Abstract
Purpose
The human WD repeat-containing protein 46 (WDR46; also known as C6orf11), located at the disease-relevant centromere side of the class II major histocompatibility complex region, is hypothesized to be associated with risk of aspirin-exacerbated respiratory disease (AERD) as well as a decline in forced expiratory volume in the first second (FEV1), an important diagnostic marker of asthma.
Methods
To investigate the association between WDR46 and AERD, five single-nucleotide polymorphisms (SNPs) were genotyped in 93 AERD cases and 96 aspirin-tolerant asthma controls of Korean ethnicity. Three major haplotypes were inferred from pairwise comparison of the SNPs, and one was included in the association analysis. Differences in the frequency distribution of WDR46 SNPs and haplotype were analyzed using logistic and regression models via various modes of genetic inheritance.
Results
Depending on the genetic model, the logistic and regression analyses revealed significant associations between rs463260, rs446735, rs455567, rs469064, and WDR46_ht2 and the risk of AERD (P=0.007-0.04, Pcorr=0.01-0.04) and FEV1 decline after aspirin provocation (P=0.006-0.03, Pcorr=0.01-0.03). Furthermore, functional analysis in silico showed that the G>A allele of rs463260 located in the 5' untranslated region potentially matched a nucleotide sequence within an upstream open reading frame of WDR46.
Conclusions
These findings show for the first time that WDR46 is an important genetic marker of aspirin-induced airway inflammation and may be useful for formulating new disease-management strategies.
Keywords: Aspirin exacerbated respiratory disease, WDR46, FEV1, haplotype, single-nucleotide polymorphism
INTRODUCTION
Aspirin-exacerbated respiratory disease (AERD) is characterized by asthma, the presence of polyps in nasal passages, and sensitivity to aspirin and other non-steroidal anti-inflammatory drugs (NSAIDs).1 Although previous research has attributed AERD pathophysiology to inhibition of the cyclooxygenase-2 (COX-2) pathway, resulting in decreased synthesis of prostaglandin E2 (PGE2) and overproduction of cys-leukotrienes (cysLTs),2 recent reports have suggested that several genes may play a predisposing role, implicating other pathways in AERD pathogenesis; these include major histocompatibility complex (MHC), class II, DP beta 1 (HLA-DPB1), solute carrier family 6 (neurotransmitter transporter, betaine/GABA) member 12 (SLC6A12), complementary component 6 (C6), D-tyrosyl-tRNA deacylase (DTD1), and emilin/multimerin domain-containing protein 2 (EMID2).3-7
The MHC, a group of genes that determines the immune response to pathogens, is central to the activation of the innate immune system. The human WD repeat-containing protein 46 (WDR46, also called C6orf11 and BING4; NM_005452) gene is located on chromosome 6p21.3 at the centromere side of the class II MHC region, and encodes a protein that is 610 amino acids in length8 that shows homology to Saccharomyces cerevisiae and Caenorhabditis elegans proteins.9 In particular, WDR46 contains a class I type aminotransferase pyridoxal phosphate attachment site motif, a class I aminoacyl-tRNA synthetase adenylate binding site motif, and G-beta (WD40) repeats found in the β-subunits of G-proteins.10 A previous study of metastatic cancer patients reported high expression levels of WDR46 mRNA in melanoma cell lines, which correlated with lymphocyte recognition, resulting in secretion of additional cytokines. Furthermore, one of the MHC class II genes, HLA-DPB1, is a genetic marker for AERD in the Korean population, suggesting that WDR46 may be a candidate gene for disease susceptibility. However, little of the structure of WDR46 has been reported, and its function remains unknown.
Because genes at the centromere end of the MHC are often associated with several human diseases including asthma,3 and recruitment and activation of T-lymphocytes with increased cytokine production has been implicated in aspirin-induced airway inflammation,11,12 WDR46 is hypothesized to be associated with the risk of AERD. Thus, we conducted an association analysis on a Korean asthma cohort.
