Abstract
Anthocyanins are antioxidants and are among the natural products synthesized via the flavonoid biosynthesis pathway. Anthocyanins have been recommended for dietary intake in the prevention of cardiovascular diseases, cancer, and age-related conditions such as Alzheimer's disease or dementia. With an increasingly aging population in many parts of the world, strategies for the commercial production of in vitro synchronized red cell cultures as natural antioxidants will be a significant contribution to human medicine. Red pigmented fruits such as grapes (Vitis sp.) are a major source of bioavailable anthocyanins and other polyphenols. Since the level of antioxidants varies among cultivars, this study is the first one that phytochemically and genetically characterizes native grape cultivars of North America to determine the optimal cultivar and berry cells for the production of anthocyanins as antioxidants. Using real-time PCR and bioinformatics approaches, we tested for the transcript expression of the chalcone synthase (CHS) gene, an enzyme involved in the flavonoid and anthocyanin biosynthesis pathway, in different parts of physiologically mature grape berries and in vitro synchronized red cells. A low level of expression was recorded in berry flesh, compared with an elevated expression in berry skins and in vitro synchronized red cells, suggesting increased production of flavonoids in skin and cell cultures. This preliminary study demonstrates the potential of functional genomics in natural products research as well as in systematic studies of North American native grapes, specifically in muscadine (Vitis rotundifolia).
Introduction
Flavonoids are antioxidants that form various natural products such as anthocyanin, proanthocyadin, and phylobaphene pigments. They are involved in many aspects of plant growth and development, such as pathogen resistance, pigment production, UV light protection, pollen growth, and seed coat development (Harbone, 1986). Red pigmented fruits such as grapes are a major source of anthocyanins, as the latter is responsible for the red pigment. The pigments of anthocyanin dictate the degree of redness of the berry skin in different cultivars of grapevine, and this applies to wines fermented in the presence of red skins. The red color is crucial to the wine industry, and wine redness has valuable commercial and economic relevance—premium red wine is considered to be of high quality, which is also a primary factor in visual characterization. Red grapes and wines are particularly rich in bioavailable anthocyanins that are rapidly absorbed as intact molecules (Bitsch et al., 2004) and delivered into the brain within minutes of ingestion (Passamonti et al., 2005).
Due to their high antioxidant capacity, there is increasing evidence which suggests that flavonoids are health-protecting components in the human diet (Sugihara et al., 1999; Dugas et al., 2000). Anthocyanins have been used in therapy against cardiovascular diseases, cancer (Trevisanato and Kim, 2000), and age-related conditions such as Alzheimer's disease or dementia (Commenges et al., 2000). Due to increasing health problems in many parts of the world, ways for the commercial production of antioxidants in functional foods will be a significant contribution to human health.
Native grapes of North America (Muscadine or Vitis rotundifolia) are considered the most important cultivated Vitis species in the Southeastern part of the United States due to their possession of a high level of antioxidants. However, the lack of genomic studies and the phytochemistry of muscadine has limited the rapid discovery of genes that influence specific traits within the species. Available information indicates that muscadine grape genotypes contain several unique polyphenolic compounds that are beneficial in the human diet. Apart from being the only cultivar producing ellagic acid, muscadine contains important genetic resources that are not utilized in grape breeding (Samuelian et al., 2009b).
Chalcone synthases (CHS) is the first committed enzyme in the flavonoid pathway that facilitates the stepwise synthesis of chalcone. The function of CHS is believed to be a regulatory focal point of substrate flow between the flavonoid pathway and the other competing directions of the phenylpropanoid pathway. This leads to the diversity of phenolic compounds such as lignins and phytoalexins (Winkel-Shirley, 2001; Szalma et al., 2002). Genes encoding CHS constitute a multigene family in which the copy number varies among the plant species, and functional divergence appears to have repeatedly occurred (Tuteja et al., 2004; Yang et al., 2004). The expression of the CHS gene in fruit tissue is developmentally regulated and associated with fruit coloring (Aharoni and O'Connell, 2002; Kumar and Ellis, 2003). CHS is encoded by a single gene in Arabidopsis thaliana and by a large multiple gene family in other plants such as soybean and petunia (Koes et al., 1994). In soybean, there are nine members of the CHS gene families sharing high nucleotide sequence identity within their coding regions (Tuteja and Vodkin, 2008). Many genes for the anthocyanin biosynthesis pathway have been isolated and characterized in Vitis vinifera grapes. However, only one member of the CHS gene family (CHS1) has been reported in muscadine (Samuelian et al., 2009a). It is not clear whether there are other CHS gene families available in M. rotundifolia genome.
