Abstract
Cytological studies of fluorescent proteins are rapidly yielding insights into chromatin structure and dynamics. Here we describe the production and cytological characterization of new transgenic maize lines expressing a fluorescent histone fusion protein, H2B-mCherry. The transgene is expressed under the control of the maize ubiquitin1 promoter, including its first exon and intron. Polymerase chain reaction–based genotyping and root-tip microscopy showed that most of the lines carrying the transgene also expressed it, producing bright uniform staining of nuclei. Further, plants showing expression in root tips at the seedling stage also showed expression during meiosis, late in the life cycle. Detailed high-resolution three-dimensional imaging of cells and nuclei from various somatic and meiotic cell types showed that H2B-mCherry produced remarkably clear images of chromatin and chromosome fiber morphology, as seen in somatic, male meiotic prophase, and early microgametophyte cells. H2B-mCherry also yielded distinct nucleolus staining and was shown to be compatible with fluorescence in situ hybridization. We found several instances where H2B-mCherry was superior to DAPI as a generalized chromatin stain. Our study establishes these histone H2B-mCherry lines as new biological reagents for visualizing chromatin structure, chromosome morphology, and nuclear dynamics in fixed and living cells in a model plant genetic system.
Introduction
Chromosome research has a rich history in cytology, and advances in optical microscopy together with recent interests in epigenetics and chromatin-templated processes have highlighted the importance of using core nuclear proteins tagged with fluorescent marker proteins (for reviews see Herman, 1998; Pawley, 2006; Allis, 2007; Day and Davidson, 2009; Figueroa and Bass, 2010; Craig and Wong, 2011).
A fundamental structural unit of chromatin is the nucleosome, which is made up of an octamer of histone proteins known as the core particle with 150–200 bp of DNA wrapped around the octamer (Kornberg, 1974). The octamer is composed of two each of the four core histone proteins—H2A, H2B, H3, and H4—which are highly conserved proteins. Nucleosomes are conserved across eukaryotes (reviewed by Kornberg and Lorch, 1999; Andrews and Luger, 2011). Although the fundamental biochemical and genetic properties of plant chromatin are known to be similar to those of animals and fungi (Spiker, 1985), advances in plant chromatin research have been made primarily in the areas of molecular genetics (Chaubet et al., 1986; Koning et al., 1991; Razafimahatratra et al., 1991; Joanin et al., 1992; Chaboute et al., 1993; Sundas et al., 1993; Riggs, 1994; Huh et al., 1995; Reichheld et al., 1995; Kanazin et al., 1996; Okada et al., 2005; Ingouff and Berger, 2010). In contrast, relatively few cytological studies of core nucleosomal histone proteins in plants have been reported.
Fluorescent protein (FP) tags are useful cytological markers for localizing and tracking proteins within fixed or living cells (reviewed by Day and Davidson, 2009). This approach originated more than 50 years ago, when the green fluorescent protein (GFP) was first isolated from the jellyfish Aequorea victoria (Shimomura et al., 1962; reviewed by Shimomura, 2005). Since then, derivatives with different spectral properties and variants from other bioluminescent marine organisms have been characterized and adapted for use as cell markers (Shaner et al., 2004).
Among the most widely used FPs for localization studies in plants are GFP (reviewed by Stewart, 2001; Haseloff and Siemering, 2006), red fluorescent protein (RFP) (Jach et al., 2001), and yellow fluorescent protein (YFP) (Mohanty et al., 2009). The ability to use stable or transient transformation to direct fluorescent fusion proteins to different targets within plant cells has proven useful for plant cell biology (Sparkes et al., 2006; DeBlasio et al., 2010).
Given the recent increased interest in plant chromatin research together with the advances in FP and imaging technologies, we developed a constitutive histone fused to the FP mCherry. The successful development of histone H2B fluorescent fusion proteins in mammalian systems (Kanda et al., 1998; Hadjantonakis and Papaioannou, 2004) prompted us to produce comparable maize lines expressing a histone H2B-mCherry transgene under the control of the maize ubiquitin promoter. The RFP is reported to be among the most photostable of all the FP variants, and one in particular, mCherry, stands out due to its superior photostability and is therefore the preferred variant for cell imaging (Shaner et al., 2004, 2005, 2007; and Day and Davidson, 2009). Here, we describe the production of maize histone H2B-mCherry-expressing plants, the localization of H2B-mCherry in somatic and meiotic cells, and discuss several of their useful properties.
Materials and Methods
Transgene design and plant transformation
We designed the H2B-mCherry fusion protein by fusing the gene for histone H2B.5 (GenBank U08226, UniGene accession Zm.20180) to that for mCherry. The clone was synthesized with maize codon preferences (GenScript USA), and the resulting gene fusion was obtained in plasmid form and subcloned into the T-DNA construct pPTN290 (Howe et al., 2006); GUSPlus was replaced with our histone H2B-mCherry. The resulting plasmid pPTN828, harboring the H2B-mCherry-containing T-DNA construct, was used to transform maize (Plant Transformation Core Research Facility, University of Nebraska-Lincoln, Lincoln, NE). Agrobacterium-mediated transformation of maize was carried out as previously described (Sattarzadeh et al., 2010). T0 plants from neomycin-resistant transformation events were outcrossed as males to inbred B73 females, and the resulting T01 seed was examined by polymerase chain reaction (PCR) and microscopy for the presence and expression of the H2B-mCherry gene. Events (individual lines) were named ZH2B-mC01 through ZH2B-mC18 (e.g., the line ZH2B-mC01 refers to Zea mays histone H2B fused to mCherry in line number 01). Here, these are designated by the shorter suffix-only names, C01–C18.
Polymerase chain reaction
Two primers were designed to detect plants carrying the H2B-mCherry sequence; Tm2_F (forward primer, 5′-GGAGTCCAGACTTCCGTGA-3′), located 81 bp before the end of the histone H2B sequence, and Tm2_R (reverse primer, 5′-AGCTGCATGGCTTCTTAGC-3′), located near the middle of the mCherry sequence. This primer pair was expected to generate a 695-bp PCR product. Control primers for ZmSUN4_F (forward primer, 5′-GCACGGAGTGAGAACACAGA-3′) and ZmSUN4_R1 (reverse primer, 5′-CCTTAGCCCCTTTAGATGCC-3′) and ZmGAPDH_F1 (forward primer, 5′-CCTTGCTCCCCTTGCTAAGG-3′) and ZmGAPDH-R1 (reverse primer, 5′-TGCCACCTCTCCAGTCCTTG-3′) were also used. ZmSUN primers were from SP Murphy (gift, unpublished) and the ZmGAPDH primers were from Brown et al. (2011). Leaf punches were aseptically collected into 1.7-mL microfuge tubes with a sterile hole punch. DNA was prepared from the leaf punches, and 1 μL (of 25 μL/sample) was used for a standard 35-cycle, 30-μL PCR with Platinum PCR SuperMix (Invitrogen). The PCR products from individuals representing eight different events (C03, C05, C06, C07, C08, C09, C10, and C11) were analyzed by agarose gel electrophoresis and visualized by ethidium bromide staining.
Harvesting and fixation of plant tissues for imaging
For the collection of root tissues, kernels were germinated in small plastic pots with maize soil mix (2 parts Fafard 3, 1 part course sand, with high-nitrogen Osmocote) under natural plus supplemental light in a greenhouse (King Life Sciences Building and Mission Road Research Facility, FSU, Tallahassee, FL). Half-inch sections of root tips were cut off and fixed in 3 mL meiocyte buffer A (MBA) with 4% paraformaldehyde for 1 h at room temperature in the dark as described for anther fixation by Howe et al. (2012). Fixed root tips were stored in MBA at 4°C for up to several months before imaging.
For collection of meiotic cells, tassels were harvested before emergence from greenhouse- and field-grown plants. Tissues were kept hydrated on damp paper towels while intact florets were microdissected to yield whole anthers, which were fixed in MBA with formaldehyde exactly as described previously (Howe et al., 2012). All other tissues were fixed in a similar manner and stored at 4°C in the dark for up to 1 year.
