Abstract
Many strains of the soil bacterium Bacillus subtilis are capable of producing and being resistant to the antibiotic sublancin because they harbor the Spβ prophage. This 135 kb viral genome is integrated into the circular DNA chromosome of B. subtilis, and contains genes for the production of and resistance to sublancin. We investigated the role of SigY in sublancin production and resistance, finding that it is important for efficient maintenance of the Spβ prophage. We were unable to detect the prophage in mutants lacking SigY. Additionally, these mutants were no longer able to produce sublancin, were sensitive to killing by this factor, and displayed a delay in sporulation. Wild-type cells with normal SigY activity were found to partially lose the Spβ prophage during growth and early sporulation, suggesting a mechanism for the bistable outcome of sibling cells capable of killing and of being killed. The appropriate regulation of SigY appears to be essential for growth as evidenced by the inability to disrupt the gene for its putative antisigma. Our results confirm a role for SigY in antibiotic production and resistance, as has been found for other members of the extracytoplasmic function sigma factor family in B. subtilis, and shows that this role is achieved by affecting maintenance of the Spβ prophage.
Introduction
SigY is a member of a family of sigma factors, the extracytoplasmic function (ECF) sigmas (Lonetto et al., 1994; Missiakas and Raina, 1998). Members of this family are characterized by their ability to respond to environmental stress, and to initiate transcription of genes necessary to adapt to the stress. This includes the gene that encodes for the ECF sigma itself (i.e., autotranscription) (Helmann, 2002, 2006). In Bacillus subtilis the ECF sigmas have been shown to respond to a number of stressors, and to primarily regulate functions associated with antibiotic production and resistance (Huang and Helmann, 1998; Huang et al., 1999; Cao and Helmann, 2002, 2004; Cao et al., 2002; Pietiainen et al., 2005). In its natural soil environment the coexpression of killing factors and resistance proteins by B. subtilis is thought to be an important survival strategy in response to starvation. This response kills competing bacteria and creates additional nutritional resources, while protecting the cells that produce the killing factors by the concomitant synthesis of resistance proteins (Stein, 2005). For example, proteins responsible for both killing and resistance (i.e., ABC transporters) are responsible for processing the mature killing factor during export, and conferring resistance to the mature antibiotic by keeping it out of the cell (Havarstein et al., 1995; Higgins, 2001). However, subpopulations of cells unable to produce these proteins will die in the presence of cells capable of their production.
While antibiotic production and resistance in B. subtilis has largely been described as a process that targets competing bacteria, the bistable expression of these functions leads to subpopulations of sibling cells capable of killing and of being killed. For example, in response to starvation, two killing factors (Skf and SdpC) are produced by a subpopulation of cells that are also resistant to them, but are not produced by another subpopulation that is sensitive to killing by these factors (González-Pastor et al., 2003; Ellermeier et al., 2006; González-Pastor, 2011). This bistable outcome is achieved through differential phosphorylation of Spo0A, the master regulator of sporulation (Grossman, 1995; Stragier and Losick, 1996; Perego and Hoch, 2002; Veening et al., 2005). In sporulating cells Spo0A is active because adequate levels of phosphorylated Spo0A have been attained, whereas in nonsporulating cells Spo0A is inactive due to insufficient amounts of phosphorylated Spo0A. Because both sporulation and Skf production and resistance are coupled to the activity of Spo0A, sporulating cells kill the nonsporulating sibling cells. In this way the subpopulation of cells destined to die, due to their inability to sporulate because of insufficient phosphorylated Spo0A, are a nutritional resource for the remaining cells and delay sporulation (González-Pastor et al., 2003; Ellermeier et al., 2006; González-Pastor, 2011).
In addition to Spo0A, ECF sigmas play a role in antibiotic production and resistance in B. subtilis (Helmann, 2002, 2006). In previous work we confirmed the biochemical activity of SigY as a sigma factor, and found that it maintained hallmark features characteristic of the ECF sigma family. Like other ECF sigmas, the activity of SigY is upregulated by stress (i.e., low nutrients), and it is responsible for transcribing its own operon (Cao et al., 2003). In addition to sigY, the gene that encodes for SigY, this operon contains five genes of unknown function, yxlC, yxlD, yxlE, yxlF, and yxlG. A sixth gene immediately downstream of the sigY operon, yxlH, appears to be subject to antisense regulation by the transcript produced from the sigY operon (Tojo et al., 2003). The last two genes in the sigY operon, yxlF and yxlG, are predicted to encode components of an ABC transporter, and the predicted protein product of yxlH is a multidrug efflux transporter. The functions predicted for these SigY-regulated gene products suggest a role for SigY in antibiotic production and resistance consistent with studies that demonstrate a broad role for the ECF sigmas of B. subtilis in these activities (Helmann, 2002, 2006). In this study we find that SigY is required for maintenance of the Spβ prophage that contains genes necessary to produce and resist killing by the antibiotic sublancin.
Materials and Methods
Bacterial strains and media
All B. subtilis strains used in this study are listed in Table 1. Bacterial strains were streaked out on Luria-Bertani (LB) plates and incubated at 37°C the night before the experiment. Bacterial cultures were grown in LB media or 2×Schaefer Growth (SG)-rich medium at 37°C with vigorous shaking. The LB medium contained 25 g of Difco LB per liter and the 2×SG medium was first prepared as a base containing 16 g of Difco nutrient broth, 2 g of KCl, and 0.5 g of MgSO4.7H2O per liter that was stored in the dark and used within 2 weeks. The following supplements were added to the 2×SG base medium on the day of experiment to make it complete: 1 mM Ca(NO3)2, 0.1 mM MnSO4, 1 μM FeSO4, 0.1% glucose, 100 mg of tryptophan, 100 mg of phenylalanine, and antibiotics when appropriate. The antibiotics used in both liquid and solid media were the following: chloramphenicol (7 μg/mL) and erythromycin (1 μg/mL). Bacterial growth was measured using a spectrophotometer at 600 nm (optical density [OD]600).
Table 1.
Strains, Plasmids, and Primers Used
| Strain or plasmid | Genotype or description | Derivation (source and/or reference)a |
|---|---|---|
| Bacillus subtilis | ||
| CU1065 | Spβ−trpC2 | (D. Zeigler) (Zahler et. al., 1977) |
| CU1065 sigY | Spβ− trpC2 sigY::mls | Transform [CU1065:LMB276, MLSr]a (This study) |
| LMB7 | trpC2 pheA1 | JH642b (J. Hoch) |
| LMB10 | trpC2 pheA1 sigD::pLM5 | CB100b (M. Chamberlin) (Marquez et al., 1990) |
| LMB276 | Spβ−trpC2 pheA1 sigY::mls | Transform [LMB7:HB0009 (J. Helmann), MLSr]a (Perez, 2006) |
| LMB276Sc | Spβ+ trpC2 pheA1 sigY::mls | Transform [LMB7:LMB276, MLSr]a (This study) |
| LMB307 | trpC2 pheA1 skfABCDEF::tet | Transform [LMB7:EG168 (J.E. González-Pastor]a (This study) |
| LMB309 | trpC2 pheA1 spo0A::cat | Transform [LMB7:AG475 (A. Grossman)]a (This study) |
| Plasmid | ||
| pCLP03 | Disruptional plasmid for yxlC | Perez (2006) |
| Primers | Sequence | |
| BETAFd | Primers for empty Spβ site | TATTCCCCAAAGAGGTGGTG |
| BETARd | AGGTTGCCCCATTCATACAG | |
| yonRFd | Primers for amplification of yonR | TTTTTGCTTCTTTTTGTTTACGA |
| yonRRd | TTCATTTCCCCTTTTTACTCCA | |
| sigDF | Primers for amplification of sigD | TGCCGCTTGTCACATATC |
| sigDR | GTTTTCCCCGTCATCTTG | |
Transformations are shown as follows: [recipient strain: chromosomal DNA from listed strain used for transformation, and selection for specified resistance].