With an increasingly aging population in many parts of the world, strategies for the commercial production of natural antioxidants from in vitro synchronized red cells of muscadine will be a significant contribution to human medicine. The purpose of this study was to investigate the expression patterns of the muscadine CHS gene in the physiologically mature berries and in vitro synchronized red cell cultures of North American native grapes. This is because our goal is to phytochemically and genetically characterize the native grape cultivars of North America and to determine the tissues that express high anthocyanin levels as potential candidates for the commercial production of anthocyanins as antioxidants.
Materials and Methods
Plant materials and cell growth
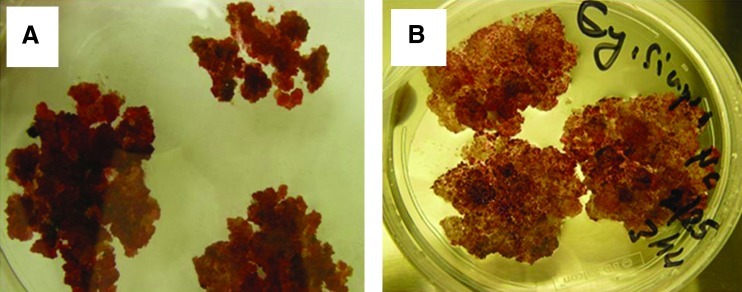
In vitro synchronized red cell cultures (Fig. 1A, B) established from super-epidermal cells of red berries (Fig. 2A, B) of North American native grapes: muscadinia “Noble” var. and aestivalis “Cynthiana” var. by Colova, (2011) (patent publication no. US 2011/0054195 A1) were used in this study. The cells were grown in a growth chamber at 23°C under a white light (150 μE m−2S−1) with a 16 h light/8 h dark cycle. The callus developed produces anthocyanin, and it is red in color. Figure 2A and B shows the two varieties that were used to initiate the in vitro cell lines at the physiological maturity stage. They are grown at the experimental vineyard of the Florida A&M University in Tallahassee, Florida.
FIG. 1.
In vitro synchronized red cell cultures established from super-epidermal cells of red berries of North American native grapes: muscadinia Noble var. (A) and aestivalis Cynthiana var. (B) (patent publication no. US 2011/0054195A1). Color images available online at www.liebertonline.com/dna
FIG. 2.
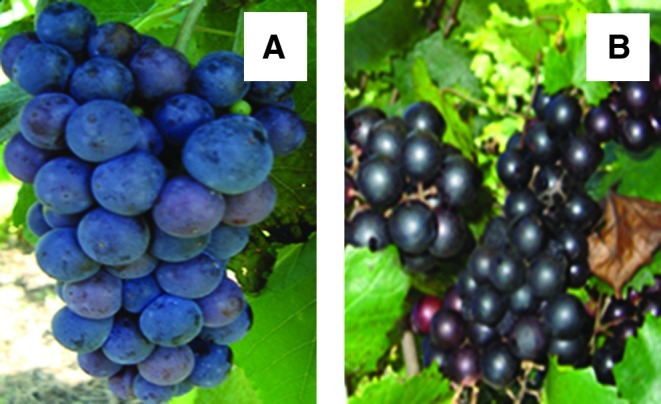
Physiologically mature berries from Cynthiana var. (A) and Noble var. (B) used to analyze CHS expression profile. CHS, chalcone synthase. Color images available online at www.liebertonline.com/dna
Maintenance of cell cultures
The cells were maintained in B-5 media in petri plates, as described by Gollop et al. (2002). The cultures were transferred to a fresh medium every 30 days or when thickness and the required color were attained. To transfer the cells, light forceps were gently used to scoop off the top layers of the cells. The cells were gently spread onto new culture media in 3 medium-sized layers in each plate in order to give them enough room to multiply.
Protein three-dimensional structure prediction
The Muscadine CHS Structural model was obtained from its amino-acid sequence by using the SWISS MODEL (http://swissmodel.expasy.org) (Kiefer et al., 2009) and Protein Homology/analogy Recognition Engine (PHYRE) (www.sbg.bio.ic.ac.uk/phyre) prediction servers (Altschul et al., 1997, Bennett-Lovsey et al., 2008). The model obtained was classified according to identity percentage.
