Abstract
The chemical modification of histones at specific DNA regulatory elements is linked to the activation, inactivation and poising of genes. A number of tools exist to predict enhancers from chromatin modification maps, but their practical application is limited because they either (i) consider a smaller number of marks than those necessary to define the various enhancer classes or (ii) work with an excessive number of marks, which is experimentally unviable. We have developed a method for chromatin state detection using support vector machines in combination with genetic algorithm optimization, called ChromaGenSVM. ChromaGenSVM selects optimum combinations of specific histone epigenetic marks to predict enhancers. In an independent test, ChromaGenSVM recovered 88% of the experimentally supported enhancers in the pilot ENCODE region of interferon gamma-treated HeLa cells. Furthermore, ChromaGenSVM successfully combined the profiles of only five distinct methylation and acetylation marks from ChIP-seq libraries done in human CD4+ T cells to predict ∼21 000 experimentally supported enhancers within 1.0 kb regions and with a precision of ∼90%, thereby improving previous predictions on the same dataset by 21%. The combined results indicate that ChromaGenSVM comfortably outperforms previously published methods and that enhancers are best predicted by specific combinations of histone methylation and acetylation marks.
INTRODUCTION
The differential regulation of genes allows cells to respond to a number of changing external and internal stimuli that eventually determine a cell's developmental fate, its function as part of a complex tissue or its ability to respond to invading pathogens. Genetic information can be regulated at many levels from DNA transcription to a vast array of protein post-translational modifications (1). However, the regulation of gene transcription appears to be the primary and most important level of control, as Derman and colleagues suggested over 30 years ago (2).
The regulation of a gene is an exceedingly complex process controlled by interacting proximal and distal DNA sequence elements usually placed in a cis configuration. The proximal element is the basal promoter where the general transcription machinery assembles. A promoter is always located in close proximity to the 5′-end of a gene and is necessary but not sufficient for its transcription (3). Distal elements are either enhancers or silencers. Enhancers are thought to be composed of binding sites for transcription factors (TF) that upon recruitment to the enhancer loop over to the promoter, thus activating the transcription of the target gene. Enhancers may also be transcribed into non-coding RNAs that together with cohesin are thought to control specific long-range enhancer–promoter interactions (4). The functional mechanism of enhancers seems to be independent of their location and orientation, and enhancers are known to work at a great distance. For instance, a key enhancer of the Ssh gene lies within another gene (Lmbr1) located 1 Mb away from the Ssh promoter, and its disruption causes a limb malformation known as preaxial polydactyly (5). In another example, a 2.1 kb enhancer, located 1.1 Mb upstream of the male sexual development SOX9 gene, has been reported to regulate its expression (6). Silencers, on the other hand, negatively regulate the activity of target promoters and are much more difficult to characterize as their study requires a more complex and sophisticated experimental design (3).
Enhancers have traditionally been studied using experimental techniques such as electrophoretic mobility shift assays (EMSA), molecular cloning with a reporter gene and mutation analyses. The observation that a number of experimentally well-defined enhancers share sequence conservation with orthologous regions in other mammalian genomes led to the assumption that regulatory sequences are under negative evolutionary selection (7–11). This concept spearheaded the global identification of putative enhancers by computational means alone. In practice, however, this approach is limited for three reasons: (i) even if we could identify bona fide enhancers by such means alone, we would not know when, where or under what conditions such enhancers will be active; (ii) conservation might be indicative of function of many sorts (e.g. matrix attachment regions) and thus may not be necessarily indicative of enhancer activity; and (iii) we would miss rapidly evolving enhancers that are not found in evolutionarily conserved regions. In fact, the subsequent targeted deletion of four independent ultra-conserved elements of the mouse genome (12) had no obvious phenotype under the detection assays applied.
The experimental investigation of the proteins and chemical modifications associated with enhancers and promoters, especially by ChIP-chip (13), and later by ChIP-seq (14), showed that the post-translational modification of the histones, including phosphorylation, acetylation and methylation, is linked to specific events, including transcriptional activation, silencing, heterochromatin formation (15–19), DNA damage sensing and repair (20) and chromosomal segregation (21). In fact, the acetylation and methylation of specific histones is of particular interest in the field of gene regulation as these effects determine the activation, inactivation and poising of cis-acting regulatory DNA elements, such as promoters, enhancers and insulators, which in turn control gene expression programs both in tissue-specific and temporal manners (22–25). Genome-wide chromatin epigenetic maps have shown that enhancers, but not promoters, display the largest variability in their activation states across diverse cell types (26). Therefore, enhancers must be responsible for the development and differentiation of the many different cell types in the body by activating cell type-specific gene expression programs. This is clearly illustrated in haematopoiesis, where specific TFs are known to direct developmental fates, and the various haematopoietic progenitors are characterized by distinct gene expression programs (27,28).
The recent discovery of a broad domain of histone H3 lysine 4 monomethylation (H3K4me1) specific to enhancers, combined with low amounts of trimethylation on the same amino acid residue (H3K4me3) (25), has encouraged the genome-wide identification of enhancers. This approach involves the identification of chromatin methylation patterns by ChIP-seq, followed by the application of pattern recognition algorithms trained with specific chromatin profile signatures.
Although peak discovery tools perform well at identifying specific TF binding events in ChIP-seq libraries, the accurate identification of functional enrichment regions in chromatin modification maps has demanded the implementation of sophisticated pattern recognition algorithms (25,29–31). In these methods, the aligned tag counts from ChIP-seq libraries are processed into profiles of specified window sizes with positive profiles centered at enhancer marker-enrichment regions, and background profiles are generated at random loci. Classifiers are then implemented to discriminate functional from non-functional profiles. These methods are nevertheless limited as they either consider only a small number of epigenetic marks, or need far more marks to make accurate predictions than are experimentally viable for most laboratories. The profile method (PM) (25) and the hidden Markov model method (HMM) (29) only explored datasets using a limited number of chromatin modifications, mainly focusing on H3K4 methylations, whereas the artificial neural network method CSI-ANN (31) combined ∼40 datasets of methylation and acetylation signatures. To overcome these limitations, we have developed a novel method for chromatin state detection combining support vector machines (SVM) with genetic algorithm (GA) optimization (ChromaGenSVM). ChromaGenSVM automatically selects the types of histone epigenetic marks that best characterize active enhancers. The GA optimizes the window size of the epigenetic profiles and the SVM hyperparameters. ChromaGenSVM was initially trained with a small set of ChIP–chip chromatin maps from the pilot ENCODE region in untreated HeLa cells (25). ChromaGenSVM successfully predicted 88.0% of the experimentally supported enhancer regions in IFN-γ treated HeLa cells (independent test set). Our method managed to recover higher numbers of supported functional regions both in untreated and IFN-γ-treated HeLa cell libraries in a parsimonious way.
In a second exercise, ChromaGenSVM was trained with 38 distinct chromatin marks derived from genome-wide (ChIP-seq) histone methylation and acetylation maps done in human CD4+ T cells. Our method selected an optimum combination of only five epigenetic marks that accurately characterize active enhancers. These marks include both activating and repressive methylations and acetylations that combine in putative enhancers in various ways. This suggests that well-trained signal detection algorithms are in principle better at locating enhancers than simpler methods that look for the presence or absence of specific marks. About 90% of the enhancers predicted in human CD4+ T cells were supported by at least one type of experimental evidence. This demonstrates ChromaGenSVM's high sensitivity and specificity for the identification of active enhancers from specific combinations of chromatin epigenetic marks.
MATERIALS AND METHODS
Datasets
Our first SVM model was built to recognize enhancers found in the pilot ENCODE region in untreated HeLa cells (25). This was done in order to compare the performance of ChromaGenSVM to previously published methods (PM, HMM and CSI-ANN), and also to demonstrate its applicability to ChIP-chip libraries. For this, the six distinct ChIP-chip chromatin modification maps reported by Heintzman et al. (25), both in untreated and treated HeLa cells, were used. These include the core histone H3 (H3), the acetylation of lysines 9 and 14 of histone H3 (H3Ac), the acetylation of lysines 5, 8, 12 and 16 of histone H4 (H4Ac), the mono-methylation of lysine 4 of histone H3 (H3K4Me1), the di-methylation of lysine 4 of histone H3 (H3K4Me2) and the tri-methylation of lysine 4 of histone H3 (H3K4Me3).
From the original ENCODE data, we derived a positive class set (called E1) with epigenetic profiles centered at TSS-distal regions (≥ 2.5 kb upstream and downstream of the TSS) that were enriched in p300 binding. p300 is a transcriptional coactivator that works by binding to transcription factor activation domains to then position histone acetyltransferases (HATs) near specific nucleosomes in target gene promoter regions (32) and found to localize to many active enhancers (25), but not all. The E1 set includes 74 TSS-distal high-confidence peaks as described in Heintzman et al. (25) (Supplementary Table S1).