Preparation of slides for microscopy
Somatic cells from fixed, microdissected organs were stained with 3 μg/mL DAPI in 1×PBS for 10 min. Meiotic and postmeiotic cells from the fixed anthers were embedded in acrylamide as described previously (Howe et al., 2012). Slides were mounted with VectaShield, covered with 22×30×1.5-mm glass coverslips (Thomas Red Label Micro), sealed with clear nail polish, and stored at −20°C for up to several months.
Three-dimensional acrylamide FISH
Three-dimensional acrylamide FISH was carried out as first described by Bass et al. (1997) and later detailed by Howe et al. (2012). The fluorescent oligonucleotide probes for the detection of telomeres or centromeres have been previously described (Bass et al., 1997; Koumbaris and Bass, 2003). Slides were DAPI stained and mounted as described previously.
Microscopic imaging
For three-dimensional (3D) imaging, iterative deconvolution microscopy (Chen et al., 1995) was used, and the resulting multiple-wavelength sets of 3D data were collected and displayed essentially as described by Howe et al. (2012). For live root imaging, we adapted the method of Shaw (2006) using agarose plates with minor modifications, and imaged tissues on a fluorescence dissecting microscope. Seeds were sprouted on sterile water-soaked KimWipes treated with the fungicide Captan. Two days later, when the primary root was 5–10 mm in length, the seedlings were transferred to imaging plates (35-mm Glass Bottom Dishes No. 1.5 Uncoated; MatTek Corporation) and positioned so that the root could continue to grow across the coverslip. Agarose (0.8%) was dissolved in artificial pond water (0.1 mM NaCl, 0.1 mM KCl, and 0.1 mM CaCl2), cooled, and poured around the seed, filling the plate to about one-third capacity. The plates were stored in the dark at room temperature until the root tip reached the center of the coverslip.
Results
Production of H2B-mCherry fusion protein transgenic plants
Our objective in this study was to produce and characterize a new reagent for plant chromatin cytology, a fluorescent line of maize based on histone H2B-mCherry. H2B was selected because of its expected widespread integration into plant chromatin. mCherry was selected for its photostability and spectral properties for epifluorescence imaging with conventional microscopic filters sets, such as rhodamine, Cy3, or Texas Red, and because it does not excite in the DAPI channel. Choice of the H2B-mCherry combination was further strengthened by the successful development of similar fusions in mammalian cells (reviewed by Day and Davidson, 2009).
Several histone H2B genes are known or predicted to exist in the maize B73 genome (Joanin et al., 1992; Bokhari Riza et al., 1994; Alexandrov et al., 2009). A multiple sequence analysis of the conserved histone H2B gene family in maize is shown in Figure 1. From these, we selected the Histone H2B.5 sequence to make the fusion protein. The histone H25B.5 (GenBank U08226, UniGene Zm.20180) shows EST expression data for multiple different tissues and it has >90% amino acid sequence ID with the other histone H2B proteins.
FIG. 1.
Multiple sequence alignment of maize histone H2B proteins. Multiple protein sequence alignment; histone identification is indicated before each sequence with a suffix to indicate chromosome number (1–10) and gene order on each chromosome (a–c). The top line is the fusion protein H2B-mCherry, and the bottom line is the consensus sequence. Dashes represent gaps introduced by the sequence-alignment software (ClustalW, v1.7). Stop codons (*) are followed by chromosome number, genomic coordinate (Mbp), maize gene ID, and protein length (amino acid number). One gene model (GRMZM2G442555) with histone H2B-like motif was omitted from the alignment because it was predicted to encode a long variant with <50% amino acid sequence identity to the others in the alignment.
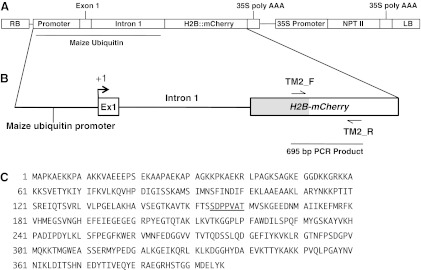
The H2B-mCherry transgene in the T-DNA construct is shown in Figure 2. The DNA sequence of the entire open-reading frame was optimized with maize codon preference, and the synthetic fusion, H2B-mCherry, was inserted into a T-DNA vector. The H2B-mCherry was placed under the control of a moderate and widely expressed promoter and consisting of the promoter, first exon (5′ UTR), and first intron of the maize ubiquitin gene, Ubi-1 (Christensen et al., 1992).
FIG. 2.
T-DNA construct, transgene structure, and protein sequence for the fusion protein H2B-mCherry. (A) Diagram of the T-DNA; RB, right border; LB, left border; histone H2B-mCherry transgene, with promoter, Exon 1, and Intron 1 of the maize ubiquitin (Ubi-1) gene; NPTII, Neomycin phosphotransferase gene. (B) Diagram of fusion protein, with maize ubiquitin promoter showing transcription start (+1), locations of primers (TM2_F and TM2_R), and length of the PCR product (695-bp PCR product). (C) Protein sequence of H2B-mCherry, indicating the linker sequence (underlined) separating the H2B and mCherry. PCR, polymerase chain reaction.
Presence and expression of H2B-mCherry in maize transgenic plants
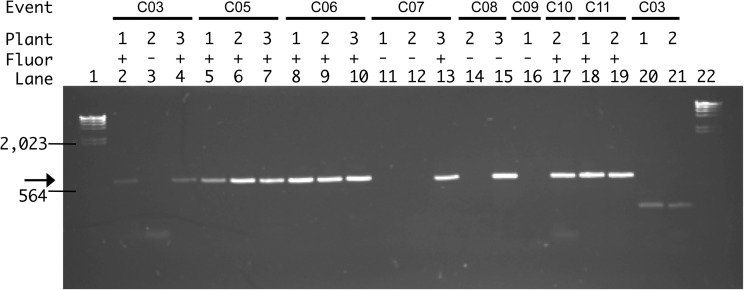
The PCR primers used to check for the presence of H2B-mCherry in the genomic DNA are shown in Figure 3. Control PCRs designed to amplify endogenous sequences (GAPDH and SUN4, lanes 20 and 21, respectively; Fig. 3) ruled out false negatives. The expected 695-bp PCR product was detected in most of the samples, indicating that a majority of these plants carry the transgene in their genome. Microscopic expression analysis, described below in Figure 4, revealed a nearly perfect correlation (12 of 13) between detection of the H2B-mCherry sequence by PCR and detection of the protein gene product by microscopy. Only one of the 13 plants (C08-3, lane 15) was positive for the PCR assay but negative for the microscopic expression assay.
FIG. 3.
PCR detection of transgene. PCR evaluating transgene incorporation; individual transformation event lines, corresponding progeny, H2B-mCherry expression, and lane number are indicated along the top of gel. Molecular size markers (λ HindIII) are in lanes 1 and 22, and the position of two marker bands (2023 and 564) is indicated at left. Expression of transgene was verified by microscopy (+or−) for all PCR products. PCRs designed to amplify fragments of H2B-mCherry (lanes 2–19) or control nontransgene sequences—SUN (lane 19) or GAPDH (lane 20)—were carried out with primer sequences described in the Materials and Methods section. Position of the H2B-mCherry 695-bp PCR product is indicated by the arrow.
