 min prior to addition of the enzyme. The reaction mixture was incubated either at 37°C for 1 h or at 16°C overnight. The reaction was stopped by heating to 80°C for 10 min. The DNA splint was removed by the addition of DNase I (0.5 U/µl) and incubated at 37°C for 15 to 30 min, followed by 10 min of heat inactivation at 80°C or phenol extraction (for samples subjected to click reaction). The reaction mixture was purified by ethanol precipitation. Reactions were analyzed by 15% seqPAGE. Shifted bands, representing ligated RNA were quantified against the bands representing non-ligated sequences.
CuAAC with internally N3-modified RNA
The CuAAC with Alexa Fluor 647 alkyne or Alexa Fluor 488 alkyne was carried out with 250 nM ligated RNA, 2 mM Alexa Fluor alkyne, 500 µM CuSO4, 2.5 mM THPTA and 5 mM Na-ascorbate. In a different approach, the ligated RNA was first annealed to a partially reverse complementary DNA (DNA3, in 40-fold excess over ligation product) that forces the modified position into a 9-nt bulge-loop, and carried out as described above. RNA was then purified by phenol extraction in the presence of EDTA (10 mM) and Na-acetate (300–600 mM, pH 5.5) followed by ethanol precipitation. Then, DNA2 was digested with DNase I in DNase I buffer with MgCl2 [10 mM Tris–HCl (pH 7.5 at 25°C), 2.5 mM MgCl2, 0.1 mM CaCl2] for 15 min, and the enzyme was heat-inactivated for 10 min at 65°C. Reactions were analyzed by 15% seqPAGE. Shifted bands, representing clicked RNA, were quantified against the bands representing non-clicked sequences.
RESULTS
Screening of nucleotidyl transferases and modified nucleotides
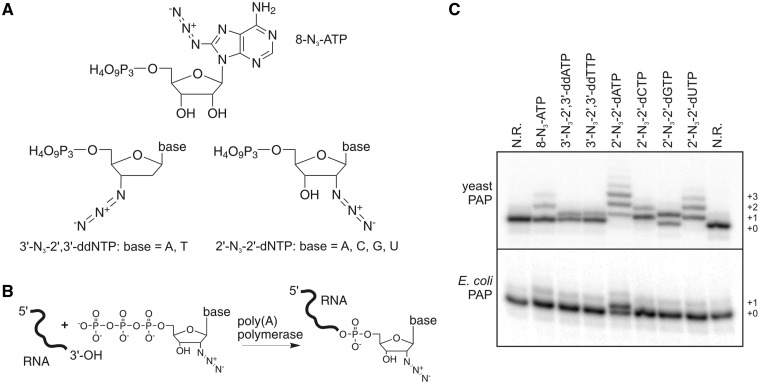
To achieve both, base- and backbone modifications, we employed NTPs bearing the azide modification at position C-2′, C-3′ or C-8 (see Figure 1A for an overview). We determined which of the four nucleotidyl transferases (yeast and E. coli PAP, Cid 1 PUP and TdT) are best suited to efficiently transfer the different N3-modified nucleotides to the 3′-termini of RNA (shown schematically in Figure 1B for modification of RNA with 2′-modified nucleotides and PAP). The results are summarized in Table 3.
Figure 1.
Incorporation of different N3-modified NTPs by different PAPs. (A) Overview of the modified NTPs employed in this study. (B) Schematic representation of the addition of a 2′-N3-modified nucleotide to the 3′-terminus of an RNA sequence by PAP. (C) Addition of various N3-modified nucleotides by yeast and E. coli PAP to the 3′-terminus of RNA1. N.R.: no reaction control. Analysis by 18% seqPAGE. Radioactive bands are shown. Bandshifts indicate the incorporation of one or more nucleotides.
Table 3.
Results of the first screening of nucleotidyl transferases for incorporation of N3-nucleotides
| Modified NTP | No. of residues added |
|||
|---|---|---|---|---|
| Yeast PAP | Escherichia coli PAP | Cid 1 PUP | TdT | |
| 8-N3-ATP | 1–2 | – | 0–2a | – |
| 2′-N3-2′-dATP | Multiple | 1(-2)b | 0–mult.a | – |
| 2′-N3-2′-dCTP | 1–2 | – | 0–mult.a | – |
| 2′-N3-2′-dGTP | 1–2 | – | 0–mult.a | 0–1a |
| 2′-N3-2′-dUTP | 1–2 | – | 0–mult.a | – |
| 3′-N3-2′,3′-ddATP | 0–1 | – | – | ND |
| 3′-N3-2′,3′-ddTTP | 0–1 | – | – | ND |
Generally, reactions were carried out with commercial buffers, 500 µM NTP, at 37°C for 60 min (TdT: 90 min). Enzyme-specific conditions were the following: 0.2 µM RNA, 24 U/µl yeast PAP; 0.2 µM RNA, 0.08 U/µl Cid1 PUP; 0.25 µM RNA, 0.25 U/µl E. coli PAP, 0.25 µM RNA, 1 U/µl TdT. No heat inactivation.
aMostly 0.
bOnly at long incorporation times or with MnCl2.
For TdT, the efficiency of N3-nucleotide addition was very low (Supplementary Figure S1A), which was expected since TdT usually modifies DNA (53) and is known to have a very low efficiency on RNA (54).
Cid 1 PUP accepted 2′-N3-2′-dNTPs with moderate yield, but no significant incorporation of NTPs modified at positions C-3′ or C-8 could be detected (Supplementary Figure S1B and S1C). Additionally, this enzyme produced heterogeneous tails in most cases.
Yeast PAP exhibited the highest incorporation activity for all modified NTPs (Figure 1C). In accordance with the published data (55), one to two C-8 modified nucleotides were incorporated. We made similar observations for NTPs modified at positions C-2′, except for 2′-N3-2′-dATP, where we observed the addition of multiple nucleotides. For NTPs modified at position C-3′, the addition of a single nucleotide is possible with limited efficiency.
With E. coli PAP, only the incorporation of 2′-N3-2′-dATP proceeded with a good efficiency, leading to the addition of one nucleotide (Figure 1C).
Taking together these results, we focused on yeast and E. coli PAP for more detailed investigations.
Yeast PAP reaction with the base-modified 8-N3-ATP
For the base-modified ATP analog 8-N3-ATP, yeast PAP added either one or two nucleotides to the RNA, after which further nucleotide additions did not occur, irrespective of RNA concentration and reaction time. The reaction was more efficient at an RNA concentration of 0.2 µM than at higher concentrations (0.8 µM and 4 µM), and the fraction of modified RNA sequences increased at longer reaction times (120 min) (Supplementary Figure S2). Using 8-N3-ATP, it is thus possible to modify RNA sequences with one to two moieties that can react in further CuAAC so that the length of the RNA is only slightly altered. The extent of modified sequences can, to a certain degree, be controlled by varying the RNA concentration and reaction time.
Optimization of single incorporation for sugar-modified nucleotides
For some applications, it is crucial that only a single modification is added to each oligonucleotide. While only a single nucleotide can be incorporated in case of 3′-modified (and therefore 3′-blocked) NTPs, for 2′-modified NTPs that carry a free 3′-OH, multiple incorporation is, in principle, possible. However, having observed the relatively short tails generated by yeast PAP with 2′-N3-2′-dCTP, -GTP and -UTP (Figure 1C), we attempted to control the reaction, so as to specifically achieve the addition of a single modified nucleotide.