A second SVM model was implemented to predict enhancers from 20 and 18 genome-wide ChIP-seq histone methylation and acetylation maps done in human CD4+ T cells (33) (Supplementary Table S2). The methylation and acetylation maps were combined to build the enhancer predictor. A set of positive class examples (called E2) was built with regions that were also enriched in p300 binding in CD4+ T cells, as described by Wang et al. (33). The E2 dataset includes 527 TSS-distal p300 peaks (Supplementary Table S3). The peak centers were selected from p300 regions spanning less than 1 kb.
For both SVM models, the epigenetic profiles of the positive class examples were computed at various window sizes (1, 2.5, 5, 7.5, 10, 12.5 and 15 kb). Tag reads were averaged in 100 bp and 200 bp bins for each window size. A background dataset (negative class example) was built for each SVM model, with a size 10-fold the number of p300 peaks in E1 and E2. Background profiles were centered at random chromosomal positions.
Support vector machines
SVMs are a machine learning method of broad applicability to many types of pattern recognition problems. Since an excellent introduction to SVMs exists (34), here we will briefly describe SVMs as applied to our specific case. In SVMs, the input vectors are first mapped onto one feature space (possibly with a higher dimension) by means of a kernel function. Then, a hyperplane is built to separate the positive and negative examples within this feature space. Only relatively low-dimensional vectors in the input space and dot products in the feature space will evolve by a mapping function. SVMs have been designed to minimize structural risk whereas other machine learning methods, such as artificial neural networks (ANN), are based on the minimization of empirical risk. Therefore, SVMs are less vulnerable to the over-fitting problem, and so they can typically deal with a large number of features. There are several important parameters in an SVM, including the kernel function (and its specific parameters) and the regularization parameters. Neither the kernel function nor the regularization parameters can be defined from the optimization problem but must be manually tuned. This can be done by applying Vapnik-Chervonenkis bounds, cross-validation, an independent optimization set, or Bayesian learning (35). GA was used to automate the selection of optimum SVM hyper-parameters through cross-validation. Since nucleosomes are dynamically allocated and their positions oscillate in small locus ranges, it is possible that the state of chromatin shifts around at certain loci (36). Thus, we try to improve profile/peak detection by adding a shift term to the Gaussian kernel. In this way, the similarity between profiles was computed by comparing profiles at different shift steps from 100 to 1000 bp. SVMs were implemented using the Python programming language and the SHOGUN toolbox (37).
ChromaGenSVM
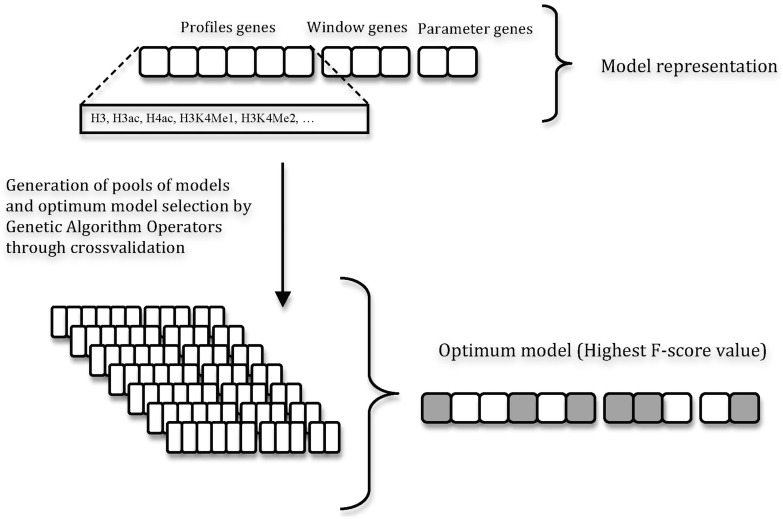
SVM models were first trained to recognize the histone modification profiles associated with putative enhancers of the ENCODE region in untreated HeLa cells, and then in human CD4+ T cells. For each dataset, the most discriminating epigenetic marks and profile sizes were selected using the GA strategy described in Figure 1. GAs are stochastic optimization methods that have been inspired by evolutionary biology principles, and as such are governed by the rules of natural selection (38). The most relevant aspect of GAs is their ability to search for many possible solutions simultaneously, each of which explores different regions in parameter space (39).
Figure 1.
ChromaGenSVM workflow. Different SVM models are trained using different epigenetic marks, profile window sizes and SVM parameters generated by GA rules. The optimum SVMs are selected by cross-validation after 100 GA runs.
The GA output was also used to tune both the SVM regularization and the Gaussian kernel parameters. For this, an initial population of SVM models was first generated with regularization and kernel parameters that were randomly selected from a pool of adequate values (ranging from 10−4 to 103). Next, SVM models were trained with combinations of unique chromatin modification profiles randomly selected from a pool of epigenetic profiles computed at seven fixed window sizes (1, 2.5, 5, 7.5, 10, 12.5 and 15 kb). In the GA framework, a model was represented as a fixed-length bit string. Each binary ‘gene’ in the bit string encoded the inclusion (1) or exclusion (0) of an epigenetic mark in the training data. This bit string was concatenated with two other bit strings encoding the window size and the SVM hyper-parameters. The fitness or cost function of each model was computed as the F-score of enhancer over background classification in three-fold-out (TFO) cross-validation tests. Furthermore, crossover and mutation operators were applied to the top-ranked predictors in a reproduction step to create a new population of models. The crossover operator combines the information from two parent models to generate children models. On the other hand, the mutation operator takes a single parent model to generate a child by randomly changing part of the information derived from the parent. Seventy percent (70%) of the new generation was created by binary crossover of the bit strings of progenitor pairs and the rest by mutating the single parent ‘genes’. A copy of the best-ranked model in the new population was also kept. Based on experimental and previous computational evidence, we set to penalize combinations of more than five marks during GA optimization. This means that models including more than five epigenetic marks needed to increase their accuracy by at least 0.01 AUC units per extra epigenetic mark to rank on top of five-mark models during evolution. The reproductive cycle continues until the best fitness score remains unchanged for 90% of the generations, or the maximum number of generations is reached. This algorithm was run 100 times training 100 models in each population for a maximum of 100 generations. The best model of each run was selected from the population of the final generation. The statistical analysis of the most informative epigenetic marks was done by histogram density plots of the chromatin methylations in the models of the final generation. The algorithm was implemented in Python using the SHOGUN toolbox (37) and the Pyevolve module (40).
Performance evaluation
A trained SVM model returns a vector of scores between 0 and 1 for a combined epigenetic profile. These scores are then transformed to a binary state indicating a ‘regulatory’ or ‘non-regulatory’ region by choosing a cut-off. For each combination of profiles, the existence of a regulatory element is considered positive (P) or negative (N) otherwise. True (T) means that the predicted and observed functional states are identical, and false (F) implies otherwise. The notations TP, FP, TN and FN combine these labels to return the number of data points (combined profile) in each category. These values correspond to a cut-off at which SVM analog values are transformed into binary predictions. The predicted functional scores are transformed into binary predictions by using different cut-offs yielding sensitivity and specificity over the entire score range. The F-score in Equation 4 was used as the fitness score or cost function in the GA optimization. ROC plots display the FP (1-specificity) values on the x-axis, and the TP (sensitivity) values on the y-axis. ROC plots show the direct relationship between the FP and TP rates. The total AUC (area under the curve) for ROC plots was used as a measure of the prediction performance of our method:
| (1) |
| (2) |
| (3) |
| (4) |
Positive predicted value (PPV) (same as precision)
Recall (same as sensitivity)
A prediction located within a number of kb equal to the profile window size of an experimental mark of enhancers was recorded as a TP hit, as done by Heintzman et al. (25). Three types of experimental evidence were used to classify predictions as TP hits. These include (i) DNase I hypersensitivity regions (DHS), which are indicative of an ‘open’ chromatin state (41); (ii) p300 binding sites; and (iii) regions marked with the mediator complex component TRAP220 (42,43). DHS, p300 and TRAP220 regions are all known to mark subsets of enhancers. Our predictions were further validated computationally by assessing their localization within evolutionarily conserved regions and the local clustering of TF binding sites (TFBS). Predicted regions were considered to be computationally supported when they either (i) overlapped with regions with normalized PhastCons (44) scores ≥0.5 and/or (ii) overlapped with regions of TFBS clusters according to the PReMod database (45). The PhastCons (44) scores measuring the evolutionary conservation across 17 vertebrate genomes were extracted from the UCSC Genome Browser. Genome regions with TFBS clusters were downloaded from the PReMod database (45) available at http://genomequebec.mcgill.ca/PReMod/.
