Abstract
Objectives
To investigate the prospects of newly available benchtop sequencers to provide rapid whole-genome data in routine clinical practice. Next-generation sequencing has the potential to resolve uncertainties surrounding the route and timing of person-to-person transmission of healthcare-associated infection, which has been a major impediment to optimal management.
Design
The authors used Illumina MiSeq benchtop sequencing to undertake case studies investigating potential outbreaks of methicillin-resistant Staphylococcus aureus (MRSA) and Clostridium difficile.
Setting
Isolates were obtained from potential outbreaks associated with three UK hospitals.
Participants
Isolates were sequenced from a cluster of eight MRSA carriers and an associated bacteraemia case in an intensive care unit, another MRSA cluster of six cases and two clusters of C difficile. Additionally, all C difficile isolates from cases over 6 weeks in a single hospital were rapidly sequenced and compared with local strain sequences obtained in the preceding 3 years.
Main outcome measure
Whole-genome genetic relatedness of the isolates within each epidemiological cluster.
Results
Twenty-six MRSA and 15 C difficile isolates were successfully sequenced and analysed within 5 days of culture. Both MRSA clusters were identified as outbreaks, with most sequences in each cluster indistinguishable and all within three single nucleotide variants (SNVs). Epidemiologically unrelated isolates of the same spa-type were genetically distinct (≥21 SNVs). In both C difficile clusters, closely epidemiologically linked cases (in one case sharing the same strain type) were shown to be genetically distinct (≥144 SNVs). A reconstruction applying rapid sequencing in C difficile surveillance provided early outbreak detection and identified previously undetected probable community transmission.
Conclusions
This benchtop sequencing technology is widely generalisable to human bacterial pathogens. The findings provide several good examples of how rapid and precise sequencing could transform identification of transmission of healthcare-associated infection and therefore improve hospital infection control and patient outcomes in routine clinical practice.
Article summary
Article focus
To investigate the prospects of newly available benchtop sequencers to provide rapid whole-genome data in routine clinical practice.
In particular to investigate the potential of such technology for identification of transmission events of healthcare-associated pathogens.
Key messages
We demonstrate benchtop sequencing can enhance hospital infection control through high precision support and rejection of transmission using genetic data.
Whole-genome data provided additional genetic resolution over existing genetic typing strategies.
We also show this technology offers turnaround times of under a week in a format that, in contrast to molecular typing, is organism independent.
Strengths and limitations of this study
The case studies presented provide several good examples of how rapid and precise sequencing could transform identification of transmission of healthcare-associated infection.
Given this is a pilot study, further evaluations of the impact of this technology on hospital infection control are required. However, this study provides a clear rationale for future work undertaking formal comparisons of benchtop sequencing with existing local and national typing schemes.
Background
Uncertainty about the exact route and timing of transmission is a major impediment to management of healthcare-associated infection, particularly for endemic pathogens that are also carried commensally, such as Staphylococcus aureus and Clostridium difficile. This problem impairs the development and implementation of effective evidence-based measures for infection control.1
High-throughput methods using next-generation sequencing (NGS) are revolutionising bacterial genomics, providing sufficient resolution potentially to determine which cases within temporo-spatial clusters are likely to be related.2–5 With the advent of rapid sequencers, sources of outbreaks have been identified in clinically relevant timescales, demonstrating the potential of NGS to transform infection control practice.6–8 The latest benchtop machines offer whole-genome sequencing in a format and at a cost accessible to routine hospital laboratories,9 but the practical prospects for their use are unclear.
Here, we apply benchtop NGS in near real time to four case studies and to a surveillance reconstruction, involving two important healthcare-associated pathogens, S aureus and C difficile. We demonstrate how NGS supports identification of outbreaks of closely genetically related cases, including highlighting potential genetic links between cases not previously known to be related. In other examples, we show how NGS can refute transmission between cases that are epidemiologically linked, including between cases sharing the same strain type, indicating the additional benefit benchtop sequencing may provide over existing typing strategies.