The optimal reaction conditions for single nucleotide addition at 4 (yeast PAP) or 5 µM (E. coli PAP—only for 2′-N3-2′-dATP) RNA concentration were determined by screening over reaction times between 2 and 240 min for RNA1, RNA2 and RNA3 and are given in Table 2.
For 2′-N3-2′-dGTP, we observed the highest amount of single nucleotide addition after 2 h of reaction time. Longer incubations led to an increase of multiple nucleotide addition and thereby reduction of products containing a single modified nucleotide. Similarly, we determined the optimal reaction times to be 5 min for 2′-N3-2′-dUTP, and 20 min for 2′-N3-2′-dCTP (Supplementary Figure S3).
Since the use of yeast PAP with 2′-N3-2′-dATP led to the addition of multiple moieties, even at low NTP concentration and short reaction time (Supplementary Figure S4A), E. coli PAP seemed to be more suitable to achieve single nucleotide addition in this particular case. To facilitate the reaction, we added 2.5 mM MnCl2 to the reaction buffer (already containing 10 mM MgCl2). In the absence of MnCl2, acceptable yields were only achieved after 16 h of incubation at 37°C, using double amounts of enzyme (Supplementary Figure S4B), which led to substantial RNA degradation and severe fluctuations in reaction yield. In contrast, MnCl2 at a concentration of 2.5 mM has been reported to increase the efficiency of E. coli PAP by ∼5-fold (56). Indeed, we observed a considerable acceleration of 2′-N3-A-incorporation, so that generally reaction yields of >50% were obtained for the product containing a single modified nucleotide, after as little as 20 min reaction time (Supplementary Figure S5). Comparing initial rates of the first and second nucleotide incorporation, we found that the second incorporation proceeds with only 5–10% of the initial rate of the first incorporation, so that single nucleotide incorporation is favored after short reaction times.
Incorporation efficiencies under optimized conditions
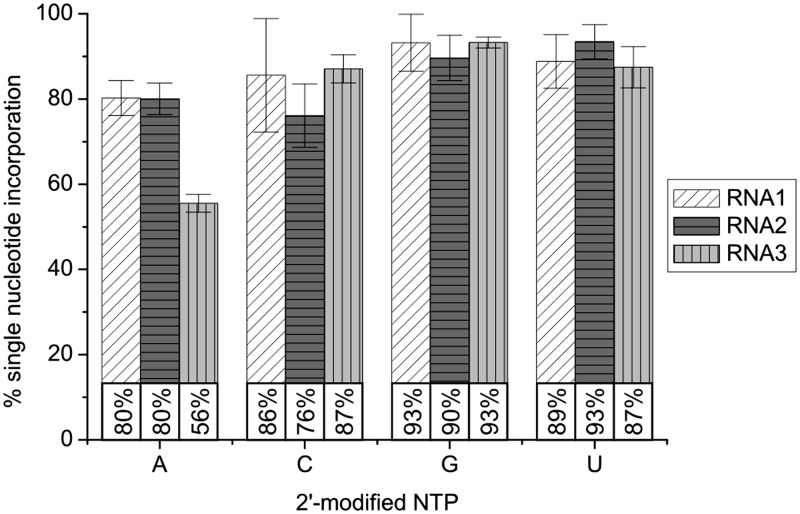
Figure 2 shows a triplicate determination of single nucleotide incorporation efficiencies under the optimized conditions (Table 2) for each of the three test sequences. In 11 out of the 12 combinations, single-addition product yields ≥75% were achieved. Altogether, we were able to identify conditions under which single nucleotide incorporation is prevalent for each of the 2′-modified NTPs, requiring relatively short reaction times and preserving the integrity of the RNA sequences.
Figure 2.
Determination of single nucleotide incorporation efficiencies for all 2′-N3-2′-dNTPs with three different RNA sequences under conditions optimized for 4 or 5 µM (only 2′-N3-2′-dATP) RNA. RNA1, 2, and 3 were subjected to PAP reactions under optimized conditions (in triplicate, except RNA3 + 2′-N3-2′-dGTP—duplicate). Analysis by 15% seqPAGE. All radioactive bands were quantified with ImageQuant software and the percentages of singly modified product were calculated for all samples. Of these, average values are shown. Error bars indicate 1 SD.
Dynamic range of PAP reaction
Depending on the application, the amount of RNA to be modified may vary considerably. While certain RNA samples from natural sources can be scarce, highly abundant natural RNAs or chemically synthesized sequences might need to be modified in bigger amounts and therefore in higher concentrations, demanding our labeling strategy to be equally efficient under very high as well as low RNA concentrations. Thus, to investigate the robustness of our approach, we sought to find out the dynamic range of the yeast PAP reaction where we varied the reaction time employing any one of the four 2′-N3-modified NTPs and RNA1 at 0.2 and 4 µM concentration (for an example, see Supplementary Figure S6), or RNA2 at 4 to 80 µM concentration in a reaction with 2′-N3-2′-dCTP and -UTP (Supplementary Figure S7). The overall reaction yield was generally lower at 0.2 µM than at 4 µM concentration. However, the reaction kinetics were similar. At elevated RNA concentrations, we found that the relative rate of the reaction was decreased. Nonetheless, efficient addition of a single nucleotide was possible at prolonged reaction times (maximally 2 h for 80 µM RNA with 2′-N3-2′-dCTP).
To further confirm the successful incorporation of a single modified nucleotide, we performed an LC–MS analysis of the purified product of a reaction containing 80 µM RNA2 and 2′-N3-2′-dCTP (Supplementary Figure S8), incubated for 2 h. In this analysis, we determined a reaction yield of ∼80%. The identity of the PAP reaction product could be confirmed according to the molecular mass.
