Abstract
Antibodies directed against the influenza hemagglutinin (HA) protein largely mediate virus neutralization and confer protection against infection. Consequently, many studies and assays of influenza vaccines are focused on HA-specific immune responses. Recombinant HA (rHA) proteins can be produced in a number of protein expression and cell culture systems. These range from baculovirus infection of insect cell cultures, to transient transfection of plants, to stably transfected human cell lines. Furthermore, the rHA proteins may contain genetic modifications, such as histidine tags or trimerization domains, intended to ease purification or enhance protein stability. However, no systematic study of these different forms of the HA protein have been conducted. It is not clear which, if any, of these different protein expression systems or structural modifications improve or diminish the biological behavior of the proteins as immunogens or antigens in immune assays. Therefore we set out to perform systematic evaluation of rHA produced in different proteins expression systems and with varied modifications. Five rHA proteins based on recent strains of seasonal influenza A and five based on influenza B HA were kindly provided by the Biodefense and Emerging Infections Reagent repository (BEIR). These proteins were evaluated in a combination of biochemical and structural assays, in vitro humoral and cellular immune assays, and in an animal vaccination model. Marked differences in the behavior of the individual proteins was evident suggesting that they are not equal when being used to detect an immune response. They were, nevertheless, similar at eliciting neutralizing antibody responses.
Keywords: Influenza virus, vaccine, hemagglutinin, B cells, antibody, immunology
1. Introduction
Influenza hemagglutinin (HA) glycoproteins are responsible for two major events in the influenza life cycle: (a) binding/attachment to sialic acid containing receptors in the target cells and (b) the fusion of the virus membrane with the endosomal membrane resulting in the release of the viral genome into the cytosol of the target cell [1, 2]. Due to the role of the HA in key aspects of the virus life cycle, and its relative abundance in the surface of the viral membrane, this protein is a relevant target for the humoral response. Indeed, a substantial portion of the antibody response following influenza infection in mice and humans is directed against the HA, with a population of these antibodies capable of neutralizing HA binding and/or fusion [3–8]. More importantly, these antibodies are sufficient for protection against live virus infection [3, 9–12] and their concentration shows a negative correlation with disease incidence in humans [12–14].
In the United States, current FDA licensed influenza vaccines have been optimized to maximize the generation of B cell and antibody responses against the HA of the strains included in the vaccine. In addition, a number of experimental vaccines, are also focused on eliciting antibody responses to the HA component of the vaccine (DNA, recombinant HA, and virus-like particles) [15–21]. Recombinant HA (rHA) proteins have been widely used to study B cell responses following vaccination and/or infection in humans and animals alike [21–23]. These proteins have been produce in an assortment of protein expression systems including yeast, bacteria, insect cells, plant cells, and human cell lines [24–30]. However, to appropriately measure responses to the HA protein of interest it is imperative that the rHA that most resembles the viral HA is chosen. In addition, there are recent efforts to license baculovirus-derived rHA vaccines for seasonal and pandemic strains of influenza [25, 31, 32]. However, while these vaccines have been shown to elicit comparable responses to conventional influenza vaccines, no systematic study of these different forms of the HA protein have been conducted. It is not clear which, if any, of these different protein expression systems improve or diminish the biological behavior of the proteins as viewed by the immune system. Therefore we set out to perform systematic evaluation of rHA produced in different protein expression systems. Five rHA proteins based on recent strains of seasonal influenza A and five based on influenza B HA were obtained through the Biodefense and Emerging Infections Research Resources Repository (BEIR). These proteins were evaluated in a combination of biochemical and structural assays, in vitro humoral and cellular immune assays, and in an animal vaccination model. Marked differences in the behavior of the individual proteins were evident suggesting that they are not equal in being used to detect or elicit an immune response.
2. Materials and Methods
2.1 Recombinant HA proteins
Purified rHAs from A/Brisbane/59/07 and B/Florida/04/06 listed below (and Table 1) were obtained through the NIH BEIR Repository (NIH/NIAID). For influenza A/Brisbane/59/07: truncated H1 HA protein with a C-terminal trimerization (T4 fibritin foldon) domain, thrombin cut site, and histidine tag produced in Trichoplusia ni High Five insect cells (Tni), NR-15476; truncated H1 HA protein with C-terminal histidine tag produced in human 293 cells (HEK), NR-15477; full-length H1 HA protein produced from baculovirus in Spodoptera frugiperda insect cells (Sf9), NR-15478; truncated H1 HA protein with C-terminal histidine tag produced in Nicotiana benthamiana plant cells (Nb), NR-15479; truncated H1 HA protein with a C-terminal trimerization (T4 fibritin foldon) domain, thrombin cut site, and histidine tag produced from baculovirus in Spodoptera frugiperda insect cells (Sf9 HAT), NR-15480. For influenza B/Florida/04/06: truncated HA protein with a C-terminal trimerization (T4 fibritin foldon) domain, thrombin cut site, and histidine tag produced in Trichoplusia ni High Five insect cells, NR-15481; truncated HA protein with C-terminal histidine tag produced in human 293 cells, NR-15482; full-length HA protein produced from baculovirus in Spodoptera frugiperda insect cells, NR-15483; truncated HA protein with C-terminal histidine tag produced in Nicotiana benthamiana, NR-15484; truncated HA protein with a C-terminal trimerization (T4 fibritin foldon) domain, thrombin cut site, and histidine tag produced from baculovirus in Spodoptera frugiperda insect cells, NR-15485. Available amino acid sequences for these proteins and their modifications are provided in supplementary figure 1 and 2. Detailed information regarding the proteins’ production and/or purification process has been previously reported [33–37] and is summarized in Table 1.
Table 1.
A/Brisbane/59/07 and B/Florida/04/06 recombinant hemagglutinin proteins
| Influenza Strain | Catalog number |
Production source |
Modifications | Purification | HAU/ug cRBC | HAU/ug tRBC | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ΔTM | TCS | TD | HT | NiC | AEX | LBC | |||||
| NR-15476 | Tni | + | + | + | + | + | + | − | ND | 16 | |
| NR-15477 | HEK | + | − | − | + | + | − | − | ND | ND | |
| A/Brisbane/59/07 | NR-15478 | Sf9 | − | − | − | − | − | + | + | >2048 | >2048 |
| NR-15479 | Nb | + | − | − | + | + | − | − | ND | ND | |
| NR-15480 | Sf9 HAT | + | + | + | + | + | − | − | ND | 256 | |
| NR-15481 | Tni | + | + | + | + | + | + | − | 64 | 512 | |
| NR-15482 | HEK | + | − | − | + | + | − | − | ND | ND | |
| B/Florida/04/06 | NR-15483 | Sf9 | − | − | − | − | − | + | + | >2048 | >2048 |
| NR-15484 | Nb | + | − | − | + | + | − | − | 1 | 1 | |
| NR-15485 | Sf9 HAT | + | + | + | + | + | − | − | >2048 | >2048 | |
ND = Not detectable ΔTM = Removal of transmembrane domain (38–47aa) TCS = Thrombin cut site inserted at C-terminus TD = Bacteriophage T4 Trimerization domain inserted at C-terminus HT = 6–8 Histidine residues inserted at the C-terminus NiC = Nickel Column AEX = Anion exchange column LBC = Lectin Binding Column
2.2 Size exclusion chromatography
Recombinant HA proteins (100–200µg) were analyzed by size exclusion chromatography (SEC) using a BioRad BioLogic Duo Flow and a Phenomenex BioSep-Sec s4000 column with native exclusion range of 15,000 to 2,000,000 daltons. Phosphate Buffed Saline (PBS) was used as the mobile phase. The flow rate was maintained at 1 ml/minute.
2.3 Mice
Six to eight week-old female C57BL/6 (B6) mice were purchased from Charles River Laboratories and maintained under pathogen free conditions at the University of Rochester, AAALAC approved, vivarium facilities. All experimental protocols involving the use of animals were reviewed and approved by the Institutional Animal Care and Use Committee.
2.4 Infection
A/Brisbane/59/07 (NR-12282) and B/Florida/04/06 (NR-9696) influenza viruses were obtained from BEIR. Each virus was passed once in specific pathogen free eggs (Charles River, SPAFAS) and a laboratory stock was generated. Mice, 10–12 weeks of age, were anesthetized with Avertin (2,2,2 Tribromoethanol) and inoculated intranasally with 30µl containing 150,000 egg infectious dose units (EID50) of A/Brisbane/59/07 virus or 5,000 EID50 of B/Florida/04/06. Mice were monitored for weight loss 8–10 days following infection.
2.5 Immunization
B6 female mice, 10–12 weeks of age, were immunized intramuscularly at the left flank, followed by a booster immunization at the same site 14 days later. Unless otherwise stated, each mouse received a priming dose of 5µg of rHA followed by a booster dose, containing 5µg of the same rHA used for priming, at day 14. A subset of mice were primed and boosted, as previously described, with the 2008–2009 trivalent influenza split virus vaccine (TIV). The dose of TIV was adjusted so that each mouse received 2ug of each of the HAs present in the formulation. All vaccine formulations were adjuvanted with CpG. The TIV reagent was obtained through the NIH BEIR Repository, NIAID, NIH: afluria® Influenza Virus Vaccine (2008–2009 Formula), NR-17598. A concentration of 25µg/mouse of CpG oligonucleotides (ODN-1826) was co-administered alongside the rHA as an adjuvant. All antigens were prepared in PBS and administered in a final volume of 100µl/mouse.
2.6 Blood collection
Blood from infected or vaccinated mice was drawn 21 days post infection or 21 days after booster immunization by cardiac puncture. Blood was allowed to coagulate and serum was collected. Serum samples were stored at −80°C until needed.