Methods
Setting and patients, Oxford-based case studies
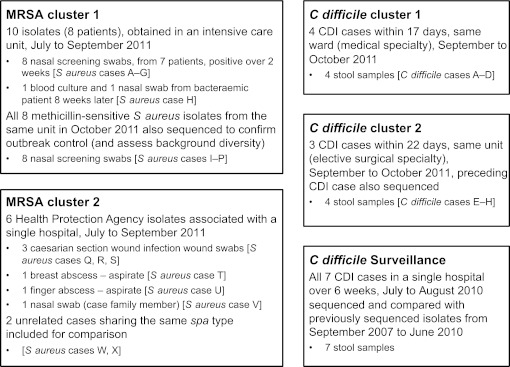
The Oxford University Hospitals NHS Trust (OUH) comprises 1600 beds across four hospitals, three in Oxford and one 35 miles north in Banbury. It provides >90% of hospital care, and all acute services, to ∼600 000 people in Oxfordshire, UK. The OUH microbiology laboratory provides all diagnostic testing for S aureus and C difficile for the region. All cases from three suspected outbreaks between July and October 2011 underwent NGS in parallel with routine infection control investigation (figure 1). A cluster of carriers of an atypical methicillin-resistant S aureus (MRSA) strain in an intensive care unit (ICU), with an associated bloodstream infection (MRSA cluster 1) was investigated. All S aureus isolates from patients in this ICU in the following month were sequenced, first to establish the level of diversity among S aureus isolates from the ICU to provide a comparison with the potential outbreak data and second to confirm control of the MRSA outbreak. Isolates were sequenced irrespective of resistance phenotype. The ICU is a 40 bedded unit offering high dependency and intensive care. Briefly, baseline infection control interventions included daily environmental cleaning, cleaning of equipment between patients and use of ‘aseptic non-touch technique’ for all line insertion, use and care. However, MRSA carriers were not routinely isolated. No other MRSA carriers were identified on the ICU in the month prior to the outbreak and only one in the 2 months following. Two clusters of C difficile infection (CDI, C difficile clusters 1 and 2) from a medical ward and an elective surgical unit were also investigated. OUH infection control policy targeting C difficile is described in a previous publication.10
Figure 1.
Clusters and samples. All clusters of cases occurred in the Oxford University Hospitals NHS Trust between July and October 2011, apart from MRSA cluster 2 where samples were obtained by the Health Protection Agency from an outbreak in southern England between July and September 2011. CDI, Clostridium difficile infection: ≥3 unformed stools in 24 h, enzyme immunoassay positive, culture positive.
Setting and patients, Health Protection Agency S aureus case study
A possible MRSA outbreak reported to the national Health Protection Agency affecting six patients in southern England between July and September 2011 was investigated (figure 1, MRSA cluster 2). All isolates possessed the lukS and lukF genes encoding Panton–Valentine leukocidin. Two additional local isolates that shared the same staphylococcal protein A (spa)-type, but without an epidemiological connection to the cluster, were also included as controls.
Setting and patients, Oxford-based surveillance reconstruction
To investigate the potential of fast turnaround benchtop sequencing as a surveillance tool, all CDI cases from one of the OUH hospitals over a 6-week period (July to August 2010) were sequenced by MiSeq. Although prepared and sequenced together, samples were analysed sequentially in the order originally sent to the routine clinical laboratory to mimic how NGS could support a real infection control investigation. Sequence data obtained on the Illumina (San Diego, California, USA) GAIIx and HiSeq platforms from 1185 of 1460 samples taken between September 2007 and June 2010 from a previously described collection of all Oxfordshire CDI cases10 were available for comparison.
Samples and sequencing
S aureus isolates were obtained from clinical samples inoculated onto MRSASelect agar (Bio-Rad, Hemel Hempstead, UK). Antimicrobial sensitivities were determined by disc diffusion and E testing as per European Committee on Antimicrobial Sensitivity Testing guidelines.11 C difficile isolates were identified following selective culture12 of toxin enzyme immunoassay-positive diarrhoeal faecal samples.
For both organisms, DNA was extracted using a commercial kit (QuickGene, Fujifilm, Tokyo, Japan), from a single colony subcultured onto a Columbia blood agar plate and incubated for 24–48 h. A combination of standard Illumina and adapted protocols was used to produce multiplexed paired-end libraries (DNA fragments with each sample's DNA tagged with a unique index). Pools of four samples were sequenced at the Wellcome Trust Centre for Human Genetics, Oxford, UK, using sequencing-by-synthesis technology,13 on the Illumina MiSeq platform, generating 150 base paired-end reads.