CuAAC at the 3′-terminus
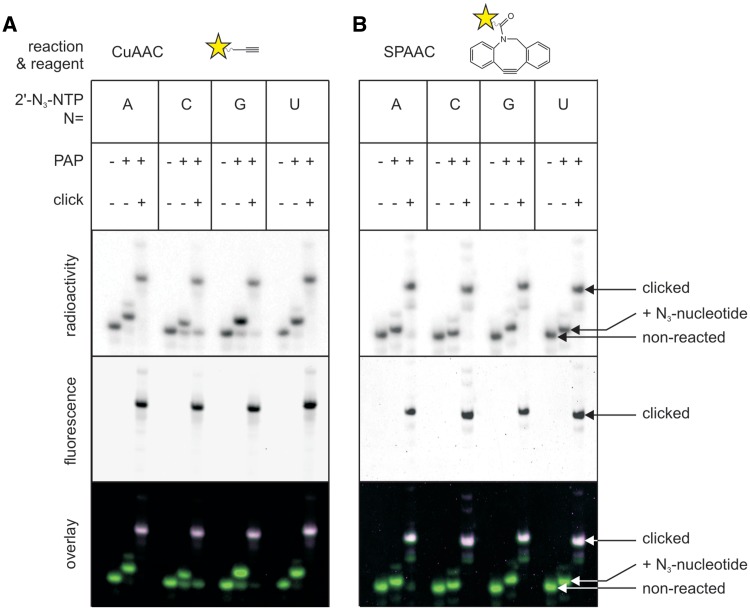
To convert the azide modification to various functional tags, we employed the CuAAC using biotin-alkyne, Alexa Fluor 488 alkyne or Alexa Fluor 647 alkyne, together with a water-soluble Cu-stabilizing ligand, THPTA (50). A Cu-stabilizing ligand is necessary to protect the biomolecule, i.e. RNA from degradation by free copper ions (57) and oxidation products of ascorbate (50). RNA modified with any one of the four 2′-N3-2′-dNTPs (Figure 3A) or with 8-N3-ATP (Supplementary Figure S9) could be efficiently conjugated with various functional moieties: fluorophores (Figure 3A) or biotin (Supplementary Figure S10). Purification of the RNA from the PAP reactions by size exclusion chromatography, ethanol or isopropanol precipitation proved to be efficient enough in removing excess azido-NTPs prior to CuAAC. Under carefully optimized conditions, we achieved high yields (from >80% up to quantitative) at RNA concentrations ranging from 50 nM to 50 µM. Yields and concentrations of the respective azide-bearing RNA, alkyne, as well as Cu(II), THPTA and ascorbate, are summarized in Table 4. In all cases, it was sufficient to incubate the reactions for 2 h at 37°C, which lead to excellent reaction yields without causing significant RNA degradation.
Figure 3.
Fluorescent labeling of RNA by CuAAC or SPAAC. RNA1, modified with each of the four 2′-N3-2′-dNTPs under optimized conditions and further conjugated with fluorescent dyes. Analysis by 15% seqPAGE. Radioactivity scan (upper panel), fluorescence scan (middle panel) and an overlay of both (green: radioactivity; magenta: fluorescence; white: both; lower panel) are given. (A) Conjugation with Alexa Fluor 647 alkyne by CuAAC. (B) Conjugation with DIBAC Fluor 488 by SPAAC.
Table 4.
Yields and corresponding reaction conditions of CuAAC at different N3-RNA concentrations
| [N3-RNA] | [alkyne] | [Cu(II)] | [THPTA] | [Asc.] | Yield |
|---|---|---|---|---|---|
| 50 nM | 50 µM | 100 µM | 500 µM | 1 mM | >80% |
| 1–10 µM | 500 µM | 100 µM | 500 µM | 1 mM | Quantitative |
| 50 µM | 2 mM | 500 µM | 2.5 mM | 5 mM | Quantitative |
General conditions: 50 mM phosphate buffer, 37°C, 2 h.
An LC–MS analysis was performed for RNA2 and RNA3, reacted with 2′-N3-2′-dCTP at an RNA concentration of 20 µM and conjugated with Alexa Fluor 488 alkyne at an RNA concentration of 10 µM. Here, the reaction products with the respective expected molecular masses were present in > 94% of overall RNA content, according to the UV absorption at 260 nm (Supplementary Figure S11 and S12).
Fluorescent labeling of RNA by SPAAC
To prove the generality of our approach, which can be extended to copper-free techniques, we performed an exemplary SPAAC reaction (Figure 3B), employing a heterocyclic aza-dibenzocyclooctyne (DIBAC) (58) previously shown to exhibit one of the highest reaction rates among the known cyclooctyne compounds (59). Similar to the case of CuAAC, RNA modified with any one of the four 2′-N3-modified nucleotides reacted quantitatively with the fluorescent DIBAC, providing thus a second possibility of high yielding RNA modification.
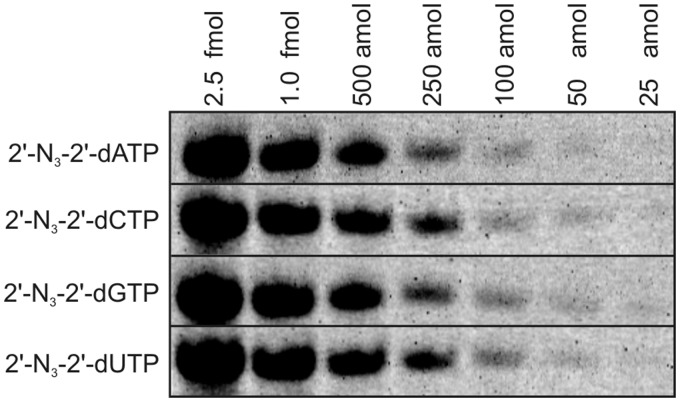
Detection limit for fluorescently labeled RNA
Figure 4 shows the determination of the detection limit for RNA that was fluorescently labeled, using our strategy, employing CuAAC. After incorporation of either one of the four 2′-N3-nucleotides (A, C, G or U) and conjugation with Alexa Fluor 647 alkyne, RNA amounts of as little as 100 amol still resulted in clearly visible bands on a 12% denaturing sequencing gel. Even lower amounts like 25 or 50 amol were still distinguishable as blurry spots. Thus, this labeling approach permits an excellent sensitivity.
Figure 4.
Detection limit for RNA1 modified with each of the four 2′-N3-2′-dNTPs and conjugated with Alexa Fluor 647 alkyne. Samples (same as in Figure 3A) were diluted and analyzed by 12% seqPAGE in amounts from 25 amol to 2.5 fmol. Fluorescence scan (Typhoon 9400, pixel size: 100 µm, PMT: 800 V, high sensitivity) is shown.
Fluorescent labeling of an RNA isolate from E. coli
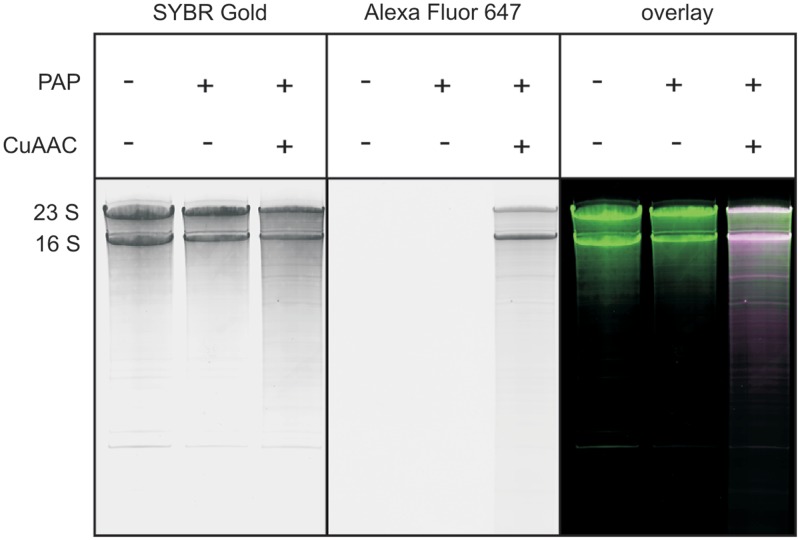
To test the robustness of our labeling approach and its applicability to RNA from biological sources, we isolated total RNA from E. coli, reacted this isolate with 2′-N3-2′-dUTP and yeast PAP. We employed a 30-min incubation time to assure that every sequence carries at least one modified nucleotide, and performed the CuAAC with Alexa Fluor 647 alkyne afterwards. To prevent degradation of the total RNA sample, both, reaction time and temperature were decreased (30 min incubation at 25°C). In the PAGE analysis (Figure 5), the appearance of fluorescent bands that perfectly overlay with the bands visible after non-specific RNA staining and the absence of degradation products indicate the applicability of this labeling approach toward long natural RNA.