Previous name of strain.
This strain likely contains a suppressor mutation that retains Spβ.
Source of BETA and yonR primers is D. Zeigler.
DNA extractions and transformation
Chromosomal DNA was extracted from donor strains and used to transform wild-type and mutant B. subtilis cells according to standard procedures (Cutting, 1990). These procedures were modified appropriately for transformation of B. subtilis strains with plasmid DNA that was extracted from Escherichia coli cells by alkaline lysis (Sambrook, 1996). When constructing the strains included in Table 1, appropriate integration in the selected transformants was verified using single-colony polymerase chain reaction (PCR) (Estacio et al., 1998).
To determine whether disruption of yxlC is essential in the wild type, but not the sigY mutant, these strains were transformed with a disruptional plasmid for yxlC (pCLP03). This plasmid contains an yxlC::cat disruptional cassette for the gene that encodes the putative antisigma factor for SigY. Ten micrograms of pCLP03 was used in each transformation, and the results were recorded as number of chloramphenicol resistant transformants normalized to total number of viable cells. In control reactions the ability of the wild-type and sigY cells to become competent and take up DNA was verified. For these control reactions 1 ng of chromosomal DNA from LMB10 was used. The DNA from LMB10 contains a sigD::cat cassette that disrupts the sigD gene (a gene unrelated to SigY function). The results of these control reactions were recorded as number of chromosomal resistance transformants per 1 ng of chromosomal DNA, normalized to total number of viable cells.
Conditioned cell-free media for killing assay
Cell-free media (CFM) were prepared by growing cells in 100 mL of 2×SG at 37°C with constant shaking (300 rpm) until reaching T1 (1 h after reaching OD600=1.2, T0). In 2×SG rich sporulation medium the break from logarithmic growth (i.e., T0) is achieved at an OD600 of 1.2. Time following this initiation of nutrient stress is marked from this breakpoint with T1 marking early events in sporulation, and T2 marking commitment to sporulation.
Cells were removed by centrifugation and the exhausted media were filtered using a 0.22 μm filter apparatus (Millipore). To enrich for antibiotic peptide the CFM was subjected to boiling and stored at −20°C before use in killing assays. These treatments denature proteins in the exhausted media that may contribute to cell death.
Killing assay
The four tester strains used were LMB7 wild-type, LMB276 sigY null, CU1065 wild-type that lacks Spβ, and CU1065 that additionally bears the sigY null mutation. Single-colony isolates from overnight plates were inoculated into 5 mL prewarmed 2×SG LB broth with appropriate antibiotic if necessary. The culture was incubated in an environmental shaker with 3000 rpm agitation for aeration until an OD600 of 0.8–1.0 (∼2 h) was reached. Cells were then collected by centrifugation and the exhausted medium was decanted. The tester strains were then resuspended in 5 mL of each CFM to an OD600 of 0.5 maintained at 37°C. Resuspended cells were incubated for 1 h in an environmental shaker at 37°C and 300 rpm. Aliquots of each culture were then serially diluted in prewarmed spizizen minimal salts (SMS) and plated in triplicate on LB plates with the appropriate antibiotic. Per liter SMS is 2 g (NH4)2SO4, 14 g K2HPO4, 6 g KH2PO4, 1 g Na3C6H5O7⊕2H2O, and 0.2 g MnSO4⊕7H2O. The number of surviving cells was determined by counting colony forming units following overnight incubation, and multiplying this number by the appropriate dilution factor. To obtain the number of cells without CFM addition (i.e., No CFM) tester strains were resuspended in SMS instead of CFM then serially diluted, plated, and counted.
PCR for detection of presence or absence of Spβ
All primers used in this study are listed in Table 1. The yonR, BETA, and sigD primers were designed by Dr. Daniel Zeigler, Dr. Leticia Márquez-Magaña, and Christy Perez, respectively.
To check for the presence of the intact Spβ prophage in wild-type and mutant strains, PCR was used to detect the yonR target gene located within Spβ. Excision of Spβ from the B. subtilis chromosome was monitored using BETA primers that amplify across the Spβ attachment site. Primers that amplify an unrelated gene, sigD (Marquez et al., 1990), were used in positive control reactions to demonstrate that the lack of a product in yonR or BETA reactions was due to lack of target and not inappropriate reaction conditions. All PCRs were carried out using a GoTaq Flexi DNA polymerase kit. PCR was carried out under the following conditions: 100 ng/μL of DNA template, 5 μL of 5×Green GoTaq Flexi Buffer, 2.5 μL of dNTP, 1.25 μL of reverse and forward primers, 1.25 μL of MgCl2, and 0.25 μL Taq DNA polymerase. PCR amplification was performed in an Eppendorf gradient cycler, using the program set to denature at 95°C for 5 min, and then to denature at 94°C for 1 min, anneal at 53°C for 45 s, and extend at 72°C for 1 min for a total of 30 cycles, with a final extension of 72°C for 1 min.
Plate and quantitative sporulation assays for timing of sporulation
Plates were prepared as previously described (González-Pastor et al., 2003). Test strains were streaked in duplicate onto plates and incubated at 37°C, and the timing of sporulation was recorded by digital camera. Sporulation is evidenced by increased opaqueness, whereas the lack of sporulation is demonstrated by translucent bacterial growth. Cells bearing a mutation in skfA were used as a positive control for accelerated sporulation, and spo0A cells were used as a negative control for sporulation.
The percent sporulation at 8, 12, 17, 21, and 24 h was determined by quantitative sporulation assay as previously described (González-Pastor et al., 2003) with the following modifications. Cells were directly collected from the sporulation plates as 10 mm plugs. Additionally, 50 μL of chloroform was used to eliminate nonsporulating cells instead of heat treatment.
Results
SigY and Spβ are required for production of and resistance to a killing factor
The known function of ECF sigmas in B. subtilis in antibiotic production and resistance (Helmann, 2002), coupled with a similarity between the sigY and skf operons, caused us to consider a role for SigY in the control of killing factor production and resistance. The sigY operon contains the genes for SigY and five proteins of unknown function encoded by yxlC, yxlD, yxlE, and yxlF (Cao et al., 2003). Two of these genes were found to be similar to genes in the skf operon. The skf operon contains the gene for the sporulation killing factor Skf and seven proteins encoded by skfB, skfC, skfD, skfE, skfF, skfG, and skfH. A subset of these proteins is required to produce the mature Skf toxin, and to provide cellular resistance by eliminating Skf from the cell. The skfE and skfF genes appear to form an ABC transporter required for export and resistance to the sporulation killing factor that is an antibiotic peptide encoded by skfA (Gonzalez-Pastor et al., 2003). A pair-wise comparison of the predicted protein products encoded in the sigY and skf operons demonstrated a 66% similarity between YxlF and SkfE that spans the entire SkfE sequence, and identified conserved motifs shared by YxlG and SkfF (data not shown). Given that yxlF and yxlG are transcribed by the SigY-holoenzyme because of their location in the autoregulated sigY operon (Cao et al., 2003) we tested whether SigY is involved in antibiotic production and resistance.