RNA isolation and gel electrophoresis
Samples were prepared from different tissues of Noble and Cynthiana grape varieties (In vitro synchronized red cell cultures, physiologically mature berry skins, and flesh). Total RNA was isolated using the RNeasy Plant Mini Kit according to the manufacturer's protocol. RNA was quantified using UV absorbance, and the inactivity was inspected by formaldehyde agarose gel electrophoresis. Purified RNA was treated with RNase-free DNAse 1, and immediately frozen to −20°C. Formaldehyde gel electrophoresis (1.2% agarose) was used to evaluate the RNA quality. The gel apparatus (including the gel tray and comb) was treated with RNase Away and rinsed with distilled water.
cDNA synthesis
Total RNA was used in primary gene expression profiling. The SuperScript First-strand Synthesis System for RT-PCR (Invitrogen) was used to synthesize cDNA in a 20 μL reaction containing 1 μg of DNase I-treated total RNA, 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 2.5 mM MgCl2, 10 mM dithiothreitol, 0.5 μg oligo (dT), 0.5 mM each of dATP, dGTP, dCTP, and dTTP, and 200 U SuperScript II Reverse Transcriptase. RNA, dNTPs, and oligo (dT) were mixed first, heated to 65°C for 5 min, and placed on ice until the addition of the remaining reaction components. The reaction was incubated at 50°C for 50 min, and terminated by heat inactivation at 85°C for 5 min. The cDNA product was treated with 1μL of Rnase H (Invitrogen) for 20 min at 37°C. An identical reaction without the reverse transcriptase was performed to verify the absence of genomic DNA (no-RT control). The cDNA was stored at −20°C until it was ready for use.
Relative-quantitative real-time PCR using SYBR green assay
Relative-quantitative real-time PCR reactions were performed in a 96-well plate with an iCycler iQ Multicolor Real-Time PCR Detection system (Bio-Rad; Vandesompele et al., 2002), using an iQ SYBR Green Supermix (Bio-Rad) to monitor cDNA amplification, according to the manufacturer's protocol. As a control, parallel amplification reactions of Actin (a housekeeping gene) were also performed. Each primer set was designed based on the 3′-end cDNA sequence of the corresponding gene. The specific primers used for real-time PCR were as follows: for CHS, 5′-TACTCGCTCAAGACGTGTCG-3′ (Forward) and 5′-CCTACCCACACACAACTTCG-3′ (Reverse), giving a product of 245 bp; for Actin 5′-TAGAAGCACTTCCTGTGGAC-3′ (Forward) and 5′-GGAAATCACTGCACTTGCTC-3′ (Reverse), giving a product of 184 bp. RT-PCR was performed using the iQ SYBR Green Supermix (Bio-Rad) on an iQ-Cycler (Bio-Rad) under the following conditions: an initial denaturating cycle (2 min at 95°C), followed by 40 cycles constituted by three steps of denaturation, annealing, and polymerization (20 s at 95°C, 30 s at 60°, and 30 s 72°C). PCR amplification was done in a 25 μL total volume, containing 3 μL of diluted cDNA (duplicate), 0.5 μm of each primer, and 11 μL of 2 × iQ SYBR Green Supermix. In each experiment, the target and the control were amplified in the same plate with a series of diluted cDNA (10−5–10−9 ng) to generate calibration-specific curves. Three independent experiments were performed for each sample. After the real-time PCR had been performed, the absence of unwanted by-products was confirmed by an automated melting curve analysis and agarose gel electrophoresis of the PCR products.
In all the experiments, three replicates for each RNA sample were included; averages were calculated, and differences in the threshold cycle (Ct) were evaluated. The comparative Ct method was used, which mathematically transforms the Ct data into the relative transcription-level genes. When comparing the expression of CHS in different tissues, the relative quantification of CHS expression was achieved by calibrating its transcription level to that of the reference gene, Actin. The expression level calculated by the formula 2−ΔΔCt represents the x-fold difference from the calibrator.
Search for muscadine CHS-related sequences
Putative CHS sequence was retrieved through Basic Local Alignment Tool (BLAST), homology, and domain searches in public domains, namely GenBank (www.ncbi.ncbi.nlm.nih.gov). The GenBank CHS protein sequence of muscadine (Accession no. ACN30003; Samuelian et al., 2009a) was used for BLAST and homology searches against other plants.
Multiple sequence alignments and phylogenetic tree construction
Multiple alignment of the putative amino-acid sequence of muscadine CHS was performed using the T-Coffee program (Notredame et al., 2000). The alignment of 14 CHS proteins was summarized using the Plotcon sequence similarity graph (http://bioweb2.pasteur.fr/docs/EMBOSS/plotcon.html), which represents the similarity along the set of aligned sequences. The molecular phylogenetic tree for CHS was built with Neighbor Joining, using p-distance as a substitution model, and Maximum Parsimony methods in MEGA Version 4.0, with 5000 iterations for calculating boostrap confidence levels (Tamura et al., 2007). The phylogenetic tree construction included the sequences for the muscadine CHS protein and/or putative CHS proteins reported in the NCBI database for 13 plants (Table 1).
Table 1.
