Abstract
Recent advances in next generation sequencing (NGS) technology now provide the opportunity to rapidly interrogate the methylation status of the genome. However, there are challenges in handling and interpretation of the methylation sequence data because of its large volume and the consequences of bisulphite modification. We sequenced reduced representation human genomes on the Illumina platform and efficiently mapped and visualized the data with different pipelines and software packages. We examined three pipelines for aligning bisulphite converted sequencing reads and compared their performance. We also comment on pre-processing and quality control of Illumina data. This comparison highlights differences in methods for NGS data processing and provides guidance to advance sequence-based methylation data analysis for molecular biologists.
INTRODUCTION
Next generation sequencing (NGS) coupled with sodium bisulphite modification of DNA has become a powerful tool to quantify DNA methylation at single nucleotide resolution (1,2). As for other NGS applications, bisulphite sequencing presents a challenge in terms of the large amount of raw data generated from the sequencing, processing, analysis and finally interpretation of the data. In particular, aligning bisulphite converted reads to a large reference genome brings substantial computational challenges.
Sodium bisulphite treatment of DNA converts unmethylated cytosines (C) to thymines (T) after subsequent PCR, but methylated Cs remain unchanged by the treatment. This method is widely used to distinguish methylated from unmethylated Cs in the DNA strands. Since C is converted to T, a T in the sequenced reads could be mapped against either C or T in the reference genome but not vice versa, and so the C to T mapping is asymmetric (3). This gives rise to the possibility of more false-positive matches between the reads and the reference genome and also increases the search space significantly, making mapping bisulphite converted reads more challenging.
As a consequence of bisulphite modification, four distinct DNA strands are created after PCR amplification. In shotgun sequencing, the reads can possibly be derived from any of the four strands. However, for our data, due to the directionality of the Illumina platform and the protocol used, reads were obtained exclusively from MspI digested 5′ CGG strands (the recognition motif of MspI is C′CGG). But in case of non-directional libraries, there is the potential to introduce bias into the methylation call, as MspI cut-sites need to be filled in by a cytosine. The sequence of the read will then depend on whether these sites are filled in with methylated or non-methylated cytosines. In either case, the cytosine might not retain the true genomic methylation state. To avoid incorrect estimation of the methylation of the initial CpG site, the filled in bases of the read should be omitted when extracting the methylation information. In the case of reads that cover the entire MspI fragment, the 3′ end of the read can be similarly affected. Furthermore, reads from non-directional libraries can map to any of the four different versions of the reference genome: (i) (bisuflite) Watson or (ii) the reverse-complement of Watson or (iii) to (bisulphite) Crick or (iv) the reverse complement of Crick.
Compared to classical Sanger sequencing, the NGS reads are shorter in length. When the read length is short, alignment against a large and complex genome (such as human) becomes more difficult. Large genomes potentially contain significant regions of repetitive sequence, and, as a result, the percentage of uniquely aligned sequence decreases significantly when the reads are short. Ensuring the quality of the data received from sequencing and appropriate computational manipulation can increase the mapping efficiency.
The processes of mapping bisulphite reads against genomic sequence not only have to manage mapping asymmetry, but also must be able to allow for a reasonable level of sequencing errors. Various computational strategies have been employed to handle these issues: CokusAlign (4) uses a tree-based lookup method in which the genomic sequence is pre-processed into a memory resident tree structure through which the sequence data for each read causes a progressive traversal to establish a genomic location. While this strategy works effectively for 36 bp reads versus the Arabidopsis thaliana genome, it does not scale well for longer reads or larger genomes. Seed lookup methods have been employed in which the genome is pre-processed into a series of hash values (seeds), which enable rapid indexing of oligomers from read sequences to genomic positions. A recently implemented, very efficient short sequence aligner Bowtie (5) uses the Burrows-Wheeler (BW) transformation to perform rapid mapping of fragments to genomic positions. Although a moderately slow prior build operation is required to generate BW transformed genomic sequences, subsequent use of this data is very rapid. The Bowtie authors distribute pre-built BW-transformed files for a number of model genomes, although this facility is not available for bisulphite genomes at present.
Since Illumina read quality deteriorates with later cycles, all aligners treat the start of each read as more reliable and are designed to work with limited errors in that portion. The reduced quality further into the reads is reflected in the aligners' permitting increased mismatches after mapping the initial seed. Potential erroneous methylation calls from misalignment events further into the reads and loss of information must be balanced against failure of unique alignment of short reads if excessive quality trimming is performed. We have analysed high-throughput Illumina data from the human genome, sequenced from reduced representation (RR), bisulphite-converted libraries enriched for CpG islands. In this article, we describe three major aligners (Bismark, BSMAP and RMAPBS) that map DNA methylation sequence data at single-base pair resolution. We compare their speed and mapping efficiency and also comment about the quality control and suggest pre-processing of Illumina reads for best possible output and the visualization of the methylation data.