RESULTS
Enhancer prediction in the pilot ENCODE region of HeLa cells from ChIP-chip histone modification maps
The ENCODE project was developed to provide a functionally informative representation of the human genome by using high-throughput methods to identify and catalog its functional elements. In its pilot phase, the project targeted ∼30 Mb of DNA, equivalent to 1% of the human genome. Of this, ∼50% of the DNA regions were manually selected whereas the other half was selected at random (46). The manually selected DNA regions included a number of well-studied loci for which comparative data in other species exist.
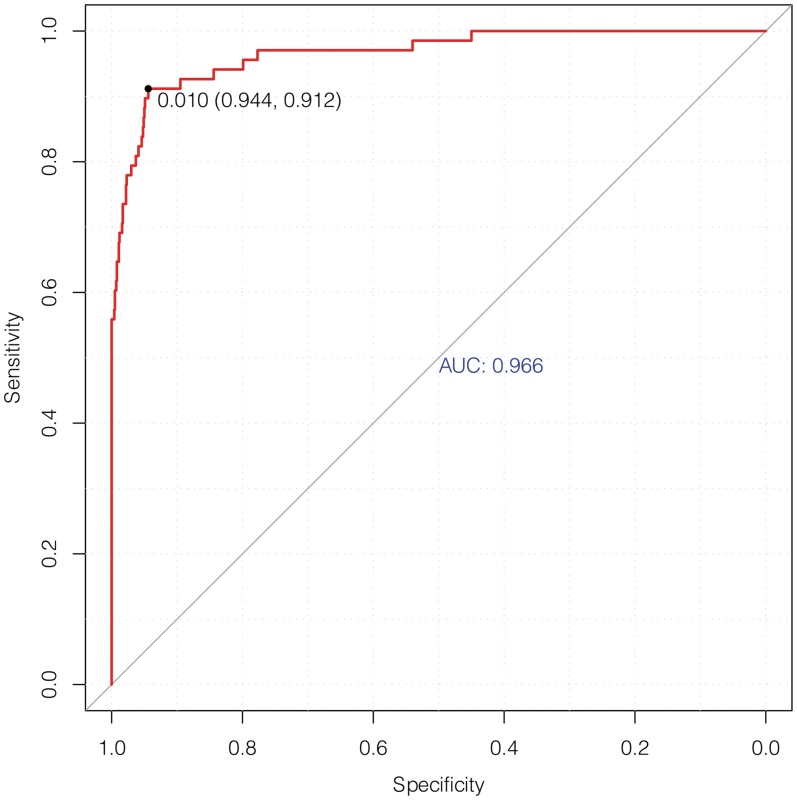
ChromaGenSVM was trained on a subset of high-confidence putative enhancers from six ENCODE ChIP-chip chromatin maps (H3, H3Ac, H4Ac, H3K4Me1, H3K4Me2, H3K4Me3) in untreated HeLa cells (25) (see Materials and Methods). The optimum ChromaGenSVM predictor for the ENCODE region combines H3, H3K4Me1 and H3K4Me3 methylation profiles generated at 5.0 kb windows with a signal shift of 400 bp. Figure 2 shows the cross-validation ROC curve for enhancer predictions in HeLa cells with a maximum F-score of 0.928, and an AUC, specificity and sensitivity of 0.97, 0.944 and 0.912, respectively.
Figure 2.
Cross-validation ROC plot of the optimum SVM model to predict enhancers in the pilot ENCODE region of HeLa cells using the H3, H3K4Me1 and H3K4Me3 epigenetic signatures.
The ENCODE genome region was scanned both in untreated and in IFN-γ-treated HeLa cells using the optimum ChromaGenSVM model at different classification thresholds. Enhancers were predicted from histone modification profiles computed at 5.0 kb windows and with a 1.25 kb resolution. If more than one region was predicted within a 5.0 kb window, only the best-scoring region was recorded. The quality of our predictions was evaluated by counting the number of predicted regulatory regions lying within 2.5 kb of a DHS, p300 or TRAP220 region (‘overlap’), as explained above. The predicted regions were also characterized according to their evolutionary conservation across 17 vertebrate genomes using PhastCons scores (44) and the presence of TF binding site (TFBS) clusters from the PReMod database (45).
The definition of a non-functional genomic region (a true negative) is not straightforward, because a region may appear to be non-functional under a specific set of experimental conditions, but not in others. Therefore, the evaluation of the predictive ability of the method was determined by the recovery of regions whose functionality is inferred from external experimental datasets or computational analyses (true positives). The precision of our method was thus calculated from the number of predictions that presented either experimental or computational support and the sensitivity as the proportion of supported enhancer regions recovered by our predictions.
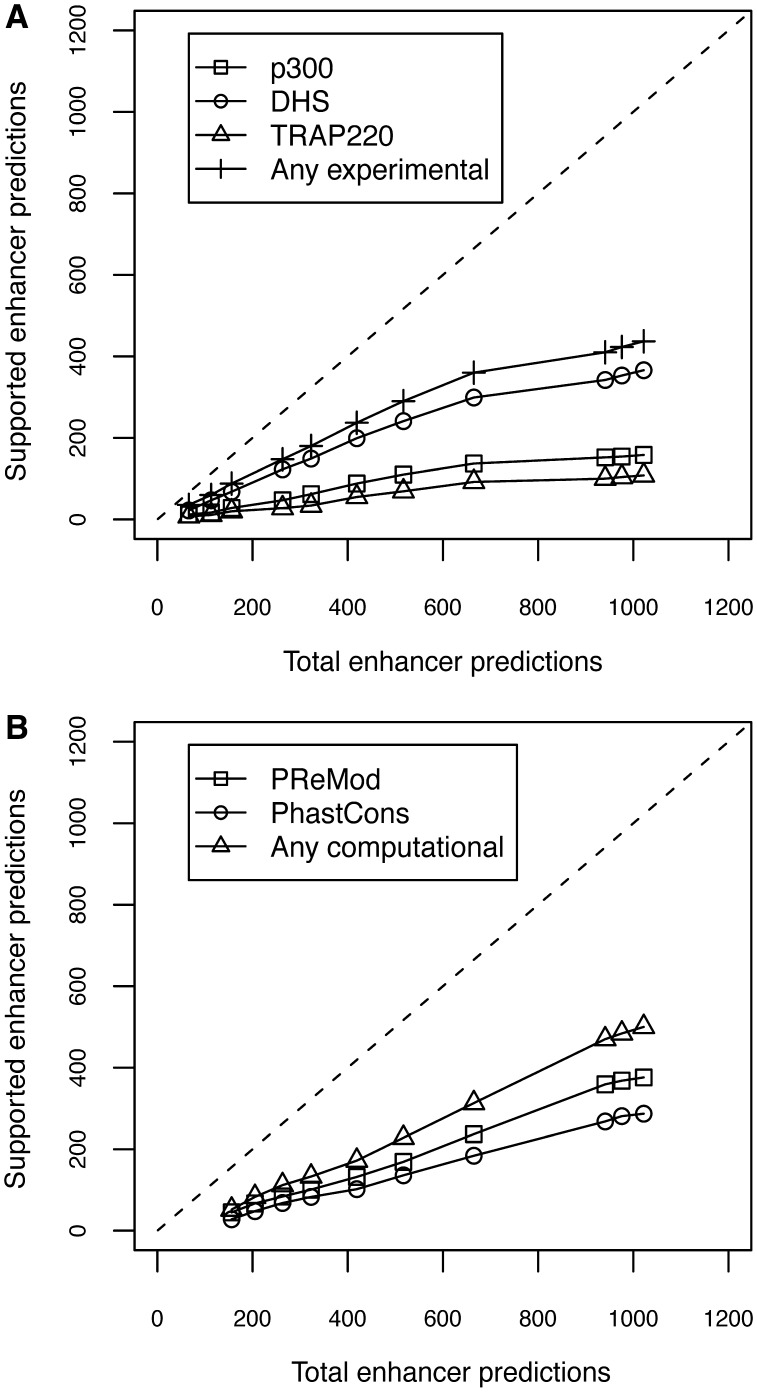
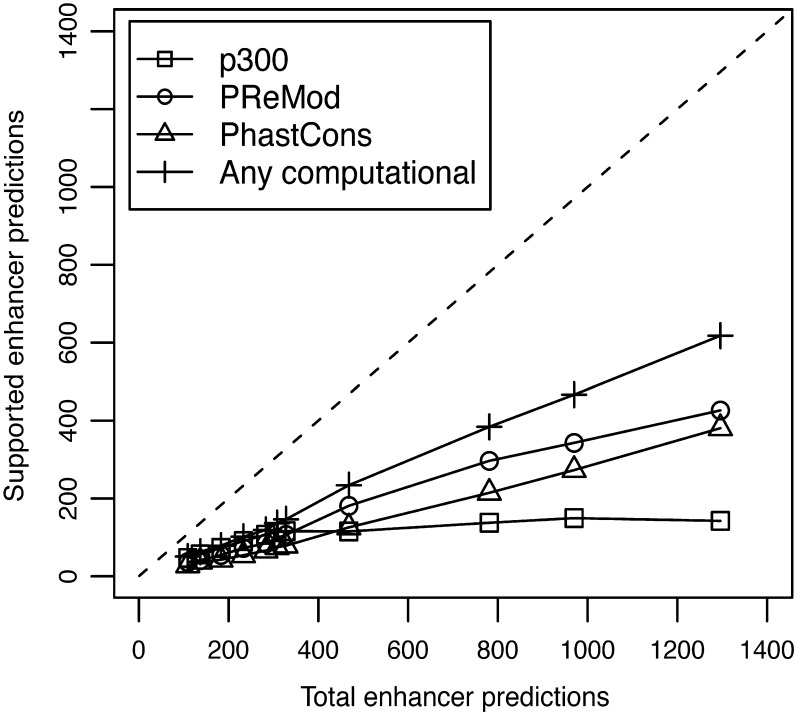
The enhancers predicted both in untreated and in IFN-γ-treated HeLa cells were found to overlap extensively with experimental and computational enhancer marks characteristic of functional regulatory regions. Figures 3 and 4 show the plots of total predictions versus predicted regions overlapping different enhancer marks and functional evidences (supported predictions), both in untreated and in IFN-γ-treated HeLa cells, respectively.