2.7 Hemagglutination assay
Recombinant HA proteins were diluted to a concentration of 2µg/100µl of PBS and 50µl were placed in column 1 of a round bottom 96-well plate. Recombinant HA samples were diluted two-fold along the length of the plate and 50µl of 1% chicken red blood cells (CRBC) in PBS were added to each well. Samples in a duplicate plate received 50µl of 1% turkey red blood cells (TRBC) in PBS. The plates were incubated at room temperature for one hour and agglutination was recorded. The highest dilution in which agglutination was observed was used to calculate the amount of hemagglutination units per microgram of rHA.
2.8 Hemagglutination inhibition (HAI) assay
Sera from infected and/or vaccinated mice were heat inactivated at 56°C for 45 minutes. The sera was allowed to return to room temperature and it was then diluted 1:5 in 1% CRBC/PBS to adsorb nonspecific agglutination activity in the sera. This mixture was incubated at room temperature for 30 minutes, mixing the contents by inversion at 10-minute intervals. Following incubation the CRBCs were pelleted by centrifugation and 100µl of cleared serum were added to row of a 96-well plate. Two-fold serial dilution of the sera was performed followed by incubation with 4HAU of either A/Brisbane/59/07 or B/Florida/04/06 virus. The serum/virus mixture was incubated for one hour at 37°C. 1% CRBCs were added to serum/virus mixture and the hemagglutination inhibition (HAI) activity was recorded following one hour of incubation at room temperature. A back titration of the stock virus preparations used in the assay was performed to confirm 4 HAU. The HAI titer was determined as the highest dilution in which agglutination was inhibited.
2.9 Human sample collection and processing
Blood was collected at days 7 and 28 following administration of the 2009–2010 trivalent influenza vaccine formulation. Day 7 samples were collected into CPT tubes (Becton-Dickinson, Franklin Lakes, NJ) and the lymphocytes separated per the manufacturer’s protocol. PBMC were cryopreserved using a modified Immune Transplantation Network protocol. Briefly, isolated PBMC were resuspended in 90% human AB serum (Gemini Bio-Products, Woodland, CA) 10% high-grade DMSO (Sigma, St. Louis, MO) and frozen to −80° C at a density of 107 per ml using an isopropanol-filled, controlled-rate freezing device. After 24–48h at −80°C, vials were transferred into liquid nitrogen for long-term storage. Prior to immune assay, cells were rapidly thawed, then slowly diluted by adding cold RPMI + 10% human AB serum drop wise. Viability and recovery (>80%) were assessed by counting cells in a hemacytometer in the presence of trypan blue dye. Serum was separated from day 28 blood samples and subsequently stored at −80°C until used.
2.10 Enzyme-linked immunosorbent (ELISA) assay
Mouse and human HA-specific immunoglobulin G (IgG) antibodies were detected by ELISA. 96 well flat-bottom plates (Nunc #422404) were coated with a concentration of 0.05µg per well of rHA or control antigen. The plates were blocked with 1% PBS/BSA, serial dilutions of the serum were performed, and 50µl of each dilution were added in duplicate to the antigen-coated wells. Plates were incubated overnight at 4°C. Antigen specific IgG was detected by incubating the plates for 1 hour with 50µl of a goat anti-mouse IgG antibody conjugated to horseradish peroxidase (Southern Biotech 1030-05) or goat anti-human IgG antibody conjugated to horseradish peroxidase (Southern Biotech 2040-05) diluted according to manufacture’s recommendations. The assay was developed by adding 75µl of TMB substrate (Biolegend Cat#421101) for 7–10 minutes and stopped by adding equal volume of 2N H2SO4. The optical density was read at 450nm. ELISA end point titers for mouse assays are expressed as the logarithm of the reciprocal of the highest dilution at which the optical density is above 3 times the average of the mean of a similarly diluted negative control.
2.11 Enzyme-linked immunospot (ELISPOT) assay
Sterile 96-well nitrocellulose plates (Millipore, Bedford, MA) were coated overnight with 50µl of HA antigens in PBS at concentration of 0.1 µg per well. Plates were washed and blocked for 2 hours in 10% complete MEM (Cellgro cat. No. 10-010-CV). Mediastinal lymph nodes were collected from infected mice (Day 12) and pooled (2 LN/pool). Single cell suspensions were prepared and diluted two-fold down the plate. The plates were incubated for 6–8 hours at 37°C. HA-specific IgG was detected by a goat anti-mouse IgG antibody conjugated to alkaline phosphatase (Southern Biotech 1030-04). Spots were developed by alkaline phosphatase substrate kit III (Vector labs SK-5300) and counted on a CTL scanner. Human B cell ELISPOTs were performed from freshly thawed PBMCs by serially diluting them down the plate and incubating for 6 hours in the presence of a phosphatase labeled goat anti-human IgG detection antibody (KPL 4751-1006). Development and assay analysis was performed as described above for the mouse ELISPOT.
3. Results
3.1 Physical properties of recombinant influenza HA proteins
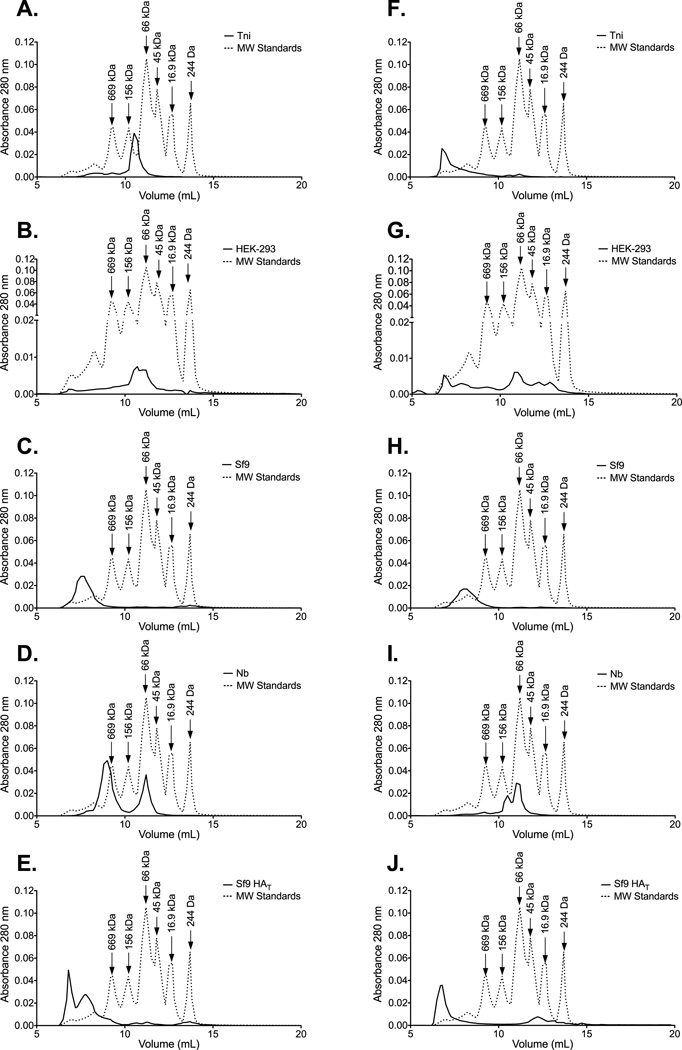
HA purified from the surface of influenza virions aggregates into structures containing 6–8 HA trimers [38]. However, expressed rHA proteins can be observed as oligomers, trimers, and/or monomers [16, 39]. In order to determine the predominant form of the rHA in these preparations we performed size exclusion chromatography. Protein preparations derived from Sf9 cells were primarily composed of high molecular weight oligomers (HMWO) (Figure 1C, 1E, 1H, and 1J). These oligomers display molecular weights greater than 669KDa suggesting that they are composed of at least 3 HA trimers. In contrast, the elution profile of HEK- and Nb- derived HA proteins exhibited preparations with a more heterogeneous range of molecular weights (Figure 1B, 1D, 1G, and 1I). In these preparations, monomeric HA was readily observed (peak at approximately 66KDa). It is worth noting that the detection of HMWO in the HEK-derived HA from B/Florida/04/06 was not consistent across experiments. Interestingly, the elution profile of the Tni-derived HAs was the most different between the influenza A and B proteins. Here the A/Brisbane/59/07 preparation contained a homogeneous population of protein that eluted between the 156KDa and the 66KDa standards (Figure 1A). In contrast, the bulk of the B/Florida/04/06 rHA was arranged into HMWO (Figure 1F).
Figure 1. Influenza rHA proteins produced in various protein expression systems differ in their ability to aggregate into HMWO.
SEC reveals the profile of the oligomeric species contained in the (A) Tni-, (B) HEK-, (C) Sf9-, (D) Nb-, and (E) Sf9 HAT- derived HA preparations from A/Brisbane/59/07. Likewise, the oligomeric species contained in the Tni-, HEK-, Sf9-, Nb-, and Sf9 HAT- derived HA preparations from B/Florida/04/06 are displayed in panels F-J. Molecular weight standards (dotted line) are included as a reference. Fractions containing monomeric HA elute at volumes that correspond to the 66 kDa standard. HMWO fractions (> 669 kDa) contain aggregates that are composed of at least 9 HA monomers.
Virus-derived HA forms oligomers with a rosette-like structural organization [38]. Rosettes are readily observed when full-length rHA proteins are produced by baculovirus expression vector systems [40]. To determine the structural organization of the HA in the various rHA preparations we performed TEM. HMWO arranged into rosette formation, like those previously reported [38, 40], are predominantly observed in the Sf9-derived full-length HA. Rosette structures were not as apparent in the other rHA preparations (Supplementary Figure 3). These data suggests that the structural organization of rHA proteins is more a result of the cell culture system, the genetic modifications in the HA, and/or the purification method employed than the strain of virus from which the HA is derived.