MATERIALS AND METHODS
Study patients
Asthma patients enrolled in nine hospitals in Korea affiliated with the Asthma Genome Research Center were recruited. Diagnosis of asthma and tests measuring total immunoglobulin E (IgE), atopy, and individuals' reactions to inhalant allergens were performed. Oral aspirin challenge (OAC) was performed using increasing doses.13,14 Briefly, patients with a history of aspirin hypersensitivity were given 30 mg orally. Respiratory and nasal symptoms, blood pressure, external signs (urticaria and angio-oedema), and FEV1 were documented every 30 minutes for 1.5 hours. In the absence of any symptom or sign suggesting an adverse reaction after 1.5 hours, increasing aspirin doses (60, 100, 300, and 400 mg) were administered until the patient developed a reaction; subsequently the same measurements were repeated hourly. Those with no history of aspirin hypersensitivity were started on 100 mg aspirin, which was increased to 200, 350, and 450 mg until a reaction developed. If no reaction occurred 4 hours after the final dose, the test was deemed negative. Aspirin-induced bronchospasm, reflected by a decline (%) in FEV1, was calculated as the pre-challenge FEV1 minus the post-challenge FEV1, divided by the pre-challenge FEV1. OAC reactions were categorized into three groups: (1) a 15% or greater decrease in FEV1 or nasal reactions (AERD), (2) less than a 15% decrease in FEV1 without naso-ocular or cutaneous reactions (aspirin-tolerant asthma [ATA]), or (3) less than a 15% decrease in FEV1 with cutaneous reactions (aspirin-induced urticaria). Written informed consent was obtained from each patient, and the study protocol was approved by the Institutional Review Board of each participating hospital.
Selection and genotyping of single-nucleotide polymorphisms
Using the tagging approach, candidate single-nucleotide polymorphisms (SNPs) in WDR46 were screened from the International HapMap database (version release #27; http://www.hapmap.org) based on linkage disequilibrium (LD) status in the Asian population (Han Chinese and Japanese) and location within the gene. Genotypes were derived by TaqMan assay15 using an ABI prism 7900HT sequence detection system (Applied Biosystems, Foster City, CA, USA). Evaluation of LDs between screened SNPs was carried out using the Haploview software,16 and SNPs with a minor allele frequency (MAF)>0.05 and those tagged because several polymorphisms had an LD>0.98, were selected for association analysis.
Statistical analyses
The association between differences in genotype frequency distribution of WDR46 variation and the risk of AERD and airway obstruction was analyzed according to previous methods.7 The statistical power of single associations was determined using the Power for Genetic Association Analyses software,17 and multiple testing corrections were calculated using the effective number of independent marker loci (Meff), that accounts for the eigenvalue spectral decomposition (SpD) of all of the genotypes represented in the correlation matrix extracted from the SNPSpD program.18 Furthermore, functional analyses of WDR46 SNPs were carried out in silico using the UTRScan program19 and the European Molecular Biology Laboratory-European Bioinformatics Institute (EMBL-EBI) splice site prediction software (http://www.ebi.ac.uk/asd-srv/wb.cgi?method=2).
RESULTS
Categorization and clinical characteristics
Based on individual reactions to aspirin, the asthma patients (n=189) were stratified into 93 AERD cases (males, 32; females, 61) and 96 ATA controls (males, 24; females, 72) (Table 1). The aspirin-induced decrease in FEV1 (AERD, 23.61 vs. ATA, 0.94), positivity rate of aspirin side effects including external signs of urticaria and angioedema (AERD, 26.67% vs. ATA, 8.42%), and positive history of aspirin intolerance (AERD, 63.86% vs. ATA, 29.27%) were significantly different between cases and controls (P<0.0001; Table 1). No significant differences for any other diagnostic factors were observed.
Table 1.