Sequence assembly and analysis
Sequence read data were analysed and assembled using a pipeline developed specifically for bacterial sequencing, as follows: to measure genome-wide sequence similarity, the full set of properly paired reads from each isolate was mapped using Stampy14 V.1.0.11 (without BWA pre-mapping, using an expected substitution rate of 0.01), to either S aureus MRSA252 (GenBank: NC_002952) 15 or C difficile 630 (GenBank: AM180355), CD630.16 Single nucleotide variants (SNVs) were identified across all mapped non-repetitive sites using SAMtools17 mpileup, after parameter tuning based on bacterial sequences. Known mobile genetic elements in CD630 were excluded from the analysis, as they account for 11% of the genome,16 and exchange of a single element may result in multiple SNVs in one event. A consensus of at least 75% was required to support an SNV, and calls were required to be homozygous under a diploid model. Only SNVs supported by at least five reads, including one in each direction, which did not occur at sites with unusual depth and were not within 12 bp of other variants, were accepted. Maximum likelihood trees were estimated from the mapped whole genomes using PhyML18 using a Jukes–Cantor model. To identify variation in gene content, sequence reads were assembled de novo using Velvet V.1.0.18, 19 with parameter values optimised to maximise n50 (50% of the assembly in contigs equal to or larger than this value). These assemblies were used to identify antibiotic resistance genes for S aureus and to determine in silico multilocus sequence types (MLST)12 for C difficile.
To assess both MiSeq data quality and its comparability with existing HiSeq data, the MRSA252 and CD630 references and one other isolate of each organism were sequenced on both platforms. Duplicates of two of the S aureus isolates from MRSA cluster 1 and an MRSA252 sample were run on MiSeq as controls.
Ethics statement
Ethical approval for sequencing S aureus and C difficile isolates from routine clinical samples and linkage to patient data without individual patient consent was obtained from Berkshire Ethics Committee (10/H0505/83) and the UK National Information Governance Board (8-05(e)/2010). The Health Protection Agency has Patient Information Advisory Group approval to hold and analyse surveillance data for public health purposes under Section 60 of the Health and Social Care Act 2001.
RESULTS
Twenty-six S aureus isolates (from 24 patients) and 15 C difficile isolates (from 15 patients) (figure 1) were sequenced using the MiSeq benchtop sequencer. Mean read-depths for S aureus and C difficile were 77.6 and 50.4, respectively, and reference genome coverages were 82.7% and 79.5%, respectively, after quality filtering (see supplementary figures 1–3 for regions called; uncalled regions include repetitive regions which, in contrast to Sanger sequencing, 150 bp reads cannot cover, and mobile elements, as well as other non-core genome). The entire process from commencing DNA extraction to measuring genomic relatedness for each set of sequences was completed within five working days of culture. No sequence differences were detected in the two pairs of replicates: between CD630 and MRSA252 and published reference sequences or between four samples sequenced both with MiSeq and earlier with HiSeq.
NGS supports a ward outbreak, despite discordant antimicrobial and strain-typing data (MRSA cluster 1)
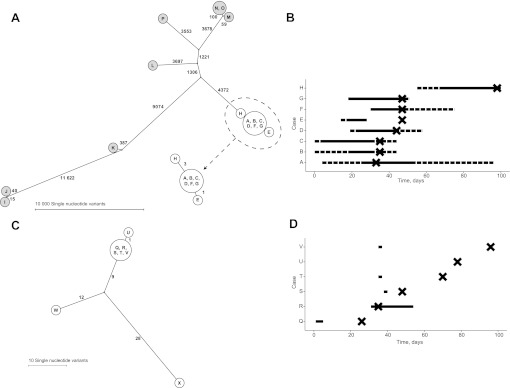
Ten MRSA isolates obtained from eight patients from the same ICU over 4 months belonged to the same spa-type (t5973) and were indistinguishable by pulsed-field gel electrophoresis (PFGE) (figure 2A,B). Ward stays and the first positive isolate per patient are shown in figure 2B. The extreme rarity of this spa-type in the UK (no t5973 isolates held by the National Reference Laboratory), and the fact they exhibited indistinguishable PFGE types, would normally be considered sufficient evidence for an outbreak. However, the isolates showed methicillin heteroresistance (which impaired initial detection of the outbreak) and differed in penicillin and tetracycline sensitivity (table 1); no common source was identified, casting doubt on the connection between the first seven cases and with the bloodstream infection, which occurred after a further 8 weeks, but in the absence of any contemporaneous patient MRSA isolates.
Figure 2.