Because the rHA proteins were produced by independent entities we further analyzed their conformation and biological activity to confirm that the proteins retained the properties reported in their respective product sheets. SDS-PAGE analysis of the rHA preparations from A/Brisbane/59/07 and B/Florida/04/06 shows that the bulk of the HA protein contained in all five of the rHA preparations is in an HA0 or uncleaved configuration (Supplementary Figure 4). The rHAs derived from HEK cells lack the transmembrane (TM) domain (38–47aa) but are highly glycosylated, which is reflected in a slightly higher molecular weight when compared to the Sf9- and Nb-derived HAs (Supplementary Figure 5). In addition, a considerable shift in molecular weight is observed upon enzymatic cleavage of the carbohydrate groups with PNGase F (Supplementary Figure 5).
The biological activity of HA proteins can be determined by their ability to aggregate red blood cells (RBC) through binding to sialic acids on the cell surface [41–43]. The hemagglutination activity of the rHA preparations against turkey and chicken RBC is shown in Table 1. Agglutination activity was only observed for the rHA (both A/Brisbane/59/07 and B/Florida/04/06) preparations derived from insect cell lines and correlates for the most part with the presence of HMWO (Figure 1). Agglutination activity to either of the avian RBCs was not detectable in any of the HEK-derived HAs. Agglutination of TRBCs by the B/Florida/04/06 Nb-derived rHA was not consistent across experiments. Furthermore, agglutination activity could not be restored by enzymatic removal of carbohydrate groups (data not shown). These differences in biological activity between rHA preparations recapitulate the information provided in the product specification sheets. These data suggest that the structural differences imparted to the rHA, by the production and/or purification methods employed in their generation, affect the biological activity of the HA protein independent of the HA being expressed.
3.2 Recombinant HA antigenicity measured by ELISA
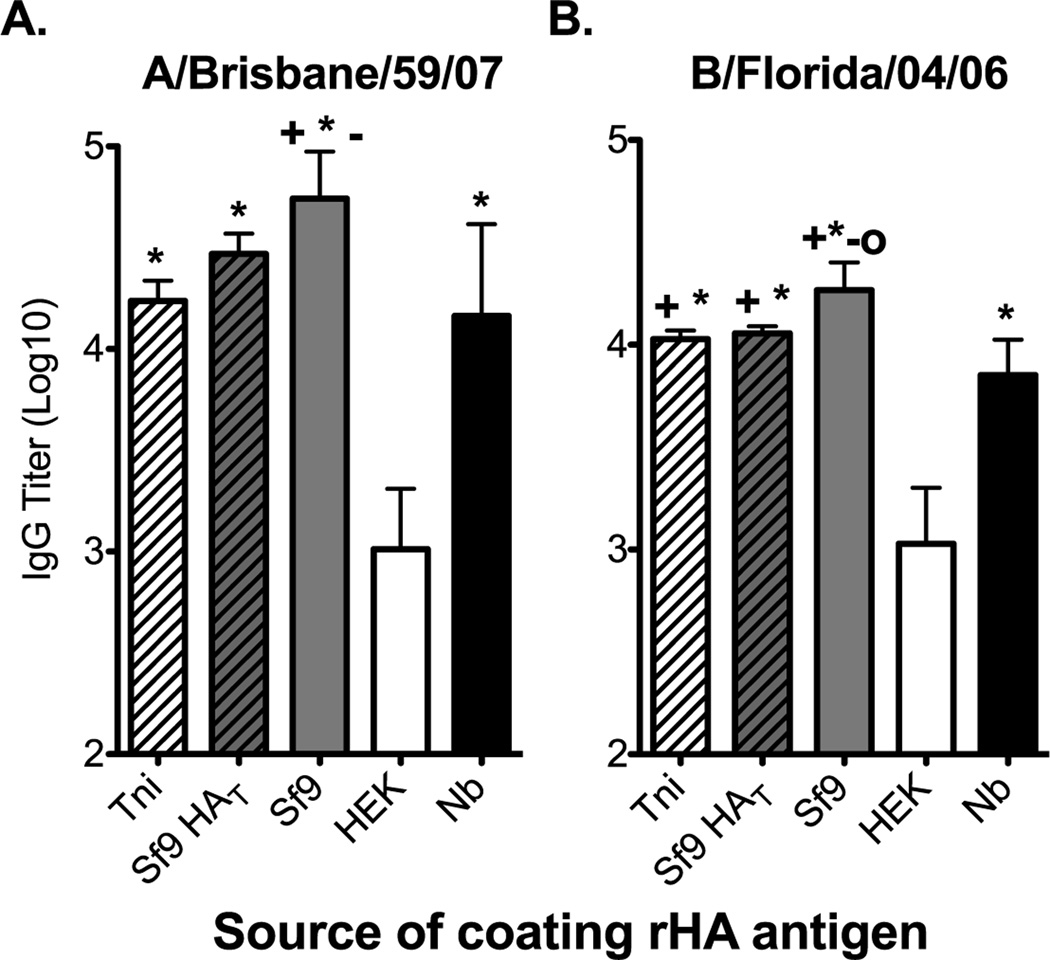
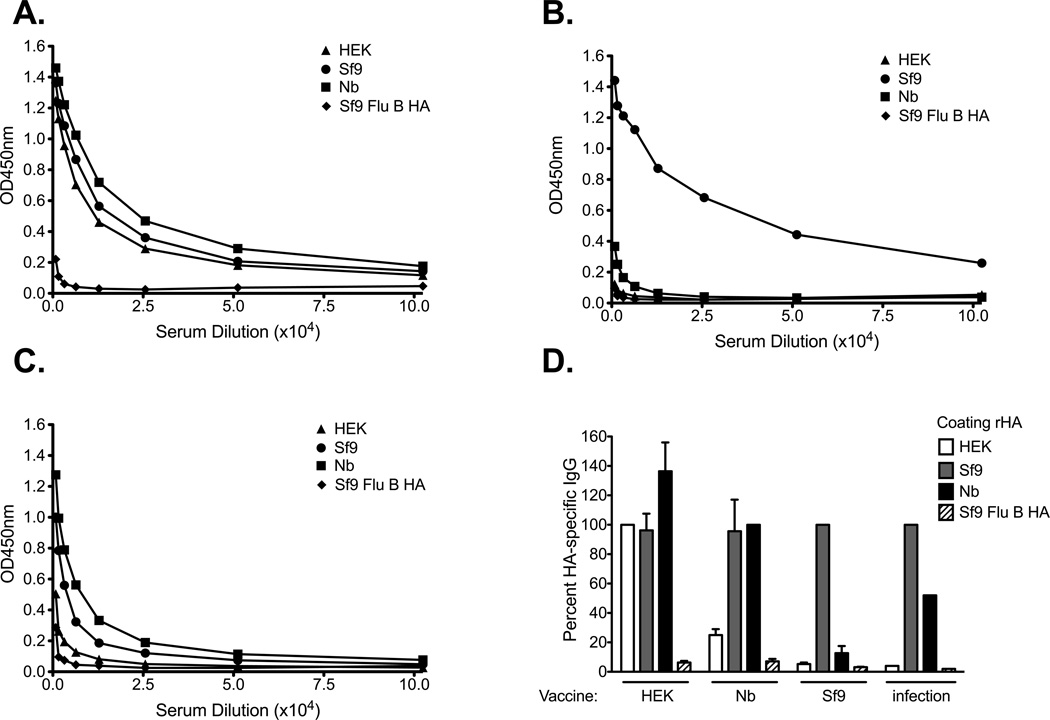
To determine if the differences in the physical and biological properties of the rHA proteins translated into differences in antigenicity in an ELISA we infected B6 mice with either A/Brisbane/59/07 or B/Florida/04/06 influenza viruses. The serum antibodies generated following virus infection did not bind the rHAs equally (Figure 2A and B). A/Brisbane/59/07 immune sera bound the Sf9-derived HA the best, while binding to the HEK-derived HA was 54-fold and 14-fold lower when compared to that observed with the Sf9- and Nb-derived HAs, respectively. Only a 4-fold, albeit significant, decrease in serum IgG binding was observed between the Sf9- and Nb-derived HAs (Figure 2A). Likewise, a small reduction in HA-specific IgG binding was observed when the Tni- (3-fold) or the Sf9 HAT (1.8-fold) proteins were used as coating antigens (Figure 2A). Similar results were observed with B/Florida/04/06 immune sera (Figure 2B). Here we observed a 17-fold difference in binding of immune serum between the HEK-derived HA and the Sf9-derived HA. Again, a slight, but significant decrease (2.6-fold) in binding was observed when the Nb-derived HA was used as coating antigen. Binding of HA-specific IgG to the Tni- and the Sf9 HAT proteins was only reduced less than 1.7-fold. However, binding to the Tni, Nb-, or Sf9 HAT proteins was greater than 6.6-fold higher than that obtained against the HEK-derived HA (Figure 2B). Due to the small differences in binding observed between the insect cell-derived rHAs we choose to perform all remaining experiments, unless otherwise stated, with the Sf9-, Nb-, and HEK-derived HAs. It is worth noting that we did not observe differences in the ability of these proteins to saturate the wells at the coating concentrations used for these experiments. Furthermore, up to an 8-fold increase in the coating concentration of the HEK and Nb-derived HAs did not significantly increased the binding of virus-specific sera against these proteins (data not shown).
Figure 2. The rHA production process significantly affects its antigenicity in a mouse IgG ELISA assay.
B6 female mice were infected intranasally with 150,000 EID50 of (A) A/Brisbane/59/07 or (B) 5000 EID50 of B/Florida/04/06 and serum was harvested 21 days post infection. ELISA plates were coated overnight with equal concentrations of Tni (white hatched bars), Sf9 HAT (grey hatched bars), Sf9 (grey bars), HEK (white bars), or Nb (black bars) derived rHAs (0.05µg/well). Convalescent serum was diluted two-fold and added to the rHA-coated plates. IgG titer is defined as the highest dilution in which the optical density is greater than 3 times the average of an equally diluted negative (naïve) control. Mean IgG titer and standard deviation are presented on the graph. Mann-Whitney (t-test) statistical analysis performed with n=14/group (A) and n=9/group (B). Statistically significant comparisons against HEK (*), Nb(+), Tni (−), and Sf9 HAT (O) proteins are denoted by the corresponding symbols. The respective p-values for the statistically significant comparisons are as follows for panel A + = p<0.0213, * = p<0.0003, and − = p<0.004. For panel B + = p<0.0463, * = p<0.0004, − = p<0.0113, and O = p<0.0170.