Figure 3.
Plots of total predictions versus supported predictions in untreated HeLa cells using the H3, H3K4Me1 and H3K4Me3 epigenetic signatures. The dashed line represents an ideal predictor. (A) Experimental evidences of functional regions: square (p300), circle (DHS), triangle (TRAP200) and cross (any experimental); (B) Computational evidences of functional regions: square (PReMod), circle (PhastCons) and triangle (any computational).
Figure 4.
Plot of total predictions versus supported predictions in IFN-γ treated HeLa cells using the H3, H3K4Me1 and H3K4Me3 epigenetic signatures. The dashed line represents an ideal predictor. Evidences of functional regions: square (p300), circle (PReMod), triangle (PhastCons) and cross (any computational).
The number of supported predictions increases as we augment the total number of predictions with the corresponding decrease in the cut-off value. The selection of the optimum cut-off for a binary classifier is a trade-off between a predictor's specificity and its sensitivity. Depending on the experimental datasets, predictions can be computed at different recall (or sensitivity) and specificity levels. So far, experimental evidences for enhancers cover only a fraction of the total number of existing enhancers, and the vast majority of the distal functional regions most likely remain unknown. For this reason, we selected the cut-off of the maximum F-score to ensure a good coverage of putative enhancers. A total of 1022 distal regulatory elements were predicted in untreated HeLa cells at this cut-off value. ChromaGenSVM successfully recovered 85.1% of the high-confidence p300 peaks reported by Heintzman et al. (25) in untreated HeLa cells. Our predicted regions overlap with 38.2% of the DHS sites and 71.1% of the TSS-distal regions enriched in the mediator complex component TRAP220 (Supplementary Figure S1). Additional computational evidences supported our predicted set of enhancers. About 30% of the predicted enhancers are found within ENCODE regions that are conserved across vertebrate genomes according to PhastCons scores (44) and 34% of these regions contain computationally predicted TFBS clusters according to the PReMod database (45). Finally, ∼66% of the enhancers predicted in untreated HeLa cells were supported by at least one type of experimental or computational evidence (Supplementary Figure S1).
The ChromaGenSVM scan of the ENCODE ChIP-chip regions of IFN-γ-treated HeLa cells yielded a set of 1001 putative enhancers (Supplementary Figure S2). These predicted regions overlap with 88.0% of the distal p300 binding sites found in these stimulated cells. Although p300 is the only bona fide enhancer mark available in the IFN-γ-treated HeLa cell dataset, the predicted regions were well supported by other types of computational evidence. Twenty-seven percent of the enhancers predicted in IFN-γ-treated HeLa cells are conserved across vertebrates, and TFBS clusters are found in 40% of these regions. Overall, 55% of the enhancers predicted in IFN-γ-treated HeLa cells were supported by at least one type of computational evidence.
Performance comparison of ChromaGenSVM with other published methods
Table 1 summarises a comparative analysis of the performance of our method and three other predictive approaches on the same ENCODE region of HeLa cells. Using statistical criteria, the PM method reported a total of 389 and 324 predictions in untreated and IFN-γ-treated HeLa cells, respectively. The HMM and CSI-ANN methods reported the same number of predictions for the sake of comparison. In this regard, we also produced a reduced set of 391 and 325 enhancers in untreated and IFN-γ-treated HeLa cells. In our top 391 predictions in untreated HeLa cells, our model recovered ∼70% of the p300 peaks, ∼26% of the DHS regions and ∼53% of the TRAP220 binding regions. We consider the lower sensitivity of our model in the training data as a sign of a higher parsimony rather than evidence of underperformance. Our optimum SVM was set to yield optimum predictions in cross-validation experiments rather than simple fitting of the training data. Regardless of the lower sensitivity, our model had the second highest precision (57%) in untreated HeLa cells, outperforming the PM and HMM methods by 4% and 3%, respectively. CSI-ANN reported the highest precision in untreated HeLa cells (66.3%) but did not report enhancer predictions in the independent dataset of IFN-γ-treated HeLa cells. The implementation of CSI-ANN that is publicly available can only be trained and tested on the same epigenetic library, thus hampering the analysis of the prediction performance of this method in an independent dataset. In contrast, our method predicted enhancers in IFN-γ-treated HeLa cells with a higher sensitivity of 72.0%, very similar to that of the HMM method. This result indicates a high sensitivity for the prediction of enhancers in an independent dataset that is yet 4% lower than that of the PM method. However, it is worth mentioning that the PM method predicts enhancers within 10 kb windows whereas ChromaGenSVM defines functional regions twice as precisely within 5.0 kb regions.
Table 1.
Comparative performance analysis of the enhancer predictions in the pilot ENCODE region of untreated and IFN-γ treated HeLa cells showing the number of regions recovered over the number of total predictions and sensitivity values in parentheses
| Cell | Method Marker | PM(%) | HMM(%) | CSI-ANN(%) | ChromaGenSVM(%) |
|
|---|---|---|---|---|---|---|
| Adapted cut-off | Optimum cut-off | |||||
| Untreated HeLa cells | P300 (n = 94) | 77/389 (81.9) | 82/389 (87.2) | 79/389 (84.0) | 66/391 (70.2) | 80/1022 (85.1) |
| DHS (n = 587) | 165/389 (28.1) | 179/389 (30.5) | 243/389 (41.4) | 152/391 (25.9) | 224/1022 (38.2) | |
| TRAP220 (n = 76) | 43/389 (55.8) | 47/389 (61.0) | 54/389 (71.0) | 40/391 (52.6) | 54/1022 (71.1) | |
| p300 or DHS or TRAP220 | 206/389 (52.9)* | 213/389 (54.8)* | 258/389 (66.3)* | 223/391 (57.0)* | 299/1022 (30.1)* | |
| IFN-γ treated HeLa cells | P300 (n = 151) | 116/324 (76.8) | 109/324 (72.2) | – | 109/325 (72.0) | 133/1001 (88.0) |
Note. ChromaGenSVM also reports the number of predictions in untreated HeLa cells at the cut-off of maximum F-score.
*Precision [TP/(TP + FP)] in brackets (Equation 3).
Table 1 also reflects the prediction performance of ChromaGenSVM at the optimum cut-off of the maximum F-score, yielding 1022 predicted enhancers in untreated HeLa cells. ChromaGenSVM improved the discovery of p300 enrichment regions by 3% and 1% with respect to the PM and CSI-ANN methods, respectively. We also observed that the number of predictions by the PM and HMM methods that were supported by DHS evidence were surpassed by ChromaGenSVM by ∼10% and ∼8%, respectively. Moreover, the number of TRAP220 regions recovered in untreated HeLa cells is greater than those of the PM and HMM methods by ∼15% and ∼10%. The performance analysis for the 1001 predicted enhancers in IFN-γ treated HeLa cells shows that ChromaGenSVM makes an improvement of ∼12% and ∼16% over the PM and HMM methods for the recovery of p300-enriched regions.
Genome-wide enhancer prediction in human CD4+ T cells from ChIP-Seq histone modification maps
ChromaGenSVM was implemented to predict enhancers in human CD4+ T cells by exploring their chromatin epigenetic landscape. ChromaGenSVM is likely to yield more accurate and robust enhancer predictions when exploring a more diverse epigenetic landscape (i.e. with a larger number of epigenetic maps). We can fully exploit the power of GA to unveil robust functional relationships from complex information spaces (38). In this context, a vast collection of ChIP-seq epigenetic maps (including 20 chromatin methylations and 18 acetylations, Supplementary Table S2) profiled in human CD4+ T cells (33) was explored in order to predict enhancers genome-wide.