Clinical profiles of the study patients
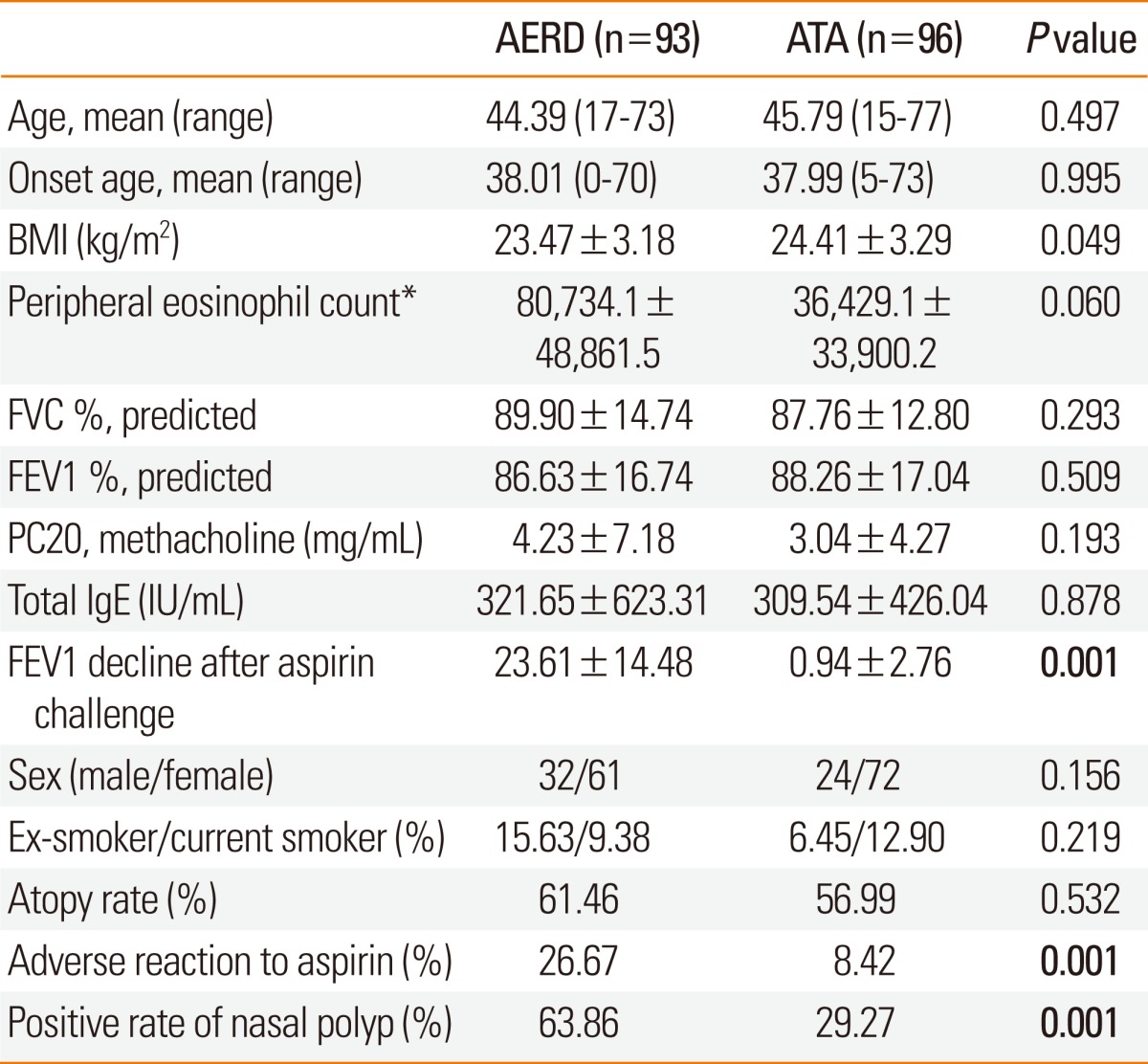
Values are mean±SE.
AERD, aspirin-exacerbated respiratory disease; ATA, aspirin-tolerant asthma; BMI, body mass index.
*Blood eosinophil (%): 6.29±5.80 (AERD group), 4.88±4.19 (ATA group).
Distribution of WDR46 variants
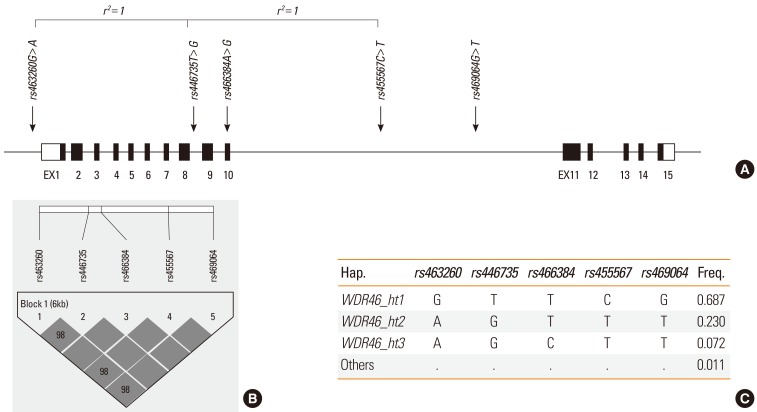
With high rates of concordance in duplicates (>99%), 5 WDR46 SNPs, rs463260 in the 5' untranslated region (UTR), rs466384 in the exon, and rs446735, rs455567, and rs469064 in the introns (Figure A and Supplementary Table S1), were successfully genotyped. The genotype counts and MAFs of each SNP are summarized in Supplementary Table S1. Furthermore, none of the variants deviated from the Hardy-Weinberg Equilibrium (HWE). Pairwise comparison of the genotyped SNPs showed a tight LD in the study population (Figure B) and produced three major haplotypes (MAF>0.05; Figure C). However, because WDR46_ht1 and WDR46_ht3 were equivalent to some SNPs, only WDR46_ht2 was included in the association analysis.