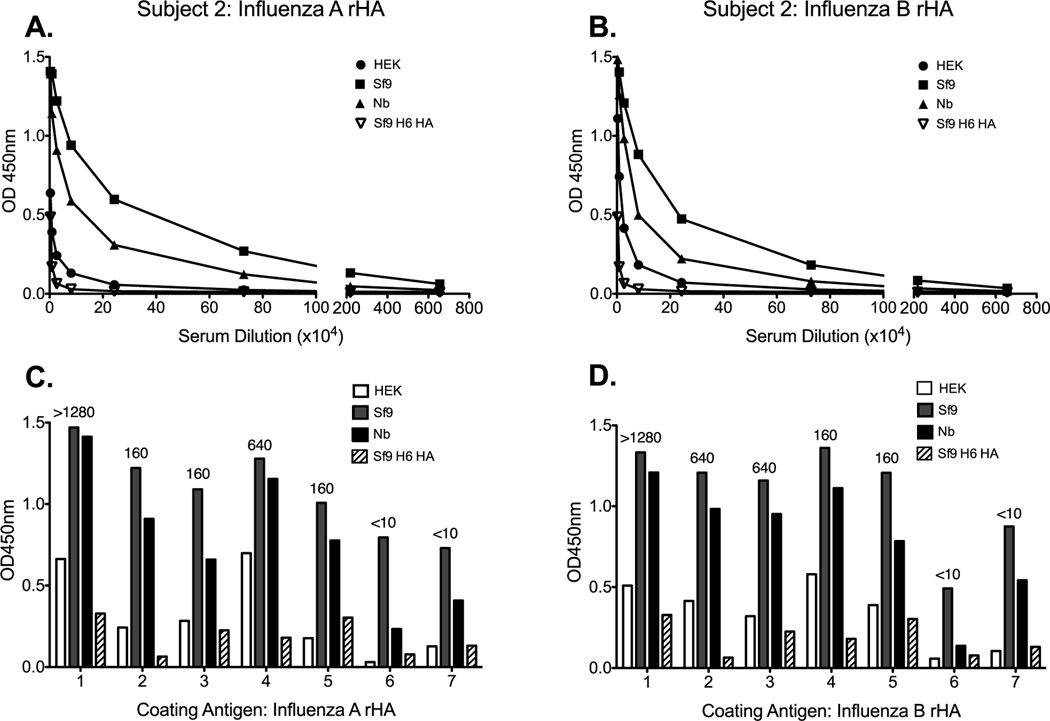
Next we tested sera from seven human subjects vaccinated with the 2009–2010 TIV formulation to determine if the observed binding pattern could be replicated in another species. Indeed, antibodies elicited following vaccination with TIV bound Sf9-derived HA from both influenza A and B more readily than they did Nb or HEK counterparts (Figure 3A and B). The reduced antibody binding to the HEK-derived HA, when compared to the Sf9-derived HA, was pronounced and it was consistent across all subjects. However, the extent of antibody binding to the Nb-derived HA was variable between subjects and in occasions it was comparable to the Sf9-derived proteins (Figure 3C and D). Next we wanted to know if there was a relationship between hemagglutination inhibition (HAI) titers and the pattern of binding to the different versions of the rHA proteins from influenza A and B viruses. HAI titers were obtained against the 2009–2010 vaccine reference H1N1 and influenza B antigens and are displayed above the corresponding subjects IgG titer bar (Figure 3C and D). As expected, a higher HAI titer was associated with higher binding to the HAs. However, we did not observe any relationship between the extent of HAI activity and the pattern of binding to the different versions of the influenza A and B proteins (Figure 3C and D). Taken together, these data suggest that the rHA production process significantly affects its ability to detect antibodies induced during live-virus infection or TIV vaccination as measured by binding to these proteins in an IgG ELISA assay.
Figure 3. The rHA production process significantly affects its antigenicity in a human IgG ELISA assay.
Human sera from seven subjects vaccinated with the 2009–2010 trivalent influenza vaccine were tested for antibody binding against the rHA antigens from influenza A and B. A representative plot of antibody binding against HEK- (circle), Sf9- (square), and Nb- (triangle) derived rHA, or control Sf9-derived H6 HA (upside down triangle) is presented in panels A (A/Brisbane/59/07) and B (B/Florida/04/06). The antibody binding profile, for each individual subject, to the various versions of rHA from A/Brisbane/59/07 and B/Florida/04/06 are presented in C and D respectively. HAI titers were obtained against the vaccine reference antigens for the corresponding influenza strain. Numbers above the bars correspond to the HAI titer obtained for each subject in panels C and D.
3.3 Recombinant HA antigenicity in B cell ELISPOT assays
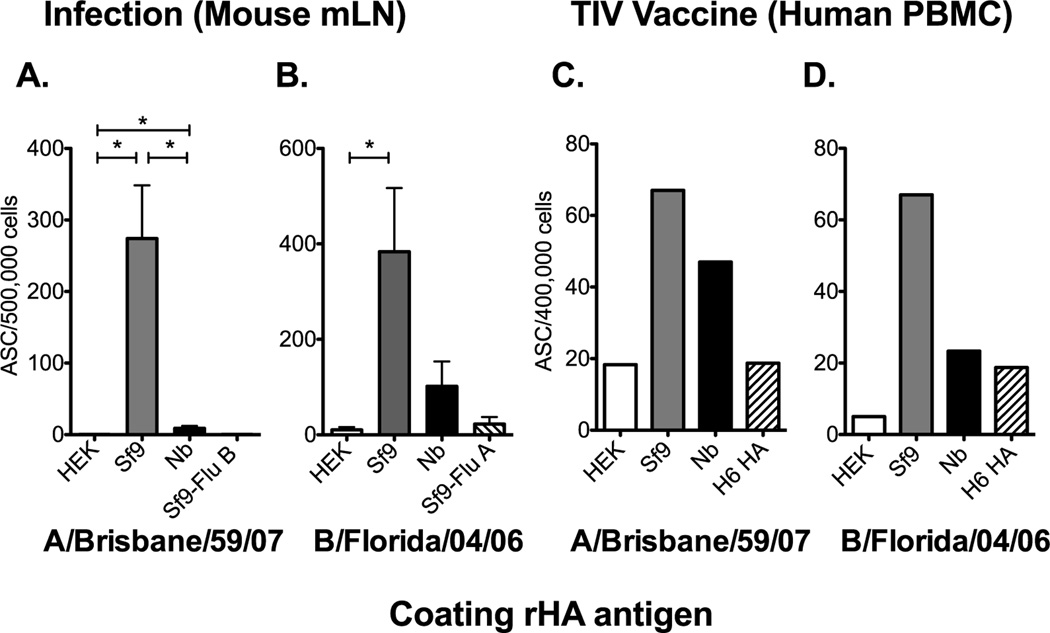
To further characterize the antigenicity of these rHA proteins in routine assays used to study responses to influenza infection and vaccination we performed B cell ELISPOT assays. Here we assessed the generation of HA-specific antibody (IgG) secreting cells (ASC) in the mediastinal lymph node (mLN) following live virus infection of B6 mice. HA-specific ASC were readily detected from the mLN of A/Brisbane/59/07 infected mice when the Sf9-derived rHA was used as the coating antigen (Figure 4A). The detection of HA-specific ASC was reduced or not detectable when the ELISPOT plates were coated with Nb- or HEK-derived rHAs respectively. Similar results were obtained following live virus infection of mice with B/Florida/04/06 (Figure 4B). In addition, the detection of the ASC response in the peripheral blood following vaccination in one human subject confirms the increase in detection when the Sf9-derived rHA was used as the coating antigen (Figure 4C and D). However, the ASC responses against the HEK-derived HA were at or below background levels. Taken together these data suggest that the rHA production process can significantly affect the detection of HA-specific ASC induced during live virus infection or TIV vaccination in an IgG B cell ELISPOT assay.
Figure 4. The rHA production process significantly affects its antigenicity in a B cell ELISPOT assay.
(A) B6 female mice were infected intranasally with 150,000 EID50 of A/Brisbane/59/07 and the mLN was harvested 12 days post infection. Mediastinal LNs from 12 mice were pooled in groups of 2 and processed into single cell suspensions. (B) B6 mice were infected with 6000 EID50 of B/Florida/04/06 and the mLN were harvested at day 12 days post infection. Single cell suspensions from individual mice were added into the plate. Mean ASC frequencies and standard error of the mean are presented in panel A and B. PBMCs, collected 7 days post seasonal influenza vaccination, from one vaccinated human subject were plated (4×105 cells/well) and diluted two-fold down the plate. HA-specific IgG secreting cells against the rHA from A/Brisbane/59/07 or B/Florida/04/06 are presented in C and D respectively. Mean ASC frequencies are plotted in C and D. Error bars represent the standard deviation between normalized wells. For panel A and B, Mann-Whitney (t-test) statistical analysis was performed with n = 5–6 and * = p<0.002.