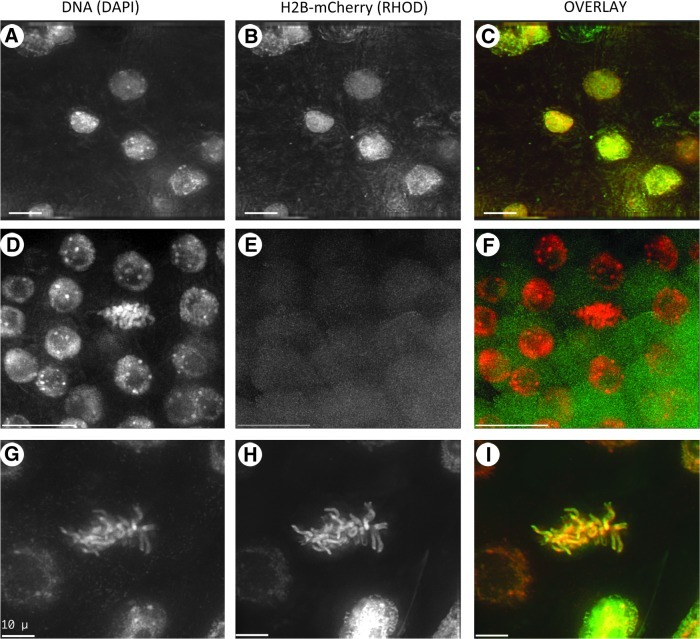
FIG. 4.
Fluorescent images of root tissue from plants transformed with H2B-mCherry. Microscopic analysis of transgene expression in root sections containing fixed nuclei subjected to 3D imaging. Three-dimensional data sets are shown as maximum-intensity projections. Image projections for a given field of view are shown in each row; DNA (DAPI), DAPI image; H2B-mCherry (RHOD), rhodamine image; OVERLAY, pseudocolor overlay image showing DAPI in red and rhodamine in green to maximize visualization of primary data (H2B-mCherry). (A–C) Several interphase nuclei expressing H2B-mCherry. (D–F) Several interphase nuclei that are not expressing the transgene. (G–I) Metaphase cell expressing H2B-mCherry. Scale bars are 10 μm. 3D, three dimensional.
Detection of histone H2B-mCherry in nuclei and metaphase chromosomes
Figure 4 shows images of formaldehyde-fixed root tips stained with DAPI. A representative example of a line expressing H2B-mCherry is shown (line C11, Fig. 4B) for comparison with a line that does not show H2B-mCherry expression (line C09, Fig. 4E). The expressing line shows general colocalization of fluorescence in the nuclei of several cells (OVERLAY, Fig. 4C). Little or no fluorescence was observed in other channels (FITC, not shown), confirming the specificity of mCherry signal for the rhodamine channel. Plants lacking red fluorescent nuclei (Fig. 4E) probably do not carry the transgene, as might be expected from seed of outcrossed T01 plants.
Having established that the H2B-mCherry was in the nucleus, we next wanted to determine whether the fusion protein was being incorporated into the chromatin or just localized to the nucleoplasm compartment of the cell. For this question, we imaged cells in mitosis, a stage when the nuclear envelope should be broken down or absent, leaving soluble nuclear proteins to diffuse into the cytoplasm. Images of a representative metaphase cell (Fig. 4G–I) revealed that the DAPI and mCherry staining was limited to chromosome fibers, with little or no cytoplasmic staining. We conclude from these observations that most of the protein is in the chromatin, with a relatively small pool of unincorporated protein.
A summary of expression levels for each of the 11 primary lines is shown in Table 1. The majority of the nuclei showed strong expression, whereas some plants showed no obvious fluorescent nuclei in any of the cells examined. Several of the families with stable expression were outcrossed to several different maize genotypes (B73, A344, and KYS) to produce T02 plants, most of which continued to show expression of H2B (Table 1).
Table 1.
Summary of Transgenic H2B-mCherry Maize Lines
| Line name | Source event ID | T01 expressiona | T02 expression | Image examples (figure numbers) |
|---|---|---|---|---|
| C01 | SS 585-2-1-4 | Yes | Yes | 5B, 6A–C, 7A–C, 8A–C, 9E–H |
| C02 | SS 585-1-1-2 | Yes | Yes | |
| C03 | SS 585-2-1-3 | Yes | — | |
| C04 | SS 585-2-1-1 | Yes | Yes | 5A |
| C05 | SS 583-1-1-4 | Yes | Yes | |
| C06 | SS 589-2-1-4 EL#8 | Yes | Yes | |
| C07 | SS 589-2-1-3 | Yes | No | |
| C08 | SS 589-2-2-3 | No | — | |
| C09 | SS 585-2-1-2 | No | — | 4E |
| C10 | SS 589-2-2-4 | No | — | |
| C11 | SS 589-2-2-5 | Yes | — | 4A |
“Expression” refers to detection by microscopic assay for red fluorescent nuclei.
“—” indicates lines for which outcrosses were not made.
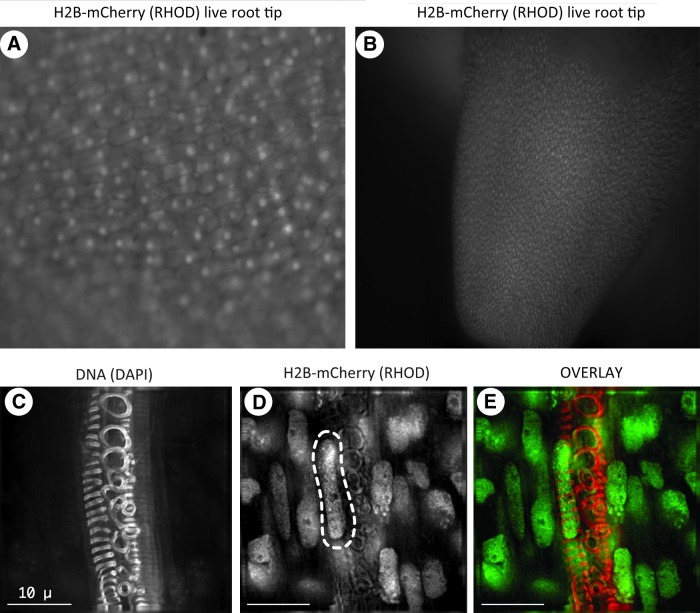
The ability to image living cells is valuable for study of complex and dynamic processes that occur during cell growth and differentiation (Shaw, 2006). Representative live root images of H2B-mCherry within cells of living plants are shown in Figure 5, taken at a low magnification with a fluorescence dissection microscope. Individual nuclei can be seen (Fig. 5A) in the cells, and their uniformity of expression over a large field of view (Fig. 5A, B) is evident.
FIG. 5.
Fluorescent images of root tissue from plants transformed with H2B-mCherry. (A, B) Live imaging of root from plants expressing H2B-mCherry, imaged with rhodamine fluorescence channel on a dissecting microscope; shown at high (A) and low (B) magnifications. (C–E) Xylem tissue fixed, imaged, and displayed as described in Figure 4. Autoflourescence in DAPI channel obscures somatic nuclei in this tissue, such as the individual nucleus (D, dashed outline) inside a xylem cell. Scale bars are 10 μm.
Another notable observation made during the screening of plants for red fluorescent nuclei was that, in some cell types, such as xylem, the autofluorescence in the DAPI channel prevents routine DAPI imaging of nuclei within cells. An example is evident in the images of xylem cells shown in Figure 5C–E. H2B-mCherry fluorescence revealed the location of an elongated nucleus (dashed line, Fig. 5D) that could not be visualized with DAPI because of the cell wall autofluorescent rings that are characteristic of this cell type (Fig. 5C).