Figure.
Physical map, linkage disequilibrium, and haplotypes of WDR46. (A) Schematic gene map and single-nucleotide polymorphisms (SNPs) of WDR46 on chromosome 6p21.3 (10.11 kbp). Black blocks represent coding exons and white blocks represent 5' and 3' untranslated regions. The first base of translation sites are denoted as nucleotide +1. SNPs in absolute linkage are indicated by brackets (r2=1). (B) LD coefficient (|D'|) among WDR46 SNPs in a Korean population. (C) Haplotypes of WDR46.
Association analysis of WDR46 variants with a risk for AERD and FEV1 decline
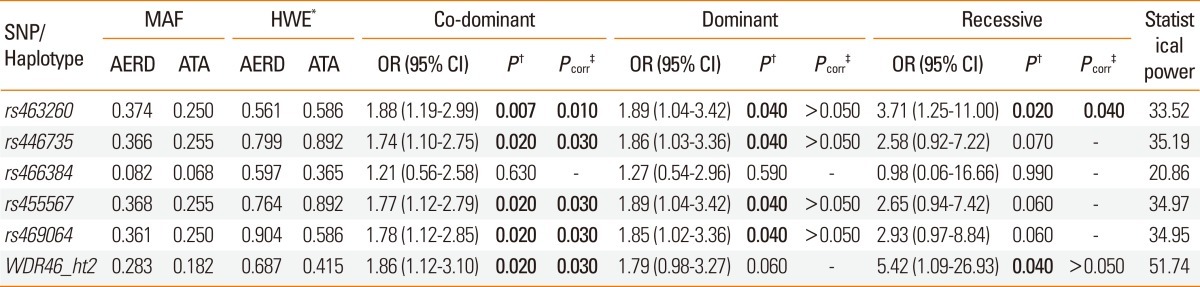
The results of logistic analysis controlling for age, sex, smoking status, atopy, and body mass index (BMI) revealed that the majority of the analyzed WDR46 variants (rs463260, rs446735, rs455567, rs469064, and WDR46_ht2) were associated with a risk for AERD (P=0.007-0.04, odds ratio [OR]=1.88-1.89 depending on the genetic model; Table 2). The association signals maintained significance under co-dominant and recessive models after multiple testing corrections (Pcorr=0.01-0.04; Meff=2.2767; Table 2).
Table 2.
Association analysis of variation in WDR46 with AERD
Logistic analysis controlling for age, sex, smoking status, atopy, and body mass index.
*P values of Hardy-Weinberg equilibrium (HWE).
†P values at 0.05 levels of significance. ‡P values after multiple testing corrections (Meff=2.2767). Significant values are shown in bold.
WDR46, human WD repeat-containing protein 46; AERD, aspirin-exacerbated respiratory disease; SNP, single-nucleotide polymorphism; MAF, minor allele frequency; ATA, aspirin-tolerant asthma; OR, odds ratio; CI, confidence interval.