MATERIALS AND METHODS
RR bisulphite sequencing
A RR human genome was generated according to published protocols (6–8). The RR library is enriched for CpG islands and is predicted to include 84% of the CpG islands in the human genome and ∼3.4 million unique CpG sites (6,7). In brief, the genomic DNA was digested with MspI (New England Biolabs, Ipswich, MA) followed by end repair and addition of 3′ A overhangs. Methylated adaptors (Illumina, San Diego, CA) with a 3′ T overhang were then ligated with the generated fragments. Following adaptor ligation, DNA fragments ranging from 40 to 220 bp (preligation size) were cut from a 3% (w/v) NuSieve GTG agarose gel (Lonza, Basel, Switzerland) and subsequently bisulphite modified using the EZ DNA methylation kit (Zymo Research, Irvine, CA). The final library was amplified by PCR. The resulting library was sequenced on an Illumina GAIIX platform with a single-ended, 100 bp run. Data from a single lane, from which 18 490 898 sequence reads were obtained, were used for subsequent analysis.
Quality check of the sequenced data
The Illumina base calling program converts the captured signal images into sequence, and a technical concern of this process is that the base calling accuracy decreases with increased read length, since the sequential chemistry results in progressive decline of the signal and the corresponding increase in background noise results in less reliable base calls.
SolexaQA (9) was used to evaluate the quality of the data. SolexaQA is a Perl application using the R statistics package and the matrix2png program to generate a graphical representation of data quality and is in the public domain. It samples entries in the FASTQ file and uses the quality score for each cycle to generate a graphical ‘heat-map’, which indicates the quality of the reads generated from a lane. One axis represents the base, and the other represents each individual tile in a flow cell: the darker the colour of a box, the lower the quality of that tile for that cycle (Supplementary Figure S1).
In addition, SolexaQA generates a plot of the quality of every tile along the reads, in which the probability of bases being called in error is plotted against the read position (Supplementary Figure S2). SolexaQA further plots the distribution of read lengths where the x-axis represents the length of contiguous sequence (with a base-calling error rate ≤5%) and the y-axis maps the proportion of all the reads. From these data, we can work out the proportion of reads with full length sequence (Supplementary Figure S3). Together, these graphs give a good indication of the quality of the run.
Based on the various quality indicators, it is possible to establish where in the read cycles the base-call reliability has declined beyond a reasonable level and, hence, where the reads should be trimmed before further processing. On one hand, trimming the reads discards some information while, on the other, less reliable sequence towards the ends of reads may contribute to misalignment or failure of alignment. While the trim length is, to a degree, arbitrary our experience is that the quality indicators discussed above allow it to be set with reasonable confidence. Our reads were 100 bp but after evaluation of their quality we hard-trimmed to 75 bp for further processing. This not only ensures better quality data for further analysis but also it reduces the rate of mismatches during mapping with the reference genome. Some tests were performed with reads hard-trimmed to 60 bp. The SolexaQA application also identifies potentially bad tiles (Supplementary Figure S1), the sequences from which were eliminated to reduce the risk of artefacts from less reliable reads.
Scan for adaptor contamination
Contamination by adaptor sequences in the data set was also assessed. For this purpose, a program cleanadaptors was developed. The program scans the reads, identifying sections, which show 75% or higher matching with any of a series of adaptor sequences (the threshold is adjustable). The scanning was performed against 100, 75 and 60 bp data sets to estimate the amount of adaptor sequence contamination in the reads (Supplementary Table S1).
Dynamic trimming
We used the program fastq_quality_trimmer (v 0.0.13 http://hannonlab.cshl.edu/fastx_toolkit/) to perform dynamic trimming of the original 100 bp and the 75 bp data sets to a Phred quality level of 30 (=0.001 probability of a base-call error as assessed by the Illumina base calling pipeline).
Mapping the reads
The 75 and 60 bp data sets (created by hard trimming the original data) were mapped against the human reference genome (build GRCh37) to assess the effect of trimming on mapping efficiency. The mapping was performed with three aligners described below. With a purpose-written program mkrrgenome, we also created an in silico RR genome based on MspI fragments in our 40–220-bp size range and mapped the data sets against this RR genome as well (Supplementary Figure S5).