Percent Similarity of Muscadine Chalcone Synthase Amino-Acid Sequence with Chalcone Synthase Amino Sequence from Other Plant Species
| Plant species | NCBI accession no. | Sequence similarity (%) | |
|---|---|---|---|
| 1 | Vitis rotundifolia | ACN30003 | 100 |
| 2 | Vitis vinifera | BAA31259 | 100 |
| 3 | Camellia japonica | BAI66465 | 98 |
| 4 | Gossypium hirsutum | AEO96985 | 98 |
| 5 | Camellia chekiangoleosa | ADW11243 | 98 |
| 6 | Camellia sinensis | AAO13091 | 98 |
| 7 | Nelumbo nucifera | ADD74168 | 98 |
| 8 | Abelmoschus manihot | ACE60221 | 98 |
| 9 | Rhododendron simsii | CAC88858 | 98 |
| 10 | Pyrus communis | ABI79305 | 98 |
| 11 | Humulus lupulus | CAK19317 | 98 |
| 12 | Citrus maxima | ACX37403 | 99 |
| 13 | Glycine max | XP_003553986 | 98 |
| 14 | Nicotiana alata | ACS12837 | 98 |
Results
RT-PCR primers used were specific to the target gene
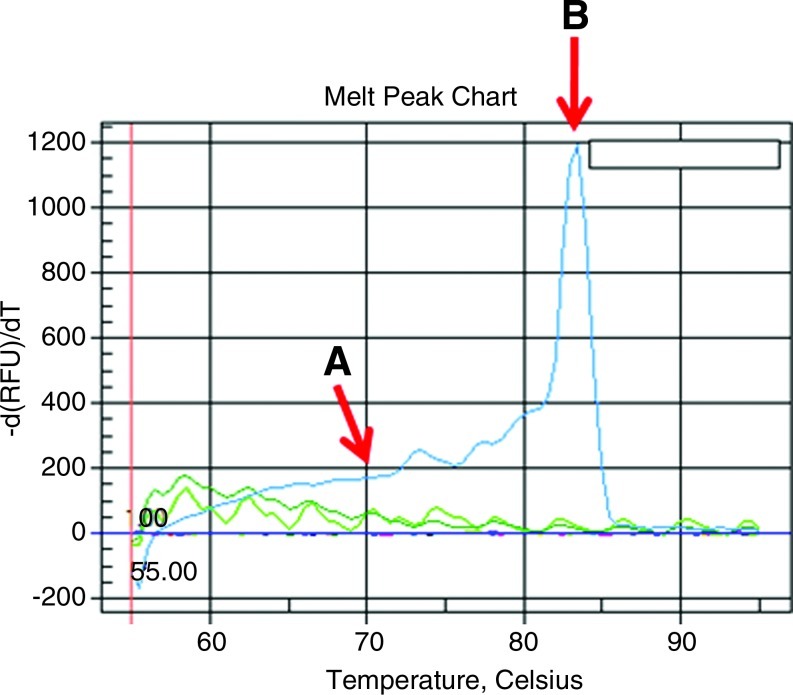
The CHS fragment showed a sharp peak for the PCR product of the quantitative standard sample, Cynthiana, and Noble at 88°C (Fig. 3). A single melting peak at the same melting temperature was produced for the PCR product of the cDNA samples. Every PCR product also generated prominent bands with the expected sizes in the gel electrophoresis. These results indicated that nonspecific PCR products were not detected in the analyzed temperature range. The fidelity of the real-time PCR assay is also demonstrated by the lack of specific signals for the no-RT and H2O controls and by the Ct determined for each gene.
FIG. 3.
Melting curve indicating temperature verses fluorescence [-d(RFU)/dT] for the CHS gene. All reactions with the cDNA template showed one sharp and fully overlapping melting peak, indicating the specificity of primers designed from a partial gene sequence. (A) indicates the melting curve with no peaks for water and no RT controls. (B) indicates the melting temperature of specific products in these reactions. Color images available online at www.liebertonline.com/dna
cDNA encoding muscadine CHS amplified
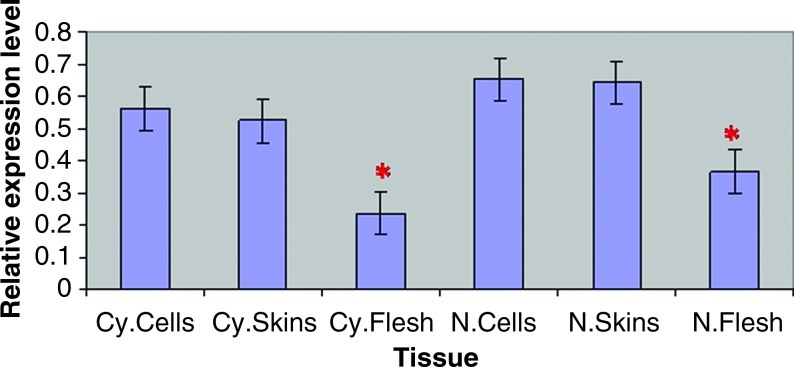
The results showed that the CHS transcript accumulated in the skin, flesh, and in vitro synchronized cell cultures of Noble and Cynthiana. However, the relative expression levels varied significantly (Fig. 4). CHS showed the highest transcript levels in the skins, and in vitro red cell cultures, and low levels in the flesh of physiologically mature berries. Our study confirms, for the first time, the expression of CHS in in vitro synchronized red cell cultures of Noble and Cynthiana varieties after the cells have gone through the tissue culture process, which involves different hormone (Auxins and Cytokines) stimuli. The RT-PCR analysis of total RNA from the in vitro synchronized red cells of Noble and Cynthiana varieties indicated that there was a slight difference in the expression of the CHS gene transcripts in both varieties.