Killing assays were performed to test whether or not SigY plays a role in producing and conferring resistance to an antibiotic. In these assays CFM from either the wild-type strain or the isogenic sigY mutant that lacks SigY activity (see Table 1) was collected following nutrient stress. CFM was collected 1 h after the break from logarithmic growth in rich medium because sigY expression was found to be maximal at the end of logarithmic growth in this medium (data not shown). CFM was also collected from a wild-type strain that lacks Spβ (CU1065). The collected CFM was tested for killing activity by using it to resuspend four different strains: the wild-type strain (LMB7), the sigY mutant (LMB276), a wild-type strain that lacks Spβ (CU1065), and an isogenic derivative that lacks both Spβ and SigY (CU1065 sigY). The latter two strains were included because the work of others demonstrated that the role of the ECF sigma factor SigW in antibiotic resistance is masked in a strain bearing the Spβ prophage (Butcher and Helmann, 2006). The Spβ prophage contains genes necessary for synthesis and export of the antibiotic sublancin, which is expressed at high levels and masks the expression of other antibiotics and resistance proteins expressed at low levels. Thus, inclusion of the strains lacking Spβ in the killing assay was predicted to unmask the role of SigY in the production of antibiotics that are produced at low levels (like other ECF sigmas in B. subtilis), but instead demonstrated the role of both SigY and Spβ in the production of and resistance to a killing factor.
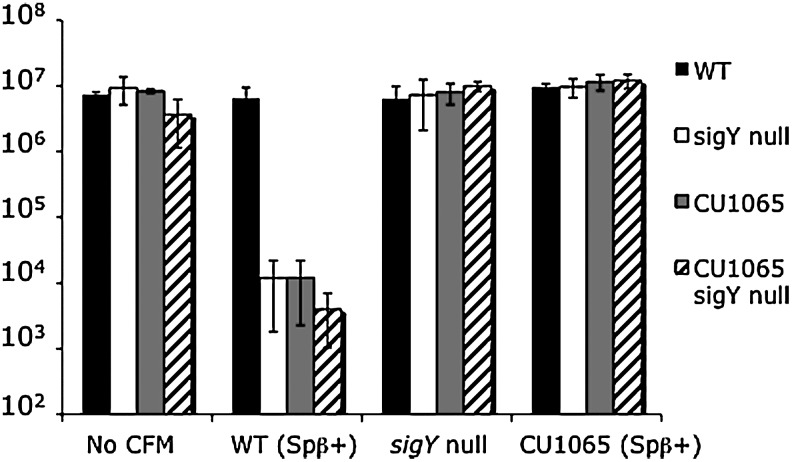
The cell survival results shown in Figure 1 demonstrate that only CFM from a strain that contains both SigY and Spβ is capable of killing sensitive strains that lack a functional sigY gene and/or the Spβ prophage. CFM from the wild-type strain that contains Spβ (Spβ+) led to significant killing of sensitive strains: only 1:10,000 cells survive treatment with this CFM. Conversely, CFM from strains that lack a functional sigY gene (sigY) and/or the Spβ prophage (Spβ−) failed to cause significant killing: the number of surviving cells is comparable with the number of viable cells determined in the absence of CFM treatment (i.e., No CFM). Further, CFM from the wild-type strain that maintains a functional sigY and the Spβ (Spβ+) is only capable of killing strains that lack sigY and/or the Spβ prophage. Thus, resistance to the killing factor found in this CFM requires both SigY and Spβ. Taken together, our results show that both SigY and Spβ are necessary to produce and confer resistance to a killing factor found in the medium of growing cells subjected to nutrient stress.
FIG. 1.
Results of killing assays in the absence of CFM (i.e., No CFM) or CFM obtained from wild-type strain containing the Spβ prophage, WT(Spβ+); sigY null; or different wild-type strain that lacks the prophage, CU1065 (Spβ−). Source of CFM is listed on the x-axis, and number of cells surviving treatment is given on the y-axis. Survival of WT that contains Spβ (black bars), sigY null (white bars), CU1065 that lacks Spβ (gray bars), and CU1065 that lacks both Spβ and sigY (hatched bars) following 1 h incubation in the indicated CFM. The results presented are an average of three experiments, and the standard error is presented. CFM, cell-free media; WT, wild type.
Loss of Spβ in SigY mutant and wild-type strain
Given that both SigY and Spβ are required for the killing activity shown in Figure 1, which is presumably due to production of mature sublancin, we studied the sunA gene that encodes for the precursor of sublancin in the sigY mutant. Unexpectedly, we found that this gene was absent in the sigY null mutant (Perez, 2006). The sunA gene is located on the Spβ prophage (Paik et al., 1998) so the absence of sunA in the sigY null led us to propose that the prophage is missing in this strain.
To test the postulate that Spβ is missing in the sigY null mutant used in the killing assays, LMB276, PCR was used to detect whether or not the insertion site for the prophage was empty or occupied. The BETA primers amplify a 1 kb fragment of DNA if the prophage is absent, and fail to generate a product if the 135 kb Spβ genome is inserted. Conversely, the yonR primers amplify a 0.5 kb fragment if the prophage is present in the bacterial genome because they are complementary to the yonR gene that resides on Spβ. Absence of product using these primers indicates loss of the prophage or inappropriate reaction conditions for successful PCR. To demonstrate that the reaction conditions were appropriate to support successful PCR, a positive control was used. Primers specific to the sigD gene that is unrelated to SigY function were used in this positive control.
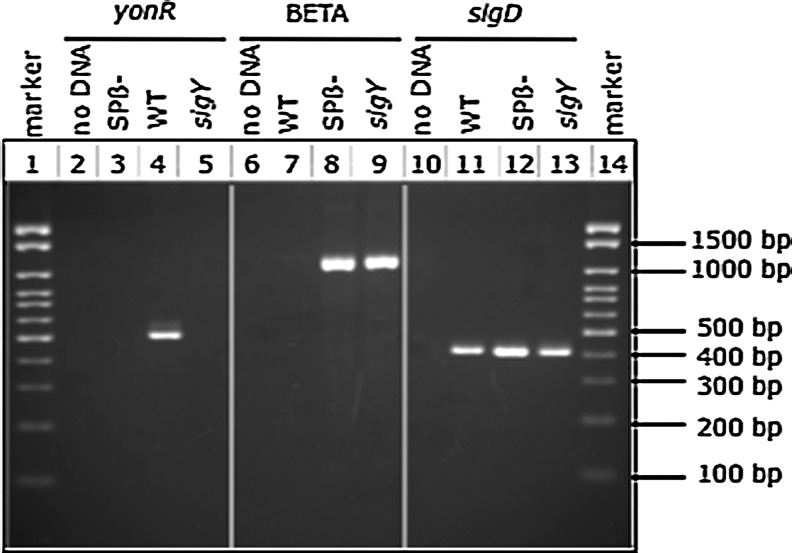
The results shown in Figure 2 demonstrate that the Spβ prophage is missing in the sigY null mutant used in the killing assays. PCR using template DNA extracted from this strain fails to yield the 0.5 kb yonR product, but supports amplification of the 1.0 kb product using the BETA primers. Conversely, the wild-type strain retains Spβ. DNA extracted from this strain yields the 0.5 kb yonR product, and fails to amplify the 1.0 kb product indicative of an empty insertion site. The positive control for PCR amplification, sigD, is amplified using DNA from all strains, demonstrating that the absence of product is not attributable to inappropriate reaction conditions. Additional controls show no amplification in the absence of DNA, and show amplification using the BETA primers only in strains known to lack the prophage (i.e., CU1065).