Figure 5.
Fluorescent labeling of a natural RNA sample. A total RNA isolate from E. coli was reacted with 2′-N3-2′-dUTP and yeast PAP, and further subjected to CuAAC with Alexa Fluor 647 alkyne. Analysis by 8% denaturing PAGE. SYBR Gold scan (left panel), Alexa Fluor 647 scan (middle panel) and overlay (green: radioactivity; magenta: fluorescence; white: both; right panel) are given. The two main bands represent 23S and 16S ribosomal RNA.
Enzymatic reactions involving the azido-modified 3′-termini of PAP reaction products
Since the azido-modifications at the base or the sugar moieties of the nucleotides employed in this study only constitute minor changes with respect to their natural analogs, we assumed that they would not interfere with certain enzymatic downstream reactions. To test this assumption, we investigated the feasibility of (i) further tailing these RNAs with yeast and E. coli PAP employing the enzyme′s preferred substrate ATP; (ii) carrying out an adapter ligation to add a short DNA strand; and (iii) a splinted ligation in order to add a second RNA strand to the modified 3′-terminus.
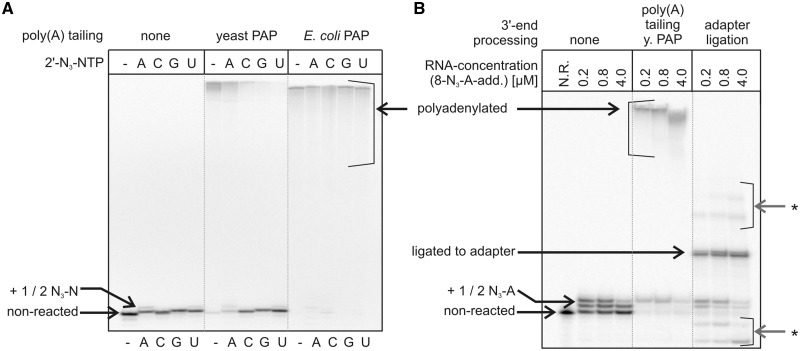
The results of polyadenylation and adapter ligation are summarized in Table 5. While yeast PAP could not efficiently polyadenylate most of the templates containing a 2′-N3-modified nucleotide at their 3′-termini, the polyadenylation reaction proceeded quantitatively when carried out with E. coli PAP (Figure 6A). For RNA containing 8-N3-A at the 3′-terminus (Figure 6B), near-quantitative polyadenylation by yeast PAP was observed for those sequences that contained a single 8-N3-A moiety. Interestingly, the polyadenylation reaction was inhibited when two 8-N3-A moieties were present.
Table 5.
Enzymatic manipulation of 3′-termini of RNA previously modified in PAP reactions
| 3′-terminal modified nt | Polyadenylationa by |
Adapter ligation | |
|---|---|---|---|
| Yeast PAP | Escherichia coli PAP | ||
| None | + (∼93%) | + (>95%) | + (>95%)c |
| 2'-N3-2'-dA | + (∼65%) | + (>95%) | + (∼75%)c |
| 2'-N3-2'-dC | − (<5%)b | + (>95%) | + (∼90%)c |
| 2'-N3-2'-dG | − (<5%) | + (>95%) | + (∼85%)c |
| 2'-N3-2'-dU | − (<5%) | + (>95%) | + (∼80%)c |
| 3'-N3-2',3'-ddA | Not possible | Not possible | Not possible |
| 3'-N3-2',3'-ddT | Not possible | Not possible | Not possible |
| (1×) 8-N3-A | + (∼84–89%) | ND | + (∼50–80%)c |
| (2×) 8-N3-A | − (<5%) | ND | (+) (∼8–40%)c |
aIn presence of 500 µM ATP, 1 h, 37°C.
bA higher fraction of polyadenylated sequences was attributed to remaining, non-modified RNA.
cConsumption of starting material (RNA).
Figure 6.
Enzymatic manipulation of N3-modified 3′-termini. (A) Polyadenylation of RNA1 modified with each of the four 2′-N3-nucleotides by yeast PAP and E. coli PAP. RNA1, reacted with each of the four 2′-N3-NTPs under optimized conditions, and further reacted either with yeast PAP or with E. coli PAP in the presence of ATP. Analysis by 12% seqPAGE. (B) RNA1 modified with 8-N3-ATP at the indicated concentrations, and further subjected to polyadenylation with yeast PAP or adapter ligation. N.R.: no reaction control. Analysis by 15% seqPAGE. Asterisk indicates bands that appear as artifacts, due to the presence of a monophosphate at the 5′-terminus of the radiolabeled RNA, which was doped into the reaction for visualization. Lower running bands represent circularized RNA, while higher running bands represent products of RNA–RNA ligation (with or without adapter added). Both radioactive scans shown here have been scaled. A non-scaled representation is given in Supplementary Figure S13.
We also investigated the possibility of ligating pre-adenylated DNA adapters to the modified 3′-termini, employing the ligases RNL1 and RNL2, tr (Figure 6B, for reaction yields see Table 5). The RNA strands containing one 8-N3-A were accepted almost as well as the unmodified RNA, whereas there was a strong bias against sequences with two 8-N3-A modifications. For 2′-N3-nucleotide-containing sequences, such a preference was not observed (Supplementary Figure S14). In addition to the desired RNA-adapter ligation product, we observed some concatenated (running higher) and circularized RNAs (running lower) in the 32P scans of the gels. Both of these are due to the presence of a monophosphate at the 5′-terminus of the radiolabeled RNAs (≤2%), as observed by others [e.g. (60)]. Non-labeled RNA sequences do not contain a 5′-monophosphate and can thus not form these adducts, which amount to <0.35% of the total RNA in any of the samples.
Encouraged by these results, we investigated the possibility of joining a second RNA strand to RNA carrying a 2′-N3-nucleotide at the 3′-terminus, employing splinted ligation. In this method, the use of a splint oligonucleotide enables the correct ligation of the sequences of choice in high yield (61). In our case, this would convert the 3′-terminal N3-modification into an internal modification within a defined RNA sequence and considerably extend the scope of the approach.