FIG. 4.
Expression profile of CHS in different tissues collected from the muscadine and hybrid berries. A real-time PCR analysis was performed using total RNA from tissues of Cynthiana var. such as red cells (Cy. Cells), skins of physiologically mature berries (Cy. skins), and flesh of physiologically mature berries (Cy. Flesh). Real-time PCR analysis was also performed using total RNA from tissues of Noble var. such as red cells (N. Cells), skins of physiologically mature berries (N. Skins), and flesh of physiologically mature berries (N. Flesh). Actin and ubiquitin were used as an internal control. Data are mean values±SDs from the relative expression of CHS in cells, skins, and flesh analyzed from two independent varieties. *Significant difference between flesh and other tissues (p<0.05). Color images available online at www.liebertonline.com/dna
Multiple alignments and phylogenetic analysis of the CHS protein
The BLAST search for CHS-related sequences in the NCBI Genebank database showed that homologous protein sequences are present in other plants, as indicated in Table 1. The alignment of the deduced amino-acid sequences indicated that muscadine CHS shares the most identity with V. vinifera CHS, showing 100% identity. Furthermore, the alignment of muscadine CHS with other plants revealed that there are conserved amino-acid substitutions between CHS from muscadine and CHS from other plants mentioned in Table 1. The same conservation pattern was observed when the corresponding nucleotide sequences were aligned (>99% identity). Similarity percentages were found in the multiple amino-acid alignment of other plant species within the genus such as Citrus maxima (99%), Camellia japonica (98%), Pyrus communis (98%), and Glycine max (98%), showing that CHS is highly conserved at the genus level.
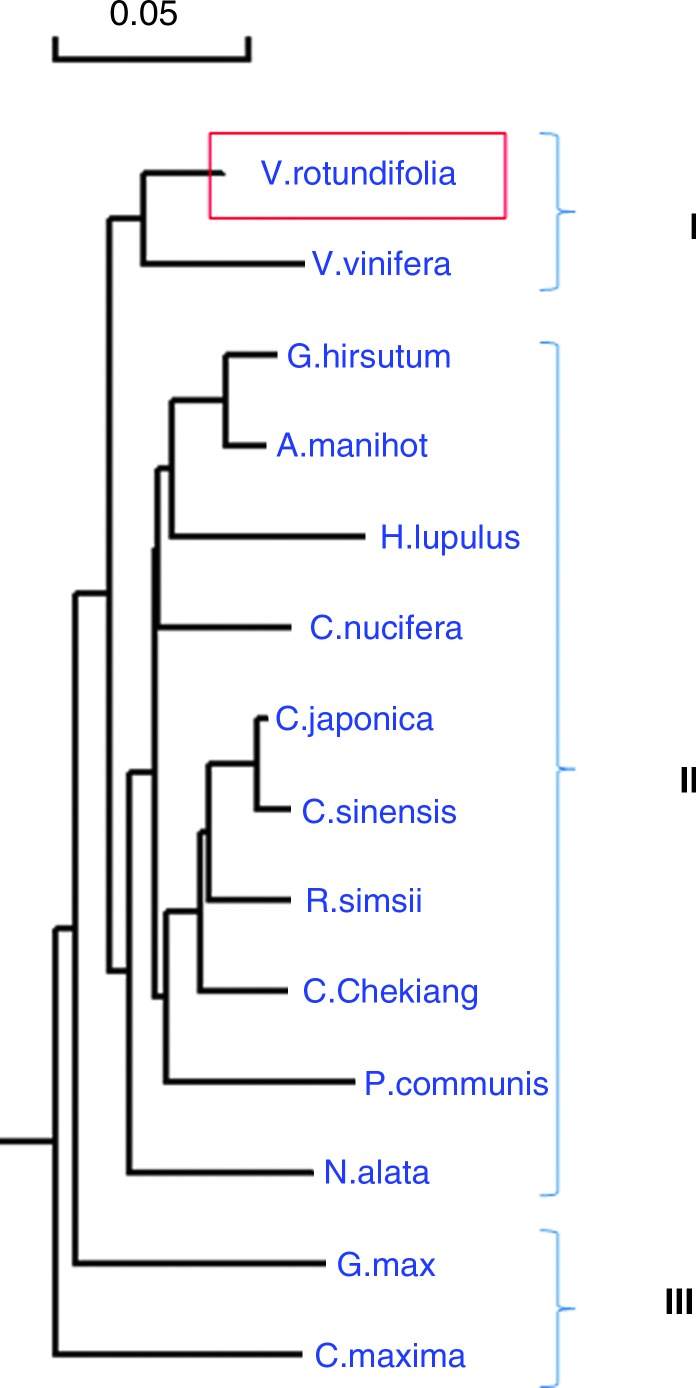
The three-dimensional (3D) structure of muscadine CHS (Fig. 5) shared an 84.3% similarity with the template, which further facilitated the positive identification of muscadine CHS. The cluster tree of the alignment of CHS proteins and the cladogram confirmed the close relationship between muscadine CHS and V. vinifera CHS (Fig. 6I). The phylogenetic tree was grouped into three distinct clades (Fig. 6I–III). Muscadine CHS was located in cluster I Fig. 6, while cluster II was further divided into several subgroups based on the plant species. For example, CHS proteins from Gossypium hirsutum and Abelmoschus manihot (both in Malvaceae family) appeared in the same subgroup and those from C. japonica, C. Sinensis, and C. chekiangoleosa (all in the Theaceae family) were nested in another subgroup (Fig. 6). Our results concur with those by Zhou et al. (2011), which suggest that CHS is well conserved among plants of different groups and has distinct species specificity.