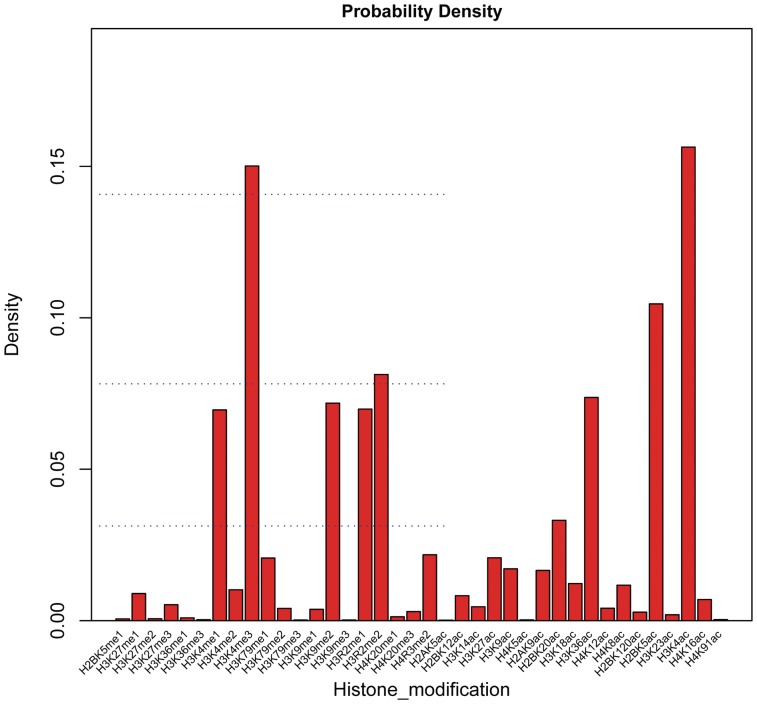
ChromaGenSVM was run on the combined histone methylation and acetylation maps. Then, we analyzed the relative relevance of each epigenetic signature to characterize the chromatin state across the entire genome of human CD4+ T cells. The probability density histogram in Figure 5 depicts the frequency density of every epigenetic mark in the pool of models in the final generations from 100 independent ChromaGenSVM runs. The acetylation and tri-methylation of the lysine 4 residue of histone H3 (H3K4Ac and H3K4Me3) were the most frequent profiles in the top-ranked models. In addition, the acetylation of the lysine 5 residue of the histone H2B (H2BK5Ac) was found to be the next most frequent profile. ChromaGenSVM yielded other interesting epigenetic features of enhancers including the activating mono-methylation of the lysine 4 residue of histone H3 (H3K4Me1).
Figure 5.
Probability density histogram of the epigenetic signatures in the pool of top-ranked SVM predictors selected after 100 GA runs for the prediction of distal regulatory elements in human CD4+ T cells. The most frequent signatures in the top-ranked predictors were H3K4Ac, H3K4Me3 and H2BK5Ac.
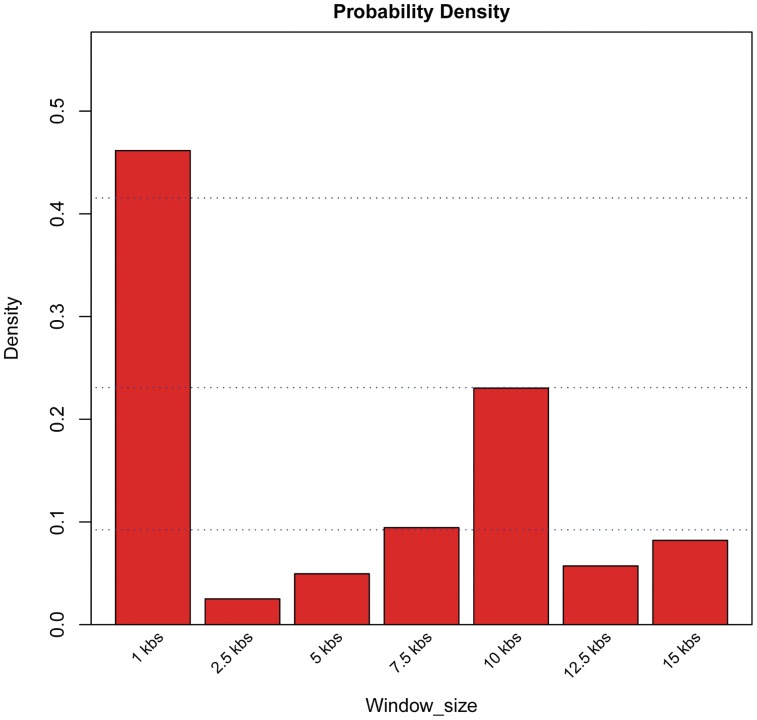
Similarly, the probability density histogram of the optimum window size is depicted in Figure 6. The extension and boundaries of the unique epigenetic landscape associated with functional regions in human CD4+ T cells were best encoded in epigenetic profiles spanning 1.0 kb.
Figure 6.
Probability density histogram of profile window sizes in the pool of top-ranked SVM predictors selected after 100 GA runs for the prediction of distal regulatory elements in human CD4+ T cells. The most frequent window size in the top-ranked predictors was 1.0 kb.
In fact, several types of histone modifications coexist as shown by the preliminary inspection of the histone acetylation and methylation data from human CD4+ T cells. This analysis uncovered strong inter-correlations among the epigenetic profiles associated with the putative p300 enhancers of the training set (Supplementary Figure S3). This suggests that several, overlapping, combinations of epigenetic marks could yield similar prediction performances. Therefore, it has been difficult to assess the nature and number of necessary marks that uniquely define the functional state of a particular genomic region. In our computational analyses, we control the number of chromatin marks in a single model by penalizing predictors combining more than five histone methylations.
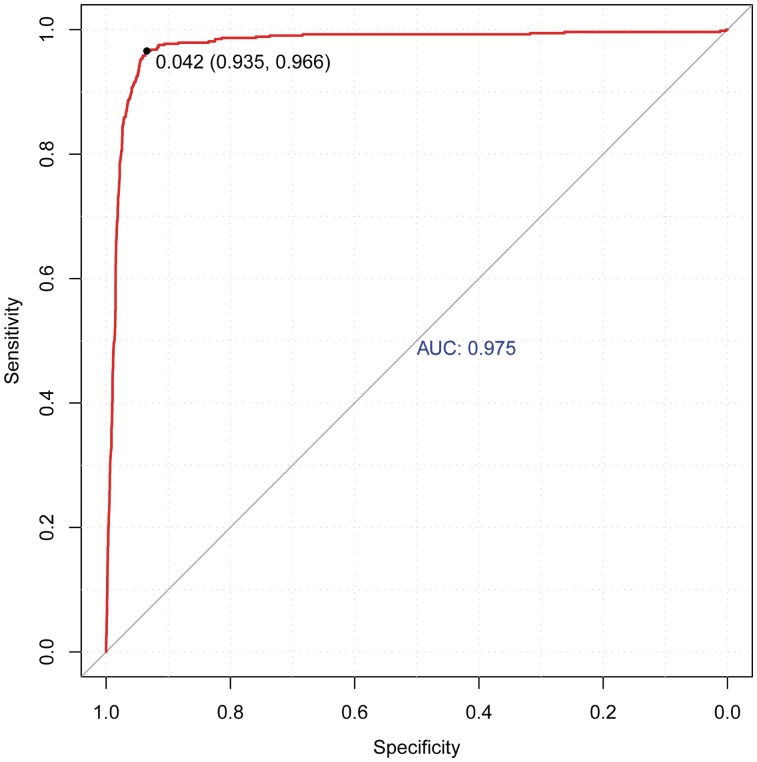
Figure 7 depicts the ROC of the optimum SVM model generated by ChromaGenSVM that recognizes enhancers in human CD4+ T cells with an AUC, specificity and sensitivity of 0.975, 0.935 and 0.966, respectively, and a maximum F-score of 0.950. This predictor was trained with three methylations and two acetylations marks computed at 1.0 kb windows: H3K4Me1, H3K4Me3, H3R2Me2, H4K8Ac and H2BK5Ac. The optimum Gaussian kernel has a shift parameter of 400 bp. This optimum combination of methylation and acetylation marks includes two of the most frequent marks (H2BK5Ac, H3K4Me3) accompanied by two other methylation marks (H3K4Me1, H3R2Me2) that complement the epigenetic landscape at TSS distal regulatory regions along with another acetylation mark (H4K8Ac).
Figure 7.
Cross-validation ROC plot of the optimum SVM model to predict enhancers in human CD4+ T cells using the histone modification maps H3K4Me1, H3K4Me3, H3R2Me2, H4K8Ac and H2BK5Ac.