Detection of histone H2B-mCherry during meiotic prophase
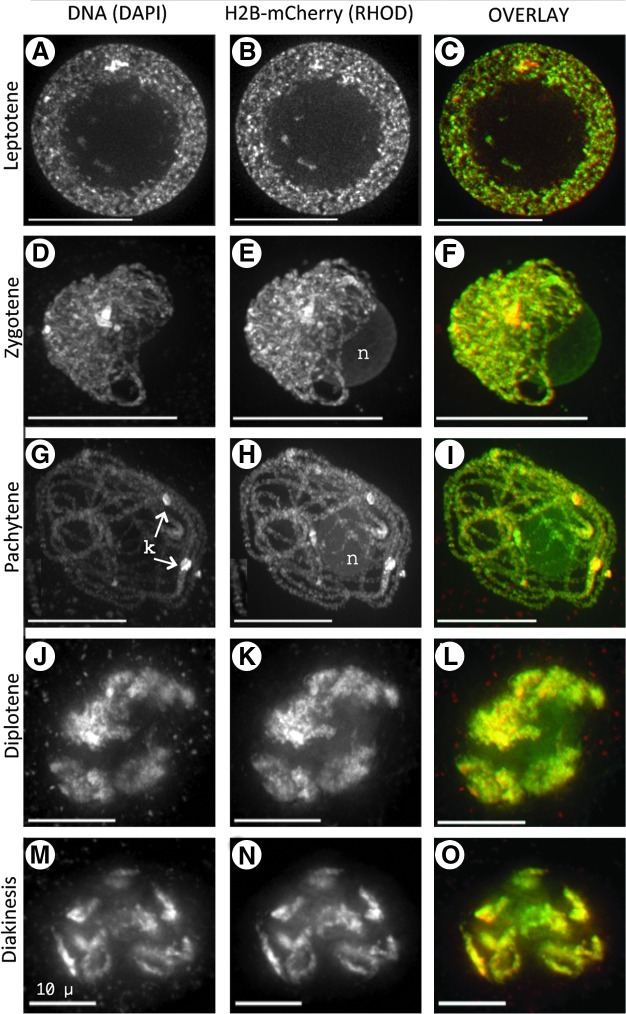
We found H2B-mCherry fluorescence in all stages of meiotic prophase, as shown in Figure 6. These large cells with dense cytoplasm are easily recognized as cells going through meiosis (DNA column, Fig. 6). At the first stage of meiotic prophase (Leptotene, Fig. 6A–C), typical patterns of chromatin condensation and a centralized nucleolus are evident in both the DAPI and the rhodamine images. Small segments that appear to be parts of chromosome fibers are seen within the enlarged nucleolus (arrows, Fig. 6A, B). The colocalization of DAPI and H2B-mCherry signals indicates that these are indeed chromatin fibers. At the zygotene stage, the condensation of the chromatin is more apparent (Fig. 6D–F). The nucleolus exhibits a distinct staining pattern in the rhodamine channel, less bright than the chromosome fibers but brighter than anything else in the cell. This observation, seen in many cell types, suggests that some of the histone H2B-mCherry fusion protein is localized throughout the nucleolus. For comparison, the DAPI image of the nucleolus is typically understained. The difference in nucleolar staining between DAPI and H2B-mCherry is most clearly seen in the pseudocolored overlay image (Fig. 6F).
FIG. 6.
H2B-mCherry in meiotic prophase of pollen mother cells. Meiocytes (A–O) from fixed anthers were imaged in 3D and displayed as described in Figure 4. Each row shows a single nucleus at the stage indicated on the left. Within nuclei, the locations of the nucleoli (n) and knobs (k) are indicated. Scale bars are 10 μm.
The pachytene-stage nuclei (Fig. 6G–I) contain fully synapsed chromosome fibers. The H2B-mCherry fluorescence is nearly identical in morphological detail to the DAPI fluorescence at this stage, indicating that the marker protein is uniformly distributed even at pachytene and that its presence does not appear to disrupt the progression of meiotic chromosome synapsis. Chromomere substructure (Fig. 6H, zoom inset) in the rhodamine channel is remarkably clear, and the whole-cell images are not obscured by staining of cytoplasmic structures, as seen with DAPI. Later in prophase, at the diplotene stage (Fig. 6J–L) and at diakinesis (Fig. 6M–O), the H2B-mCherry signal is bright and similar to that of DAPI. Overall, the coincidence of DAPI and H2B-mCherry staining can be seen as yellow in the red–green overlay images. A notable exception is the nucleolus, which shows relatively little staining in DAPI but uniform, largely diffuse staining in rhodamine and appears green in the overlay images.
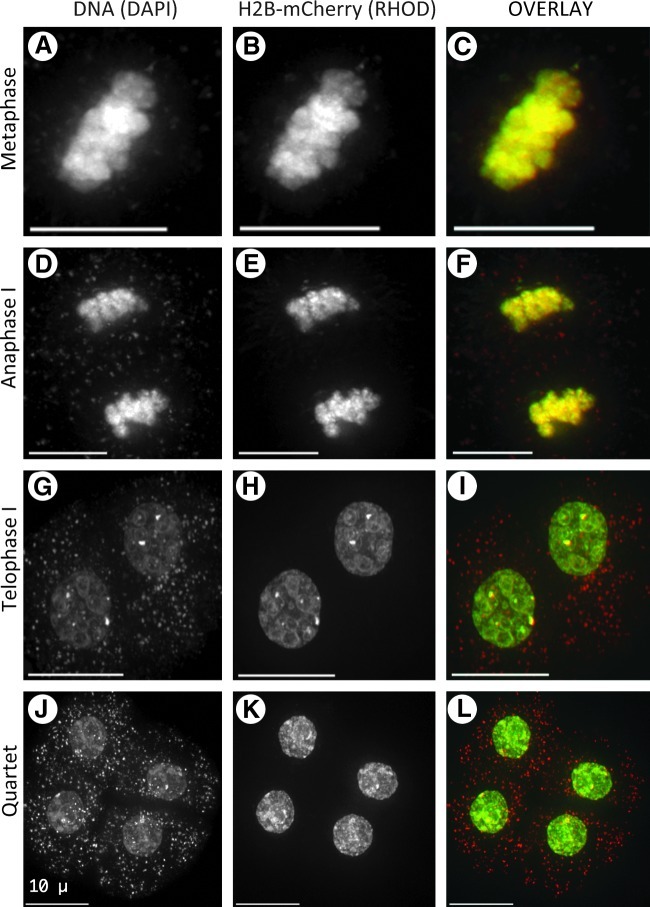
Detection of histone H2B-mCherry in meiosis I and II
Following meiotic prophase in male meiocytes, the cells go through a relatively rapid progression to complete meiosis I and meiosis II, resulting in tetrad-stage haploid cells. Given that no DNA replication takes place between meiosis I and II, the H2B-mCherry observed at the end of meiotic prophase should persist throughout the remainder of meiosis. Data consistent with this idea are shown in Figure 7. The DAPI and rhodamine images give similar staining patterns in the nuclei of metaphase, anaphase, and telophase cells (Fig. 7A–I). Cells exiting meiosis II can be seen as the four uniformly sized haploid daughter cells (Quartet, Fig. 7J–L). The greater clarity of nuclear imaging with H2B-mCherry versus DAPI is especially evident in the image projections from cells at the end of the first (Fig. 7G, J) and second (Fig. 7H, I) meiotic divisions (see Supplementary Movies S1 and S2; also see Appendix Table 1; Supplementary Data are available online at www.liebertonline.com/dna).
FIG. 7.
H2B-mCherry in later stages of meiosis I and II of pollen mother cells. Meiocytes (A–L) from fixed anthers were imaged in 3D and displayed as described in Figure 4. Each row shows a single nucleus at the stage indicated on the left. Scale bars are 10 μm.