FIG. 2.
Presence or absence of Spβ in a strain lacking the prophage (CU1065; Spβ−), wild-type (LMB7; WT), and sigY mutant (LMB276) as determined by PCR using yonR or BETA primers, respectively. Lanes 2–5, products obtained using yonR primers that amplify part of the yonR gene on the prophage and template DNA from control and test strains; lanes 6–9, products obtained with BETA primers that amplify across the Spβ attachment site; lanes 10–13, products obtained using primers that amplify a region of the sigD gene as a positive control for PCR amplification. Molecular weights of some DNA markers in lanes 1 and 14 are provided. PCR, polymerase chain reaction.
Having found that Spβ is missing in the sigY null mutant used in the killing assays, we sought to determine how quickly the prophage is lost when the sigY mutation is introduced by plasmid transformation. The disruptional plasmid pCLP03 was transformed into wild-type cells. PCR amplification with yonR and BETA primers was then used to monitor the presence or absence of the prophage in the transformants. This analysis showed that 83% of the transformants studied (5/6) lacked the Spβ, and that 17% (1/6) retained the prophage. Additional studies find that on average 75% of wild-type cells transformed with pCLP03 lose the prophage when analyzed immediately following transformation, and that those that retain Spβ often lose the prophage after multiple rounds of growth (data not shown). However, we were unable to detect loss of the Spβ detected in the original transformant (i.e., 1/6).
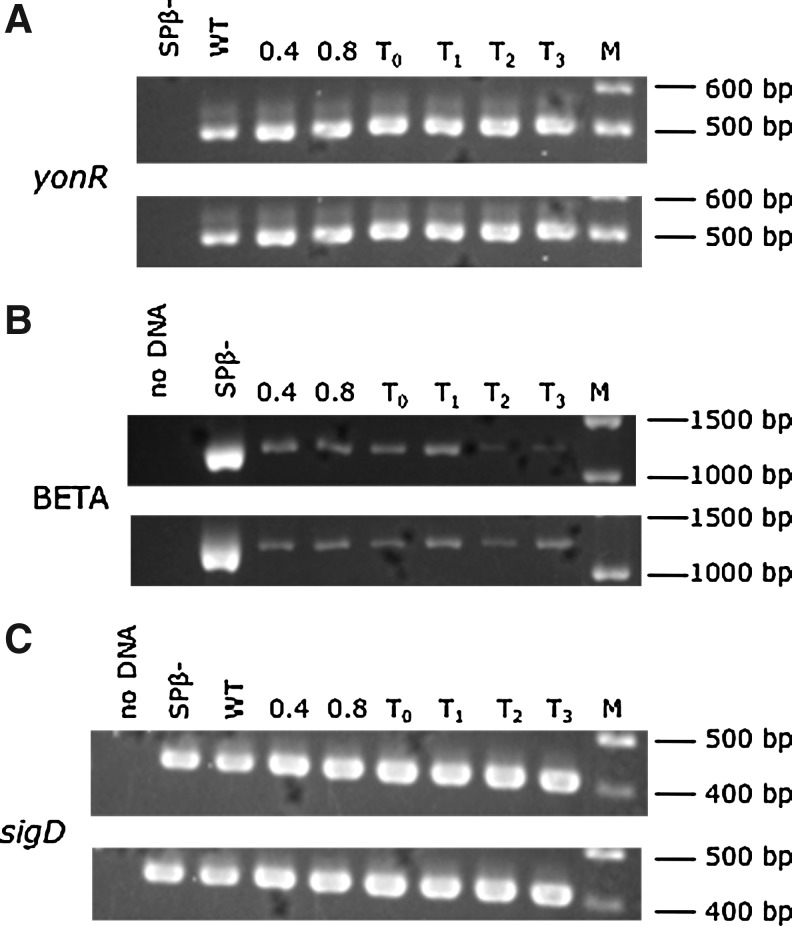
To monitor loss of Spβ in the original transformant (i.e., 1/6) during growth, the strain was grown in rich medium and cells were collected during logarithmic growth, and following the initiation of nutrient stress (i.e., T0). Wild-type cells were similarly grown and cells were collected at the identical time points. The yonR and BETA primers were used to monitor the presence and absence of the Spβ prophage, respectively, and primers complementary to sigD were used as a positive control for amplification. The results presented in Figure 3 show that cells with the newly inserted sigY mutation (lower panel for each target shown) retain Spβ as evidenced by the amplification of yonR. However, a subpopulation of these cells lose the prophage as demonstrated by amplification using the BETA primers. This strain was found to retain the prophage after additional rounds of growth, and is predicted to contain a suppressor mutation that has not been characterized. This strain has been designated LMB276S.
FIG. 3.
Presence or absence of Spβ throughout growth in the wild-type (top panel), or a sigY null mutant strain, LMB276S, that retains the prophage likely due to a suppressor mutation (bottom panel). (A) PCR products obtained using yonR primers, (B) products obtained using BETA primers, and (C) products obtained using sigD primers as a positive control for amplification. The first two lanes in each panel contain the results of negative and positive control reactions for each of the primer sets used in the PCR.
More importantly, the results obtained using DNA from the wild-type strain show that the loss of Spβ in a subpopulation of cells is apparently a normal process. In fact, Dr. Daniel Zeigler who is the curator of the Bacillus Genetic Stock Center finds that subpopulations of other wild-type strains similarly lose the prophage (Zeigler, personal communication). Our results suggest that in wild-type cells Spβ is lost at greater amounts during logarithmic growth and exhibits the greatest levels of loss at T1 (Fig. 3, upper panel of BETA amplification) when we have found SigY-dependent killing activity to be maximal (Perez, 2006). However, the results presented in Figure 3 are not quantitative. Nonetheless, it appears that the pattern of Spβ loss found in the wild-type strain is absent in the sigY mutant LMB276S.
Negative regulation of SigY activity appears to be essential
Our studies demonstrating that subpopulations of wild-type cells either lack or retain Spβ during growth (Fig. 3), coupled with the finding that cells lacking the prophage are sensitive to killing by cells with Spβ (Fig. 1), suggest that the subpopulation lacking the prophage is killed during growth. This cell death phenotype is likely subjected to complex regulation, and we sought to better understand regulation of this process by studying the putative antisigma factor for SigY. The paradigm for ECF sigma regulation dictates that the antisigma negatively controls ECF sigma activity and is encoded by the gene immediately downstream of the structural gene for the ECF sigma (Missiakas and Raina, 1998; Helmann, 2002). The gene immediately downstream of sigY is yxlC.