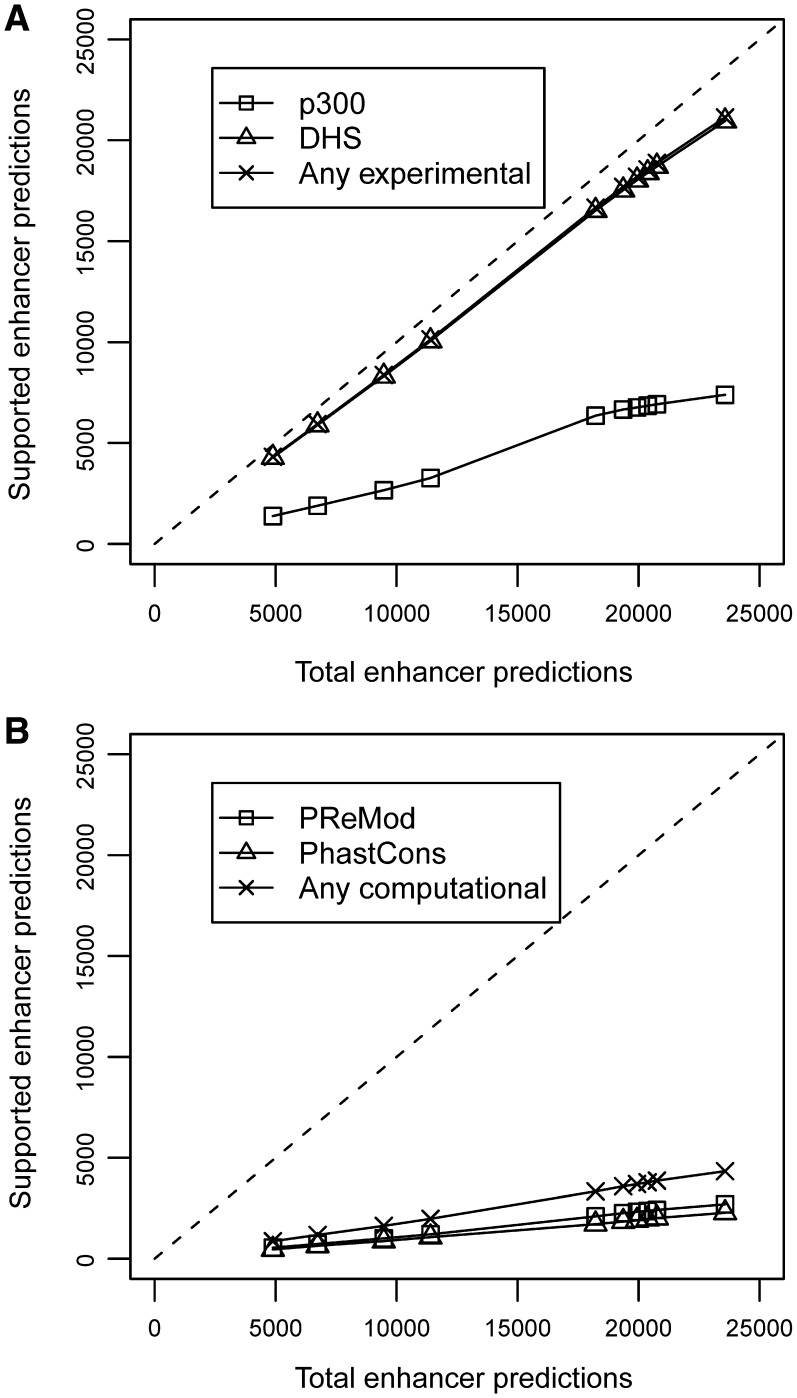
Using this optimum combination of five epigenetic marks, we predicted enhancers genome-wide in human CD4+ T cells. Epigenetic signals were scanned using 1.0 kb windows with a resolution of 400 bp, retaining only the highest scoring element within every 1.0 kb region. Here we report 23 574 predicted enhancers in human CD4+ T cells (Supplementary Table S4). The quality of the predictions in CD4+ T cells was evaluated by counting the overlap with 72 646 DHS regions and 3989 p300 binding sites. The clustering of TFBS and the evolutionary conservation of the predicted regions were also analyzed. Figure 8 shows the plots of the total number of predictions versus supported predictions in a range from 5000 to 24 000 predicted enhancers. Substantial overlaps with DHS regions and p300 binding sites (in the ranges 88–91% and 28–34%) were observed for different subsets of predicted enhancers. Taken these data together, 88–91% of the predictions were supported by at least one of the experimental lines of evidence. Moreover, 10–12% of the predicted regions were found to be evolutionarily conserved according to the PhastCons scores (44), and TFBS clusters were detected in a similar fraction of these regions according to the PReMod database (45). About 19% of the predictions were at least supported by one of the computational lines of evidence.
Figure 8.
Plots of total predictions versus supported predictions in human CD4+ T cells using the histone modification maps of H3K4Me1, H3K4Me3, H3R2Me2, H4K8Ac and H2BK5Ac. The dashed line represents an ideal predictor. (A) Experimental evidences of functional regions: square (p300), triangle (DHS) and cross (any experimental); (B) Computational evidences of functional regions: square (PReMod), triangle (PhastCons) and cross (any computational).
A large set of 20 953 enhancers, selected by ChromaGenSVM at the optimum cut-off of the maximum F-score, had a very good rate of supporting evidence. Eighty-nine percent of the enhancers in this predicted dataset are located in open chromatin regions according to DHS evidence and 31% overlap with p300-binding regions. TFBS clusters were found in 11% of these loci and 10% were found to be conserved across 17 vertebrates genomes.
Performance comparison of the genome-wide enhancer predictions by ChromaGenSVM with other published methods in human CD4+ T cells
Table 2 shows a comparative performance analysis between the 23 574 enhancers predicted by our method (Supplementary Table S4) at the optimum cut-off and the 36 769 enhancers predicted by CSI-ANN. Larger fractions of our predictions overlapped with experimental evidences. We predicted a total of 20 953 enhancers located at open chromatin regions versus the 23 017 reported by CSI-ANN. The number of p300-associated regulatory regions increased by 2414, indicating an improvement of 50% over CSI-ANN. Moreover, 90% of ChromaGenSVM's predictions (versus 69% of CSI-ANN's predictions) overlapped with at least one of the experimental evidences, demonstrating that our method represents an improvement in precision of 21% as determined by the presence of experimental marks. The evolutionary conservation and presence of TFBS clusters in our predicted enhancers were slightly smaller than those the reported by CSI-ANN (31). However, these types of evidence have been shown to overlap weakly with cell-type specific and environment-dependent enhancer activation (47).
Table 2.
Comparative performance analysis of the genome-wide enhancer predictions in human CD4+ T cells according to experimental evidences
| Method | CSI-ANN Total number of predictions: 36 769 |
ChromaGenSVM Total number of predictions: 23 574 |
||
|---|---|---|---|---|
| Enhancer marker | S.P. | Precision (%) | S.P. | Precision (%) |
| p300 | 4964 | 13.5 | 7378 | 31.3 |
| DHS | 23 017 | 62.6 | 20 953 | 88.9 |
| p300 or DHS | 25 444 | 69.2 | 21 122 | 89.6 |
| PReMod | 9037 | 24.6 | 2695 | 11.4 |
| PhastCons | 8124 | 22.1 | 2274 | 9.6 |
| PReMod or PhastCons | – | – | 4337 | 18.4 |
S.P.: Supported predictions; Precision [TP/(TP + FP)] (Equation 3).
Enhancers have tissue-specific activity, and therefore we expected that our predicted set of enhancers possess specific functions in human CD4+ T cells. According to the gene ontology (GO) analysis using the GREAT tool (48), the genes closest to the predicted regions are significantly (P < 10−4) related to immune system processes and cell types (Supplementary Figure S4). This analysis reflects that a significant set (P < 10−4) of the discovered enhancers are involved in the regulation of differentiation and activation of T cells. Thus, we would also expect that our set of enhancers is associated with genes having T-cell-specific expression compared to non-specific genes. In this regard, we found that the genes associated with our predicted set of enhancers (by proximity) were also differentially regulated in CD4+ T cells according to the analysis of the data in the Gene Expression Atlas (49). We found that 112 genes associated with the discovered enhancers belong to the top 1000 upregulated genes in CD4+ T cells, whereas only 73 would be randomly expected (P < 10−4). Meanwhile, the top 1000 downregulated genes included 145 of the genes associated with the predicted enhancers in comparison to 182 randomly expected (P < 10−4). This represents a 1.5-fold enrichment in upregulated genes and a 1.3-fold decrease in downregulated genes in comparison to what would be randomly expected (Supplementary Figure S4).
DISCUSSION
A large body of evidence suggests that genetic programs and cellular states are tightly controlled by the chemical modification of histones and other proteins that package the genome (43,50,51). Accordingly, the chromatin states for a wide variety of cell types and environmental conditions have been mapped, allowing the computational modeling of these epigenetic landscapes. ChromaGenSVM was designed to predict enhancers by combining the minimum possible number of epigenetic marks. In the tests reported here, ChromaGenSVM yielded optimum combinations of three and five histone modification marks that best predict enhancers in the pilot ENCODE region of HeLa cells and in human CD4+ T cells (genome-wide), respectively.