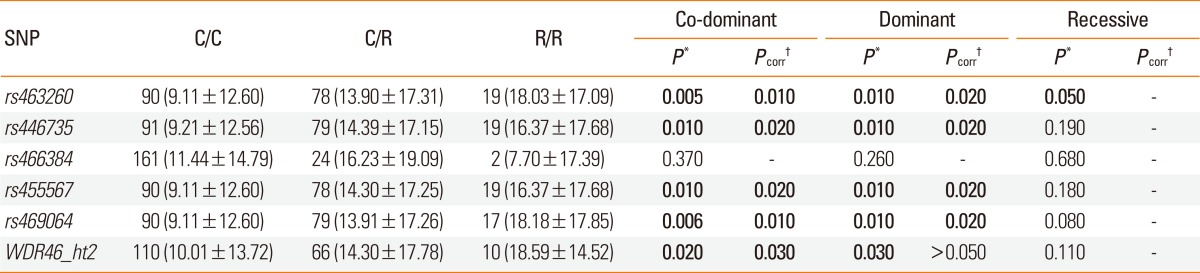
The results of regression analysis after adjustment for age, sex, smoking status, atopy, and BMI showed a significant association between a similar set of WDR46 variants (rs463260, rs446735, rs455567, rs469064, and WDR46_ht2) and decreased FEV1 in asthma patients (P=0.006-0.03 under co-dominant and dominant models; Table 3), even after multiple comparisons (Pcorr=0.01-0.03 in co-dominant and dominant models; Table 3).
Table 3.
Regression analysis of variation in WDR46 with decrease in FEV1 rate due to aspirin ingestion
Co-dominant, dominant, and recessive models of regression analysis controlling for age, sex, smoking status, atopy, and body mass index.
C/C, C/R, and R/R refer to major homozygote, heterozygote, and minor homozygote, respectively.
*P values at 0.05 levels of significance. †P values after multiple testing corrections (Meff=2.2767). Significant values are shown in bold.
Functional analyses in silico
Functional estimation of SNPs in intronic regions of WDR46 revealed that none of the analyzed variants act as branch point sites for alternative splicing (data not shown). However, results of another computer simulation analysis showed that the sequence containing the G>A allele of rs463260 located in the 5' UTR matched a nucleotide sequence within a UTRSite signal of WDR46, more specifically, an upstream open reading frame (uORF; Supplementary Table S2).
DISCUSSION
Although ample progress has been made in understanding the pathogenesis of AERD, the observed association between it and genes that were originally thought to play no relevant role in aspirin-induced airway bronchospasm suggests that the exact mechanisms of AERD development remain to be elucidated. The current findings provide important insight into the etiology of aspirin-hypersensitive asthma by reporting, for the first time, a significant association between the WDR46 variants rs463260, rs446735, rs455567, rs469064, as well as the major haplotype WDR46_ht2, and the risk of AERD. Furthermore, with a marginal increase in association signals, similar variants were found to induce significant decreases in FEV1, an important diagnostic marker of respiratory obstruction, among AERD patients compared to ATA controls. The reported association maintained significance under the strict criteria of multiple testing corrections. In particular, the rs463260 polymorphism exhibited a consistently robust association signal with the risk of AERD (P=0.007, Pcorr=0.01 in a co-dominant model) and FEV1 (P=0.005, Pcorr=0.01 in a co-dominant model) decline even after multiple comparisons. The association of WDR46 with risk of AERD was suggested via co-dominant and dominant modes of genetic inheritance in a Korean population.
Despite the dearth of previous reports, the positioning of the WDR46 gene in the MHC region is suggested to play a role in AERD development as well as decreased FEV1 due to aspirin ingestion. In further analysis, a uORF was detected in the 5' UTR of WDR46 and the G>A allele of the rs463260 polymorphism was found within the start and end positions of the UTRSite signal. The predicted uORF in the 5' UTR is speculated to be among the nine potential ORFs previously identified within WDR46.8 Although the role of uORFs is not yet fully understood, the 5' UTR harbors cis/trans-acting elements that play a crucial role in the regulation of gene expression by mediating translation efficiency and mRNA stability.19 Thus, the results of our in silico analyses suggest that the relevance of the significant SNP, rs463260G>A, in AERD susceptibility may be a manifestation of cis-regulated gene expression.
Examination of expressed sequence tags suggested the presence of alternative splicing within the WDR46 transcript.9 Although polymorphisms located in the non-coding intronic regions of the gene were originally thought to have no role in protein function, these intronic variants may also affect alternative splicing, splicing efficiency, or mRNA turnover as has been reported for other disease-causing genes.20 Thus, an in silico analysis was carried out to functionally characterize the intronic WDR46 polymorphisms. None of the analyzed variants were predicted to be branch point sites for alternative splicing, refuting our hypothesis.