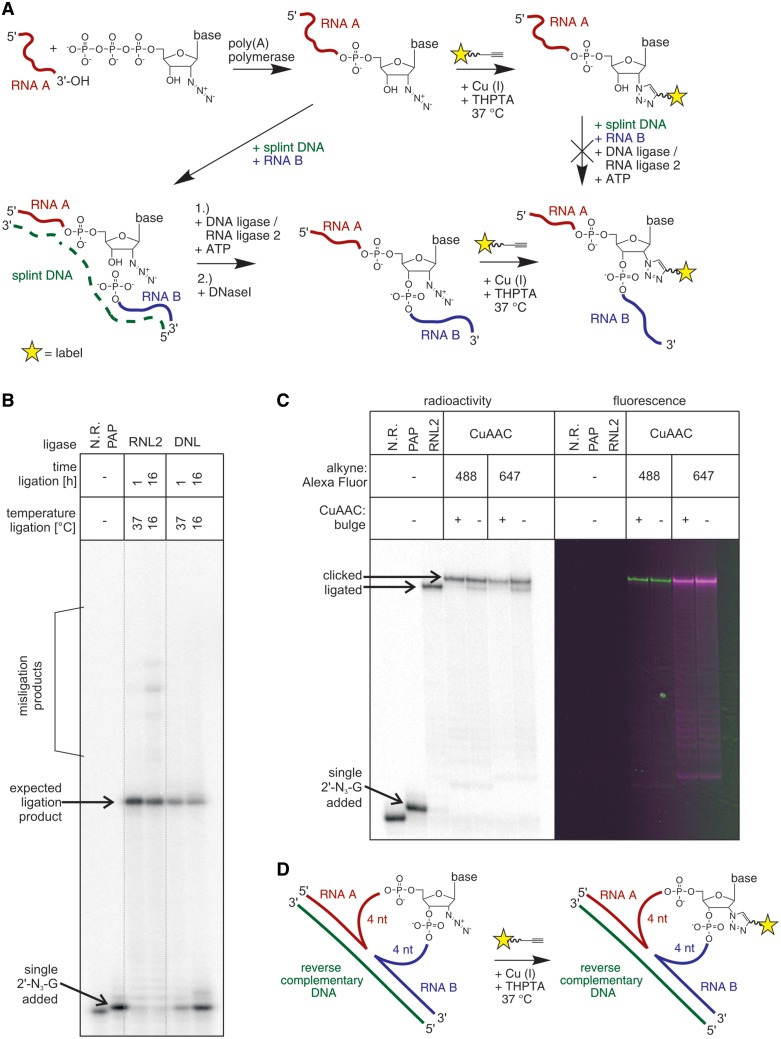
An RNA oligonucleotide modified with 2′-N3-2′-dGTP was subjected to splinted ligation (as shown schematically in Figure 7A), demonstrating the feasibility of the approach. Both, DNL and RNL2 accept the modified 3′-terminus as substrate (Figure 7B), with RNL2 showing better efficiencies than DNL. For both enzymes, an incubation for 1 h at 37°C produced significantly higher yields than 16 h at 16°C. With a 5-fold excess of the second ligation fragment and incubation times of 1 h at 37°C, ∼50% ligation efficiency could be achieved with DNL, while the ligation with RNL2 had an excellent efficiency of ∼ 80% of desired product. In contrast to this, as expected, ligation was not possible any longer when a bulkier modification like a fluorophore was attached by CuAAC prior to the ligation (Supplementary Figure S15). In this case, the modification at the 2′-position of the 3′-terminal nucleotide is much bulkier than just an azide and inhibits the ligase.
Figure 7.
Creating an internal fluorescent label. (A) General reaction scheme for creation of terminal (upper right corner) or internal modifications (lower right corner). An internal modification can be created by first adding an N3-modified nucleotide to the 3′-terminus of the RNA sequence, connecting this RNA to a second RNA sequence via splinted ligation, and subjecting the product, with an internal N3-modification to CuAAC. (B) Splinted ligation of RNA1 and RNA4, employing different ligases (RNL2 and DNL) under different reaction conditions (time and temperature). Analysis by 15% seqPAGE. (C) Addition of 2′-N3-guanosine to RNA1, followed by splinted ligation to RNA4 (using RNL2), and CuAAC with Alexa Fluor 488/647 alkyne, with or without the use of a helper DNA that forces the modified position into a 9-nt bulge loop. Analysis by 15% seqPAGE. Radioactivity scan (left panel), and an overlay of Alexa Fluor 488 scan (green, middle two lanes) and Alexa Fluor 647 scan (magenta, right two lanes) are shown. (D) Formation of 9 nt bulge loop to assist CuAAC. N.R.: no reaction control.
CuAAC of internally azido-modified RNA
After ligation, the former terminal modification has been converted to an internal one. As a side effect of this, the accessibility of the azide for both, alkyne reactants and copper-catalyst is reduced during CuAAC. Depending on whether or not the modified position is situated in a base-paired region or in a more flexible loop, the cycloaddition can be hampered, which has also been observed for click reactions of 5-ethynyl-deoxyuridine in double-stranded DNA (36). Indeed, in our test system, where the modified nucleotide is situated in a partially base paired structure, we observed a decrease in cycloaddition efficiency under standard conditions. In order to improve the labeling yields, we successfully applied forced conditions involving 5-fold higher Cu concentrations and ≥10-fold higher alkyne concentrations, which led to efficiencies in the range of 75–85%. A further increase in labeling efficiency could be achieved by hybridizing a DNA strand with partial reverse complementarity to the ligated sequence that forced 4 nt both, up- and downstream of the modification site into a non-paired bulge loop structure (Figure 7C and D). Here, we again observed near-quantitative reaction yields (>92%) as observed before with 3′-terminal modifications.
DISCUSSION
In our modular RNA labeling approach, we employ enzymes (PAPs) to incorporate a convertible nucleotide at the 3′-terminus of the RNA, in order to provide a reactive handle (an azide) which is further converted to a modification of interest in a bio-orthogonal click reaction, namely CuAAC or SPAAC. The azide is not stable under the standard conditions of solid-phase chemical oligonucleotide synthesis (46), and can therefore not be incorporated easily with these methods, making enzymatic reactions an attractive alternative, which has the additional advantage of not being limited to short sequences.
As we have shown, yeast PAP can readily add one (or sometimes more) azido-modified nucleotide(s) (modified at C-8 or C-2′) to the 3′-terminus of RNA but can, in most cases, not easily extend this reaction product, even with the preferred PAP substrate ATP. In a reaction mixture containing the above-mentioned modified NTPs, this leads to the dominance of RNA containing a single (or sometimes two) modified nucleotide(s).
Similarly, E. coli PAP can well accept 2′-N3-2′-dATP as an NTP-substrate, and can also easily extend RNA substrates containing a 2′-N3-modification at their 3′-termini with standard ATP. However, when both, the NTP and the 3′-terminus of the RNA-substrate are 2′-N3-modified, the E. coli PAP reaction is considerably slowed down (by 10- to 20-fold), which again makes it possible to achieve a prevalence of singly modified RNA sequences.
Unlike most other RNA 3′-labeling approaches, we provide a method that is capable of introducing each of the four (A, C, G, U) nucleotides in a chemically modified form. We took advantage of kinetic control of the enzymatic reaction to ensure single label incorporation. This enables the 3′-terminal modification of sequences from any origin (synthetic or natural), causing only minimal mutation, and thereby preserving function.
Both, the PAP reactions, and the CuAAC can be adjusted to a wide range of RNA concentrations (at least 200 nM to 80 µM for PAP, at least 50 nM to 50 µM for CuAAC), thereby making our labeling strategy applicable for very little as well as large amounts of RNA. Even at the lowest RNA concentration (50 nM), CuAAC reaction yields of ≥ 80% could be achieved under optimized conditions. For such low concentrations of nucleic acids carrying single reactive handles, no previous reports exist for CuAAC modification. At higher concentrations, we routinely observed quantitative reaction yields.