Description of the aligners
Bismark
Bismark (10) v0.2.3 is a Perl application, which works by calling the Bowtie fragment aligner (5). Genome files are pre-processed in a separate step to generate CT- and GA-converted files, which are then scanned with parallel invocations of Bowtie. By default, Bismark will map all reads, directional or non-directional against all four conversions, introducing possible mismapping of directional reads. This distortion can be suppressed with recent versions (0.2.3 or later) of the Bismark package by using the directional switch. The output contained genomic and read sequences for each match from which methylated CpGs were determined either with the script methylation_extractor or directly by the SeqMonk visualization application.
BSMAP
BSMAP (3) is a C++ application based on a modified version of the SOAP aligner (11) in which the reference genome is converted to a series of typically octamer seeds on which hashing and fast lookup methods can be applied to attain efficient performance. BSMAP generates C/T and G/A converted seeds for the reference genome in which all possible methylation patterns exist for each seed. A bit-mapping strategy is applied within the program to highlight mismatches from methylation and sequencing errors. Further processing permits a user-specified degree of mismatching in each read, and the algorithm expends significant effort in resolving multiple or conflicting mapping of reads. BSMAP output was a list for each read of its matching status (unmatched, uniquely or multiply matched) with suggested methylation positions for unique matches.
Initially, we used BSMAP v1.02 and subsequently v1.2. In some cases, we have contrasted the performance of both versions.
RMAPBS
RMAPBS v2.05 (12) is a C++ application based on the RMAP program for mapping single-ended bisulphite reads. The RMAP algorithm uses an advanced seed and hashing strategy, related to that for BSMAP, to locate partial matches for reads while permitting mismatches and methylation. RMAPBS output was a BED format file giving the chromosome, position, the read identifier, a quality parameter and the strand.
Data visualization
SeqMonk (http://www.bioinformatics.bbsrc.ac.uk/projects/seqmonk) was used to view methylation data. It is a graphical Java application, which is distributed for a wide range of computer platforms (Windows, Linux, MacOS X and as Java source code). SeqMonk is pre-configured with a significant set of genomic sequences and their annotations and is configured to check those in use for any updates at each invocation as well as checking for any updates to the application itself. When a genome is loaded, either de novo or as a project, the displays are capable of showing genes, mRNAs and exons, but the displayed information is widely configurable depending on requirements (Supplementary Figure S4).
SeqMonk is capable of importing mapping information in a variety of mapping formats or as a tab-delimited list. Methylation data from Bismark (from the same authors as SeqMonk) were imported directly from Bismark files, but each CpG was treated as a single-mapped entity with methylated positions indicated as 5′ mappings and unmethylated as 3′. Further quantification of methylation was possible by generating ‘probes’ on a residue-by-residue basis. Despite the significant overheads of working on the human genome and tens of millions of CpG positions, SeqMonk performed acceptably fast, although the memory requirements were large. SeqMonk could display bisulphite mapping data from more than one treatment or pipeline in the same window, enabling visual comparison of the methods. Feature reports could be created in which ‘probe’ data can be related to genomic annotations.
The importing of RMAPBS and BSMAP methylation data into SeqMonk was not automatic, as for Bismark, but was possible by pre-processing the various output files into tab-delimited lists of methylated and unmethylated CpG positions, which were then imported as raw data. We have written rmapbsbed2cpg (a C command line application) to do this pre-processing for both (Supplementary Figure S5).
RR genome
A purpose written program mkrrgenome was used to generate a RR genome in silico. The program scans for MspI recognition sites (C′CGG) saving only those fragments that fall in the specified size range of 40–220 bp. The output was a series of FASTA files one for each chromosome. The program could return statistics on CpG sites for each chromosome and the fragment positions.
Computing resource
The mapping runs were performed on a Mac Pro with 64 bit duo quad core Intel Xeon processors and with 22 Gb RAM running MacOS 10.6.
Distribution of new programs
The software written to support this work is distributed as a shell archive (meth_progs_dist.shar) along with the supplementary data and, while developed on a MacOS X platform, has successfully been compiled with gcc on various Linux distributions. We have also included a test data set containing 5000 sequenced reads along with chromosome 22 and a file (test_progs_readme.txt) describing the set, as a compressed shell archive (meth_progs_test.shar.gz) in the supplementary data.
RESULTS
Despite the difference in the algorithms, all three programs efficiently mapped the sequence reads. Following trimming to 75 bp, 42.2, 58.9 and 65.1% of the reads were uniquely aligned by using BISMARK, BSMAP and RMAPBS, respectively (Table 1). We found that RMAPBS and Bismark were able to map the reads much faster than BSMAP v1.02. Bismark has a speed of 1642 reads/sec and RMAPBS maps 119.6 reads/sec. Despite being a single-threaded application, RMAPBS is relatively fast in operation. BSMAP v1.02 mapped the reads with a speed of 5.6 reads/sec taking 38 days when run on 6 CPU cores (Table 1). The performance of BSMAP v1.2 was much faster than BSMAP v1.02, and the mapping was completed in 22.2 h with a speed of 231.2 reads/sec (Table 1).