Disruption of yxlC in our wild-type strain had been very difficult and three independent strategies failed before we were able to replace the coding sequence of yxlC with an erythromycin cassette in collaboration with the Helmann Laboratory. SigY expression was not upregulated in this yxlC mutant and we concluded that YxlC is not the antisigma for SigY (Cao et al., 2003). However, in subsequent work, YxlC was found to bind to SigY in a yeast two-hybrid study, supporting its predicted role as the antisigma factor (Yoshimura et al., 2004). Further, yxlC was found to be essential for growth in one study (Tojo et al., 2003), but was not determined to be an essential gene in a comprehensive analysis of these genes in B. subtilis (Kobayashi et al., 2003).
As a first attempt toward testing the role of the putative antisigma factor YxlC in SigY-dependent killing and Spβ maintenance, we studied the nature of the yxlC::mls mutation described in our published work (Cao et al., 2003). The lack of increased SigY activity in the strain bearing this mutation, HB0915, had caused us to conclude that YxlC is not the antisigma factor. We now considered the possibility that the yxlC::mls mutation that had been so difficult to integrate in HB0915 was made possible by a suppressor mutation in sigY. The sequence of sigY in the yxlC::mls strain was obtained and compared with the sequence of sigY found in a wild-type strain. A deletion of eight nucleotides was found near the middle of the sigY open-reading frame leading to a frame-shift mutation (Perez, 2006). This result explains the lack of increased SigY activity in HB0915 and it has caused us to reconsider the role of YxlC as the antisigma factor for SigY. It also supports the postulate that yxlC is an essential gene in B. subtilis.
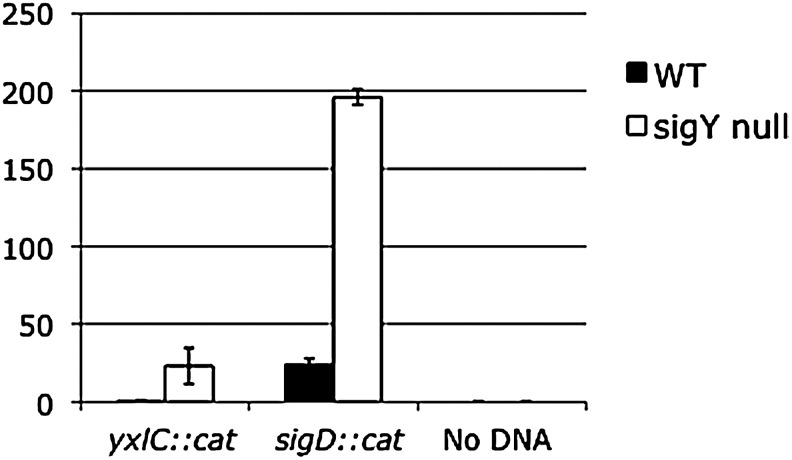
While others concluded that yxlC is essential due to their inability to disrupt the gene, these investigators failed to test whether the lethal phenotype was dependent on the presence of a functional copy of sigY (Tojo et al., 2003). Further, the suppressor mutation found in the sigY gene of HB0915 is located in a strain that lacks Spβ. Therefore, we sought to test the postulate that yxlC is essential only in the presence of a functional sigY gene in our wild-type strain that contains Spβ. We tested this prediction by attempting to transform wild-type and sigY null mutant strains with a disruptional plasmid for yxlC (pCLP03). If yxlC is essential only in the presence of a functional sigY gene, then its disruption would only be possible in the sigY null mutant and pCLP03 transformants obtained only in this strain. In parallel transformations, wild-type and sigY null mutant cells were made competent and the disruptional plasmid pCLP03 was introduced. Chloramphenicol-resistant transformants were obtained in the sigY null mutant (Fig. 4), but only one transformant was obtained in the wild-type strain and is suspected to contain a suppressor mutation in sigY. The virtual lack of pCLP03 transformants in the wild-type strain is not due to a lack of competence. In control transformations using chromosomal DNA from LMB10 that contains a chloramphenicol-resistant cassette integrated at an unrelated gene (sigD::cat), the wild-type strain was found to be competent to take up this DNA. Therefore, our inability to disrupt the yxlC gene in the wild-type strain supports our postulate that it is essential in the presence of a functional copy of sigY. This result also supports the speculation that inappropriate upregulation of SigY activity in the absence of YxlC leads to cell death.
FIG. 4.
Relative number of transformants obtained for wild-type and sigY mutant strains following introduction of a plasmid that disrupts yxlC and introduces the gene for choramphenicol resistance, yxl::cat. Transformants obtained for the wild-type strain (black bars) and a sigY null mutant (white bars). Chromosomal DNA from a strain bearing a plasmid insertion that disrupts sigD and introduces the gene for chloramphenicol resistance, sigD::cat, was used as a positive control for transformation, and no DNA was added in the negative control. The results for three transformations using 10 μg plasmid and 1 ng chromosomal DNA are presented along with the standard error. The number of transformants obtained for each amount of DNA has been normalized to total number of viable cells to obtain the relative number of transformants. The relative number of transformants is presented on the y-axis.
Loss of SigY leads to sporulation delay
While the apparent negative regulation of SigY by its putative antisigma factor appears to be essential for overall survival, we sought to determine the importance of SigY-dependent killing in sporulation. In previous work by others it had been demonstrated that killing of sibling cells, through a process these investigators termed cannibalism, delayed sporulation (González-Pastor et al., 2003). It was found and later confirmed that a subpopulation of cells fail to produce, and are sensitive to killing by, the SkfA and SdpC factors exported by sibling cells (Ellermeier et al., 2006; González-Pastor, 2011). This results in the killing of the subpopulation of cells incapable of producing these antibiotics, and the release of additional nutrients that delay sporulation. We suspected that a similar phenomenon is found in wild-type cells that exhibit subpopulations of cells that retain or lose Spβ (Fig. 3), and is absent in sigY mutant cells that lack Spβ (Fig. 2). Thus, we predicted that sporulation of the sigY null mutant that lacks the prophage (Fig. 2) and the associated killing activity (Fig. 1) would be accelerated with respect to wild-type cells due to their inability to obtain additional nutrients from the killing of sibling cells.
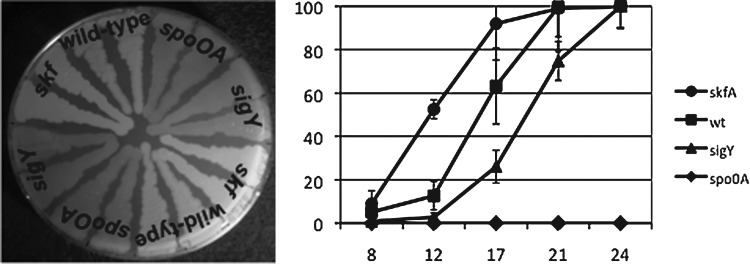
The results of the sporulation assays presented in Figure 5 show that sporulation is delayed in the sigY null mutant compared with sporulation in the wild-type strain. This result is inconsistent with our prediction, and does not align with the results obtained with the skf mutant. As first shown by others (González-Pastor et al., 2003), the skf mutant exhibited accelerated sporulation. At 17 h the skf strain displays opaque growth on sporulation plates (Fig. 5, left panel) indicative of efficient sporulation, and an average sporulation rate of 92% (Fig. 5, right panel). At this time the wild-type strain also displays opaque growth, but a lower average sporulation rate of 63%. At 17 h the sigY mutant has not efficiently sporulated. The growth of the sigY strain on sporulation plates is less opaque than skf and wild-type strains (Fig. 5, left panel), and the average sporulation rate is 26% (Fig. 5, right panel). Further, the results of the quantitative sporulation assay (Fig. 5, right panel) show that the skf strain achieves >90% sporulation in 17 h, the wild-type strain at 21 h, and the sigY mutant at 24 h. The negative control for sporulation, the spo0A strain, fails to sporulate. Thus, the loss of SigY and Spβ in the sigY mutant delays sporulation, suggesting that one or both play a role in the timely initiation of this process.