Since both reactions proceed with high yield and without significant degradation, the overall efficiency of our two-step functionalization strategy is very high. As a consequence, an outstanding sensitivity can be achieved upon fluorescent labeling, enabling the detection of RNA amounts <100 amol, which exceeds the only comparable report of RNA detection sensitivity after CuAAC labeling (32) by more than three orders of magnitude.
Another major advantage of our labeling protocol arises from the kinetic control over the PAP reactions that we employ in order to achieve the incorporation of a single label, instead of using a 3′-protected, and thus blocked nucleotide (lacking a 3′-hydroxyl function), as in the case of the recently reported RNA labeling using E. coli PAP and 3′-N3-2′,3′-ddATP (32). RNA that is end-modified with a 3′-blocked nucleotide cannot be subjected to further enzymatic manipulations of that modified 3′-end. Contrary to this, in our experimental setup, the modified 3′-terminus bears a free 3′-hydroxyl group, which is available for further enzymatic downstream processing, such as polyadenylation (with non-modified ATP) by E. coli PAP or ligation employing a number of different ligases. Thereby, the terminal azido-modification can be converted into an internal modification, which is still able to undergo CuAAC. To achieve near-quantitative labeling, also with the sterically challenging internal backbone modifications, we had to further optimize the CuAAC conditions. In order to circumvent the problems of sterical clashes in base-paired regions, we introduced a simple but efficient strategy. Using a helper oligonucleotide that forces the modified nucleotide into an available position, we were able to recover full reactivity.
Noteworthy, due to the possibility of attaching the nucleotide of choice (A, C, G or U) prior to ligation, we are capable of targeting any internal position within a de novo synthesized RNA, without mutating its sequence. This should help preserving the integrity of functional RNAs that can be studied after labeling.
To even further broaden the scope of azide incorporation into oligonucleotides, other click reactions can be employed to convert azides, which is a subject of ongoing research in our laboratory. The copper-free reactions include the Staudinger ligation, which has already been successfully applied to RNA (62) and the SPAAC (41), which has also been applied to nucleic acids (63–66). For the latter, we exemplarily show the fluorescent labeling of N3-modified RNA with a DIBAC-fluorophore. Similar to our observations with CuAAC, the reaction proceeded with excellent efficiency. Copper-free reactions are more compatible with living cells than the CuAAC and may therefore allow for new in vivo applications. In our decision in favor of incorporating azides instead of alkynes, this was a crucial factor.
In conclusion, we provide a simple method for chemoenzymatic RNA functionalization, which does not require any special skills or lab equipment for chemical synthesis. Since all components are commercially available, the technique should be readily available to prospective users from different disciplines. Our method can be applied to many different RNA substrates, at variable concentrations, to create either terminal or internal modifications with an ever-growing number of possible labels. Owing to the modular nature of the approach, a set of only four different modified NTPs is sufficient to target RNA for conjugation with a multitude of different labels. Therefore, comparatively little optimization is needed, while high yields can be expected even at concentrations that are relatively high for standard enzymatic reactions. The only requirement for a sequence to be modified is that it is a substrate of the respective PAP. Due to the popularity of copper-catalyzed, as well as metal-free click reactions, a growing number of possible labels are commercially available. Therefore, the technique is open to a broad range of applications, e.g. in vitro studies of RNA structure and dynamics, RNA imaging, as well as the modulation of RNA properties through chemical modification.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online: Supplementary Figures 1–15, Supplementary Table 1 and Supplementary Methods.
FUNDING
Deutsche Forschungsgemeinschaft (Ja 794-3); Hartmut Hoffmann-Berling International Graduate School of Molecular and Cellular Biology (HBIGS) to M.-L.W. Funding for open access charge: Institutional budget.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Juliane Schoch for assistance with LC–MS experimentation and data analysis, and Hannah R. Hudson for experimental support.
REFERENCES
- 1.Weil TT, Parton RM, Davis I. Making the message clear: visualizing mRNA localization. Trends Cell Biol. 2010;20:380–390. doi: 10.1016/j.tcb.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wachowius F, Höbartner C. Chemical RNA modifications for studies of RNA structure and dynamics. Chembiochem. 2010;11:469–480. doi: 10.1002/cbic.200900697. [DOI] [PubMed] [Google Scholar]
- 3.Strauss B, Nierth A, Singer M, Jäschke A. Direct structural analysis of modified RNA by fluorescent in-line probing. Nucleic Acids Res. 2012;40:861–870. doi: 10.1093/nar/gkr733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rouault TA, Hentze MW, Haile DJ, Harford JB, Klausner RD. The iron-responsive element binding protein: a method for the affinity purification of a regulatory RNA-binding protein. Proc. Natl Acad. Sci. U SA. 1989;86:5768–5772. doi: 10.1073/pnas.86.15.5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vilfan ID, Kamping W, van den Hout M, Candelli A, Hage S, Dekker NH. An RNA toolbox for single-molecule force spectroscopy studies. Nucleic Acids Res. 2007;35:6625–6639. doi: 10.1093/nar/gkm585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kellner S, Seidu-Larry S, Burhenne J, Motorin Y, Helm M. A multifunctional bioconjugate module for versatile photoaffinity labeling and click chemistry of RNA. Nucleic Acids Res. 2011;39:7348–7360. doi: 10.1093/nar/gkr449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wower J, Rosen KV, Hixson SS, Zimmermann RA. Recombinant photoreactive tRNA molecules as probes for cross-linking studies. Biochimie. 1994;76:1235–1246. doi: 10.1016/0300-9084(94)90054-x. [DOI] [PubMed] [Google Scholar]
- 8.Shim MS, Kwon YJ. Efficient and targeted delivery of siRNA in vivo. FEBS J. 2010;277:4814–4827. doi: 10.1111/j.1742-4658.2010.07904.x. [DOI] [PubMed] [Google Scholar]