Although the results from power calculations revealed a 35.24% average statistical power to detect effect sizes with the current sample (Table 2), indicating insufficient sample size, the allele frequency of the analyzed variants in our Korean population was similar to that of other Asian ethnic groups (Japanese and Han Chinese; Supplementary Table S1). However, while our findings successfully elucidated the association of WDR46 variation with aspirin-induced airway inflammation, the lack of an in-depth molecular investigation may be a limitation of this preliminary report. Furthermore, differences in the effects of genetic variation with regard to geographical location and/or ethnicity should also be taken into consideration. The present findings would, therefore, benefit from future studies addressing these limitations. Moreover, the advent of next-generation sequencing (NGS) with one-base ultra-resolution and high-density chip microarrays makes it possible to investigate the association of rare WDR46 variants with the risk of AERD.
In conclusion, our data provide convincing evidence that WDR46 variants are associated with the risk for both AERD and a relevant phenotype, FEV1 decline, in a Korean asthma cohort. Because the function of WDR46 in human diseases remains to be elucidated, our data represent a marked contribution to the knowledge on the role of this gene in airway inflammation.
ACKNOWLEDGMENTS
This work was supported by Korea Science and Engineering Foundation (KOSEF) funded by the Korea government (MEST) (No. 2009-0080157), and a grant by the Korea Healthcare Technology R&D Project, Ministry for Health, Welfare & Family Affairs, Republic of Korea (A010249). The DNA samples were generously provided by Soonchunhyang University, Bucheon Hospital Biobank, a member of the National Biobank of Korea, supported by the Ministry of Health, Welfare and Family Affairs, Republic of Korea.
Footnotes
There are no financial or other issues that might lead to conflict of interest.
Supplementary Table
References
- 1.Lee RU, Stevenson DD. Aspirin-exacerbated respiratory disease: evaluation and management. Allergy Asthma Immunol Res. 2011;3:3–10. doi: 10.4168/aair.2011.3.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bennett A. The importance of COX-2 inhibition for aspirin induced asthma. Thorax. 2000;55(Suppl 2):S54–S56. doi: 10.1136/thorax.55.suppl_2.S54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choi JH, Lee KW, Oh HB, Lee KJ, Suh YJ, Park CS, Park HS. HLA association in aspirin-intolerant asthma: DPB1*0301 as a strong marker in a Korean population. J Allergy Clin Immunol. 2004;113:562–564. doi: 10.1016/j.jaci.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 4.Pasaje CF, Kim JH, Park BL, Cheong HS, Chun JY, Park TJ, Lee JS, Kim Y, Bae JS, Park JS, Yoon SH, Uh ST, Choi JS, Kim YH, Kim MK, Choi IS, Cho SH, Choi BW, Park CS, Shin HD. Association of SLC6A12 variants with aspirin-intolerant asthma in a Korean population. Ann Hum Genet. 2010;74:326–334. doi: 10.1111/j.1469-1809.2010.00584.x. [DOI] [PubMed] [Google Scholar]
- 5.Pasaje CF, Kim JH, Park BL, Cheong HS, Kim MK, Choi IS, Cho SH, Hong CS, Lee YW, Lee JY, Koh IS, Park TJ, Lee JS, Kim Y, Bae JS, Park CS, Shin HD. A possible association of EMID2 polymorphisms with aspirin hypersensitivity in asthma. Immunogenetics. 2011;63:13–21. doi: 10.1007/s00251-010-0490-8. [DOI] [PubMed] [Google Scholar]
- 6.Pasaje CF, Bae JS, Park BL, Jang AS, Uh ST, Kim MK, Koh IS, Kim JH, Park TJ, Lee JS, Kim Y, Park CS, Shin HD. Association analysis of DTD1 gene variations with aspirin-intolerance in asthmatics. Int J Mol Med. 2011;28:129–137. doi: 10.3892/ijmm.2011.669. [DOI] [PubMed] [Google Scholar]