Table 1.
Comparison of mapping performance of the different packagesa
| Programme | Aligner | Number of readsb | Uniquely mapped readsc (%) | Multiple mapping | Cores used | CPU time taken | Reads/sec |
|---|---|---|---|---|---|---|---|
| Bismark | Perl application uses Bowtie | 18 490 898 | 42.2 | 7.7 | 4 | 3 h 7 min | 1642 |
| BSMAP v1.02 | SOAP modified | 18 471 799 | 58.9 | 14.0 | 6 | 38 days | 5.6 |
| BSMAP v1.2 | SOAP modified | 18 490 898 | 55.5 | 10.9 | 6 | 22.2 h | 231.2 |
| RMAPBS | RMAP modified | 18 458 028 | 65.1 | 16.8 | 1 | 42 h 48 min | 119.6 |
aThe reads were mapped against the complete human genome GRCh37. bBSMAP v1.02 and RMAPBS rejected a proportion of lower quality reads. cThe reads were hard trimmed to 75 bp for better alignment.
We observed that shortening the read length improves the percentage of uniquely aligned sequences. All three programs showed improved mapping efficiency with shorter read length, a consequence of the poorer quality sequence towards the ends of Illumina reads. Bismark showed an increase of 12.1% in unique mapping when 75 bp reads were trimmed to 60 bp (Table 2). A similar trend was seen for alignments against the RR genome. However, further trimming of the data set did not improve mapping efficiency significantly (Table 4), implying that the reads are of sufficient quality up to 60 bp. Shorter bisulphite reads are more challenging to map and give an increased proportion of multiple alignments. In their original description of RR bisulphite sequencing (RRBS), Meissner et al. (6) found that a significant proportion of RRBS reads did not align to the reference genome even after allowing up to six mismatches in the mapping, which was attributed to repetitive sequence and sequencing process artifacts. We showed that quality control and pre-processing of the data set improve the alignment efficiency (Table 2).
Table 2.
Comparison of mapping against RR genome and full length genome and different read lengths
| Programme | Read length | % Uniquely mapped sequence against RRa | CPU time (h) | % Uniquely mapped sequence against full genomeb | No. of CpG sites in size selected region | % of CpG sites in size selected region |
|---|---|---|---|---|---|---|
| Bismark | 75 | 42.0 | 1.30 | 42.2 | 23 415 803 | 82.9 |
| 60 | 53.2 | 0.85 | 54.3 | 27 795 960 | 84.5 | |
| RMAPBS | 75 | 59.1 | 8.65 | 65.1 | 44 104 796 | 91.4 |
| 60 | 64.0 | 4.37 | 65.2 | 36 700 533 | 82.8 | |
| BSMAP v1.02 | 75 | 49.3 | 19.37 | 58.9 | 29 829 964 | 86.7 |
| 60 | 58.8 | 24.03c | 64.0 | 25 976 326 | 91.8 | |
| BSMAP v1.2 | 75 | 49.3 | 1.52 | 55.5 | 21 453 738 | 81.7 |
| 60 | 58.7 | 0.58 | 65.0 | 35 642 607 | 83.9 |
aRR genome (40–220 bp). bComplete human genome GRCh37. cThe longer time taken for the 60 bp reads must reflect more time spent resolving potential mismatches in comparison with that necessary for 75 bp reads.
Table 4.
Effect of sequence trimming on alignment efficiency and methylation percentage
| Program | Total number of reads | Uniquely mapped reads (%) | Total methylation percentage against RRa | Methylated CpG sites against RRa |
|---|---|---|---|---|
| 50 bp data set | ||||
| Bismark | 18 490 898 | 52.5 | 36.6 | 11 238 582 |
| RMAPBS | 18 458 028 | 62.5 | 34.7 | 12 813 410 |
| BSMAP v1.02 | 18 471 799 | 58.2 | 35.3 | 13 074 829 |
| BSMAP v1.2 | 18 490 898 | 59.5 | 35.4 | 13 044 161 |
| 40 bp data set | ||||
| Bismark | 18 490 898 | 53.2 | 35.4 | 9 635 443 |
| RMAPBS | 18 458 028 | 62.9 | 33.4 | 10 651 296 |
| BSMAP v1.02 | 18 471 799 | 59.1 | 34.7 | 10 933 808 |
| BSMAP v1.2 | 18 490 898 | 58.0 | 34.8 | 10 805 047 |
| 36 bp data set | ||||
| Bismark | 18 490 898 | 52.7 | 34.9 | 8 824 724 |
| RMAPBS | 18 458 028 | 62.6 | 32.9 | 9 700 349 |
| BSMAP v1.02 | 18 471 799 | 54.0 | 34.1 | 9 960 586 |
| BSMAP v1.2 | 18 490 898 | 57.1 | 34.2 | 9 836 729 |
aThe runs were performed against our RR genome (40–220 bp).