Staphylococcus aureus cluster genetic and epidemiological relationships. (A and B) MRSA cluster 1 and related MSSA isolates. (C and D) MRSA cluster 2. Panel A shows all isolates from MRSA cluster 1 (white circles) and all S aureus isolates from the following month (grey circles, all methicillin sensitive). The left panels show the genetic relationships between cases as maximum likelihood trees, labelled with the number of single nucleotide variants at which samples differ. Genetically indistinguishable samples are shown in the same circle. The right panels show time spent on the same ward as a horizontal line for each case in both clusters. In panel B, the dashed line indicates time on the same ward and the solid line time in the same bay. In panel D, the solid line indicates time spent on the same ward. The timing of the first positive sample for each case is indicated with a cross. Case H subsequently developed a bloodstream infection.
Table 1.
MRSA cluster 1, comparison of antibiotic susceptibility and associated genetic elements
| Patient | Sample date | Antibiotic susceptibility |
Gene presence/absence |
||||
| Penicillin |
Tetracycline |
blaZ | tetK | ||||
| DD | MIC | DD | MIC | ||||
| A | 25/7 | R | 2 | S | 0.094 | + | − |
| B | 27/7 | R | 2 | R | 24 | + | + |
| C | 27/7 | R | 4 | S | 0.094 | + | − |
| D (i) | 5/8 | R | 3 | R | 24 | + | + |
| D (ii) | 10/8 | R | 2 | R | 24 | + | + |
| E | 8/8 | R | 4 | R | 32 | + | + |
| F | 8/8 | S | 0.047 | S | 0.064 | − | − |
| G | 8/8 | R | 3 | S | 0.094 | + | − |
| H (i) | 28/9 | R | 4 | R | 24 | + | + |
| H (ii)* | 29/9 | R | 2 | R | 24 | + | + |
Screening swabs were obtained from patient D on two separate dates. All isolates were methicillin heteroresistant, appearing susceptible on routine testing, despite detection of mecA by PCR and sequencing. This explains why isolate F appeared phenotypically penicillin susceptible on DD and E testing. The mechanism of heteroresistance for these isolates has not yet been fully elucidated, although the penicillin-susceptible methicillin-resistant phenotype has been described.20
The second isolate from patient H is from a positive blood culture.
DD, disc diffusion (R, resistant; S, susceptible); MIC, minimum inhibitory concentration (milligrams per litre).
No sequence differences were detected in the mapped genomes between isolates from six of the carriage cases (A, B, C, D, F and G); case E differed at a single site. The two isolates (nasal swab and blood culture) from the bloodstream infection case (H) were indistinguishable and differed by three SNVs from the other cases. Given previous estimated rates of short-term S aureus evolution of 9.6 (95% CI 7.3 to 11.6) 4 and 7.9 (95% credibility interval 4.8–12.8) 21 SNVs/genome/year, these data are consistent with recent acquisition from a common source. In contrast, the mean number of SNVs between all pairs of S aureus isolates from the same unit from the following month was 7541 (figure 2A). All the outbreak cases (A–H) were mecA positive by PCR and sequencing; however, two plasmids not present in MRSA252, encoding the blaZ and tetK genes, were detected in seven and five of the isolates, respectively. These genotypic findings matched the phenotypic susceptibility results (table 1).
As sequencing data provided support for an outbreak, this directly led to implementation (and escalation with subsequent cases) of intensive infection control supervision of the unit with visits up to four to five times per day. Observations made resulted in retraining for medical and nursing staff covering administration of intravenous medication and taking blood cultures. Additionally, isolation of all MRSA carriers was implemented and reinforced.
NGS supports transmission of isolates during short periods of shared ward exposure (MRSA cluster 2)
Six Panton–Valentine leukocidin-positive MRSA isolates were identified over 3 months (figure 2C,D). Five had been inpatients on the same ward (Q, R, S, T and V) (with overlapping stays of 1–2 days in four cases) and one (U) was a relative of V. All were spa-type t657, which, although relatively rare, has occurred sporadically in this region. The isolates were also indistinguishable by PFGE. Therefore, given the prolonged timescale, it was unclear whether these cases reflected a genuine outbreak or background circulation of related organisms. In fact, only one SNV was detected among all six samples, indicating a recent common source, whereas two isolates originating from the same geographical area and with the same spa and susceptibility profiles, but with slightly different PFGE types and no known epidemiological link, differed from the main cluster at 21 and 37 sites, respectively. The sites were distributed throughout the genome ruling out a single recombination event accounting for all the differences (supplementary figure 1). Additionally, no variant sites were detected within the mutS, mutS2 and mutL genes associated with hypermutation.22 The SNV differences are therefore consistent with a shared ancestry 2–5 years earlier.