- 9.Lönnberg H. Solid-phase synthesis of oligonucleotide conjugates useful for delivery and targeting of potential nucleic acid therapeutics. Bioconjug. Chem. 2009;20:1065–1094. doi: 10.1021/bc800406a. [DOI] [PubMed] [Google Scholar]
- 10.Singh Y, Murat P, Defrancq E. Recent developments in oligonucleotide conjugation. Chem. Soc. Rev. 2010;39:2054–2070. doi: 10.1039/b911431a. [DOI] [PubMed] [Google Scholar]
- 11.Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin CT, Coleman JE. T7 RNA polymerase does not interact with the 5'-phosphate of the initiating nucleotide. Biochemistry. 1989;28:2760–2762. doi: 10.1021/bi00433a002. [DOI] [PubMed] [Google Scholar]
- 13.Hirao I. Unnatural base pair systems for DNA/RNA-based biotechnology. Curr. Opin. Chem. Biol. 2006;10:622–627. doi: 10.1016/j.cbpa.2006.09.021. [DOI] [PubMed] [Google Scholar]
- 14.Oh BK, Pace NR. Interaction of the 3'-end of tRNA with ribonuclease P RNA. Nucleic Acids Res. 1994;22:4087–4094. doi: 10.1093/nar/22.20.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hansske F, Seela F, Watanabe K, Cramer F. Modification of the rare nucleoside X in Escherichia coli tRNAs with antigenic determining, photolabile, and paramagnetic residues. Methods Enzymol. 1979;59:166–171. doi: 10.1016/0076-6879(79)59078-8. [DOI] [PubMed] [Google Scholar]
- 16.England TE, Uhlenbeck OC. 3'-terminal labelling of RNA with T4 RNA ligase. Nature. 1978;275:560–561. doi: 10.1038/275560a0. [DOI] [PubMed] [Google Scholar]
- 17.Igloi GL. Nonradioactive labeling of RNA. Anal. Biochem. 1996;233:124–129. doi: 10.1006/abio.1996.0016. [DOI] [PubMed] [Google Scholar]
- 18.Hausch F, Jäschke A. Libraries of multifunctional RNA conjugates for the selection of new RNA catalysts. Bioconjug. Chem. 1997;8:885–890. doi: 10.1021/bc9701151. [DOI] [PubMed] [Google Scholar]
- 19.Rosemeyer V, Laubrock A, Seibl R. Nonradioactive 3'-end-labeling of RNA molecules of different lengths by terminal deoxynucleotidyltransferase. Anal. Biochem. 1995;224:446–449. doi: 10.1006/abio.1995.1068. [DOI] [PubMed] [Google Scholar]
- 20.Martin G, Keller W. Tailing and 3'-end labeling of RNA with yeast poly(A) polymerase and various nucleotides. RNA. 1998;4:226–230. [PMC free article] [PubMed] [Google Scholar]
- 21.Baum DA, Silverman SK. Deoxyribozyme-catalyzed labeling of RNA. Angew. Chem. Int. Ed. Engl. 2007;46:3502–3504. doi: 10.1002/anie.200700357. [DOI] [PubMed] [Google Scholar]
- 22.Vauleon S, Ivanov SA, Gwiazda S, Müller S. Site-specific fluorescent and affinity labelling of RNA by using a small engineered twin ribozyme. Chembiochem. 2005;6:2158–2162. doi: 10.1002/cbic.200500215. [DOI] [PubMed] [Google Scholar]
- 23.Onizuka K, Taniguchi Y, Sasaki S. Site-specific covalent modification of RNA guided by functionality-transfer oligodeoxynucleotides. Bioconjug. Chem. 2009;20:799–803. doi: 10.1021/bc900009p. [DOI] [PubMed] [Google Scholar]
- 24.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. A stepwise huisgen cycloaddition process: copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew. Chem. Int. Ed. Engl. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 25.Tornoe CW, Christensen C, Meldal M. Peptidotriazoles on solid phase: [1,2,3]-triazoles by regiospecific copper(i)-catalyzed 1,3-dipolar cycloadditions of terminal alkynes to azides. J. Org. Chem. 2002;67:3057–3064. doi: 10.1021/jo011148j. [DOI] [PubMed] [Google Scholar]
- 26.Seo TS, Li Z, Ruparel H, Ju J. Click chemistry to construct fluorescent oligonucleotides for DNA sequencing. J. Org. Chem. 2003;68:609–612. doi: 10.1021/jo026615r. [DOI] [PubMed] [Google Scholar]
- 27.Gramlich PM, Wirges CT, Manetto A, Carell T. Postsynthetic DNA modification through the copper-catalyzed azide-alkyne cycloaddition reaction. Angew. Chem. Int. Ed. Engl. 2008;47:8350–8358. doi: 10.1002/anie.200802077. [DOI] [PubMed] [Google Scholar]
- 28.El-Sagheer AH, Brown T. Click chemistry with DNA. Chem. Soc. Rev. 2010;39:1388–1405. doi: 10.1039/b901971p. [DOI] [PubMed] [Google Scholar]
- 29.Jao CY, Salic A. Exploring RNA transcription and turnover in vivo by using click chemistry. Proc. Natl Acad. Sci. USA. 2008;105:15779–15784. doi: 10.1073/pnas.0808480105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Motorin Y, Burhenne J, Teimer R, Koynov K, Willnow S, Weinhold E, Helm M. Expanding the chemical scope of RNA:methyltransferases to site-specific alkynylation of RNA for click labeling. Nucleic Acids Res. 2011;39:1943–1952. doi: 10.1093/nar/gkq825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamada T, Peng CG, Matsuda S, Addepalli H, Jayaprakash KN, Alam MR, Mills K, Maier MA, Charisse K, Sekine M, et al. Versatile site-specific conjugation of small molecules to siRNA using click chemistry. J. Org. Chem. 2011;76:1198–1211. doi: 10.1021/jo101761g. [DOI] [PubMed] [Google Scholar]
- 32.Paredes E, Das SR. Click chemistry for rapid labeling and ligation of RNA. Chembiochem. 2011;12:125–131. doi: 10.1002/cbic.201000466. [DOI] [PubMed] [Google Scholar]
- 33.El-Sagheer AH, Brown T. New strategy for the synthesis of chemically modified RNA constructs exemplified by hairpin and hammerhead ribozymes. Proc. Natl Acad. Sci. USA. 2010;107:15329–15334. doi: 10.1073/pnas.1006447107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burley GA, Gierlich J, Mofid MR, Nir H, Tal S, Eichen Y, Carell T. Directed DNA metallization. J. Am. Chem. Soc. 2006;128:1398–1399. doi: 10.1021/ja055517v. [DOI] [PubMed] [Google Scholar]
- 35.Gierlich J, Gutsmiedl K, Gramlich PM, Schmidt A, Burley GA, Carell T. Synthesis of highly modified DNA by a combination of PCR with alkyne-bearing triphosphates and click chemistry. Chemistry. 2007;13:9486–9494. doi: 10.1002/chem.200700502. [DOI] [PubMed] [Google Scholar]
- 36.Wirges CT, Gramlich PME, Gutsmiedl K, Gierlich J, Burley GA, Carell T. Pronounced effect of DNA hybridization on click reaction efficiency. QSAR Comb. Sci. 2007;26:1159–1164. [Google Scholar]
- 37.Gramlich PM, Wirges CT, Gierlich J, Carell T. Synthesis of modified DNA by PCR with alkyne-bearing purines followed by a click reaction. Org. Lett. 2008;10:249–251. doi: 10.1021/ol7026015. [DOI] [PubMed] [Google Scholar]
- 38.Salic A, Mitchison TJ. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc. Natl Acad. Sci. USA. 2008;105:2415–2420. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guan L, van der Heijden GW, Bortvin A, Greenberg MM. Intracellular detection of cytosine incorporation in genomic DNA by using 5-ethynyl-2'-deoxycytidine. Chembiochem. 2011;12:2184–2190. doi: 10.1002/cbic.201100353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weller RL, Rajski SR. DNA methyltransferase-moderated click chemistry. Org. Lett. 2005;7:2141–2144. doi: 10.1021/ol0504749. [DOI] [PubMed] [Google Scholar]