FIG. 5.
Results of plate and quantitative sporulation assays to monitor the timing of sporulation. Left panel, photograph of a plate following 17 h of growth. Wild-type and mutant cells are included as duplicates and are labeled. Right panel, results of a quantitative sporulation assay for the four strains included in the plate assay. The percentage of chloroform-resistant spores versus total viable cells is presented on the y-axis. The percent sporulation is provided following growth on sporulation plates presented in hours on the x-axis.
Discussion
Our work demonstrates that SigY is required to efficiently maintain Spβ in the bacterial genome, thereby playing a critical role in the ability of B. subtilis to produce and shield itself from antibiotic(s) encoded in the genome of the bacteriophage. The sunA gene found in the Spβ prophage encodes for the antibiotic peptide sublancin, which is a primary mechanism for killing by B. subtilis (Paik et al., 1998; Dorenbos et al., 2002; Butcher and Helmann, 2006). The killing observed by the wild-type strain that contains both SigY and Spβ (Fig. 1) is likely due to the production of this antibiotic peptide. Antibiotic peptides are resistant to boiling and the CFM was boiled prior to use in the killing assays. Additionally, CFM collected 1 h after the initiation of nutrient stress from a mutant bearing an insertion deletion of the sunA gene displays the identical loss of killing activity that is found for the sigY mutant (data not shown). This result could be explained by SigY control of sunA transcription initiation. However, this gene was not identified as a target for SigY in previous work (Asai et al., 2003; Cao et al., 2003). Moreover, primer extension analysis of mRNA from cells subjected to nutrient stress that upregulates sigY failed to identify a SigY-dependent promoter upstream of sunA (data not shown).
Other ECF sigmas have been found to initiate transcription of the operon that contains sunA. The sunA gene is part of the sublancin operon that is subjected to complex regulation that includes the apparent existence of multiple promoters, at least one of which is recognized by the ECF sigmas, SigX and SigM (Paik et al., 1998; Luo and Helmann, 2009; Luo et al., 2010). Consistent with this finding a mutant that lacks SigX and SigM exhibits reduced sublancin production, but a mutant that lacks all seven ECF sigmas displays an even greater reduction in the production of the antibiotic (Luo et al., 2010). Our results show that this loss of sublancin production is likely caused by excision of the Spβ prophage in a sigY mutant (Fig. 2). Interestingly, in the strain that lacks all seven ECF sigmas, the conjugative transposon ICEBs1 was excised (Luo et al., 2010). Thus, loss of individual and collective ECF sigma function appears to lead to excision of foreign DNA (i.e., the Spβ prophage and conjugative transposon ICEBs1). While it is not clear why the transposon is excised in the strain that bears deletions of all seven ECF sigmas, it appears that excision of Spβ in the sigY mutant is part of an overall response to nutrient stress.
In response to nutrient stress B. subtilis cells that maintain Spβ produce antibiotic peptides, primarily sublancin, that kill competing bacteria and generate additional resources (Marahiel et al., 1993; Stein, 2005). However, loss of SigY leads to loss of Spβ (Fig. 2) and the concomitant loss of SigY-dependent killing (Fig. 1). The loss of Spβ is not limited to cells with sigY mutations. In a wild-type strain we find loss of this prophage in a subpopulation of cells growing in rich medium (Fig. 3). Given that expression of sigY is maximal at the end of logarithmic growth in this medium (data not shown), the loss of Spβ in a subpopulation of wild-type cells that begins during this period of growth is consistent with a role for SigY in the developmental excision of the prophage. We speculate that in response to nutrient stress, SigY is differentially activated in subpopulations of cells in the growing culture. This leads to an overall increase in sigY expression, and to bistable outcomes for sibling cells capable of killing and of being killed. Cells having sufficient levels of activated SigY maintain the Spβ prophage and are capable of killing sibling cells by producing sublancin. Cells having insufficient levels of active SigY and/or mutations in sigY lose the prophage (Fig. 2), fail to produce killing factor, and are sensitive to killing (Fig. 1). In effect, sibling cells incapable of producing killing factor in response to nutrient stress are “damaged” and the population is ridded of them. This process can be likened to a programmed cell death response in higher eukaryotes where the ingestion of dying plays an important role in energy recycling, and rids the organism of damaged or detrimental cells (Ameisen, 1996, 2002).
Likewise, the Spo0A-controlled production of the Skf and SdpC killing factors in B. subtilis allows for the nutritional recycling of cells unable to contribute to the overall survival of the bacterial population due to their inability to sporulate. Cells lacking adequate levels of active Spo0A are incapable of sporulating, and are sensitive to killing by these factors. Thus, death of this subpopulation of cells improves the overall survival of the remaining cells by providing additional nutrients that delay sporulation (González-Pastor, et al., 2003; González-Pastor, 2011).
While our results suggest that a subpopulation of cells that lose Spβ can provide a similar nutritional resource, we failed to identify a sporulation delay in wild-type cells compared with a sigY mutant strain. Wild-type cells with normal SigY activity effectively sporulate in 21 h, compared with 24 h for the strain that lacks SigY (Fig. 5). The delay of sporulation in the sigY compared with the wild-type strain suggests that in addition to nutritional resources, subpopulations of dying cells in wild-type cultures may release another factor(s) that promotes sporulation. We speculate that killing by the SigY-dependent killing factor leads to the release of a sporulation-inducing pheromone(s) (Perego and Hoch, 1996; Lazazzera et al., 1997). These pheromones are secreted into the extracellular medium at the end of logarithmic growth (Grossman and Losick, 1988), and act early in sporulation (Grossman, 1995). We found that the SigY-dependent killing factor is most active in CFM collected 1 h after the break from logarithmic growth consistent with this speculation, whereas Skf killing is most active 2 h after the break from logarithmic growth (Perez, 2006). Further, we saw no difference in the timing of sporulation in the CU1065 strains that do not harbor the Spβ, but that either retain normal SigY activity or lack it due to a null mutation in sigY (data not shown). Thus, normal sporulation in the wild-type strain (Fig. 5) requires both SigY and Spβ, suggesting that in strains bearing the prophage the death of a subpopulation of cells promotes normal timing of this process. However, future work is necessary to demonstrate the death of the subpopulation that loses Spβ in growing cultures of the wild-type strain (Fig. 3).