NGS refutes transmission between suspected linked cases (C difficile cluster 1)
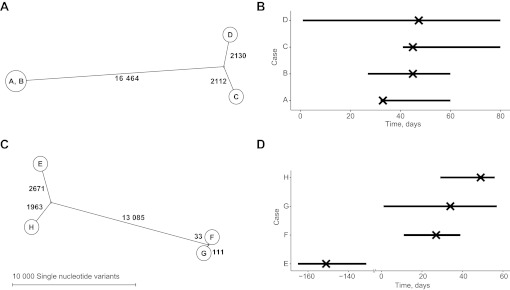
The OUH infection control service identified three CDI cases (B, C and D) occurring over 4 days among inpatients on the same ward (figure 3A,B). While UK Department of Health guidance23 states that a cluster should only be considered an ‘outbreak’ when the cases share a strain type (eg, by MLST or PCR-ribotyping), such information is slow to obtain. In practice, therefore, such clusters are treated as presumptive outbreaks requiring intensive management. Sequencing showed that all three cases had different computationally predicted sequence types (STs, ST2, ST10 and ST37) and differed at >4000 sites distributed throughout the genome (supplementary figure 2). With short-term C difficile evolution rates estimated from serial sampling of 91 patients with samples taken between 1 and 561 days apart at 2.3 SNVs/genome/year (95% credibility interval 1.6–3.0). (Xavier Didelot, personal communication 20 January 2012, manuscript under review), recent transmission between these cases can be excluded with confidence. However, a fourth case (A) that occurred 12 days previously, and was not initially included in the infection control investigation, was indistinguishable from case B; case A was found to have been in an adjacent side room to case B, strongly suggesting ward-based transmission. Presentation of this sequencing data backed transmission event to an outbreak review meeting, resulted in a detailed infection control audit, which in turn led to markedly improved cleaning of equipment.
Figure 3.
Clostridium difficile cluster genetic and epidemiological relationships. (A and B) C difficile cluster 1. (C and D) C difficile cluster 2. The left panels show the genetic relationships between cases as maximum likelihood trees, labelled with the number of single nucleotide variants at which samples differ. Genetically indistinguishable cases are shown in the same circle. The right-hand panels (B and D) show time spent on the same ward as a horizontal line for each case. The timing of the first positive sample for each case is indicated with a cross.
NGS demonstrates that isolates of the same strain type are not necessarily linked by transmission (C difficile cluster 2)
Three CDI cases (F, G and H) occurring over a 3-week period in an elective surgical unit were suggestive of transmission since the most recent previous CDI (E) had occurred 6 months before (figure 3C,D). However, isolates from the three cases were sufficiently genomically diverse to rule out transmission. Notably, two isolates shared a sequence type (ST5) under the strain-typing scheme used in OUH but differed at 144 sites distributed throughout the genome (supplementary figure 3), providing an example of the extra discriminatory power of sequencing over existing typing schemes in ruling out transmission.
NGS-based C difficile surveillance, a reconstruction
All seven CDI cases occurring in one of the OUH hospitals over 6 weeks were sequenced on the MiSeq platform and compared with each other and with an ‘historical’ sequence database comprising 1185 isolates from regional CDI cases occurring in the previous 3 years.
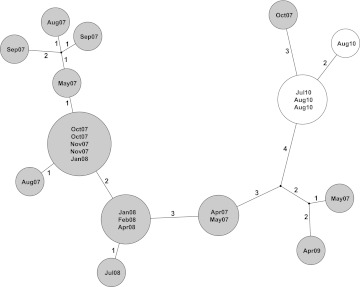
Four of the seven cases (ST3) formed a genetic cluster containing variation at only two SNVs, indicating probable transmission; these cases shared time and space on the same ward around their CDI. The genetically most similar historical CDI case differed from the four genetically clustered cases at three SNVs (figure 4); however, it occurred 3 years earlier and 30 miles way. Therefore, no direct relationship could be discerned between the historical cases and the current outbreak.
Figure 4.
Potential for whole-genome sequencing to enhance Clostridium difficile surveillance. Genetic relationships between cases are shown as a maximum likelihood tree, labelled with the number of single nucleotide variants (SNVs) at which samples differ. Genetically indistinguishable cases are shown in the same circle. Cases from an existing database of sequenced isolates are shown in grey circles with the date of isolation. Four cases arising during a 6-week surveillance reconstruction are shown in white circles. Three additional cases sequenced as part of the reconstruction differed at >3000 SNVs from the cases shown and are not shown in the figure. The maximum likelihood tree shown is consistent with data; however, homoplasy was observed at one of the variant sites, 2464157, in the CD630 reference genome.