FIG. 5.
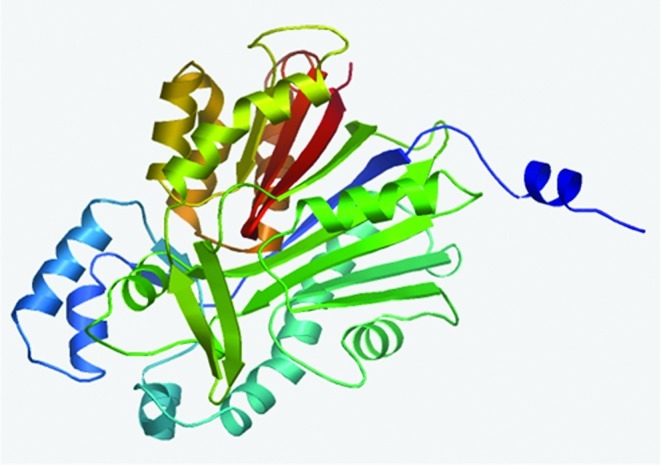
The computational modeled three-dimensional structure of muscadine CHS. Color images available online at www.liebertonline.com/dna
FIG. 6.
A phylogenetic tree based on the deduced amino-acid sequence of various CHS from other plants. The muscadine CHS protein is boxed in red. Color images available online at www.liebertonline.com/dna
Discussion
This study amplified a cDNA encoding muscadine CHS homolog from physiologically mature berry skins, flesh, and in vitro synchronized red cell cultures of North American native grapes and characterized its expression patterns. The expression analysis revealed that the CHS gene was highly expressed in the in vitro synchronized red cell cultures and the skin of the physiologically mature red berries of North American native grapes. The activation of specific sets of genes depends on plant development and response to environmental stimuli (Ban et al., 2003; Ageorges et al., 2006). Variations in expression can be explained by the fact that CHS mRNA and enzyme levels are highly regulated during development associated with tissue and cell-type-specific accumulation of flavonoid pigments, and in response to environmental stimuli for the synthesis of flavonoid or isoflavonoids involved in adaptation (Hahlbrock and Scheel, 1989).
This study has shown that strategies for the commercial production of antioxidants such as bioavailable anthocyanins and other polyphenols should target berry skins and in vitro synchronized red cells, which have in this study been shown to harbor increased production of flavonoids. Apart from giving clear solutions in natural products research, this preliminary study demonstrates the potential of functional genomics in the systematic studies of North American native grapes. The high expression of CHS in anthocyanin-pigmented tissues (Fig. 4) confirms that CHS is responsible for anthocyanin biosynthesis in red berries, as reported earlier in other species: G. hybrida (Helariutta et al., 1995), Asiatic hybrid lily (Nakatsuka et al., 2003), Dendodrobium orchid (Mudalige-Jayawickrama et al., 2005), and Paeonia suffruticosa (Zhou et al., 2011).