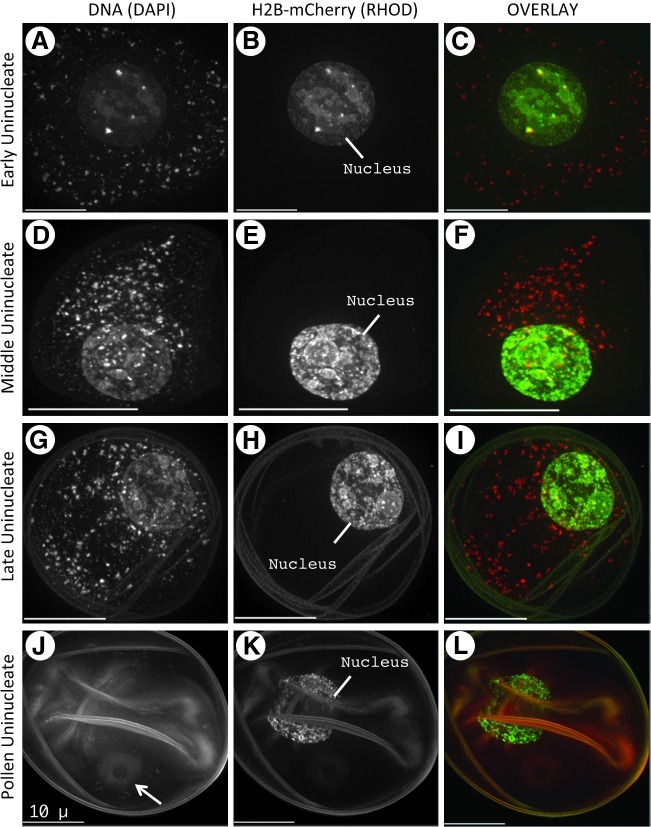
Detection of histone H2B-mCherry in postmeiotic microgametophyte cells
During maturation of the relatively short-lived male maize gametophyte into a three-nucleus pollen grain (reviewed by Chang and Neuffer, 1989), we observed excellent postmeiotic detection of H2B-mCherry, as shown in Figure 8 for early gametophyte developmental stages. In the early uninucleate cell (Fig. 8A–C), the DAPI image reveals the centrally located nucleus within the dense cytoplasm. The punctate DAPI staining outside of the nucleus probably reflects cytoplasmic structures containing nucleic acids, such as plastids, mitochondria, and possibly RNA-containing granules. In contrast, the H2B-mCherry staining is limited to the nucleus (Fig. 8B). As seen for virtually all other comparisons, the DAPI and H2B-mCherry images show similar substructure and morphology within the nuclei. At the middle uninucleate stage, the nucleus takes up an eccentric position (Fig. 8D–F). At the late uninucleate stage, the extracellular exine accumulation is evident in the form of autofluorescence in the DAPI channel (Fig. 8G–I).
FIG. 8.
H2B-mCherry in postmeiotic early microgametophyte stages leading to mature pollen. Cells (A–L) from fixed anthers were imaged in 3D and displayed as described in Figure 4. Each row shows a single nucleus at the stage indicated on the left. The DAPI image of the nucleus is obscured or difficult to visualize at some of these stages. The position of the nucleus (nucleus) is indicated in the rhodamine channel, and the annulus (J, arrow) is indicated. Scale bars are 10 μm.
At an early pollen grain stage (Fig. 8J–L), when the gametophyte is still uninucleate, the DAPI image fails to reveal the nucleus, whereas it can be clearly seen in the H2B-mCherry image (Nucleus, Fig. 8K), as can other diagnostic features associated with pollen development (arrow denoting the annulus, Fig. 8J). The ability to detect the nucleus during pollen development with H2B-mCherry highlights one of the main advantages of these chromatin marker lines, allowing for immediate nuclear detection in fresh-mounted preparations of cells where DAPI staining is unsatisfactory.
H2B-mCherry remains fluorescent after FISH
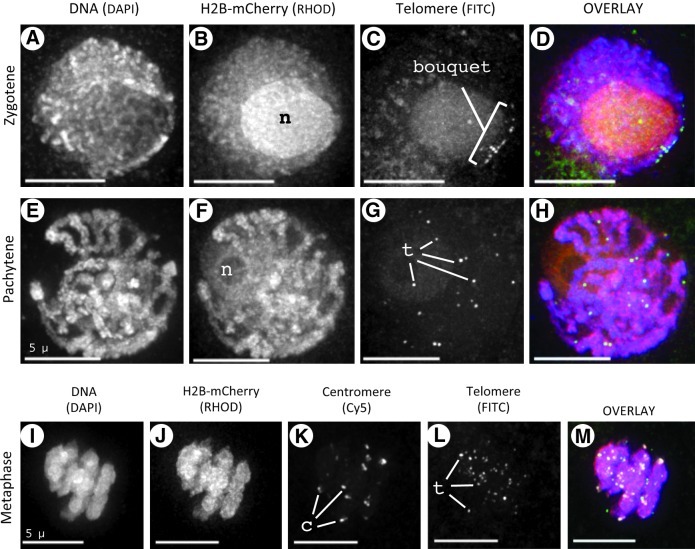
Having established the ability to detect H2B-mCherry throughout meiosis, we investigated whether the fluorescent tag was compatible with in situ hybridization procedures. We have previously established 3D FISH approaches (Howe et al., 2012) to gain insights into the organization and distribution of chromosomes, telomeres, and other specific sequences during meiosis in the pollen mother cells of maize (Bass et al., 1997, 2000, 2003).
Figure 9 shows images from three different stages of meiosis subjected to 3D acrylamide FISH with fluorescent oligonucleotide probes to localize repetitive sequences. We found the bouquet, a hallmark of meiotic prophase nuclei formed by telomere clustering on the nuclear envelope in early meiotic prophase (reviewed by Bass, 2003), appears normal at zygotene in plants expressing H2B-mCherry. At this stage, most of the telomere FISH signals show grouped localization to a small area of the nuclear periphery (bouquet, Fig. 9C). Later, during midprophase, we saw typical pachytene fibers in both the DAPI and mCherry images (Fig. 9E–H), with telomere distribution indicative of a late-pachytene-stage nucleus. Hybridization of telomere and centromere probes to meiotic metaphase cells (Fig. 9I, J) showed that the H2B-mCherry remained associated with the chromatin even after FISH, which includes a heat-denaturation step in the presence of formamide-containing buffers. We found that, in all cases, the H2B-mCherry fluorescence is retained and therefore compatible with FISH.
FIG. 9.
H2B-mCherry in meiotic cells following 3D FISH with telomere and centromere probes. Cells from fixed anthers were subjected to 3D acrylamide FISH. Each row shows a single nucleus at the stage indicated on the left. The molecule was stained and the associated wavelength used to collect the image is indicated at the top. Images from cells at early prophase (zygotene, A–D), middle prophase (pachytene, E–H), or metaphase of meiosis I (I–M) show preservation of chromosome fibers and retention of H2B-mCherry fluorescence. The positions of the nucleolus (n), the bouquet (bouquet), telomere FISH signals (t), and centromere FISH signals (c) are indicated. Pseudocolor overlay image shows DAPI (blue), FITC (green), RHOD (red), and, for (M), Cy5 (white). Scale bars are 5 μm.
Discussion
Our results show that H2B-mCherry is expressed in multiple genotypes and that the protein shows the localization pattern expected for a core histone protein. We conclude that it is stably incorporated into the chromatin, presumably as a nucleosomal component. The H2B-mCherry fluorescence closely resembled that of DAPI in that it appeared to stain bulk chromatin, which is indicative of its incorporation into nucleosomes distributed throughout the genome. Remarkably, the expression and incorporation of histone H2B-mCherry into maize chromatin did not noticeably alter the phenotype any point in development of segregating families grown under both field and greenhouse conditions (data not shown). We found that plants could be rapidly screened microscopically from sections of root tissue harvested at any stage in the life cycle. Plants that were positive for seedling root expression were also positive for expression in all other tissues examined throughout development, indicating that the root tip screen is a quick and reliable method and that H2B-mCherry expression continues during the entire plant life cycle.
Detection of the transgene through PCR
We developed PCR primers to genotype plants carrying the transgene and showed that it predicts which plants will express the FP (Fig. 3), but in rare cases, PCR-positive plants failed to exhibit red fluorescent nuclei. These exceptions could represent cases of transgene silencing, a common phenomenon in maize, often associated with the production of transgenic plants (reviewed by Matzke and Matzke, 1995; McGinnis, 2010). If reversible silencing of this transgene can occur, then this material may be an ideal expression marker for studying epigenetic regulation because its expression can be phenotyped at early stages of development from any cell type.