We observed that dynamic trimming (performed by fastq_quality_trimmer) of the data set improved mapping efficiency (Supplementary Tables S2 and S3). However, mapping efficiency was improved to a greater extent when adaptor sequences were trimmed (performed by our cleanadaptor program) especially with the longer reads (Supplementary Tables S1 and S3). An interesting corollary was that RMAPBS required all reads to be the same length and rejected reads that were padded with ‘N’s to that length after adaptor trimming. Consequently, RMAPBS could not align any reads that contained the adaptor sequence. As a result, we could not compare the performance of all the aligners with dynamically trimmed or adaptor trimmed data set. But our results strongly suggest that the trimming of adaptor sequences is an important step for improving mapping efficiency, supporting the conclusion of Gu et al. (8).
Uncertainty in the selection of fragments by size from gels poses a problem for alignments against an in silico RR genome in that sequence reads of fragments falling outside the size limits are unlikely to map correctly to it. Although this will cause the rejection of some otherwise valid sequence data, alignment against the RR genome maximizes consistency of the outputs. For the latter reason, we have opted to use an in silico RR genome restricted to the expected size range for our fragments in order that outlying fragments are rejected at the mapping stage and that experimental variation should be suppressed as a consequence. An in silico genome of 40–220 bp fragments from the GRCh37 build had a total size of 74 Mb from 647 626 MspI fragments and a total of 4 068 947 CpGs representing 13.4% of the genomic total. This corresponds to a 5.7-fold enrichment of CpGs.
The mapping against the RR genome was comparable to that for the full genome for both 75 and 60 bp trimmed reads. Bismark showed 42.0 and 53.2% unique matches against 75 and 60 bp reads and RMAPBS showed 59.1 and 64.0% unique mapping against them. Both versions of BSMAP showed similar rates of unique matches against the RR genome. For our 75 bp data set, both versions of BSMAP (v1.02 and v1.2) produced 49.3% unique mapping, and, for 60 bp data set, BSMAP v1.02 and BSMAP v1.2 showed 58.8 and 58.7% unique mapping respectively (Table 2). The RR genome is 42.5 times smaller than the full genome, and, as a consequence, alignment was much faster than for the whole genome. Bismark took 1.3 h to map 18.5 million reads (read length = 75) against the RR genome, and BSMAP v1.2 completed the run in 1.52 h, whereas RMAPBS and BSMAP v1.02 completed the run in 8.65 and 19.37 h, respectively (Table 2).
From the uniquely mapped reads against the full genome, we determined the number of CpG sites that were contained within the reduced representation (RR) genome of 40–220 bp for our data set. We observed that all the aligners mapped more than 80% of the CpG sites into the size selected region of the genome (Table 2). These results give us confidence that the RR library was well constructed as majority of the sequenced CpGs fell in the size range of 40–220 bp and that the mapping processes are producing valid alignments. Different reads vary in the number of potential CpG sites they contain, and it is apparent that different aligners perform differently in their abilities to uniquely map the bisulphite converted CpGs to a genome.
Bismark and RMAPBS produced a total methylation percentage of 44.8 and 36.9 for 75 bp data set, respectively, when aligned against the complete human genome. When aligned against the RR genome, Bismark gave 43.2% total methylation, and RMAPBS indicated 38.6% total methylation. Initially, BSMAP v1.02 showed 18.6% methylation when mapped against the whole genome (Table 3), and, for the RR genome, it found 15.0% CpG methylation (data not shown). Detailed investigation of the reads from the aligned output revealed that this anomaly is due to poorly documented trimming behaviour by an option (−c) in the program. This had unexpectedly caused a 5′ single-base truncation as well as a 10 bp 3′ truncation, causing misalignment of reads in further processing. As a result, the aligned files produced inaccurate and lower percentage of methylation. This option has been omitted from the latest version of BSMAP v1.2. Reanalysis of BSMAP v1.02 without the –c switch against the RR genome indicated 42.1% methylation for our 75 bp data set. Runs against the full genome were not repeated. On the other hand, BSMAP v1.2 showed 42.9% methylation when mapped against the whole genome (Table 3), and, for the RR genome, it found 42.1% CpG methylation. We performed similar operations on the 60 bp trimmed data set and found that the results are similar to those of 75 bp data set (Table 3) although the percent CpG methylation figures decreased slightly. The percent reductions for BSMAP and Bismark were slightly higher than for RMAPBS.