Similarly to previously published methods (25,29,31), ChromaGenSVM discovered enhancers from histone modification data by recognizing the epigenetic signals associated with p300-enrichment regions. The coactivator p300 only targets a reduced subset of enhancers and the total number of real enhancers is thus unknown (47). Thus, limiting the analysis of our method's performance to a reduced fraction of the predictions has little biological basis. Since non-functional regions (true negatives or false positives) are difficult to define, the experimentally and/or computationally supported regions (true positives) should be regarded as part of the real enhancer repertoire. In this context, the quality of a method should be better assessed by the precision [Equation (3)] rather than by its sensitivity [Equation (1)] or specificity [Equation (2)]. When the positive examples available constitute a very reduced subset of the positive space, it is crucial to avoid over-fitting of the training data. The methods HMM (29) and CSI-ANN (31) recovered large fractions (∼85%) of p300-associated enhancers in the top 389 predictions from the training data of untreated HeLa cells. Nevertheless, these performances could suggest an over-fitting of the training data while evidence of improved performance should rather be provided by the enhancer predictions in an independent dataset. This is clear from the lower sensitivity of HMM (29) predictions in IFN-γ-treated HeLa cells. The precision of our predictions in untreated HeLa cells was 57%, the second highest among all the methods under analysis. In addition, ChromaGenSVM predicted similar top sets of 391 and 324 functional regions in untreated and treated HeLa cells (with an accuracy of 72% in the latter case) and yielded 1022 well-supported enhancers at the optimum cut-off value in untreated HeLa cells. We have shown that ChromaGenSVM is a parsimonious and robust SVM implementation that surpasses previously published methods in predicting enhancers in the pilot ENCODE region.
Besides the positive and conclusive results from the modeling of ChIP-chip histone methylation maps, ChromaGenSVM also outperformed other existing methods at genome-wide enhancer prediction in human CD4+ T cells. Instead of combining all the methylation and acetylation maps available for this cell type, ChromaGenSVM automatically selected an optimum subset of (ChIP-seq) histone modification maps to identify functional regions. The discovery of regulatory regions by profiling a very large number of histone modification maps is experimentally unviable for most laboratories. CSI-ANN (31) used a feature extraction step to combine 39 histone modification marks for ANN training, thereby predicting regulatory elements using pieces of information coming from all the available epigenetic maps. Thus, ranking the epigenetic modifications according to their statistical relevance is difficult. CSI-ANN condensed the information in a smaller set of input variables to facilitate the implementation of computationally intensive machine-learning techniques such as ANNs. However, its downside is that it provides no biological insight into the role of the epigenetic landscape in transcriptional regulation. In sharp contrast, ChromaGenSVM only needed to combine five histone modifications, including three methylations and two acetylations, to predict 23 574 enhancers with a precision much higher than that of CSI-ANN (90% versus 69%). The GA feature selection identified three distinct histone methylations (H3K4Me1, H3K4Me3 and H3R2Me2), and two histone acetylations (H4K8Ac and H2BK5Ac), which points to the existence of a complex epigenetic pattern associated with TSS-distal regulatory elements. These unique methylation patterns at enhancers include the activating marks H3K4Me1 and H3K4Me3 (25), and a transcriptional silencing flag (H3R2Me2) (52). The enhancers also contain an acetylation mark (H2BK5Ac) that strongly correlates to gene expression profiles (53). The five epigenetic mark rule was implemented as a penalty function rather than as a priori cut-off: the models are actually allowed to combine more than five epigenetic marks as long as every extra mark is associated with an increase of at least 0.1 units in the AUC so that any five-mark model is substantially out-performed by a six (or more) mark model. In summary, ChromaGenSVM outperformed CSI-ANN using much less experimental information and much less computation time, making our method an ideal tool for enhancer discovery from chromatin epigenetic maps.
ChromaGenSVM: code implementation and availability
The current version of ChromaGenSVM is freely available as a stand-alone Python application. ChromaGenSVM is implemented to predict functional regions genome-wide using SVMs, and starting with (ChIP-seq) epigenetic libraries and a set of putative functional regions. Prior to the computation of the epigenetic profiles (see Materials and Methods), the script uses a modified version of the ChIP-seq enrichment discovery tool SICER (54) to filter and pre-process the epigenetic libraries with a positive control. A list of target loci can be directly provided by the user or otherwise generated by the modified version of SICER (54) from a custom library mapping functional regions (e.g. p300-enrichment regions). The epigenetic profiles associated with the target loci are labeled as positive class examples while a background set of negative class profiles are derived from randomly selected loci (see ‘Materials and Methods’ section). The program trains an SVM model with the positive and negative epigenetic profiles from all libraries. Finally, functional regions are predicted genome-wide at the cut-off of the maximum F-score from cross-validation experiments, or by using a user-defined cut-off. The script yields not only a list of putative regulatory loci at the chosen cut-off as main output, but also generates two other files listing all the scanned loci per chromosome and their corresponding scores as computed by the model. The ChromaGenSVM Python code is freely available at http://sysimm.ifrec.osaka-u.ac.jp/download/Diego/.
CONCLUSIONS
The discovery of cell type-specific enhancers is ideally accomplished by the computational modeling of the chromatin epigenetic landscape rather than by evolutionary conservation sequence analysis. From a pool of epigenetic marks, the GA part of ChromaGenSVM systematically trained SVMs with relevant chromatin epigenetic marks associated with enhancers. ChromaGenSVM is the first successful implementation of SVMs to discover functional regulatory regions from histone methylation maps with an excellent performance, as indicated below:
The enhancers predicted in the pilot ENCODE region using three histone methylations maps recovered 85.1% and 88.0% of the p300-enrichment sites in untreated (training set) and IFN-γ-treated (test set) HeLa cells, respectively.
When predicting enhancers in IFN-γ-treated HeLa cells (test set), ChromaGenSVM improves on the PM and HMM methods by ∼12% and ∼16% for the recovery of p300 regions, with the added advantage that ChromaGenSVM defines functional regions more precisely (within 5.0 kb windows).
In a second exercise, the automatic selection of relevant epigenetic marks in human CD4+ T cells yielded a very select combination of only five histone methylation and acetylation marks (from a pool of 38 histone epigenetic maps) encoding the activation, inactivation, transcription and silencing of the DNA. Several types of histone modifications coexist at regulatory regions.
ChromaGenSVM predicted 23 574 enhancers in human CD4+ T cells (within only 1.0 kb windows, 400 bp resolution), of which ∼90% overlapped with at least one type of experimental evidence. This is an improvement in precision of 21% over the CSI-ANN method as determined by the presence of experimental marks, and using much less experimental information or computation time.
Our set of 23 574 predicted enhancers is specifically associated with genes involved in the differentiation and/or regulation of T-cell activation, and the expression of these genes is differentially regulated in such cells.
ChromaGenSVM is a parsimonious and robust SVM implementation that surpasses previously published methods for enhancer prediction from chromatin epigenetic maps.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–4 and Supplementary Figures 1–4.
FUNDING
The World Premier International (WPI) Research Center Initiative, the Kishimoto Foundation, the ETHZ-JST Japanese-Swiss Cooperative Program, and the Japan Society for the Promotion of Science (JSPS). Funding for open access charge: Japan Society for the Promotion of Science (JSPS).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
MF would like to thank the Kishimoto Foundation for funding his post-doctoral studies through a Kishimoto Foundation Fellowship.