- 7.Pasaje CF, Bae JS, Park BL, Cheong HS, Jang AS, Uh ST, Kim MK, Koh IS, Kim JH, Park TJ, Lee JS, Kim Y, Park CS, Shin HD. Association analysis of C6 genetic variations and aspirin hypersensitivity in Korean asthmatic patients. Hum Immunol. 2011;72:973–978. doi: 10.1016/j.humimm.2011.05.022. [DOI] [PubMed] [Google Scholar]
- 8.Rosenberg SA, Tong-On P, Li Y, Riley JP, El-Gamil M, Parkhurst MR, Robbins PF. Identification of BING-4 cancer antigen translated from an alternative open reading frame of a gene in the extended MHC class II region using lymphocytes from a patient with a durable complete regression following immunotherapy. J Immunol. 2002;168:2402–2407. doi: 10.4049/jimmunol.168.5.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herberg JA, Beck S, Trowsdale J. TAPASIN, DAXX, RGL2, HKE2 and four new genes (BING 1, 3 to 5) form a dense cluster at the centromeric end of the MHC. J Mol Biol. 1998;277:839–857. doi: 10.1006/jmbi.1998.1637. [DOI] [PubMed] [Google Scholar]
- 10.Johnson EN, Druey KM. Heterotrimeric G protein signaling: role in asthma and allergic inflammation. J Allergy Clin Immunol. 2002;109:592–602. doi: 10.1067/mai.2002.122636. [DOI] [PubMed] [Google Scholar]
- 11.Akahoshi M, Obara K, Hirota T, Matsuda A, Hasegawa K, Takahashi N, Shimizu M, Nakashima K, Cheng L, Doi S, Fujiwara H, Miyatake A, Fujita K, Higashi N, Taniguchi M, Enomoto T, Mao XQ, Nakashima H, Adra CN, Nakamura Y, Tamari M, Shirakawa T. Functional promoter polymorphism in the TBX21 gene associated with aspirin-induced asthma. Hum Genet. 2005;117:16–26. doi: 10.1007/s00439-005-1285-0. [DOI] [PubMed] [Google Scholar]
- 12.Varga EM, Jacobson MR, Masuyama K, Rak S, Till SJ, Darby Y, Hamid Q, Lund V, Scadding GK, Durham SR. Inflammatory cell populations and cytokine mRNA expression in the nasal mucosa in aspirin-sensitive rhinitis. Eur Respir J. 1999;14:610–615. doi: 10.1034/j.1399-3003.1999.14c21.x. [DOI] [PubMed] [Google Scholar]
- 13.Kim BS, Park SM, Uhm TG, Kang JH, Park JS, Jang AS, Uh ST, Kim MK, Choi IS, Cho SH, Hong CS, Lee YW, Lee JY, Choi BW, Park HS, Park BL, Shin HD, Chung IY, Park CS. Effect of single nucleotide polymorphisms within the interleukin-4 promoter on aspirin intolerance in asthmatics and interleukin-4 promoter activity. Pharmacogenet Genomics. 2010;20:748–758. doi: 10.1097/FPC.0b013e3283402155. [DOI] [PubMed] [Google Scholar]
- 14.Nizankowska-Mogilnicka E, Bochenek G, Mastalerz L, Swierczyńska M, Picado C, Scadding G, Kowalski ML, Setkowicz M, Ring J, Brockow K, Bachert C, Wöhrl S, Dahlén B, Szczeklik A. EAACI/GA2LEN guideline: aspirin provocation tests for diagnosis of aspirin hypersensitivity. Allergy. 2007;62:1111–1118. doi: 10.1111/j.1398-9995.2007.01409.x. [DOI] [PubMed] [Google Scholar]
- 15.Livak KJ. Allelic discrimination using fluorogenic probes and the 5' nuclease assay. Genet Anal. 1999;14:143–149. doi: 10.1016/s1050-3862(98)00019-9. [DOI] [PubMed] [Google Scholar]
- 16.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 17.Menashe I, Rosenberg PS, Chen BE. PGA: power calculator for case-control genetic association analyses. BMC Genet. 2008;9:36. doi: 10.1186/1471-2156-9-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004;74:765–769. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grillo G, Turi A, Licciulli F, Mignone F, Liuni S, Banfi S, Gennarino VA, Horner DS, Pavesi G, Picardi E, Pesole G. UTRdb and UTRsite (RELEASE 2010): a collection of sequences and regulatory motifs of the untranslated regions of eukaryotic mRNAs. Nucleic Acids Res. 2010;38:D75–D80. doi: 10.1093/nar/gkp902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maquat LE. The power of point mutations. Nat Genet. 2001;27:5–6. doi: 10.1038/83759. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.