The three remaining cases in the group of seven reconstruction cases (representing ST1, ST11 and ST13) differed from the other four cases and from each other at >3000 SNVs. The overall mean pairwise SNV difference between all reconstruction cases was 13 012 SNVs. One individual (ST1, ribotype-027), diagnosed on the day of admission and last admitted 8 months previously, yielded a C difficile sequence indistinguishable from 11 previous CDI cases, including local strains from 6 months previously. Since this patient had not shared inpatient time with any of the other cases and most of the genetically related cases had occurred outside of OUH hospitals, this may represent previously unsuspected community-based transmission, which could have been investigated had sequence information been available in 2010. No plausible hospital or community patient source could be identified for the other two cases; however, previous cases differing from them at four to 10 SNVs were identified in the previous 6 months to 3 years, consistent with a local strain origin.
Discussion
This study provides strong evidence in two major healthcare-associated pathogens, S aureus and C difficile, that benchtop sequencing can enhance hospital infection control through high precision support and rejection of transmission using genetic data. We also show this technology offers turnaround times of under a week in a format that, in contrast to molecular typing, is organism independent.
The results obtained in this study were obtained quickly enough to influence cluster investigations and in the outbreaks described were used to inform the hospital's response. Where suspected transmission events were supported by sequencing data in two of the outbreaks infection control supervision of measures to prevent case-to-case spread was significantly enhanced. In contrast, increases in incidence without transmission between infected patients still merit a response, for example, clusters of genetically unrelated C difficile cases on wards have prompted a review in our hospitals of antibiotic use and guidance. If such clusters were identified in patients with S aureus infection, this might prompt, for example, review of line care or perioperative care. Had the set of cases in the C difficile surveillance reconstruction been sequenced in real time, an outbreak would have become apparent after the second case, and it is possible that subsequent transmissions might have been prevented, particularly as compliance with infection control measures was incomplete at the time of the outbreak. Clearly formal evaluation of the use of the technology in an appropriately controlled trial is needed to determine the extent to which the control of these outbreaks was, or would have been, enhanced by the availability of sequence data.
Whole-genome sequencing provides the ultimate resolution of genetic relationships. This offers two clear benefits for inference of transmission events. First, putative transmission events between genetically very distinct isolates can be refuted with confidence. This is of particular value by comparison with widely used current typing strategies that are unable to distinguish isolates belonging to a prevalent strain type; for example, PCR-ribotype-027 (NAP-1) accounts for ∼35% of C difficile strains in UK and North American hospitals.24 25 Second, close genetic relationships combined with clinical and epidemiological evidence can provide strong evidence in favour of a putative transmission event, justifying infection control intervention, as detailed above. Notably, a genetic match in the absence of an obvious epidemiological link may legitimately prompt investigation of new routes of transmission, such as the possible community transmission identified in our C difficile surveillance reconstruction. However, close genetic links cannot be used in isolation to confirm transmission. For example, we identified genetically similar cases separated by both time and space, emphasising the importance of analysing genetic data alongside epidemiological information. The main limitation of this study is it is not large enough to provide a formal comparison with existing technology including a health economic evaluation. Additionally, although we used the Illumina MiSeq platform, other similar benchtop technology exists, for example, Ion Torrent (Life Technologies, Guildford, Connecticut, USA), or is under development, for example, Oxford Nanopore (Oxford Nanopore Technologies, Oxford, UK).