H2B-mCherry expression and localization in plants
The overall localization pattern of the H2B-mCherry can be described as nuclear, largely absent from the cytoplasm, and specifically associated with chromatin (Figs. 4, 5, 7, and 8). This pattern is that expected for a fluorescent histone marker protein. Interestingly, we observed that the intensity of H2B-mCherry fluorescence differed in different cells from the same tissue (Fig. 4B, H). The basis for this variation is unknown, but possibilities include cell-type-specific variation in protein accumulation, the degree of incorporation of the modified histone into the nucleosomes, and DNA content per nucleus, which varies during the cell cycle. We also observed that the heterochromatic knobs in DAPI images were not always evident in the rhodamine channel (Fig. 4), but this distinction was not seen in the meiotic and postmeiotic nuclei (Figs. 6–8). Determining the distribution of H2B-mCherry in relation to the endogenous histone H2B variants will be interesting.
In planta imaging
We sprouted seeds on agarose imaging plates to explore the possibility of using this material for imaging of live tissue. Normal organ growth and cell morphology were observed for 2–4 days in root tissue (Fig. 5). During this time frame, the H2B-mCherry fluorescence remained stable without apparent photobleaching, even after repeated imaging over multiple days. These observations suggest that these lines hold promise for time-lapse imaging of intact organs or even single-nucleus experiments, such as fluorescence recovery after photobleaching (reviewed by Day and Davidson, 2009).
Fluorescent histones for meiosis research
A major and exciting observation is that our fluorescent histone marker was present in the chromatin before, during, and after meiosis. These observations demonstrate that this transgene, controlled by the maize ubiquitin promoter, is suitable for both somatic and meiotic studies. We do not know whether the H2B-mCherry is transcriptionally active during meiosis or whether the fluorescence results from installation of the protein in the chromatin before onset of meiosis. In either case, the ability to image fluorescent chromatin will create additional opportunities for imaging the dynamic processes that occur during meiotic prophase in living cells. This long-standing challenge in plant cell biology has recently been successfully met by means of 2-photon confocal microscopy with DAPI-stained meiotic prophase cells imaged within cultured anthers (Sheehan and Pawlowski, 2009). Similarly, this material appears ideal for live cytology of chromosome and nuclear dynamics at developmental stages following meiotic prophase (Figs. 7 and 8), such as the analysis of neocentromere behavior (Yu et al., 1997), microsporogenesis (Chang and Neuffer, 1989), and pollen biology (Slotkin et al., 2009).
Incorporation of H2B-mCherry into meiotic chromatin also did not appear to alter or disrupt the complex progression of chromosome condensation, telomere clustering, synapsis, or any other aspect of meiotic nuclear architecture that has been previously described with molecular cytology of maize (Bass et al., 1997, 2000, 2003; and reviewed by Murphy and Bass, 2012). Further, the fusion protein allowed direct visualization of the nucleolus (Fig. 6E, H; see Supplementary Movies S3–S8 for pachytene-stage nucleus) in meiotic prophase. This property is advantageous because this organelle's position is usually inferred only from lack of staining in DAPI images (Fig. 6D, G). The nucleolar fluorescence is uniform throughout the entire organelle, possibly reflecting a pool of nonnucleosomal histones. Of interest would be to determine whether this staining reflects a normal pathway for histone accumulation before nucleosome assembly or an accumulation of extra protein resulting from the introduction of the transgene.
From our analysis of meiotic prophase cells, we conclude that the H2B-mCherry expression is evident and that the protein is uniformly distributed throughout the chromatin. These observations further indicate that the H2B-mCherry does not appear to undergo any sort of global removal at premeiotic S phase or at the onset of meiotic prophase. These findings establish that these H2B-mCherry lines represent robust and novel red fluorescent chromatin markers for the analysis of meiotic chromosome dynamics in plants.
Exceptional staining of male gametophyte nuclei with H2B-mCherry
In plants, the microgametophyte stage of the life cycle is difficult to study using fluorescence microscopy because it occurs within the sporophyte. Sporophytic cells have exine, which produces autofluorescence and blocks the entry of traditional dyes, such as DAPI. We found that many of these problems are alleviated by H2B-mCherry, which yields clear images of nuclei within the gametophyte (OVERLAY, Fig. 8; see Supplementary Movies S9–S18, for Fig. 8G–L). In addition, during this stage of plant development, a number of dynamic and specialized events occur in the haploid nuclei, including nuclear division and generation of vegetative and sperm nuclei, each with different properties. H2B-mCherry allows better cytological access to these stages and is expected to be similarly valuable for examination of the female gametophyte and processes associated with fertilization.
Prospects for widespread chromatin markers in plants
Our study has revealed several experimentally useful qualities of H2B-mCherry. First, the uniform staining of the chromatin makes it a good general chromosome marker for live- or fixed-cell imaging. Second, the visualization of chromosome fiber detail during meiotic prophase was superb; it provided bright, crisp staining of chromosomes during the pairing, synapsis, and recombination stages of meiosis. Third, the nucleolus was clearly stained throughout meiosis. This organelle is difficult to visualize, and H2B-mCherry allows detailed 3D analysis. Fourth, H2B-mCherry was stable through FISH, allowing spatial analysis of specific sequences in individual nuclei. Fifth, its staining of nuclei was stable throughout prolonged live imaging. Finally, the rhodamine images are not compromised by UV-excited autofluorescence, which can be particularly problematic in some plant cell types, such as pollen grains and vascular tissue (see Supplementary Movies S19–S22 [for xylem, Fig. 5C–E] and S15–S18 [for pollen, Fig. 8J–L]). In summary, H2B-mCherry represents an exciting new resource for cytological analysis of chromosomes in a classic genetic organism. It creates new opportunities for the investigation of nuclear architecture and chromatin structure and dynamics in a model eukaryotic organism.
Supplementary Material
Appendix
Appendix Table 1.
Supplementary 3D Deconvolution Image Data of Histone H2B-mCherry
| Supplementary Movie number | Movie file name (.mov files) | Cell type | Figure | Description |
|---|---|---|---|---|
| Supplementary Movie S1 | H2BmC01_Xylem_Dapi | Somatic, with xylem | 5C | Gray-scale sections, DAPI (DNA) |
| Supplementary Movie S2 | H2BmC01_Xylem_Rhod | ” | 5D | Gray-scale sections, RHOD (H2B-mCherry) |
| Supplementary Movie S3 | H2BmC01_Xylem_DapiRhod | ” | 5E | Color sections, DAPI/RHOD=red/green |
| Supplementary Movie S4 | H2BmC01_Xylem_DapiRhod_360 | ” | 5E | Color, spinning projections, DAPI/RHOD=red/green |
| Supplementary Movie S5 | H2BmC01_Pachytene_Dapi | Male meiotic prophase, pachytene | — | Gray-scale sections, DAPI (DNA) |
| Supplementary Movie S6 | H2BmC01_Pachytene_Dapi_360 | ” | — | Gray-scale, spinning projections, DAPI (DNA) |
| Supplementary Movie S7 | H2BmC01_Pachytene_Rhod | ” | — | Gray-scale sections, RHOD (H2B-mCherry) |
| Supplementary Movie S8 | H2BmC01_Pachytene_Rhod_360 | ” | — | Gray-scale, spinning projections, RHOD (H2B-mCherry) |
| Supplementary Movie S9 | H2BmC01_Pachytene_DapiRhod | ” | — | Color sections, DAPI/RHOD=red/green |
| Supplementary Movie S10 | H2BmC01_Pachytene_DapiRhod_360 | ” | — | Color, spinning projections, DAPI/RHOD=red/green |
| Supplementary Movie S11 | H2BmC01_TeloI_Dapi_360 | Male meiosis, telophase I | 7G | Gray-scale, spinning projections, DAPI (DNA) |
| Supplementary Movie S12 | H2BmC01_TeloI_Rhod_360 | ” | 7H | Gray-scale, spinning projections, RHOD (H2B-mCherry) |
| Supplementary Movie S13 | H2BmC01_LateUniNuc_Dapi | Gametophyte, late uninucleate | 8G | Gray-scale sections, DAPI (DNA) |
| Supplementary Movie S14 | H2BmC01_LateUniNuc_Dapi_360 | ” | 8G | Gray-scale, spinning projections, DAPI (DNA) |
| Supplementary Movie S15 | H2BmC01_LateUniNuc_Rhod | ” | 8H | Gray-scale sections, RHOD (H2B-mCherry) |
| Supplementary Movie S16 | H2BmC01_LateUniNuc_Rhod_360 | ” | 8H | Gray-scale, spinning projections, RHOD (H2B-mCherry) |
| Supplementary Movie S17 | H2BmC01_LateUniNuc_DapiRhod | ” | 8I | Color sections, DAPI/RHOD=red/green |
| Supplementary Movie S18 | H2BmC01_LateUniNuc_DapiRhod_360 | ” | 8I | Color, spinning projections, DAPI/RHOD=red/green |
| Supplementary Movie S19 | H2BmC01_PollenUniNuc_Dapi | Early pollen, uninucleate | 8J | Gray-scale sections, DAPI (DNA) |
| Supplementary Movie S20 | H2BmC01_PollenUniNuc_Rhod | ” | 8K | Gray-scale sections, RHOD (H2B-mCherry) |
| Supplementary Movie S21 | H2BmC01_PollenUniNuc_DapiRhod | ” | 8L | Color sections, DAPI/RHOD=red/green |
| Supplementary Movie S22 | H2BmC01_PollenUniNuc_DapiRhod_360 | ” | 8L | Color, spinning projections, DAPI/RHOD=red/green |
These movies are derived from five different multi-wavelength image data sets (from Howe, Clemente, & Bass).