Table 3.
Comparison of methylation mapping between different aligners
| Aligners | Total methylation percentage against complete genome | Methylated CpG sites against complete genome | Total methylation percentage against RRa | Methylated CpG sites against RRa |
|---|---|---|---|---|
| 75 bp data set | ||||
| Bismark | 44.8 | 12 646 435 | 43.2 | 13 184 924 |
| RMAPBS | 36.9 | 8 557 383 | 38.6 | 17 997 752 |
| BSMAP v1.02 | 18.6b | 6 395 684b | 42.1 | 16 196 332 |
| BSMAP v1.2 | 42.9 | 11 251 307 | 42.1 | 16 212 461 |
| 60 bp data set | ||||
| Bismark | 40.2 | 13 227 156 | 39.4 | 13 579 291 |
| RMAPBS | 36.3 | 16 115 692 | 36.1 | 15 634 090 |
| BSMAP v1.02 | 12.3b | 3 484 013b | 38.0 | 15 374 907 |
| BSMAP v1.2 | 38.7 | 16 439 913 | 38.0 | 15 369 712 |
aRR genome constructed in the size range of 40–220 bp. bSee text.
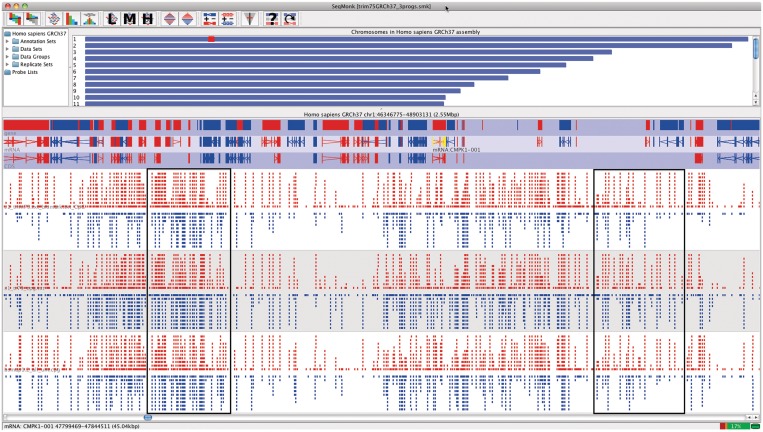
We have visualized and compared the methylation tracks for three aligners in SeqMonk. Regions showing extensively methylated and unmethylated CpGs are generally comparable between the aligners. However, closer examination of some regions revealed large differences between the outputs from the aligners. Figure 1 shows CpG methylation tracks produced by three different aligners in a 2.55 Mbp region of chromosome 1 (a section chosen at random), which documents this behaviour.
Figure 1.
SeqMonk display of differential methylation from different aligners. About 75 bp trimmed read data for 18 × 106 reads were aligned against the Human genome GRCh37 build by Bismark. v0.23, RMAPBS v2.05 and BSMAP v1.2 for which the methylation is displayed, respectively, from top to bottom below the gene, mRNA and CDS panes. Methylated CpG positions are shown in the red panes for each aligner, and unmethylated CpGs are in the blue panes. The display is of a randomly selected 2.55 Mbp region of chromosome 1. The black boxes indicate some regions of significant difference in methylation.
Furthermore, to assure that the mapping was performed only with high-quality reads and misalignment events did not affect the results, we further trimmed the data set to 50, 40 and 36 bp and compared the mapping statistics of all the aligners against them. This step would have removed virtually all of the adaptor contamination and sequencing errors from the data set. The results from these runs were summarized in Table 4. As mentioned earlier, we did not find notable improvement in the mapping efficiency for these trimmed data sets compared to the 60 bp data set.
Hard trimming resulted in a small proportional drop in percentage of methylation (Tables 3 and 4), but, otherwise, no significant change was observed.