3.4 Available B cell epitopes are not the same across rHA produced in different expression systems
Due to the observed differences in the rHA physical properties it is possible that B cell epitope availability is not the same across these proteins. To study this possibility we turned to a mouse model of rHA prime/boost vaccination. Here, mice received two intramuscular doses of 5µg of rHA protein, adjuvanted with CpG, 14 days apart. This vaccination strategy consistently elicited high titers of HA-specific antibodies (data not shown). Serum was collected 21 days post boost and tested for binding activity against each of the influenza A/Brisbane/59/07 rHAs. As expected, vaccination with either Sf9- or Nb-derived rHAs, elicited IgG antibodies that reacted primarily against the immunizing HA (Figure 5B circles and 5C squares). However, the extent of cross-reactivity across the various rHA versions varied depending on the rHA used for immunization. To compare the extent of antibody cross reactivity, against the other versions of the protein, we selected one dilution (1:3200) that provided the best signal to noise ratio and normalized the data to the optical density obtained by coating with the immunizing rHA. Due to the lack of a virus -purified HA the binding activity from virus immune serum was normalized to that of the Sf9-derived HA. This HA was selected because virus immune sera had the highest reactivity to it in an ELISA based assay (Figure 2A). Binding reactivity of sera from immunized mice (3–4 mice/group) is presented figure 5D. Limited antibody cross reactivity was observed to the HEK- and Nb-derived HAs when mice were immunized with the Sf9-derived protein. Here, the elicited antibodies were more likely to bind to the immunizing HA than to the HEK (5%) and Nb (12%) derived rHA antigens (Figure 5D). This cross reactivity profile is not unlike that observed for antibodies elicited following A/Brisbane/59/07 infection suggesting similar presentation of epitopes between the HA produced during a viral infection and the Sf9-derived recombinant (Figure 2 and Figure 5D). In contrast, antibodies elicited by immunization with Nb-derived HA displayed cross reactivity against the Sf9- (95%) and HEK-derived (25%) proteins (Figure 5D). The high cross reactivity against the Sf9-derived HA suggests that the majority of the epitopes, to which an antibody response was elicited following vaccination with Nb-derived rHA, are present in the Sf9-derived HA (Figure 5D). However, 88% percent of the antibody response to Sf9-derived HA appears to be directed against epitopes not presented by the other two rHA versions (Figure 5D). Interestingly, the antibodies elicited by immunization with HEK-derived HA displayed a similar degree of cross reactivity towards all 3 versions of the A/Brisbane/59/07 HA (Figure 5D). Taken together these data suggest that, from an immunological standpoint, the Sf9-derived rHA displays a similar array of potentially conformation-dependent B cell epitopes to the HA presented during a live virus infection.
Figure 5. Recombinant HA IgG binding profile following vaccination with various rHAs from A/Brisbane/59/07.
B6 female mice were immunized intramuscularly (left flank) with 5ug of rHA adjuvanted with CpG. 14 days after priming, the mice received a booster immunization consisting of the same rHA preparation (5µg rHA and CpG). Serum from mice immunized with (A) HEK-, (B) Sf9-, or (C) Nb- derived HA was collected 21 days after boost. ELISA plates were coated overnight with equal concentrations of each of the rHA proteins (0.05ug/well). Serum was diluted two-fold and added to the rHA-coated plates. A representative plot for one mouse is presented in panels A–C. In panel D the antibody binding was normalized against the rHA administered as vaccine. The percentage of IgG detected to each version of the rHA protein is plotted in panel D. Error bars represent the standard error of the mean for n= 3–4 mice/group. Pooled convalescent serum from A/Brisbane/59/07 was normalized against the Sf9-derived HA and is presented as a control (D). The proportion of IgG detected to each version of the rHA protein is plotted in panel D.
3.5 Recombinant HA proteins produced by different expression systems differ in their ability to induce antibodies with HAI activity
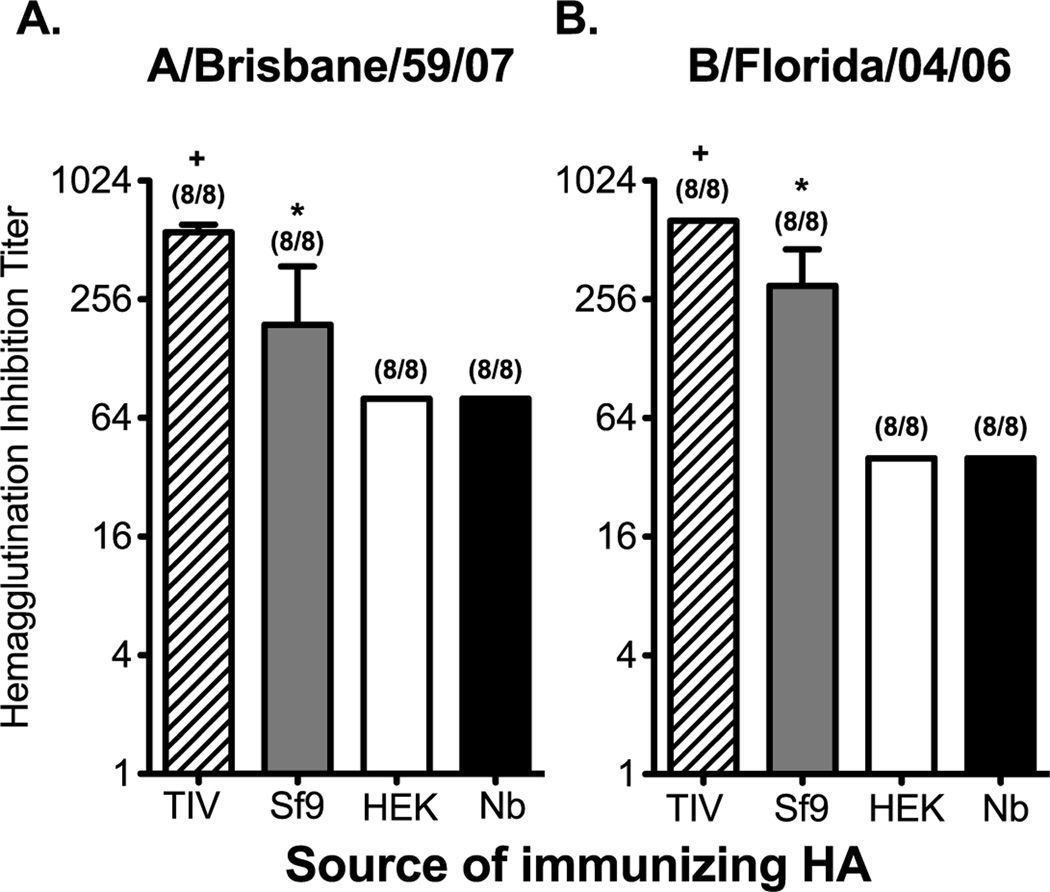
To test whether the protein source affects the induction of antibody responses with HAI activity we used a mouse model of rHA prime/boost vaccination. It is worth noting that, due to the lack of agglutination activity on the rHAs produced in HEK and Nb protein expression systems, we were unable to normalize the dose for rHA vaccination in the context of hemagglutination units (Table 1). Instead, mice were immunized with equal concentrations of the corresponding rHA protein form both influenza A and influenza B (2µg of each rHA/mouse). A separate cohort of mice was vaccinated in a similar fashion with the 2008–2009 TIV formulation (2ug of each HA/mouse). In the field of influenza vaccination an HAI titer of 1:40 has been used the standard correlate of protection [9, 14]. While a standard for protection has not been established in mice, protective antibody responses are generally observed when animals have HAI titers above 1:40 [27, 44]. Following prime/boost vaccination, serum was collected 21 days post boost, and tested for HAI activity against the influenza A/Brisbane/59/07 and B/Florida/04/06 viruses. Independent of the source of the HA used for vaccination, 100 percent of the mice generated antibodies with HAI titers over 1:40. However, vaccination with the traditional egg-derived vaccine elicited the highest titers of antibodies with HAI activity. In comparison, Sf9-derived proteins elicited titers that were 2- to 3-fold lower than TIV. Nonetheless, mean HAI titers induced by Sf9-derived rHAs (A/Brisbane/59/07) were at least 2-fold higher when compared to rHA’s produced by other expression systems (Figure 6A). Likewise, a 7-fold increase in the induction in HAI titer against B/Florida/04/06 was observed when vaccinating with Sf9-derived rHA (Figure 6B). In a similar vaccination experiment we did not observed significant differences in HAI titers against influenza A and B between mice vaccinated with Tni-, Nb-, Sf9 HAT, and HEK-derived rHA proteins (Supplementary Figure 6). The data presented here suggests that the protein expression method used to generate rHAs does not affect the ability of these proteins to elicit a functional antibody response, however the magnitude of such a response can be significantly affected by the source of rHA. Unfortunately, vaccine-elicited protection could not be assessed as the corresponding non-mouse adapted influenza A and influenza B viruses generated mild infections in mice, even at the highest dose possible that could be administered.
Figure 6. The induction of antibodies with HAI activity is affected by the source of the rHA protein.
B6 female mice were immunized intramuscularly (left flank) with a preparation containing 2µg of each Sf9-, HEK, or Nb-derived HA proteins from A/Brisbane/59/07 and B/Florida/04/06. Another group of mice received 2008–2009 TIV. The dose of TIV was adjusted so that each mouse received 2ug of each of the HAs present in the formulation. All vaccine formulations were adjuvanted with CpG. Fourteen days later the mice received a booster immunization containing the same concentration of HA and CpG used during priming. Serum from immunized mice was collected 21 days after boost. Serum was diluted and incubated with (A) A/Brisbane/59/07 or (B) B/Florida/04/06. HAI titer is defined as the highest dilution in which RBC agglutination was observed. Mean HAI titer and standard deviation are plotted on the graphs. Mann-Whitney (t-test) statistical analysis performed with n=8 (A) and n=8 (B). Numbers at the top of each bar represent the fraction of mice that antibodies with HAI activity greater that 1:40. For panels A and B statistically significant comparisons against HEK-, Nb-, and Sf9- derived proteins are denoted by the (+) symbol. Likewise, statistically significant comparisons against HEK- and Nb-derived proteins are denoted by the (*) symbol. The respective p-values for the statistically significant comparisons are as follows for panel A + = p<0.003 and * = p<0.0118. For panel B + = p<0.001 and * = p<0.0003.