NGS technology is widely generalisable to human bacterial pathogens26 and has been used previously to investigate transmission of infectious disease.2–5 Although the benefits of rapid sequencers have been shown in high-profile national outbreaks,6–8 we provide the first demonstration of rapid sequencing in a benchtop format applied to routine patient care and healthcare-associated pathogens. Existing typing schemes, such as spa, PFGE, PCR-ribotyping and MLST, have established a framework in the application of genetic data in outbreak investigation. However, the organism-specific application of these strain-typing methods, often requiring isolates to be sent to a specific reference laboratory, in practice means that many infection clusters remain untyped. Benchtop sequencing builds upon existing typing expertise, offering rapidly available and increased resolution data from an organism-independent platform. This means that a single technology will provide the capacity for individual hospital laboratories to support outbreak investigation for a range of pathogens and allow infection control personnel to efficiently target resources to genuine outbreaks. Sequencing costs are falling rapidly,27 28 with the prospect within 12 months of being able to obtain complete accurate pathogen sequences in hours, and for as little as US$10 per sequence.29 It is therefore likely that benchtop sequencing will soon be comparable in price to existing molecular typing while offering consider additional benefits. As well as identifying transmissions, the reproducible and digital nature of locally generated sequence data make these ideal for sharing in national and potentially international surveillance, for sequence-based resistance prediction and for precise generic species identification.26
The case studies in this pilot study provide a clear rationale for future work undertaking formal comparisons of benchtop NGS with existing local and national typing schemes. Such comparisons will need to include formal health economic assessments that account for the capital expense of establishing benchtop NGS equipment and expertise in a laboratory, as well as the potential cost-savings associated with more focused cluster investigation, and infection control interventions. The improved accuracy in identifying within-hospital transmission should also lead to better metrics of hospital infection control performance and provide an opportunity for further reductions in the incidence of healthcare-associated infections and hence improvements in patient outcomes.
Supplementary Material
Acknowledgments
We thank Margot Nicholls and Angela Iversen from Surrey and Sussex Health Protection Unit for providing epidemiological details. We thank David Griffiths and Alison Vaughan for assistance with sample preparation and the Oxford MRC High-Throughput Sequencing Hub team.
Footnotes
To cite: Eyre DW, Golubchik T, Gordon NC, et al. A pilot study of rapid benchtop sequencing of Staphylococcus aureus and Clostridium difficile for outbreak detection and surveillance. BMJ Open 2012;2:e001124. doi:10.1136/bmjopen-2012-001124
Contributors: All authors were involved in critical review of the manuscript and have seen and approved the final version. Specific contributions as follows: study conception and design: DWC, TEAP, ASW, PJD, RB, MW and JP; sample acquisition: LO, RL, NCG, AK, AS and JP; sample sequencing: PP and DB; sequence data processing pipeline: RB, TG, EMB and CLCI; analysis of epidemiological and sequence data: DWE, TG, NCG, DJW, XD, TEAP, ASW and DWC; drafting the manuscript: DWE, NCG, TG, ASW, TEAP and DWC. The following two groups of authors contributed equally to this article, DWE, TG, NCG and RB; and TEAP, ASW and DWC. All authors had full access to all the study data and take responsibility for the integrity of the data and the accuracy of the data analysis. DWC is the guarantor.
Funding: This study was supported by the National Institute for Health Research (NIHR) Oxford Biomedical Research Centre and the UKCRC Modernising Medical Microbiology Consortium, the latter funded under the UKCRC Translational Infection Research Initiative supported by Medical Research Council, Biotechnology and Biological Sciences Research Council and the NIHR on behalf of the Department of Health (grant G0800778) and the Wellcome Trust (grant 087646/Z/08/Z). We acknowledge the support of Wellcome Trust core funding (grant 090532/Z/09/Z). TEAP and DWC are NIHR Senior Investigators. DWE is an NIHR Doctoral Research Fellow.
Competing interests: All authors have completed the Unified Competing Interest form at http://www.icmje.org/coi_disclosure.pdf (available on request from the corresponding author). The institution of DWC and TEAP received per-case funding from Optimer Pharmaceuticals to support fidaxomicin trial patient expenses. DWC and TEAP also received honoraria from Optimer Pharmaceuticals for participation in additional meetings related to investigative planning for fidaxomicin. MHW has received honoraria for consultancy work, financial support to attend meetings and research funding from bioMerieux, Optimer, Novacta, Pfizer, Summit, The Medicines Company, Viropharma and Astellas.
Ethics approval: Ethical approval for sequencing S aureus and C difficile isolates from routine clinical samples and linkage to patient data without individual patient consent was obtained from Berkshire Ethics Committee (10/H0505/83) and the UK National Information Governance Board (8-05(e)/2010). The Health Protection Agency has Patient Information Advisory Group approval to hold and analyse surveillance data for public health purposes under Section 60 of the Health and Social Care Act 2001.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: The sequences reported in this paper have been deposited in the European Nucleotide Archive Sequence Read Archive under study accession number ERP001413 (http://www.ebi.ac.uk/ena/data/view/ERP001413).