Sequence analysis and comparison of the novel muscadine CHS revealed that the ORF was 1525 bp in length and putatively encoded a polypeptide of 393 amino acids, which had high similarities (98% to 100%) with CHSs from other plant species (Table 1), with a predicted molecular mass of 42.8 kDa and a pI of 6.01. Sequence analysis of the putative amino acids revealed that muscadine CHS contained the family signature and the active amino-acid residues that are highly conserved among all CHS sequences. In addition, the results of the 3D structural modeling and phylogenetic analysis indicated that muscadine CHS and CHS from V. vinifera are closely identical (Fig. 5 and 6). These findings indicate that the muscadine CHS characterized in this study is a homolog of the CHS gene, and its protein is a typical CHS protein.
Conclusion
In this study, we amplified and characterized muscadine CHS and by expression analysis confirmed that CHS is involved in anthocyanin biosynthesis in the in vitro synchronized red cell cultures, skins, and flesh of Noble and Cynthiana. To our knowledge, this is the first time that CHS has been analyzed in the in vitro synchronized red cell cultures of North American native grapes. The amplification, characterization, and expression of other genes related to flavonoid accumulation in North American native grapes may pave the way to elucidate the molecular basis of berry pigmentation and can also facilitate and increase the development of in vitro red cell cultures of muscadine with more neutraceuticals compounds by manipulating the flavonoid structural and regulatory genes through functional genomics. The commercial production of in vitro synchronized red cell cultures as natural antioxidants will be a significant contribution to human medicine.
Acknowledgments
The authors are grateful to the Florida A&M University Dean's Office College of Agriculture and Food Science for providing Gina Davis, an undergraduate student, with a scholarship to do this work. They also thank Ms. Julian Bourne for her technical assistance. The research work was done with the financial support of the USDA/NIFA/AFRI Plant Biochemistry Program Grant # 2009-03127.
Disclosure Statement
No competing financial interests exist.
References
- Ageorges A. Fernandez L. Vialet S. Merdinoglu D. Terrier N. Romieu C. Four specific isogenes of the anthocyanin metabolic pathway are systematically co-expressed with the red colour of grape berries. Plant Sci. 2006;170:372–383. [Google Scholar]
- Aharoni A. O'Connell A.P. Gene expression analysis of strawberry achene and receptacle maturation using DNA microarrays. J Exp Bot. 2002;53:2073–2087. doi: 10.1093/jxb/erf026. [DOI] [PubMed] [Google Scholar]
- Altschul S.F. Madden T.L. Schaffer A.A. Zhang J. Zhang Z. Miller W. Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban T. Ishimaru M. Kobayashi S. Shiozaki S. Goto-Yamamoto N. Horiuchi S. Abscisic acid and 2,4-dichlorohenoxyaceic acid affect the expression of anthocyanin biosynthetic pathway genes in ‘Kyoho’ grape berries. J Horticultural Sci Biotechnol. 2003;78:586–589. [Google Scholar]
- Bitsch R. Netzel M. Frank T. Strass G. Bitsch I. Bioavailability and biokinetics of anthocyanins from red grape juice and red wine. J Biomed Biotechnol. 2004;24:293–298. doi: 10.1155/S1110724304403106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett-Lovsey R.M. Herbert A.D. Sternberg M.J. Kelley L.A. Exploring the extremes of sequence/structure space with ensemble fold recognition in the program Phyre. Proteins. 2008;70:611–625. doi: 10.1002/prot.21688. [DOI] [PubMed] [Google Scholar]
- Colova V. Synchronized strains of subepidermal cells of muscadine (muscadine sp.) grapevine pericarp for use as a sourse of flavonoids (nutraceuticals) Patent Publication No. US 2011/0054195 A1 2011.