4. Discussion
Long lasting immunity to influenza, whether it is induced by infection or vaccination, depends on the ability of the humoral arm of the immune system to respond to the virus HA protein. Indeed, a substantial portion of the antibody response following influenza infection in mice and humans is directed against the HA. Because of this, understanding the responses against the HA is one of the main focuses of influenza research. Recombinant versions of influenza HAs are widely available as reagents and can be purchased commercially from a number of vendors, or requisitioned from the NIH BEIR. However, until now, a systematic approach to compare the antigenicity and immunogenicity of a particular HA when it is produced across different protein expression systems was not available. In this study, we report that the production and/or purification process of rHA proteins affects their biological behavior and structural organization ultimately influencing the way in which the immune system recognizes them.
There are various factors that can potentially influence the structural and antigenic properties of rHA proteins. These can be divided into the following groups: 1) the protein expression system, 2) the purification methods, and 3) the nature of the protein’s modifications. For instance, the complexity of the N-glycans in a recombinant protein can vary according to the protein expression system employed in its production [45, 46]. The glycosylation state of viral glycoproteins have been associated with immune evasion [47]. The nature of the HA glycosylation can influence epitope availability as well as the range of the HA-receptor specificity [48]. The role that the chosen protein expression system could have on epitope masking and HA receptor specificity remains to be tested. However, our observations suggest that the state of rHA glycosylation is not responsible for the apparent lack of agglutination activity observed in the HEK- and Nb-derived rHA proteins. We have not extensively examined the binding of rHA proteins to RBCs from other species, which contain a different glycan profile on their surface.
Protein purification is just as important as the expression system in the manufacturing process of a recombinant protein. The major purification methods used to recover the rHAs studied in this report consist of purification tags (His-tag), lectin binding, and/or anion exchange. rHA proteins produced in Sf9, and purified in tandem by lentil-lectin binding and anion exchange, appeared organized into well-defined structures with a homogeneous range of sizes (Figure 1 and Supplementary Figure 3). In addition, these preparations contained biologically active rHA (Table 1). In contrast, rHAs purified by the binding of the His-tag to an immobilized metal ion affinity column (IMAC), were less organized and displayed reduced binding to avian RBC (Table 1, Figure 1, and Supplementary Figure 3). However, the agglutination activity and the presence of rosettes have been reported in full-length rHAs produced in transiently transfected human cell lines and plant-based protein expression systems [16, 40, 49–53]. In these reports, the rHAs were recovered from the membrane of transfected cells by detergent solubilization and submitted to anion exchange, gel filtration, and lentil lectin columns. Thus, it is possible that the added purification step may be crucial in the selection of properly folded and biologically active rHA. For instance, HEK-derived HA’s purified by His-tag/IMAC are not assembled into rosettes and fail to agglutinate avian RBCs. Likewise, HEK-derived rHA from A/California/07/09, purified by His-tag/IMAC, failed agglutinate human erythrocytes [54]. In contrast, rHAs produced in HEK-293 cells have been shown to organize into rosette-like oligomers when the HA is purified using the His-tag/IMAC method in conjunction with anion exchange chromatography and gel filtration [16]. Thus, it is possible that His-tag/IMAC purification does not discriminate between conformationally intact trimers and denatured or misfolded protein. A proportion of misfolded protein in the preparation could either interact or interfere with the hydrophobic interactions required for proper oligomerization [55, 56], effectively diluting the concentration of bioactive HA. This could potentially explain the presence of heterogeneous oligomeric structures, and the lack of avian RBC agglutination, in rHAs purified by these means (Table 1, Figure 1, and Supplementary Figure 3). However, rHAs produced in bacterial expression systems and purified by IMAC can retain their biological activity [26, 39, 54]. It is possible that the protein refolding process prior to IMAC purification improves the recovery of conformationally sound and bioactive HA. In addition, it is worth noting that the trimeric versions of the rHAs were purified by IMAC. Contrary to the HEK-derived HAs, these proteins displayed some degree of agglutination and organized oligomerization. Thus the added stability provided by the inclusion of a trimerization domain may improve the recovery of properly folded rHA.
Recombinant HA proteins, through genetic manipulation of the HA gene, have been produced with various truncations and/or modifications to improve either protein stability, immunogenicity, or increase production yields [7, 16, 26, 57–59]. Of particular relevance to this report are the addition of trimerization domains and the removal of the TM domain. The SEC profiles of HEK-derived rHAs preparations, which lack the TM domain, suggest that the proteins are present as monomers (Figure 1). One possible explanation is that the removal of TM domain influences HA trimer stability and oligomerization. Trimer destabilization by removal of the TM or HA2 domains has been previously reported [60, 61]. However, functional oligomers of HA1 subunit have been observed [26]. At the moment it is unclear what the role of the TM domain is in rHA oligomerization. It has been reported that the addition of a trimerization domain to the HA provides stability to the trimeric conformation and enhancing HA immunogenicity [16, 26, 39, 58]. Here we observed that the both of the Sf9 HAT-derived proteins were able to aggregate into HMWO and agglutinate avian RBCs (Figure 1 and Table 1). These observations mirror those made by Khurana et al. [39] using a trimeric form of the rHA from the pandemic H1N1 strain A/California/07/09.
Recombinant HA preparations derived from baculovirus infection of Sf9 cells are well recognized by the antibody and the B cell compartment of mice infected with either A/Brisbane/59/07 or B/Florida/04/06. Conversely, these compartments poorly recognize the HEK-derived rHA when compared to the other recombinant versions of the protein (Figures 2, 3, and 4). These differences in antigenicity were reproducible across two different sets of rHAs, two different assays that measure antibody binding, and across two species. It is possible that a portion of the antibodies generated against the HA are directed against the TM domain. However, it has been reported that only a 4-fold reduction in antibody binding to HA is observed when the HA2 subunit, which is comprised of approximately 230 amino acids, is not present [15]. Thus it is unlikely that the removal of the 38–47 amino acids comprising the TM domain will account for a 14 to 54-fold reduction in antibody binding to rHAs generated in HEK-293 cells. In addition, binding reactivity of sera from immunized mice suggests, 88% percent of the antibody response to Sf9-derived HA appears to be directed against epitopes not presented by the other rHA versions (Figure 5). Alternatively, the instability or lack of a quaternary structure in the HEK-derived protein preparations may decrease the availability of epitopes dependent on such a conformation [58, 62, 63]. In addition, the lack of oligomeric organization could result in the loss of relevant epitopes when the protein is adsorbed into plastic [62]. Nevertheless, baculovirus infection of Sf9 insect cells yields rHA proteins that are organized into functional oligomers, and that from an immunological standpoint, display a similar array of potentially conformation-dependent B cell epitopes to the HA presented during a live virus infection. These data establish inter-dependence between rHA structural organization, concentration of functional protein, and the extent of antigenicity. However, the status of the rHA antigenicity is not reflective on the ability of these proteins to elicit a functional neutralizing antibody response. All of the tested rHAs induced antibodies with HAI activity, and while the titer of these antibodies was higher in mice vaccinated with Sf9-derived rHA, the percentage of responders above a 1:40 titer was identical between groups. This suggests that the immune system is able to mount a substantial response to the relevant form of the rHA, even if this form is underrepresented in the preparation. However the differences in the magnitude of the response may be influence by a number of factors. One such factor is that the Sf9-derived proteins could enhance immune stimulation via interaction of the protein’s oligomannose structures, a feature of recombinant proteins produced in insect cells, with the mannose receptor on antigen presenting cells [64–67]. However, no such enhancement was observed when immunizing with the Tni or Sf9 HAT proteins. Lastly, protein yields following histidine tag/IMAC purification are greater than those obtained following lectin binding chromatography. This is permissive for dose adjustments in order to increase the magnitude of the neutralizing antibody response. Thus, despite the observed differences in antigenicity, rHA preparations from sources other than Sf9 can be useful in the context of experimental vaccines.
The data presented here suggests that the rHA production and/or purification process can affect the protein’s structural organization and biological activity leading to marked differences in protein antigenicity. As we move towards using these proteins as vaccine reagents or as reagents to study HA-specific responses following vaccination we need to scrutinize the different production systems and purification methods. The findings presented here cannot clearly distinguish whether differences inherent to the production system or the purification method used are solely responsible for differences in antigenicity among the various HA versions. Further studies are required to address the source driving the observed differences between the various rHA protein versions. It is worth noting that our results are reflective of the state in which the proteins were received and not necessarily the quality of the product generated by the corresponding manufacturers. It is conceivable that recombinant HA proteins produced in a localized facility, and in parallel, will exhibit different antigenic and immunogenic properties than those presented here. However, the data is consistent across rHAs from influenza A and influenza B strains. We strongly encourage the careful selection of protein expression and/or purification methods that preserve the HA’s biological function, while minimizing protein degradation and denaturation overtime, as it is key to the study of HA-specific immunity.
Supplementary Material
Highlights.
rHA produced and purified by various means are tested as reagents in immune assays.
rHA produced and purified by various means are tested as immunogens in mice.
rHA production process impacts the protein’s antigenicity in immune assays.
rHA proteins elicit relevant HAI responses independent of the production process.
rHA oligomerization correlate with it’s antigenic and immunogenic properties.