REFERENCES
- 1.Deribe YL, Pawson T, Dikic I. Post-translational modifications in signal integration. Nat. Struct. Mol. Biol. 2010;17:666–672. doi: 10.1038/nsmb.1842. [DOI] [PubMed] [Google Scholar]
- 2.Derman E, Krauter K, Walling L, Weinberger C, Ray M, Darnell JE., Jr Transcriptional control in the production of liver-specific mRNAs. Cell. 1981;23:731–739. doi: 10.1016/0092-8674(81)90436-0. [DOI] [PubMed] [Google Scholar]
- 3.Alonso ME, Pernaute B, Crespo M, Gómez-Skarmeta JL, Manzanares M. Understanding the regulatory genome. Int. J. Dev. Biol. 2009;53:1367–1378. doi: 10.1387/ijdb.072428ma. [DOI] [PubMed] [Google Scholar]
- 4.Ørom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lettice LA, Horikoshi T, Heaney SJ, van Baren MJ, van der Linde HC, Breedveld GJ, Joosse M, Akarsu N, Oostra BA, Endo N, et al. Disruption of a long-range cis-acting regulator for Shh causes preaxial polydactyly. Proc. Natl Acad. Sci. USA. 2002;99:7548–7553. doi: 10.1073/pnas.112212199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bien-Willner GA, Stankiewicz P, Lupski JR. SOX9cre1, a cis-acting regulatory element located 1.1 Mb upstream of SOX9, mediates its enhancement through the SHH pathway. Hum. Mol. Genet. 2007;16:1143–1156. doi: 10.1093/hmg/ddm061. [DOI] [PubMed] [Google Scholar]
- 7.Aparicio S, Morrison A, Gould A, Gilthorpe J, Chaudhuri C, Rigby P, Krumlauf R, Brenner S. Detecting conserved regulatory elements with the model genome of the Japanese puffer fish, Fugu rubripes. Proc. Natl Acad. Sci. USA. 1995;92:1684–1688. doi: 10.1073/pnas.92.5.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loots GG, Locksley RM, Blankespoor CM, Wang ZE, Miller W, Rubin EM, Frazer KA. Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparisons. Science. 2000;288:136–140. doi: 10.1126/science.288.5463.136. [DOI] [PubMed] [Google Scholar]
- 9.Nobrega MA, Ovcharenko I, Afzal V, Rubin EM. Scanning human gene deserts for long-range enhancers. Science. 2003;302:413. doi: 10.1126/science.1088328. [DOI] [PubMed] [Google Scholar]
- 10.Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, Minovitsky S, Dubchak I, Holt A, Lewis KD, et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- 11.Visel A, Rubin EM, Pennacchio LA. Genomic views of distant-acting enhancers. Nature. 2009;461:199–205. doi: 10.1038/nature08451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahituv N, Zhu Y, Visel A, Holt A, Afzal V, Pennacchio LA, Rubin EM. Deletion of ultraconserved elements yields viable mice. PLoS Biol. 2007;5:e234. doi: 10.1371/journal.pbio.0050234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aparicio O, Geisberg JV, Struhl K. Chromatin immunoprecipitation for determining the association of proteins with specific genomic sequences in vivo. Curr. Protoc. Cell. Biol. 2004 doi: 10.1002/0471143030.cb1707s23. Chapter 17, Unit 17.7. [DOI] [PubMed] [Google Scholar]
- 14.Park PJ. ChIP-seq: advantages and challenges of a maturing technology. Nat. Rev. Genet. 2009;10:669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 16.Nathan D, Ingvarsdottir K, Sterner DE, Bylebyl GR, Dokmanovic M, Dorsey JA, Whelan KA, Krsmanovic M, Lane WS, Meluh PB, et al. Histone sumoylation is a negative regulator in Saccharomyces cerevisiae and shows dynamic interplay with positive-acting histone modifications. Genes Dev. 2006;20:966–976. doi: 10.1101/gad.1404206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sims RJ, III, Reinberg D. Histone H3 Lys 4 methylation: caught in a bind? Genes Dev. 2006;20:2779–2786. doi: 10.1101/gad.1468206. [DOI] [PubMed] [Google Scholar]
- 18.Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim TH, Barrera LO, Qu C, Van Calcar S, Trinklein ND, Cooper SJ, Luna RM, Glass CK, Rosenfeld MG, Myers RM, et al. Direct isolation and identification of promoters in the human genome. Genome Res. 2005;15:830–839. doi: 10.1101/gr.3430605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bergink S, Salomons FA, Hoogstraten D, Groothuis TA, de Waard H, Wu J, Yuan L, Citterio E, Houtsmuller AB, Neefjes J, et al. DNA damage triggers nucleotide excision repair-dependent monoubiquitylation of histone H2A. Genes Dev. 2006;20:1343–1352. doi: 10.1101/gad.373706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grewal SI, Bonaduce MJ, Klar AJ. Histone deacetylase homologs regulate epigenetic inheritance of transcriptional silencing and chromosome segregation in fission yeast. Genetics. 1998;150:563–576. doi: 10.1093/genetics/150.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 23.Orford K, Kharchenko P, Lai W, Dao MC, Worhunsky DJ, Ferro A, Janzen V, Park PJ, Scadden DT. Differential H3K4 methylation identifies developmentally poised hematopoietic genes. Dev. Cell. 2008;14:798–809. doi: 10.1016/j.devcel.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 26.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miranda-Saavedra D, Gottgens B. Transcriptional regulatory networks in haematopoiesis. Curr. Opin. Genet. Dev. 2008;18:530–535. doi: 10.1016/j.gde.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 28.Wilson NK, Miranda-Saavedra D, Kinston S, Bonadies N, Foster SD, Calero-Nieto F, Dawson MA, Donaldson IJ, Dumon S, Frampton J, et al. The transcriptional program controlled by the stem cell leukemia gene Scl/Tal1 during early embryonic hematopoietic development. Blood. 2009;113:5456–5465. doi: 10.1182/blood-2009-01-200048. [DOI] [PubMed] [Google Scholar]
- 29.Won KJ, Chepelev I, Ren B, Wang W. Prediction of regulatory elements in mammalian genomes using chromatin signatures. BMC Bioinformatics. 2008;9:547. doi: 10.1186/1471-2105-9-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hon G, Ren B, Wang W. ChromaSig: a probabilistic approach to finding common chromatin signatures in the human genome. PLoS Comput. Biol. 2008;4:e1000201. doi: 10.1371/journal.pcbi.1000201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Firpi HA, Ucar D, Tan K. Discover regulatory DNA elements using chromatin signatures and artificial neural network. Bioinformatics. 2010;26:1579–1586. doi: 10.1093/bioinformatics/btq248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vo N, Goodman RH. CREB-binding protein and p300 in transcriptional regulation. J. Biol. Chem. 2001;276:13505–13508. doi: 10.1074/jbc.R000025200. [DOI] [PubMed] [Google Scholar]
- 33.Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cortes C, Vapnik V. Support-vector networks. Machine Learn. 1995;20:273–297. [Google Scholar]
- 35.Frohlich H, Chapelle O, Scholkopf B. Proceedings 15th IEEE International Conference on Tools with Artificial Intelligence. Washington DC, USA: IEEE Comput. Soc.; 2003. Feature selection for support vector machines by means of genetic algorithm; pp. 142–148. [Google Scholar]
- 36.Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Henschel S, Zien A, Binder A, Gehl C. The SHOGUN machine learning toolbox. J. Machine Learn. Res. 2010;11:1799–1802. [Google Scholar]
- 38.Holland H. Adaptation in natural and artificial systems. Ann Arbor, MI: The University of Michigan Press; 1975. [Google Scholar]
- 39.Fernandez M, Caballero J, Fernandez L, Sarai A. Genetic algorithm optimization in drug design QSAR: Bayesian-regularized genetic neural networks (BRGNN) and genetic algorithm-optimized support vectors machines (GA-SVM) Mol. Divers. 2011;15:269–289. doi: 10.1007/s11030-010-9234-9. [DOI] [PubMed] [Google Scholar]
- 40.Perone CS. Pyevolve: a Python open-source framework for genetic algorithms. ACM SIGEVOlution. 2009;4:12–20. [Google Scholar]
- 41.Felsenfeld G. Chromatin unfolds. Cell. 1996;86:13–19. doi: 10.1016/s0092-8674(00)80073-2. [DOI] [PubMed] [Google Scholar]
- 42.Hatzis P, Talianidis I. Dynamics of enhancer-promoter communication during differentiation-induced gene activation. Mol. Cell. 2002;10:1467–1477. doi: 10.1016/s1097-2765(02)00786-4. [DOI] [PubMed] [Google Scholar]
- 43.Wang Q, Carroll JS, Brown M. Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol. Cell. 2005;19:631–642. doi: 10.1016/j.molcel.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 44.Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ferretti V, Poitras C, Bergeron D, Coulombe B, Robert F, Blanchette M. PReMod: a database of genome-wide mammalian cis-regulatory module predictions. Nucleic Acids Res. 2007;35:D122–D126. doi: 10.1093/nar/gkl879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ghisletti S, Barozzi I, Mietton F, Polletti S, De Santa F, Venturini E, Gregory L, Lonie L, Chew A, Wei CL, et al. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity. 2010;32:317–328. doi: 10.1016/j.immuni.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 48.McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl Acad. Sci. USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 51.Surani MA, Hayashi K, Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128:747–762. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 52.Kirmizis A, Santos-Rosa H, Penkett CJ, Singer MA, Green RD, Kouzarides T. Distinct transcriptional outputs associated with mono- and dimethylated histone H3 arginine 2. Nat. Struct. Mol. Biol. 2009;16:449–451. doi: 10.1038/nsmb.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shi Y, Sun H, Bao J, Zhou P, Zhang J, Li L, Bu H. Activation of inactive hepatocytes through histone acetylation: a mechanism for functional compensation after massive loss of hepatocytes. Am. J. Pathol. 2011;179:1138–1147. doi: 10.1016/j.ajpath.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zang C, Schones DE, Zeng C, Cui K, Zhao K, Peng W. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25:1952–1958. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.