- Commenges D. Scotet V. Renaud S. Jacqmin-Gadda H. Barberger-Gateau P. Dartigues J.F. Intake of flavonoids and risk of dementia. Eur J Epidemiol. 2000;16:357–363. doi: 10.1023/a:1007614613771. [DOI] [PubMed] [Google Scholar]
- Dugas A.J. Castaneda-Acosta J. Bonin G.C. Price K.L. Fischer N.H. Winston G.W. Evaluation of the total peroxyl radical-scavenging capacity of flavonoids: structure-activity relationships. J Nat Prod. 2000;63:327–331. doi: 10.1021/np990352n. [DOI] [PubMed] [Google Scholar]
- Gollop R. Even S. Tsolova V. Perl A. Expression of the grape dihydroflavonol reductase gene and analysis of its promoter region. J Exp Bot. 2002;53:1397–1409. [PubMed] [Google Scholar]
- Hahlbrock K. Scheel D. Physiology and molecular biology of phenylpropanoid metabolism. Annu Rev Plant Physiol Plant Mol Biol. 1989;40:347–364. [Google Scholar]
- Harbone J.B. Advances in Research Since. 1st. Chapman and Hall; London: 1986. 1966. The Flavonoids. [Google Scholar]
- Helariutta Y. Elomaa P. Kotilainen M. Griesbach R.J. Schröder J. Teeri T.H. Chalcone synthase-like genes active during corolla development are differentially expressed and encode enzymes with different catalytic properties in Gerbera hybrida (Asteracea) Plant Mol Biol. 1995;28:47–60. doi: 10.1007/BF00042037. [DOI] [PubMed] [Google Scholar]
- Kiefer F. Arnold K. Kunzli M. Bordoli L. Schwede T. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 2009;37:D387–D392. doi: 10.1093/nar/gkn750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koes R.E. Quattrocchio R. Mol J.N.M. The flavonoid biosynthetic pathway in plants: function and evolution. BioEssays. 1994;16:123–132. [Google Scholar]
- Kumar A. Ellis B.E. A family of polyketide synthase genes expressed in ripening Rubus fruits. Phytochemistry. 2003;62:513–526. doi: 10.1016/s0031-9422(02)00572-1. [DOI] [PubMed] [Google Scholar]
- Mudalige-Jayawickrama R.G. Champagne M.M. Hieber A.D. Kuehnle A.R. Cloning and characterization of two anthocyanin biosynthetic genes from Dendrobium orchid. J Am Soc Hortic Sci. 2005;130:611–618. [Google Scholar]
- Nakatsuka A. Izumi Y. Yamagishi M. Spatial and temporal expression of chalcone synthase and dihydroflavonol 4-reductase genes in the Asiatic hybrid lily. Plant Sci. 2003;165:759–767. [Google Scholar]
- Notredame C. Higgins G.D. Heringa J. T-coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Passamonti S. Vrhovsek U. Vanzo A. Mattivi F. Fast access of some grape pigments to the brain. J Agric Food Chem. 2005;53:7029–7034. doi: 10.1021/jf050565k. [DOI] [PubMed] [Google Scholar]
- Samuelian S.K. Simova E.P. Colova V.M. Chalcone synthase gene (CHS1) expressed in the North American ‘Noble’ muscadinia var. during ripening 2009a.
- Samuelian S.K. Camps C. Kappel C. Simova E.P. Delrot S. Colova V. Differential screening of overexpressed genes involved in flavonoid biosynthesis in North American native grapes: ‘Noble’ muscadinia var. and ‘Cynthiana’ astevalis var. Plant Sci. 2009b;177:211–221. [Google Scholar]
- Szalma S.J. Snook M.E. Bushman B.S. Houchins K.E. McMullen M.D. Duplicate loci as QTL: The role of chalcone synthase loci in flavone and phenylpropanoid biosynthesis in Maize. Crop Sci. 2002;42:1679–1687. [Google Scholar]
- Sugihara N. Arakawa T. Ohnishi M. Furuno K. Anti- and pro-oxidative effects of flavonoids on metal-induced lipid hydroperoxidedependent lipid peroxidation in cultured hepatocytes loaded with alphalinolenic acid. Free Radic Biol Med. 1999;27:1313–1323. doi: 10.1016/s0891-5849(99)00167-7. [DOI] [PubMed] [Google Scholar]
- Tamura K. Dudley J. Nei M. Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tuteja J. H. Vodkin L.Q. Structural features of the endogenous CHS silencing and target loci in the soybean genome. Crop Sci. 2008;48:S49–S68. [Google Scholar]
- Trevisanato S.I. Kim Y.I. Tea and health. Nutr Rev. 2000;58:1–10. doi: 10.1111/j.1753-4887.2000.tb01818.x. [DOI] [PubMed] [Google Scholar]
- Tuteja J.H. Clough S.J. Chan W.C. Vodkin L.O. Tissue-specific gene silencing mediated by a naturally occurring chalcone synthase gene cluster in Glycine max. Plant Cell. 2004;16:819–835. doi: 10.1105/tpc.021352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J. De Preter K. Pattyn Poppe B. Van Roy N. De Paepe A. Speleman F. Accurate normalization of real time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B. Flavonoid, biosynthesis: a colourful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001;126:485–493. doi: 10.1104/pp.126.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J. Gu H. Yang Z. Likelihood analysis of the chalcone synthase genes suggests the role of positive selection in Morning Glories (Ipomoea) J Mol Evol. 2004;58:54–63. doi: 10.1007/s00239-003-2525-3. [DOI] [PubMed] [Google Scholar]
- Zhou L. Wang Y. Peng Z. Molecular characterization and expression analysis of chalcone synthase gene during flower development in tree peony(Paeonia suffruticosa) African J Biotechnol. 2011;10:1275–1284. [Google Scholar]