DISCUSSION
Published descriptions of the aligners used have been based on much smaller data sets. For example, RMAPBS used human chromosome 6 for evaluation of the program (13). Chromosome 6 contains only between 5.5 and 6% of the total DNA of the human genome. Similarly, BSMAP performance was compared with several other aligners using 2.9 million, 31 nt reads against the Arabidopsis genome (119 Mb) (3). The time taken to map one lane of sequenced reads against the whole genome is much greater than had been implied (14). The statistics described with the example data in the programs differs significantly from our real time data. The number of reads, the read length of the sequences and the size of the reference genome have a pivotal role to play in determining the overall mapping speed. The algorithm of the program also has a significant effect. The slower performance of earlier BSMAP versions (e.g. v1.02, see Table 1) has been noted by others on a synthetic, smaller data set (106 simulated reads aligned to human chromosome 21) (14) and can be explained by the nature of the program. The seeding/hashing and the bitwise masking (a unique feature of BSMAP) confer considerable efficiency together with multithreading, but, despite these, the extensive alignment optimizing steps made the older version of the aligner slower. In contrast, the seeding method employed by RMABPS confers substantially greater efficiency, yet returns bisulphite alignments of comparable quality, as noted elsewhere (12). The top performer is the BW Transform of the BowTie aligner called by Bismark (10) in which the significant time expended in the pre-processing step is generously compensated by the notably superior performance of the algorithm.
The manner in which aligners manage the C-T mapping asymmetry can provide an additional source of mapping differences. BSMAP and RMAPBS allow both Cs and Ts of reads to map to genomic cytosines, whereas Bismark converts all residual Cs in the read and all genomic Cs into Ts for mapping. This can enable BSMAP and RMAPBS to achieve higher mapping efficiencies (as observed in our results) but can introduce a bias by increasing the unique mapping for more highly methylated reads compared to those less methylated where the higher proportion of Ts makes multiple mapping more probable. In contrast, Bismark will generate multiple mappings of reads irrespective of their methylation status and therefore will not bias for more highly methylated reads. This might result in reduced mapping efficiency but avoids mapping bias based on methylation status of the reads.
For libraries generated from a RR genome and enzymatic fragmentation, it is recommended to construct an in silico RR genome (7) and map the reads against it. This approach allows for fragment driven mapping against the predicted MspI digested sites only. The lengthy library preparation procedure might introduce some bias in the data set. For example, over-amplification of some fragments might occur during library preparation. Imprecise gel selection may also generate fragments outside the intended size range in the final library. Mapping against a selected RR genome compensates for experimental error to a certain extent. It also heavily reduces the computational load as we have shown in that mapping against RR genome is many times faster than full genome. We have demonstrated that the RR genome demonstrated comparable mapability to that of the full genome.
We showed that quality checking of the data set is important to obtain a maximum alignment output. The choice of trimming is arbitrary: with our quality control techniques and mapping statistics, we showed 60 bp was a reasonable choice that achieved respectable mapping efficiency. Also, we showed that short NGS reads do not necessarily increase unique mapping as they are more likely to match in multiple positions of the genome. If 100 bp reads are sequenced, it is inefficient to trim them to 40 bp for alignment, given that the increasing error rate further into reads does not necessarily lead to incorrect mapping.
All the aligners analysed showed comparable overall methylation percentage for our data set. Furthermore, the overall methylation status produced against the RR genome is also in concordance with that for the full genome. The overall methylation percentage of CpG sites in the human genome is estimated to be 68.4% (15), but the majority of the CpG islands are unmethylated. Since our library is enriched for CpG islands, we expect to see less methylation in our data set than the overall methylation percentage of CpG sites in the genome. However, while visualizing the data tracks in SeqMonk, we have observed that different aligners showed differential overall CpG methylation patterns in the same region, noting that such variations are widespread (Figure 1). These differences result from variations in how the aligners work. Our choices of runtime parameters were to use defaults or those recommended by the authors, but this may result in different performance for aligners when tackling the complex issues of the asymmetric mapping of bisulphite reads onto a genome. Ideally, all aligners would create exactly the same mapping for the same reads, but underlying differences in their strategies for locating, extending and making their optimized choices will create variant results. Attempting to achieve more similar mapping by the adjustment of runtime parameters is beyond the scope of this study. Also, the percentage of uniquely mapped sequence is different in each aligner, and the number of reads mapped to a particular region can differ as a result of that. So, our results indicate that for interpreting accurate methylation, the choice of aligner is important as are careful evaluation of data quality, trimming of reads as appropriate, removal of adaptor sequences and selection of suitable parameters for analysis. Since different experiments may produce different methylation patterns, it is possible that different aligners may produce varied mappings making this choice complex. Further, the evolving computational environment may alter preferences by, for instance, the move to large-scale parallel processing favouring slower algorithms.