References
- 1.Forder A. A brief history of infection control—past and present. S Afr Med J 2007;97:1161–4 [PubMed] [Google Scholar]
- 2.Gardy JL, Johnston JC, Ho Sui SJ, et al. Whole-genome sequencing and social-network analysis of a tuberculosis outbreak. N Engl J Med 2011;364:730–9 [DOI] [PubMed] [Google Scholar]
- 3.Lieberman TD, Michel J-B, Aingaran M, et al. Parallel bacterial evolution within multiple patients identifies candidate pathogenicity genes. Nat Genet 2011;43:1275–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harris SR, Feil EJ, Holden MT, et al. Evolution of MRSA during hospital transmission and intercontinental spread. Science 2010;327:469–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reeves PR, Liu B, Zhou Z, et al. Rates of mutation and host transmission for an Escherichia coli clone over 3 years. PLoS One 2011;6:e26907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chin C-S, Sorenson J, Harris JB, et al. The origin of the Haitian cholera outbreak strain. N Engl J Med 2011;364:33–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rasko DA, Webster DR, Sahl JW, et al. Origins of the E. coli strain causing an outbreak of hemolytic-uremic syndrome in Germany. N Engl J Med 2011;365:709–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rohde H, Qin J, Cui Y, et al. Open-source genomic analysis of Shiga-toxin-producing E. coli O104:H4. N Engl J Med 2011;365:718–24 [DOI] [PubMed] [Google Scholar]
- 9.Mellmann A, Harmsen D, Cummings CA, et al. Prospective genomic characterization of the German enterohemorrhagic Escherichia coli O104:H4 outbreak by rapid next generation sequencing technology. PLoS One 2011;6:e22751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walker AS, Eyre DW, Wyllie DH, et al. Characterisation of Clostridium difficile hospital ward-based transmission using extensive epidemiological data and molecular typing. PLoS Med 2012;9:e1001172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.European Committee on Antimicrobial Susceptibility Testing Breakpoint tables for interpretation of MICs and zone diameters, version 2.0. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Disk_test_documents/EUCAST_breakpoints_v_2.0_120101.pdf (accessed 20 Jan 2012).
- 12.Griffiths D, Fawley W, Kachrimanidou M, et al. Multilocus sequence typing of Clostridium difficile. J Clin Microbiol 2010;48:770–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bentley DR, Balasubramanian S, Swerdlow HP, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 2008;456:53–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lunter G, Goodson M. Stampy: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res 2011;21:936–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holden MT, Feil EJ, Lindsay JA, et al. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc Natl Acad Sci U S A 2004;101:9786–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sebaihia M, Wren BW, Mullany P, et al. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat Gen 2006;38:779–86 [DOI] [PubMed] [Google Scholar]
- 17.Li H, Handsaker B, Wysoker A, et al. The sequence Alignment/Map format and SAMtools. Bioinformatics 2009;25:2078–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 2003;52:696–704 [DOI] [PubMed] [Google Scholar]
- 19.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 2008;18:821–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blanc DS, Petignat C, Moreillon P, et al. Unusual spread of a penicillin-susceptible methicillin-resistant Staphylococcus aureus clone in a geographic area of low incidence. Clin Infect Dis 1999;29:1512–18 [DOI] [PubMed] [Google Scholar]
- 21.Young BC, Golubchik T, Batty EM, et al. Evolutionary dynamics of Staphylococcus aureus during progression from carriage to disease. Proc Natl Acad Sci U S A 2012;109:4550–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prunier AL, Leclercq R. Role of mutS and mutL genes in hypermutability and recombination in Staphylococcus aureus. J Bacteriol 2005;187:3455–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Department of Health Clostridium difficile Infection: How to Deal with the Problem. 2009 [Google Scholar]
- 24.Dingle KE, Griffiths D, Didelot X, et al. Clinical Clostridium difficile: clonality and pathogenicity locus diversity. PLoS One 2011;6:e19993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Louie TJ, Miller MA, Mullane KM, et al. Fidaxomicin versus vancomycin for Clostridium difficile infection. N Engl J Med 2011;364:422–31 [DOI] [PubMed] [Google Scholar]
- 26.Relman DA. Microbial genomics and infectious diseases. N Engl J Med 2011;365:347–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.The Sequence Explosion. Nature 2010;464:671–2 [DOI] [PubMed] [Google Scholar]
- 28.Metzker ML. Sequencing technologies—the next generation. Nat Rev Genet 2009;11:31–46 [DOI] [PubMed] [Google Scholar]
- 29.Nature News Nanopore Genome Sequencer Makes its Debut. http://www.nature.com/news/nanopore-genome-sequencer-makes-its-debut-1.10051 (accessed 22 Feb 2012). doi:10.1038/nature.2012.10051
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.