Our work demonstrates that the SigY-dependent loss of Spβ fails to delay sporulation, but control of this process appears to be essential for cell survival. This conclusion is based on the inability to disrupt the yxlC gene for the putative antisigma of SigY. Others have suggested that yxlC is essential for growth due to their inability to disrupt the gene (Tojo et al., 2003). However, in our work we found yxlC to be essential only in the presence of a functional copy of sigY (Fig. 5). Therefore, it is the negative regulation of SigY by YxlC that is necessary for cell survival. This appears to be the case both in the presence and absence of Spβ. The inability to disrupt yxlC in this study was monitored in a wild-type strain containing the prophage. In previous work, however, it was reported that yxlC had been successfully disrupted in a strain that lacks Spβ (Cao et al., 2003). We later found a deletion of eight nucleotides near the middle of the sigY open-reading frame that results in a frame-shift mutation (Perez, 2006). The identification of this suppressor mutation further supports the postulate that negative regulation of SigY is necessary for cell survival. Given that this negative regulation appears to be essential both in the presence and absence of Spβ and that SigY is one of the seven ECF sigmas implicated in the production and resistance to antibiotics (Luo et al., 2010), we propose that YxlC plays a critical role in the timing of killing and resistance functions. Perhaps, YxlC negatively regulates SigY until the necessary resistance proteins are expressed to protect the cell from killing by SigY-dependent gene products.
Conclusions
Overall, our work implicates the SigY ECF sigma factor of B. subtilis in the production of and resistance to sublancin at the single-cell level. It suggests that differences in SigY activity lead to loss or maintenance of the Spβ genome that encodes proteins responsible for these functions in individual cells within a growing population. Loss or maintenance of the prophage in a growing population of cells is expected to give rise to subpopulations capable of killing and of being killed. Sibling cells that lose Spβ presumably due to reduced SigY activity are likely killed due to their inability to produce these proteins. This selective pressure is predicted to maintain Spβ in the genome of cells having sufficient SigY activity, and to eliminate cells with insufficient activity. In this way a subpopulation of cells with insufficient SigY activity are killed and may release factors that promote sporulation. This speculation supports our finding that a sigY mutant is delayed in sporulation. We fail to find detectable levels of Spβ in this sigY mutant rendering it incapable of producing subpopulations of cells capable of killing and of being killed. We further show an inability to disrupt the gene for the putative antisigma factor for SigY in a wild-type strain. This result suggests that loss of SigY regulation is detrimental to the cell. Future work is aimed at determining the essential role SigY regulation plays in B. subtilis, and examining the mechanism and outcomes of the differential activation of SigY in subpopulations of growing cells predicted by this study.
Acknowledgments
The authors thank Drs. José Eduardo González-Pastor and John Helmann for their review of very early drafts of this article. Further, we appreciate the guidance of Dr. Daniel Zeigler in better understanding and studying loss of Spβ in wild-type strains, and the suggestion of Dr. Chester Price to search for a suppressor mutation in sigY in the yxlC strain generated in previous work. NIH MBRS SCORE S06 GM52588 and NSF RUI MCB 0519482 grants to L.M.-M. supported this work, as well as NIH MBRS RISE GM5-59298 undergraduate fellowship to R.M. and master's level fellowships to A.G. and J.R. R.M. was also awarded undergraduate funding as a Beckman Scholar.
Disclosure Statement
No competing financial interests exist.
References
- Ameisen J.C. The origin of programmed cell death. Science. 1996;272:1278–1279. doi: 10.1126/science.272.5266.1278. [DOI] [PubMed] [Google Scholar]
- Ameisen J.C. On the origin, evolution, and nature of programmed cell death: a timeline of four billion years. Cell Death Differ. 2002;9:367–393. doi: 10.1038/sj.cdd.4400950. [DOI] [PubMed] [Google Scholar]
- Asai K. Yamaguchi H. Kang C.M. Yoshida K. Fujita Y. Sadaie Y. DNA microarray analysis of Bacillus subtilis sigma factors of extracytoplasmic function family. FEMS Microbiol Lett. 2003;220:155–160. doi: 10.1016/S0378-1097(03)00093-4. [DOI] [PubMed] [Google Scholar]
- Butcher B.G. Helmann J.D. Identification of Bacillus subtilis sigma-dependent genes that provide intrinsic resistance to antimicrobial compounds produced by Bacilli. Mol Microbiol. 2006;60:765–782. doi: 10.1111/j.1365-2958.2006.05131.x. [DOI] [PubMed] [Google Scholar]
- Cao M. Helmann J.D. Regulation of the Bacillus subtilis bcrC bacitracin resistance gene by two extracytoplasmic function sigma factors. J Bacteriol. 2002;184:6123–6129. doi: 10.1128/JB.184.22.6123-6129.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao M. Helmann J.D. The Bacillus subtilis extracytoplasmic-function sigmaX factor regulates modification of the cell envelope and resistance to cationic antimicrobial peptides. J Bacteriol. 2004;186:1136–1146. doi: 10.1128/JB.186.4.1136-1146.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao M. Salzberg L. Tsai C.S. Mascher T. Bonilla C. Wang T. Ye R.W. Marquez-Magana L. Helmann J.D. Regulation of the Bacillus subtilis extracytoplasmic function protein sigma(Y) and its target promoters. J Bacteriol. 2003;185:4883–4890. doi: 10.1128/JB.185.16.4883-4890.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao M. Wang T. Ye R. Helmann J.D. Antibiotics that inhibit cell wall biosynthesis induce expression of the Bacillus subtilis sigma(W) and sigma(M) regulons. Mol Microbiol. 2002;45:1267–1276. doi: 10.1046/j.1365-2958.2002.03050.x. [DOI] [PubMed] [Google Scholar]
- Cutting S.M. Genetic analysis. In: Hardwood C.R., editor; Cutting S.M., editor. Molecular Biological Methods for Bacillus. John Wiley & Sons; Chichester: 1990. pp. 27–74. [Google Scholar]
- Dorenbos R. Stein T. Kabel J. Bruand C. Bolhuis A. Bron S. Quax W.J. Van Dijl J.M. Thiol-disulfide oxidoreductases are essential for the production of the lantibiotic sublancin 168. J Biol Chem. 2002;277:16682–16688. doi: 10.1074/jbc.M201158200. [DOI] [PubMed] [Google Scholar]
- Ellermeier C.D. Hobbs E.C. Gonzalez-Pastor J.E. Losick R. A three-protein signaling pathway governing immunity to a bacterial cannibalism toxin. Cell. 2006;124:549–559. doi: 10.1016/j.cell.2005.11.041. [DOI] [PubMed] [Google Scholar]
- Estacio W. Anna-Arriola S.S. Adedipe M. Marquez-Magana L.M. Dual promoters are responsible for transcription initiation of the fla/che operon in Bacillus subtilis. J Bacteriol. 1998;180:3548–3555. doi: 10.1128/jb.180.14.3548-3555.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Pastor J.E. Cannibalism: a social behavior in sporulating Bacillus subtilis. FEMS Microbil Rev. 2011;35:415–424. doi: 10.1111/j.1574-6976.2010.00253.x. [DOI] [PubMed] [Google Scholar]