- 41.Agard NJ, Prescher JA, Bertozzi CR. A strain-promoted [3 + 2] azide-alkyne cycloaddition for covalent modification of biomolecules in living systems. J. Am. Chem. Soc. 2004;126:15046–15047. doi: 10.1021/ja044996f. [DOI] [PubMed] [Google Scholar]
- 42.Gierlich J, Burley GA, Gramlich PM, Hammond DM, Carell T. Click chemistry as a reliable method for the high-density postsynthetic functionalization of alkyne-modified DNA. Org. Lett. 2006;8:3639–3642. doi: 10.1021/ol0610946. [DOI] [PubMed] [Google Scholar]
- 43.Seela F, Sirivolu VR. Pyrrolo-dC oligonucleotides bearing alkynyl side chains with terminal triple bonds: synthesis, base pairing and fluorescent dye conjugates prepared by the azide-alkyne “click” reaction. Org. Biomol. Chem. 2008;6:1674–1687. doi: 10.1039/b719459e. [DOI] [PubMed] [Google Scholar]
- 44.Seela F, Sirivolu VR, Chittepu P. Modification of DNA with octadiynyl side chains: synthesis, base pairing, and formation of fluorescent coumarin dye conjugates of four nucleobases by the alkyne–azide “click“ reaction. Bioconjug. Chem. 2008;19:211–224. doi: 10.1021/bc700300f. [DOI] [PubMed] [Google Scholar]
- 45.Berndl S, Herzig N, Kele P, Lachmann D, Li X, Wolfbeis OS, Wagenknecht HA. Comparison of a nucleosidic vs non-nucleosidic postsynthetic “click” modification of DNA with base-labile fluorescent probes. Bioconjug. Chem. 2009;20:558–564. doi: 10.1021/bc8004864. [DOI] [PubMed] [Google Scholar]
- 46.Jawalekar AM, Meeuwenoord N, Cremers JS, Overkleeft HS, van der Marel GA, Rutjes FP, van Delft FL. Conjugation of nucleosides and oligonucleotides by [3+2] cycloaddition. J. Org. Chem. 2008;73:287–290. doi: 10.1021/jo702023s. [DOI] [PubMed] [Google Scholar]
- 47.Weisbrod SH, Marx A. A nucleoside triphosphate for site-specific labelling of DNA by the Staudinger ligation. Chem. Commun. 2007:1828–1830. doi: 10.1039/b618257g. [DOI] [PubMed] [Google Scholar]
- 48.Padilla R, Sousa R. A Y639F/H784A T7 RNA polymerase double mutant displays superior properties for synthesizing RNAs with non-canonical NTPs. Nucleic Acids Res. 2002;30:e138. doi: 10.1093/nar/gnf138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kwak JE, Wickens M. A family of poly(U) polymerases. RNA. 2007;13:860–867. doi: 10.1261/rna.514007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hong V, Presolski SI, Ma C, Finn MG. Analysis and optimization of copper-catalyzed azide-alkyne cycloaddition for bioconjugation. Angew. Chem. Int. Ed. Engl. 2009;48:9879–9883. doi: 10.1002/anie.200905087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thevenaz P, Ruttimann UE, Unser M. A pyramid approach to subpixel registration based on intensity. IEEE Trans. Image Process. 1998;7:27–41. doi: 10.1109/83.650848. [DOI] [PubMed] [Google Scholar]
- 52.Hafner M, Landgraf P, Ludwig J, Rice A, Ojo T, Lin C, Holoch D, Lim C, Tuschl T. Identification of microRNAs and other small regulatory RNAs using cDNA library sequencing. Methods. 2008;44:3–12. doi: 10.1016/j.ymeth.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roychoudhury R, Jay E, Wu R. Terminal labeling and addition of homopolymer tracts to duplex DNA fragments by terminal deoxynucleotidyl transferase. Nucleic Acids Res. 1976;3:863–877. doi: 10.1093/nar/3.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ratliff RL. Terminal Deoxynucleotidyltransferase. In: Boyer P, editor. The Enzymes Vol. XIV. New York: Academic Press; 1981. pp. 105–118. [Google Scholar]
- 55.Chen LS, Sheppard TL. Chain termination and inhibition of Saccharomyces cerevisiae poly(A) polymerase by C-8-modified ATP analogs. J. Biol. Chem. 2004;279:40405–40411. doi: 10.1074/jbc.M401752200. [DOI] [PubMed] [Google Scholar]
- 56.Sippel AE. Purification and characterization of adenosine triphosphate: ribonucleic acid adenyltransferase from Escherichia coli. Eur. J. Biochem. 1973;37:31–40. doi: 10.1111/j.1432-1033.1973.tb02953.x. [DOI] [PubMed] [Google Scholar]
- 57.Hermann T, Heumann H. Structure and distance determination in RNA with copper phenanthroline probing. Methods Enzymol. 2000;318:33–43. doi: 10.1016/s0076-6879(00)18042-5. [DOI] [PubMed] [Google Scholar]
- 58.Debets MF, van Berkel SS, Schoffelen S, Rutjes FP, van Hest JC, van Delft FL. Aza-dibenzocyclooctynes for fast and efficient enzyme PEGylation via copper-free (3+2) cycloaddition. Chem. Commun. 2010;46:97–99. doi: 10.1039/b917797c. [DOI] [PubMed] [Google Scholar]
- 59.Debets MF, van Berkel SS, Dommerholt J, Dirks AT, Rutjes FP, van Delft FL. Bioconjugation with strained alkenes and alkynes. Acc. Chem. Res. 2010;44:805–815. doi: 10.1021/ar200059z. [DOI] [PubMed] [Google Scholar]
- 60.Hafner M, Renwick N, Brown M, Mihailovic A, Holoch D, Lin C, Pena JT, Nusbaum JD, Morozov P, Ludwig J, et al. RNA-ligase-dependent biases in miRNA representation in deep-sequenced small RNA cDNA libraries. RNA. 2011;17:1697–1712. doi: 10.1261/rna.2799511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moore MJ, Query CC. Joining of RNAs by splinted ligation. Methods Enzymol. 2000;317:109–123. doi: 10.1016/s0076-6879(00)17009-0. [DOI] [PubMed] [Google Scholar]
- 62.Aigner M, Hartl M, Fauster K, Steger J, Bister K, Micura R. Chemical synthesis of site-specifically 2′-azido-modified RNA and potential applications for bioconjugation and RNA interference. Chembiochem. 2011;12:47–51. doi: 10.1002/cbic.201000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jayaprakash KN, Peng CG, Butler D, Varghese JP, Maier MA, Rajeev KG, Manoharan M. Non-nucleoside building blocks for copper-assisted and copper-free click chemistry for the efficient synthesis of RNA conjugates. Org. Lett. 2010;12:5410–5413. doi: 10.1021/ol102205j. [DOI] [PubMed] [Google Scholar]
- 64.van Delft P, Meeuwenoord NJ, Hoogendoorn S, Dinkelaar J, Overkleeft HS, van der Marel GA, Filippov DV. Synthesis of oligoribonucleic acid conjugates using a cyclooctyne phosphoramidite. Org. Lett. 2010;12:5486–5489. doi: 10.1021/ol102357u. [DOI] [PubMed] [Google Scholar]
- 65.Marks IS, Kang JS, Jones BT, Landmark KJ, Cleland AJ, Taton TA. Strain-promoted “click” chemistry for terminal labeling of DNA. Bioconjug. Chem. 2011;22:1259–1263. doi: 10.1021/bc1003668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shelbourne M, Chen X, Brown T, El-Sagheer AH. Fast copper-free click DNA ligation by the ring-strain promoted alkyne-azide cycloaddition reaction. Chem. Commun. 2011;47:6257–6259. doi: 10.1039/c1cc10743g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.