Acknowledgments
The authors would like to thank A.B. Thistle, D.L. Vera, A.N. Brown, S.P. Murphy, and Y.Y. Mao for critical reading and helpful comments on the article. The authors would like to thank M. Davidson for suggesting histone H2B-mCherry and providing the associated sequence information, and L. Lyons for providing access to the fluorescence dissection microscope. This work was supported by grants and fellowships to E.S.H. from Women in Science Math and Engineering, the Charles M. McAllister Scholarship, the John Mark Caffrey Scholarship, and the Beta Beta Beta National Research Foundation. The work was also supported by grants to H.W.B. from the Florida State University Council on Research and Creativity (CRC Planning Grant-2008) and the National Science Foundation (IOS-1025954).
Disclosure Statement
No competing financial interests exist.
References
- Alexandrov N.N. Brover V.V. Freidin S. Troukhan M.E. Tatarinova T.V. Zhang H. Swaller T.J. Lu Y.P. Bouck J. Flavell R.B. KFeldmann K.A. Insights into corn genes derived from large-scale cDNA sequencing. Plant Mol Biol. 2009;69:179–194. doi: 10.1007/s11103-008-9415-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allis D. Epigenetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2007. [Google Scholar]
- Andrews A.J. Luger K. Nucleosome structure(s) and stability: variations on a theme. Annu Rev Biophys. 2011;40:99–117. doi: 10.1146/annurev-biophys-042910-155329. [DOI] [PubMed] [Google Scholar]
- Bass H.W. Telomere dynamics unique to meiotic prophase: formation and significance of the bouquet. Cell Mol Life Sci. 2003;60:2319–2324. doi: 10.1007/s00018-003-3312-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass H.W. Bordoli S.J. Foss E.M. The desynaptic (dy) and desynaptic1 (dsy1) mutations in maize (Zea mays L.) cause distinct telomere-misplacement phenotypes during meiotic prophase. J Exp Bot. 2003;54:39–46. doi: 10.1093/jxb/erg032. [DOI] [PubMed] [Google Scholar]
- Bass H.W. Marshall W.F. Sedat J.W. Agard D.A. Cande W.Z. Telomeres cluster de novo before the initiation of synapsis: a three-dimensional spatial analysis of telomere positions before and during meiotic prophase. J Cell Biol. 1997;137:5–18. doi: 10.1083/jcb.137.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass H.W. Riera-Lizarazu O. Ananiev E.V. Bordoli S.J. Rines H.W. Phillips R.L. Sedat J.W. Agard D.A. Cande W.Z. Evidence for the coincident initiation of homolog pairing and synapsis during the telomere-clustering (bouquet) stage of meiotic prophase. J Cell Sci. 2000;113:1033–1042. doi: 10.1242/jcs.113.6.1033. [DOI] [PubMed] [Google Scholar]
- Bokhari Riza A. Turcich M.P. Hamilton D.A. Mascarenhas J.P. Takacs I. Characterization of histone H2A and H2B cDNA clones isolated from a maize ovule cDNA library. Maydica. 1994;39:115–118. [Google Scholar]
- Brown A.N. Lauter N. Vera D.L. McLaughlin-Large K.A. Steele T.M. Fredette N.C. Bass H.W. QTL mapping and candidate gene expression analysis of telomere length control factors in maize (Zea mays L.) G3. 2011;1:437–450. doi: 10.1534/g3.111.000703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaboute M.E. Chaubet N. Gigot C. Philipps G. Histones and histone genes in higher plants: structure and genomic organization. Biochimie. 1993;75:523–531. doi: 10.1016/0300-9084(93)90057-y. [DOI] [PubMed] [Google Scholar]
- Chang M.T. Neuffer M.G. Maize microsporogenesis. Genome. 1989;32:232–244. [Google Scholar]
- Chaubet N. Philipps G. Chaboute M.E. Ehling M. Gigot C. Nucleotide-sequences of two corn histone H-3 genes: genomic organization of the corn histone H-3 and H-4 genes. Plant Mol Biol. 1986;6:253–263. doi: 10.1007/BF00015231. [DOI] [PubMed] [Google Scholar]
- Chen H. Swedlow J.R. Grote M.A. Sedat J.W. Agard D.A. The collection, processing, and display of digital three-dimensional images of biological specimens. In: Pawley J.B., editor. Handbook of Biological Confocal Microscopy. Plenum Press; New York: 1995. pp. 197–210. [Google Scholar]
- Christensen A.H. Sharrock R.A. Quail P.H. Maize polyubiquitin genes: structure, thermal perturbation of expression and transcript splicing, and promoter activity following transfer to protoplasts by electroporation. Plant Mol Biol. 1992;18:675–689. doi: 10.1007/BF00020010. [DOI] [PubMed] [Google Scholar]
- Craig J.M., editor; Wong N.C., editor. Epigenetics: A Reference Manual. Caister Academic Press; Norfolk: 2011. [Google Scholar]
- Day R.N. Davidson M.W. The fluorescent protein palette: tools for cellular imaging. Chem Soc Rev. 2009;38:2887–2921. doi: 10.1039/b901966a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBlasio S.L. Sylvester A.W. Jackson D. Illuminating plant biology: using fluorescent proteins for high-throughput analysis of protein localization and function in plants. Brief Funct Genomics. 2010;9:129–138. doi: 10.1093/bfgp/elp060. [DOI] [PubMed] [Google Scholar]
- Figueroa D.M. Bass H.W. A historical and modern perspective on plant cytogenetics. Brief Funct Genomics. 2010;9:95–102. doi: 10.1093/bfgp/elp058. [DOI] [PubMed] [Google Scholar]
- Hadjantonakis A.K. Papaioannou V.E. Dynamic in vivo imaging and cell tracking using a histone fluorescent protein fusion in mice. BMC Biotechnol. 2004;4:33. doi: 10.1186/1472-6750-4-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseloff J. Siemering K.R. The uses of green fluorescent protein in plants. Methods Biochem Anal. 2006;47:259–284. [PubMed] [Google Scholar]
- Herman B. Fluorescence Microscopy. 2nd. Springer-Verlag; New York: 1998. [Google Scholar]
- Howe A. Sato S. Dweikat I. Fromm M. Clemente T. Rapid and reproducible Agrobacterium-mediated transformation of sorghum. Plant Cell Rep. 2006;25:784–791. doi: 10.1007/s00299-005-0081-6. [DOI] [PubMed] [Google Scholar]
- Howe E.S. Murphy S.P. Bass H.W. 3D acrylamide FISH for plant cells. In: Pawlowski W.P., editor; Greylon M., editor; Armstrong S., editor. Plant Meiosis: Methods and Protocols. Methods in Molecular Biology, Human Press; New York: 2012. [Google Scholar]
- Huh G.H. Matsuura Y. Meshi T. Iwabuchi M. Differential expression of the two types of histone H2A genes in wheat. Biochim Biophys Acta. 1995;1261:155–160. doi: 10.1016/0167-4781(95)00017-b. [DOI] [PubMed] [Google Scholar]