Acknowledgements
This work was supported by the New York Influenza Center of Excellence, part of the NIH/NIAID network of Centers of Excellence in Influenza Research and Surveillance (CEIRS), contract #HHS-N266200700008C. The rHA reagents used to conduct this research were kindly provided by the Biodefense and Emerging Infections Research Resources Repository.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wharton SA. The role of influenza virus haemagglutinin in membrane fusion. Microbiol Sci. 1987 Apr;4(4):119–124. [PubMed] [Google Scholar]
- 2.Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 3.Potter CW, Oxford JS. Determinants of immunity to influenza infection in man. Br Med Bull. 1979 Jan;35(1):69–75. doi: 10.1093/oxfordjournals.bmb.a071545. [DOI] [PubMed] [Google Scholar]
- 4.Wrammert J, Koutsonanos D, Li GM, Edupuganti S, Sui J, Morrissey M, et al. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J Exp Med. 2011 Jan 17;208(1):181–193. doi: 10.1084/jem.20101352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Throsby M, van den Brink E, Jongeneelen M, Poon LLM, Alard P, Cornelissen L, et al. Heterosubtypic Neutralizing Monoclonal Antibodies Cross-Protective against H5N1 and H1N1 Recovered from Human IgM+ Memory B Cells. PLoS One. 2008;3(12):e3942. doi: 10.1371/journal.pone.0003942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walker DL, Horsfall FL., Jr Lack of identity in neutralizing and hemagglutination-inhibiting antibodies against influenza viruses. J Exp Med. 1950 Jan 1;91(1):65–86. doi: 10.1084/jem.91.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steel J, Lowen AC, Wang TT, Yondola M, Gao Q, Haye K, et al. Influenza virus vaccine based on the conserved hemagglutinin stalk domain. MBio. 2010 Apr;1(1) doi: 10.1128/mBio.00018-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corti D, Suguitan AL, Jr, Pinna D, Silacci C, Fernandez-Rodriguez BM, Vanzetta F, et al. Heterosubtypic neutralizing antibodies are produced by individuals immunized with a seasonal influenza vaccine. J Clin Invest. 2010 May 3;120(5):1663–1673. doi: 10.1172/JCI41902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hobson RLC D, Beare AS, Ward-Gardner A. The role of serum haemagglutination-inhibiting antibody in protection against challenge infection with influenza A2 and B viruses. Journal of Hygiene. 1972;70:767–777. doi: 10.1017/s0022172400022610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia JM, Pepin S, Lagarde N, Ma ES, Vogel FR, Chan KH, et al. Heterosubtype neutralizing responses to influenza A (H5N1) viruses are mediated by antibodies to virus haemagglutinin. PLoS One. 2009;4(11):e7918. doi: 10.1371/journal.pone.0007918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simmons CP, Bernasconi NL, Suguitan AL, Mills K, Ward JM, Chau NV, et al. Prophylactic and therapeutic efficacy of human monoclonal antibodies against H5N1 influenza. PLoS Med. 2007 May;4(5):e178. doi: 10.1371/journal.pmed.0040178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Puck JM, Glezen WP, Frank AL, Six HR. Protection of Infants from Infection with Influenza-a Virus by Transplacentally Acquired Antibody. Journal of Infectious Diseases. 1980;142(6):844–849. doi: 10.1093/infdis/142.6.844. [DOI] [PubMed] [Google Scholar]
- 13.Treanor J, Wright PF. Immune correlates of protection against influenza in the human challenge model. Dev Biol (Basel) 2003;115:97–104. [PubMed] [Google Scholar]
- 14.Hannoun C, Megas F, Piercy J. Immunogenicity and protective efficacy of influenza vaccination. Virus Res. 2004 Jul;103(1–2):133–138. doi: 10.1016/j.virusres.2004.02.025. [DOI] [PubMed] [Google Scholar]
- 15.Shen S, Mahadevappa G, Oh HL, Wee BY, Choi YW, Hwang LA, et al. Comparing the antibody responses against recombinant hemagglutinin proteins of avian influenza A (H5N1) virus expressed in insect cells and bacteria. J Med Virol. 2008 Nov;80(11):1972–1983. doi: 10.1002/jmv.21298. [DOI] [PubMed] [Google Scholar]
- 16.Wei CJ, Xu L, Kong WP, Shi W, Canis K, Stevens J, et al. Comparative efficacy of neutralizing antibodies elicited by recombinant hemagglutinin proteins from avian H5N1 influenza virus. J Virol. 2008 Jul;82(13):6200–6208. doi: 10.1128/JVI.00187-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lalor PA, Webby RJ, Morrow J, Rusalov D, Kaslow DC, Rolland A, et al. Plasmid DNA-based vaccines protect mice and ferrets against lethal challenge with A/Vietnam/1203/04 (H5N1) influenza virus. J Infect Dis. 2008 Jun 15;197(12):1643–1652. doi: 10.1086/588431. [DOI] [PubMed] [Google Scholar]
- 18.Bragstad K, Martel CJ, Thomsen JS, Jensen KL, Nielsen LP, Aasted B, et al. Pandemic influenza 1918 H1N1 and 1968 H3N2 DNA vaccines induce cross-reactive immunity in ferrets against infection with viruses drifted for decades. Influenza Other Respi Viruses. 2011 Jan;5(1):13–23. doi: 10.1111/j.1750-2659.2010.00177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galarza JM, Latham T, Cupo A. Virus-like particle (VLP) vaccine conferred complete protection against a lethal influenza virus challenge. Viral Immunol. 2005;18(1):244–251. doi: 10.1089/vim.2005.18.244. [DOI] [PubMed] [Google Scholar]
- 20.Tao P, Luo M, Zhu D, Qu S, Yang Z, Gao M, et al. Virus-like particle vaccine comprised of the HA, NA, and M1 proteins of an avian isolated H5N1 influenza virus induces protective immunity against homologous and heterologous strains in mice. Viral Immunol. 2009 Jul;22(4):273–281. doi: 10.1089/vim.2009.0017. [DOI] [PubMed] [Google Scholar]
- 21.Treanor J. Recombinant proteins produced in insect cells. Curr Top Microbiol Immunol. 2009;333:211–225. doi: 10.1007/978-3-540-92165-3_11. [DOI] [PubMed] [Google Scholar]
- 22.Talaat KR, Karron RA, Luke CJ, Thumar B, McMahon BA, Chen GL, et al. An open label Phase I trial of a live attenuated H6N1 influenza virus vaccine in healthy adults. Vaccine. 2011 Apr 12;29(17):3144–3148. doi: 10.1016/j.vaccine.2011.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baer J, Santiago F, Yang H, Wu H, Holden-Wiltse J, Treanor J, et al. B cell responses to H5 influenza HA in human subjects vaccinated with a drifted variant. Vaccine. 2010 Jan 22;28(4):907–915. doi: 10.1016/j.vaccine.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuroda K, Hauser C, Rott R, Klenk HD, Doerfler W. Expression of the influenza virus haemagglutinin in insect cells by a baculovirus vector. EMBO J. 1986 Jun;5(6):1359–1365. doi: 10.1002/j.1460-2075.1986.tb04367.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cox MM, Patriarca PA, Treanor J. FluBlok, a recombinant hemagglutinin influenza vaccine. Influenza Other Respi Viruses. 2008 Nov;2(6):211–219. doi: 10.1111/j.1750-2659.2008.00053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khurana S, Verma S, Verma N, Crevar CJ, Carter DM, Manischewitz J, et al. Bacterial HA1 Vaccine against Pandemic H5N1 Influenza Virus: Evidence of Oligomerization, Hemagglutination, and Cross-Protective Immunity in Ferrets. J Virol. 2011 Feb;85(3):1246–1256. doi: 10.1128/JVI.02107-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Biesova Z, Miller MA, Schneerson R, Shiloach J, Green KY, Robbins JB, et al. Preparation, characterization, and immunogenicity in mice of a recombinant influenza H5 hemagglutinin vaccine against the avian H5N1 A/Vietnam/1203/2004 influenza virus. Vaccine. 2009 Oct 19;27(44):6234–6238. doi: 10.1016/j.vaccine.2009.07.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jabbar MA, Sivasubramanian N, Nayak DP. Influenza viral (A/WSN/33) hemagglutinin is expressed and glycosylated in the yeast Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences 1985 April 1. 1985;82(7):2019–2023. doi: 10.1073/pnas.82.7.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsai SM, Chiang YC, Chin LT, Liu HJ, Wang CY. Novel post-translational modifications of the hemagglutinin and neuraminidase proteins of avian influenza virus expressed by Kluyveromyces lactis. J Virol Methods. 2011 May 27; doi: 10.1016/j.jviromet.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 30.Yang JL, Wang HL, Wang SX, Yang P, Liu KT, Jiang CY. High-level expression, purification and characterization of codon-optimized recombinant hemagglutinin 5 proteins in mammalian cells. Chin Med J (Engl) 2010 Apr 20;123(8):1073–1077. [PubMed] [Google Scholar]
- 31.Baxter R, Patriarca PA, Ensor K, Izikson R, Goldenthal KL, Cox MM. Evaluation of the safety, reactogenicity and immunogenicity of FluBlok((R)) trivalent recombinant baculovirus-expressed hemagglutinin influenza vaccine administered intramuscularly to healthy adults 50–64 years of age. Vaccine. 2011 Jan 28; doi: 10.1016/j.vaccine.2011.01.039. [DOI] [PubMed] [Google Scholar]
- 32.King JC, Jr, Cox MM, Reisinger K, Hedrick J, Graham I, Patriarca P. Evaluation of the safety, reactogenicity and immunogenicity of FluBlok trivalent recombinant baculovirus-expressed hemagglutinin influenza vaccine administered intramuscularly to healthy children aged 6–59 months. Vaccine. 2009 Nov 5;27(47):6589–6594. doi: 10.1016/j.vaccine.2009.08.032. [DOI] [PubMed] [Google Scholar]