Whereas the originally described RRBS protocol by Meissner et al. (6) is based on 36 bp reads, current Illumina sequencing protocols can produce up to 150 bp reads and are projected to give longer read lengths in the near future. However, when libraries generated from a significant number of smaller DNA fragments are sequenced to longer reads, it is inevitable that some reads will sequence into the adaptors. The Illumina adaptors are methylated and may account for false methylation calls in further analysis if such reads align to the genome, given that the aligners used here permit some read errors in making their alignments. Alternatively, many such reads will not align and will be rejected despite having valid leading sequence. Hence, in order to avoid bias from either of these processes, it is desirable to scan for adaptor sequences in the read file and remove them before further alignment (8). Interestingly, we also tried dynamic trimming based on read quality and found that this generated less unique mapping in comparison with adaptor trimming, both for the original 100 and the 75 bp data. This probably reflects the extent to which shorter reads are more likely to map multiply (Table 4), whereas the adaptor sequence tends to prevent those reads from mapping. Lower quality trailing sequence may still map correctly and its removal may amount to the loss of useful data.
The failure of RMAPBS to align reads that contained adaptor sequence, even if these were trimmed, reduces its value as an aligner for RRBS protocols, since it loses the information from shorter reads. For our data, the newer version of BSMAP (v1.2) is considerably improved, since it performs over 40 times faster than the older version. The outputs from BSMAP and RMAPBS require further downstream processing in order to obtain unique matches and to generate CpG methylation data.
On the basis of speed, reasonable performance and ease of extracting methylation data and interfacing to SeqMonk, we would choose Bismark as our preferred aligner at this stage, but, in establishing a processing pipeline for methylation profiling, it remains desirable to monitor the performance of other aligners in order to ensure that the most appropriate choice is made. The continuing evolution of sequencing chemistry and protocols together with further improvements in computational resources and algorithms will enhance the interpretation of genome-wide methylation sequencing data.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3, Supplementary Figures 1–5, Supplementary Program and Supplementary Dataset.
FUNDING
National Research Centre for Growth and Development (NRCGD) and Health Research Council (HRC), New Zealand (Grant HRC 09/085 D). A.C. is supported by a scholarship from NRCGD. Funding for open access charge: National Research Centre for Growth and Development (NRCGD) and Health Research Council (HRC), New Zealand.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We gratefully acknowledge assistance and comments provided by Dr. Felix Krueger and Dr. Simon Andrews of the Babraham Institute, Cambridge, UK. In addition, we appreciate the comments and suggestions made by the reviewers to improve the quality of the manuscript.
REFERENCES
- 1.Hurd PJ, Nelson CJ. Advantages of next-generation sequencing versus the microarray in epigenetic research. Brief. Funct. Genom. Proteom. 2009;8:174–183. doi: 10.1093/bfgp/elp013. [DOI] [PubMed] [Google Scholar]
- 2.Voelkerding KV, Dames SA, Durtschi JD. Next-generation sequencing: from basic research to diagnostics. Clin. Chem. 2009;55:641–658. doi: 10.1373/clinchem.2008.112789. [DOI] [PubMed] [Google Scholar]
- 3.Xi Y, Li W. BSMAP: whole genome bisulfite sequence MAPping program. BMC Bioinformatics. 2009;10:232. doi: 10.1186/1471-2105-10-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith ZD, Gu H, Bock C, Gnirke A, Meissner A. High-throughput bisulfite sequencing in mammalian genomes. Methods. 2009;48:226–232. doi: 10.1016/j.ymeth.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gu H, Smith ZD, Bock C, Boyle P, Gnirke A, Meissner A. Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nat. Protoc. 2011;6:468–481. doi: 10.1038/nprot.2010.190. [DOI] [PubMed] [Google Scholar]
- 9.Cox MP, Peterson DA, Biggs PJ. SolexaQA: at-a-glance quality assessment of illumina second-generation sequencing data. BMC Bioinformatics. 2010;11:485. doi: 10.1186/1471-2105-11-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for bisulfite-Seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li R, Li Y, Kristiansen K, Wang J. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008;24:713–714. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- 12.Smith AD, Chung WY, Hodges E, Kendall J, Hannon G, Hicks J, Xuan Z, Zhang MQ. Updates to the RMAP short-read mapping software. Bioinformatics. 2009;25:2841–2842. doi: 10.1093/bioinformatics/btp533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith AD, Xuan Z, Zhang MQ. Using quality scores and longer reads improves accuracy of Solexa read mapping. BMC Bioinformatics. 2008;9:128. doi: 10.1186/1471-2105-9-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen PY, Cokus SJ, Pellegrini M. BS seeker: precise mapping for bisulfite sequencing. BMC Bioinformatics. 2010;11:203. doi: 10.1186/1471-2105-11-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Zhu J, Tian G, Li N, Li Q, Ye M, Zheng H, Yu J, Wu H, Sun J, et al. The DNA methylome of human peripheral blood mononuclear cells. PLoS Biol. 2010;8:e1000533. doi: 10.1371/journal.pbio.1000533. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.