- Ingouff M. Berger F. Histone3 variants in plants. Chromosoma. 2010;119:27–33. doi: 10.1007/s00412-009-0237-1. [DOI] [PubMed] [Google Scholar]
- Jach G. Binot E. Frings S. Luxa K. Schell J. Use of red fluorescent protein from Discosoma sp. (dsRED) as a reporter for plant gene expression. Plant J. 2001;28:483–491. doi: 10.1046/j.1365-313x.2001.01153.x. [DOI] [PubMed] [Google Scholar]
- Joanin P. Gigot C. Philipps G. Nucleotide sequence and expression of two cDNA coding for two histone H2B variants of maize. Plant Mol Biol. 1992;20:581–588. doi: 10.1007/BF00046443. [DOI] [PubMed] [Google Scholar]
- Kanazin V. Blake T. Shoemaker R.C. Organization of the histone H3 genes in soybean, barley and wheat. Mol Gen Genet. 1996;250:137–147. doi: 10.1007/BF02174172. [DOI] [PubMed] [Google Scholar]
- Kanda T. Sullivan K.F. Wahl G.M. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr Biol. 1998;8:377–385. doi: 10.1016/s0960-9822(98)70156-3. [DOI] [PubMed] [Google Scholar]
- Koning A.J. Tanimoto E.Y. Kiehne K. Rost T. Comai L. Cell-specific expression of plant histone H2A genes. Plant Cell. 1991;3:657–665. doi: 10.1105/tpc.3.7.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg R.D. Chromatin structure: a repeating unit of histones and DNA. Science. 1974;184:868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- Kornberg R.D. Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- Koumbaris G.L. Bass H.W. A new single-locus cytogenetic mapping system for maize (Zea mays L.): overcoming FISH detection limits with marker-selected sorghum (S. propinquum L.) BAC clones. Plant J. 2003;35:647–659. doi: 10.1046/j.1365-313x.2003.01829.x. [DOI] [PubMed] [Google Scholar]
- Matzke M.A. Matzke A.J.M. Homology-dependent gene silencing in transgenic plants: what does it really tell us. Trends Genet. 1995;11:1–3. doi: 10.1016/s0168-9525(00)88973-8. [DOI] [PubMed] [Google Scholar]
- McGinnis K.M. RNAi for functional genomics in plants. Brief Funct Genomics. 2010;9:111–117. doi: 10.1093/bfgp/elp052. [DOI] [PubMed] [Google Scholar]
- Mohanty A. Luo A. DeBlasio S. Ling X. Yang Y. Tuthill D.E. Williams K.E. Hill D. Zadrozny T. Chan A. Sylvester A.W. Jackson D. Advancing cell biology and functional genomics in maize using fluorescent protein-tagged lines. Plant Physiol. 2009;149:601–605. doi: 10.1104/pp.108.130146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy S.P. Bass H.W. Genetics and cytololgy of meiotic chromosome behavior in plants. In: Bass H.W., editor; Birchler J.A., editor. Plant Cytogenetics. Part 2. Vol. 4. Springer; New York: 2012. pp. 193–229. [Google Scholar]
- Okada T. Endo M. Singh M.B. Bhalla P.L. Analysis of the histone H3 gene family in Arabidopsis and identification of the male-gamete-specific variant AtMGH3. Plant J. 2005;44:557–568. doi: 10.1111/j.1365-313X.2005.02554.x. [DOI] [PubMed] [Google Scholar]
- Pawley J.B. Fundamental limits in confocal microscopy. In: Pawley J.B., editor. Handbook of Biological Confocal Microscopy. Springer; New York: 2006. pp. 20–41. [Google Scholar]
- Razafimahatratra P. Chaubet N. Philipps G. Gigot C. Nucleotide sequence and expression of a maize H1 histone cDNA. Nucleic Acids Res. 1991;19:1491–1496. doi: 10.1093/nar/19.7.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichheld J.P. Sonobe S. Clement B. Chaubet N. Gigot C. Cell-cycle-regulated histone gene expression in synchronized plant cells. Plant J. 1995;7:245–252. [Google Scholar]
- Riggs C.D. Molecular cloning of cDNAs encoding variants of meiotin-1. A meiotic protein associated with strings of nucleosomes. Chromosoma. 1994;103:251–261. doi: 10.1007/BF00352249. [DOI] [PubMed] [Google Scholar]
- Sattarzadeh A. Fuller J. Moguel S. Wostrikoff K. Sato S. Covshoff S. Clemente T. Hanson M. Stern D.B. Transgenic maize lines with cell-type specific expression of fluorescent proteins in plastids. Plant Biotechnol J. 2010;8:112–125. doi: 10.1111/j.1467-7652.2009.00463.x. [DOI] [PubMed] [Google Scholar]
- Shaner N.C. Campbell R.E. Steinbach P.A. Giepmans B.N. Palmer A.E. Tsien R.Y. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Shaner N.C. Patterson G.H. Davidson M.W. Advances in fluorescent protein technology. J Cell Sci. 2007;120:4247–4260. doi: 10.1242/jcs.005801. [DOI] [PubMed] [Google Scholar]
- Shaner N.C. Steinbach P.A. Tsien R.Y. A guide to choosing fluorescent proteins. Nat Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- Shaw S.L. Imaging the live plant cell. Plant J. 2006;45:573–598. doi: 10.1111/j.1365-313X.2006.02653.x. [DOI] [PubMed] [Google Scholar]
- Sheehan M.J. Pawlowski W.P. Live imaging of rapid chromosome movements in meiotic prophase I in maize. Proc Natl Acad Sci U S A. 2009;106:20989–20994. doi: 10.1073/pnas.0906498106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura O. The discovery of aequorin and green fluorescent protein. J Microsc. 2005;217:1–15. doi: 10.1111/j.0022-2720.2005.01441.x. [DOI] [PubMed] [Google Scholar]
- Shimomura O. Johnson F.H. Saiga Y. Extraction, purification and properties of aequorin, a bioluminescent protein from the luminous hydromedusan, Aequorea. J Cell Comp Physiol. 1962;59:223–239. doi: 10.1002/jcp.1030590302. [DOI] [PubMed] [Google Scholar]
- Slotkin R.K. Vaughn M. Borges F. Tanurdzic M. Becker J.D. Feijo J.A. Martienssen R.A. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell. 2009;136:461–472. doi: 10.1016/j.cell.2008.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes I.A. Runions J. Kearns A. Hawes C. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat Protoc. 2006;1:2019–2025. doi: 10.1038/nprot.2006.286. [DOI] [PubMed] [Google Scholar]
- Spiker S. Plant chromatin structure. Ann Rev Plant Phys. 1985;36:235–253. [Google Scholar]
- Stewart C.N., Jr. The utility of green fluorescent protein in transgenic plants. Plant Cell Rep. 2001;20:376–382. doi: 10.1007/s002990100346. [DOI] [PubMed] [Google Scholar]
- Sundas A. Tandre K. Kvarnheden A. Engstrom P. cDNA sequence and expression of an intron-containing histone H2A gene from Norway spruce, Picea abies. Plant Mol Biol. 1993;21:595–605. doi: 10.1007/BF00014543. [DOI] [PubMed] [Google Scholar]
- Yu H.-G. Hiatt E.N. Chan A. Sweeney M. Dawe R.K. Neocentromere-mediated chromosome movement in maize. J Cell Biol. 1997;139:831–840. doi: 10.1083/jcb.139.4.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.