- 33.Shoji Y, Farrance CE, Bautista J, Bi H, Musiychuk K, Horsey A, et al. A plant-based system for rapid production of influenza vaccine antigens. Influenza and other respiratory viruses. 2011 Oct 4; doi: 10.1111/j.1750-2659.2011.00295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shoji Y, Chichester JA, Jones M, Manceva SD, Damon E, Mett V, et al. Plant-based rapid production of recombinant subunit hemagglutinin vaccines targeting H1N1 and H5N1 influenza. Human Vaccines. 2011 Jan-Feb;7(Suppl):41–50. doi: 10.4161/hv.7.0.14561. [DOI] [PubMed] [Google Scholar]
- 35.Wang K, Holtz KM, Anderson K, Chubet R, Mahmoud W, Cox MM. Expression and purification of an influenza hemagglutinin--one step closer to a recombinant protein-based influenza vaccine. Vaccine. 2006 Mar 15;24(12):2176–2185. doi: 10.1016/j.vaccine.2005.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kovaleva ES, O'Connell KP, Buckley P, Liu Z, Davis DC. Recombinant protein production in insect larvae: host choice, tissue distribution, and heterologous gene instability. Biotechnol Lett. 2009 Mar;31(3):381–386. doi: 10.1007/s10529-008-9883-2. [DOI] [PubMed] [Google Scholar]
- 37.Stevens J, Blixt O, Tumpey TM, Taubenberger JK, Paulson JC, Wilson IA. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science. 2006 Apr 21;312(5772):404–410. doi: 10.1126/science.1124513. [DOI] [PubMed] [Google Scholar]
- 38.Laver WG, Webster RG. Preparation and immunogenicity of a purified influenza virus haemagglutinin and neuraminidase subunit vaccine. Postgrad Med J. 1976 Jun;52(608):373–378. doi: 10.1136/pgmj.52.608.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khurana S, Larkin C, Verma S, Joshi MB, Fontana J, Steven AC, et al. Recombinant HA1 produced in E. coli forms functional oligomers and generates strain-specific SRID potency antibodies for pandemic influenza vaccines. Vaccine. 2011;29(34):5657–5665. doi: 10.1016/j.vaccine.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilkinson BE. Recombinant Hemagglutinin Subunit Vaccine Produced in a Baculovirus Expression Vector System. Avian Diseases. 2003;47:253–262. (ArticleType: research-article / Issue Title: Special Issue, Fourth International Symposium on Avian Influenza, 1997 Proceedings / Full publication date: 2003 / Copyright ¬© 2003 American Association of Avian Pathologists, Inc.) [Google Scholar]
- 41.Stray SJ, Cummings RD, Air GM. Influenza virus infection of desialylated cells. Glycobiology. 2000 Jul 1;10(7):649–658. doi: 10.1093/glycob/10.7.649. 2000. [DOI] [PubMed] [Google Scholar]
- 42.Gottschalk A. Neuraminidase: the specific enzyme of influenza virus and Vibrio cholerae. Biochim Biophys Acta. 1957 Mar;23(3):645–646. doi: 10.1016/0006-3002(57)90389-x. [DOI] [PubMed] [Google Scholar]
- 43.Hirst GK. The Agglutination of Red Cells by Allantoic Fluid of Chick Embryos Infected with Influenza Virus. Science. 1941 Jul 4;94(2427):22–23. doi: 10.1126/science.94.2427.22. [DOI] [PubMed] [Google Scholar]
- 44.Min JY, Vogel L, Matsuoka Y, Lu B, Swayne D, Jin H, et al. A live attenuated H7N7 candidate vaccine virus induces neutralizing antibody that confers protection from challenge in mice, ferrets, and monkeys. J Virol. 2010 Nov;84(22):11950–11960. doi: 10.1128/JVI.01305-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kost TA, Condreay JP, Jarvis DL. Baculovirus as versatile vectors for protein expression in insect and mammalian cells. Nat Biotechnol. 2005 May;23(5):567–575. doi: 10.1038/nbt1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bosch D, Schots A. Plant glycans: friend or foe in vaccine development? Expert Rev Vaccines. 2010 Aug;9(8):835–842. doi: 10.1586/erv.10.83. [DOI] [PubMed] [Google Scholar]
- 47.Vigerust DJ, Shepherd VL. Virus glycosylation: role in virulence and immune interactions. Trends in microbiology. 2007 May;15(5):211–218. doi: 10.1016/j.tim.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Vries RP, de Vries E, Bosch BJ, de Groot RJ, Rottier PJ, de Haan CA. The influenza A virus hemagglutinin glycosylation state affects receptor-binding specificity. Virology. 2010 Jul 20;403(1):17–25. doi: 10.1016/j.virol.2010.03.047. [DOI] [PubMed] [Google Scholar]
- 49.Kalthoff D, Giritch A, Geisler K, Bettmann U, Klimyuk V, Hehnen HR, et al. Immunization with plant-expressed hemagglutinin protects chickens from lethal highly pathogenic avian influenza virus H5N1 challenge infection. J Virol. 2010 Nov;84(22):12002–12010. doi: 10.1128/JVI.00940-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wharton SA, Skehel JJ, Wiley DC. Studies of influenza haemagglutinin-mediated membrane fusion. Virology. 1986 Feb;149(1):27–35. doi: 10.1016/0042-6822(86)90083-8. [DOI] [PubMed] [Google Scholar]
- 51.Godley L, Pfeifer J, Steinhauer D, Ely B, Shaw G, Kaufmann R, et al. Introduction of intersubunit disulfide bonds in the membrane-distal region of the influenza hemagglutinin abolishes membrane fusion activity. Cell. 1992;68(4):635–645. doi: 10.1016/0092-8674(92)90140-8. [DOI] [PubMed] [Google Scholar]
- 52.Cross KJ, Wharton SA, Skehel JJ, Wiley DC, Steinhauer DA. Studies on influenza haemagglutinin fusion peptide mutants generated by reverse genetics. EMBO J. 2001 Aug 15;20(16):4432–4442. doi: 10.1093/emboj/20.16.4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu YC, Luo YL, Ji WT, Chulu JLC, Chang PC, Shieh H, et al. Dual expression of the HA protein of H5N2 avian influenza virus in a baculovirus system. Journal of Virological Methods. 2006;135(1):43–48. doi: 10.1016/j.jviromet.2006.01.023. [DOI] [PubMed] [Google Scholar]
- 54.Khurana S, Verma S, Verma N, Crevar CJ, Carter DM, Manischewitz J, et al. Properly Folded Bacterially Expressed H1N1 Hemagglutinin Globular Head and Ectodomain Vaccines Protect Ferrets against H1N1 Pandemic Influenza Virus. PLoS One. 2010;5(7):e11548. doi: 10.1371/journal.pone.0011548. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 55.Munch C, Bertolotti A. Exposure of hydrophobic surfaces initiates aggregation of diverse ALS-causing superoxide dismutase-1 mutants. J Mol Biol. 2010 Jun 11;399(3):512–525. doi: 10.1016/j.jmb.2010.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yan-Ling Z, Xian-Ming P, Jun-Mei Z. Surface hydrophobicity and thermal aggregation of adenylate kinase. IUBMB Life. 1998;44(5):949–960. [Google Scholar]
- 57.Yang ZY, Wei CJ, Kong WP, Wu L, Xu L, Smith DF, et al. Immunization by avian H5 influenza hemagglutinin mutants with altered receptor binding specificity. Science. 2007 Aug 10;317(5839):825–828. doi: 10.1126/science.1135165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weldon WC, Wang B-Z, Martin MP, Koutsonanos DG, Skountzou I, Compans RW. Enhanced Immunogenicity of Stabilized Trimeric Soluble Influenza Hemagglutinin. PLoS One. 2010;5(9):e12466. doi: 10.1371/journal.pone.0012466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.de Vries RP, de Vries E, Bosch BJ, de Groot RJ, Rottier PJM, de Haan CAM. The influenza A virus hemagglutinin glycosylation state affects receptor-binding specificity. Virology. 2010;403(1):17–25. doi: 10.1016/j.virol.2010.03.047. [DOI] [PubMed] [Google Scholar]
- 60.Kretzschmar E, Veit M, Brunschõn S, Kuroda K, Klenk H-D. Secretion of fowl plague virus haemagglutinin from insect cells requires elimination of both hydrophobic domains. Journal of General Virology. 1992 Apr 1;73(1992)(4):839–848. doi: 10.1099/0022-1317-73-4-839. [DOI] [PubMed] [Google Scholar]
- 61.Doyle C, Sambrook J, Gething MJ. Analysis of progressive deletions of the transmembrane and cytoplasmic domains of influenza hemagglutinin. J Cell Biol. 1986 Oct;103(4):1193–1204. doi: 10.1083/jcb.103.4.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nestorowicz A, Laver G, Jackson DC. Antigenic determinants of influenza virus haemagglutinin. X. A comparison of the physical and antigenic properties of monomeric and trimeric forms. The Journal of general virology. 1985 Aug;66(Pt 8):1687–1695. doi: 10.1099/0022-1317-66-8-1687. [DOI] [PubMed] [Google Scholar]
- 63.Khurana S, Wu J, Verma N, Verma S, Raghunandan R, Manischewitz J, et al. H5N1 Virus-Like Particle Vaccine Elicits Cross-Reactive Neutralizing Antibodies That Preferentially Bind to the Oligomeric Form of Influenza Virus Hemagglutinin in Humans. Journal of virology. 2011 Nov;85(21):10945–10954. doi: 10.1128/JVI.05406-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tate MD, Job ER, Brooks AG, Reading PC. Glycosylation of the hemagglutinin modulates the sensitivity of H3N2 influenza viruses to innate proteins in airway secretions and virulence in mice. Virology. 2011;413(1):84–92. doi: 10.1016/j.virol.2011.01.036. [DOI] [PubMed] [Google Scholar]
- 65.Liu WC, Lin SC, Yu YL, Chu CL, Wu SC. Dendritic cell activation by recombinant hemagglutinin proteins of H1N1 and H5N1 influenza A viruses. Journal of Virology. 2010 Nov;84(22):12011–12017. doi: 10.1128/JVI.01316-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen JR, Ma C, Wong CH. Vaccine design of hemagglutinin glycoprotein against influenza. Trends Biotechnol. 2011 Sep;29(9):426–434. doi: 10.1016/j.tibtech.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 67.Betting DJ, Mu XY, Kafi K, McDonnel D, Rosas F, Gold DP, et al. Enhanced immune stimulation by a therapeutic lymphoma tumor antigen vaccine produced in insect cells involves mannose receptor targeting to antigen presenting cells. Vaccine. 2009 Jan 7;27(2):250–259. doi: 10.1016/j.vaccine.2008.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.