- González-Pastor J.E. Hobbs E.C. Losick R. Cannibalism by sporulating bacteria. Science. 2003;301:510–513. doi: 10.1126/science.1086462. [DOI] [PubMed] [Google Scholar]
- Grossman A.D. Genetic networks controlling the initiation of sporulation and the development of genetic competence in Bacillus subtilis. Annu Rev Genet. 1995;29:477–508. doi: 10.1146/annurev.ge.29.120195.002401. [DOI] [PubMed] [Google Scholar]
- Grossman A.D. Losick R. Extracellular control of spore formation in Bacillus subtilis. Proc Natl Acad of Sci U S A. 1988;85:4369–4373. doi: 10.1073/pnas.85.12.4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havarstein L.S. Diep D.B. Nes I.F. A family of bacteriocin ABC transporters carry out proteolytic processing of their substrates concomitant with export. Mol Microbiol. 1995;16:229–240. doi: 10.1111/j.1365-2958.1995.tb02295.x. [DOI] [PubMed] [Google Scholar]
- Helmann J.D. The extracytoplasmic function (ECF) sigma factors. Adv Microb Physiol. 2002;46:47–110. doi: 10.1016/s0065-2911(02)46002-x. [DOI] [PubMed] [Google Scholar]
- Helmann J.D. Deciphering a complex genetic regulatory network: the Bacillus subtilis sigmaW protein and intrinsic resistance to antimicrobial compounds. Sci Prog. 2006;89:243–266. doi: 10.3184/003685006783238290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins C.F. ABC transporters: physiology, structure and mechanism—an overview. Res Microbiol. 2001;152:205–210. doi: 10.1016/s0923-2508(01)01193-7. [DOI] [PubMed] [Google Scholar]
- Huang X. Gaballa A. Cao M. Helmann J.D. Identification of target promoters for the Bacillus subtilis extracytoplasmic function sigma factor, sigma W. Mol Microbiol. 1999;31:361–371. doi: 10.1046/j.1365-2958.1999.01180.x. [DOI] [PubMed] [Google Scholar]
- Huang X. Helmann J.D. Identification of target promoters for the Bacillus subtilis sigma X factor using a consensus-directed search. J Mol Biol. 1998;279:165–173. doi: 10.1006/jmbi.1998.1765. [DOI] [PubMed] [Google Scholar]
- Kobayashi K. Ehrlich S.D. Albertini A. Amati G. Andersen K.K. Arnaud M. Asai K., et al. Essential Bacillus subtilis genes. Proc Natl Acad Sci USA. 2003;100:4678–4683. doi: 10.1073/pnas.0730515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazazzera B.A. Solomon J.M. Grossman A.D. An exported peptide functions intracellularly to contribute to cell density signaling in Bacillus subtilis. Cell. 1997;89:917–925. doi: 10.1016/s0092-8674(00)80277-9. [DOI] [PubMed] [Google Scholar]
- Lonetto M.A. Brown K.L. Rudd K.E. Buttner M.J. Analysis of the Streptomyces coelicolor sigE gene reveals the existence of a subfamily of eubacterial RNA polymerase sigma factors involved in the regulation of extracytoplasmic functions. Proc Natl Acad Sci U S A. 1994;91:7573–7577. doi: 10.1073/pnas.91.16.7573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y. Asai K. Sadaie Y. Helmann J.D. Transcriptomic and phenotypic characterization of Bacillus subtilis strain without extracytoplasmic function sigma factors. J Bacteriol. 2010;192:5736–5745. doi: 10.1128/JB.00826-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y. Helmann J.D. Extracytoplasmic function sigma factors overlapping promoter specificity regulate sublancin production in Bacillus subtilis. J Bacteriol. 2009;191:4951–4958. doi: 10.1128/JB.00549-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marahiel M.A. Nakano M.M. Zuber P. Regulation of peptide antibiotic production in Bacillus. Mol Microbiol. 1993;7:631–636. doi: 10.1111/j.1365-2958.1993.tb01154.x. [DOI] [PubMed] [Google Scholar]
- Marquez L.M. Helmann J.D. Ferrari E. Parker H.M. Ordal G.W. Chamberlin M.J. Studies of sigma D-dependent functions in Bacillus subtilis. J Bacteriol. 1990;172:3435–3443. doi: 10.1128/jb.172.6.3435-3443.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missiakas D. Raina S. The extracytoplasmic function sigma factors: role and regulation. Mol Microbiol. 1998;28:1059–1066. doi: 10.1046/j.1365-2958.1998.00865.x. [DOI] [PubMed] [Google Scholar]
- Paik S.H. Chakicherla A. Hansen J.N. Identification and characterization of the structural and transporter genes for, and the chemical and biological properties of, sublancin 168, a novel lantibiotic produced by Bacillus subtilis 168. J Biol Chem. 1998;273:23134–23142. doi: 10.1074/jbc.273.36.23134. [DOI] [PubMed] [Google Scholar]
- Perego M. Hoch J.A. Cell-cell communication regulates the effects of protein aspartate phosphatases on the phosphorelay controlling development in Bacillus subtilis. Proc Natl Acad Sci U S A. 1996;93:1549–1553. doi: 10.1073/pnas.93.4.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perego M. Hoch J.A. Two-component systems, phosphorelays, and regulation of their activities by phosphatases. In: Sonenshein A.L., editor; Hoch J.A., editor; Losick R., editor. Bacillus subtilis and Its Closest Relatives: From Genes to Cells. American Society for Microbiology; Washington, DC: 2002. pp. 473–481. [Google Scholar]
- Perez C.L. Masters Thesis. San Francisco State University; San Francisco: 2006. The physiological role of the Bacillus subtilis extracytoplasmic function (ECF) sigma factor, Sigma Y. [Google Scholar]
- Pietiainen M. Gardemeister M. Mecklin M. Leskela S. Sarvas M. Kontinen V.P. Cationic antimicrobial peptides elicit a complex stress response in Bacillus subtilis that involves ECF-type sigma factors and two-component signal transduction systems. Microbiology. 2005;151:1577–1592. doi: 10.1099/mic.0.27761-0. [DOI] [PubMed] [Google Scholar]
- Sambrook J.F. Molecular Cloning: A Laboratory Manual. 2nd. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1996. [Google Scholar]
- Stein T. Bacillus subtilis antibiotics: structures, syntheses and specific functions. Mol Microbiol. 2005;56:845–857. doi: 10.1111/j.1365-2958.2005.04587.x. [DOI] [PubMed] [Google Scholar]
- Stragier P. Losick R. Molecular genetics of sporulation in Bacillus subtilis. Annu Rev Genet. 1996;30:297–341. doi: 10.1146/annurev.genet.30.1.297. [DOI] [PubMed] [Google Scholar]
- Tojo S. Matsunaga M. Matsumoto T. Kang C.M. Yamaguchi H. Asai K. Sadaie Y. Yoshida K. Fujita Y. Organization and expression of the Bacillus subtilis sigY operon. J Biochem. 2003;134:935–946. doi: 10.1093/jb/mvg225. [DOI] [PubMed] [Google Scholar]
- Veening J.W. Hamoen L.W. Kuipers O.P. Phosphatases modulate the bistable sporulation gene expression pattern in Bacillus subtilis. Mol Microbiol. 2005;56:1481–1494. doi: 10.1111/j.1365-2958.2005.04659.x. [DOI] [PubMed] [Google Scholar]
- Yoshimura M. Asai K. Sadaie Y. Yoshikawa H. Interaction of Bacillus subtilis extracytoplasmic function (ECF) sigma factors with the N-terminal regions of their potential anti-sigma factors. Microbiology. 2004;150:591–599. doi: 10.1099/mic.0.26712-0. [DOI] [PubMed] [Google Scholar]
- Zahler S.A. Korman R.Z. Rosenthal R. Hemphill H.E. Bacillus subtilis bacteriophage SPbeta: localization of the prophage attachment site, and specialized transduction. J Bacteriol. 1977;129:556–558. doi: 10.1128/jb.129.1.556-558.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]