Abstract
Partitioning of polypeptides between protein folding and amyloid formation is of outstanding pathophysiological importance. Using yeast phosphoglycerate kinase as model, here we identify the features of the energy landscape that decide the fate of the protein: folding or amyloidogenesis. Structure formation was initiated from the acid-unfolded state, and monitored by fluorescence from 10 ms to 20 days. Solvent conditions were gradually shifted between folding and amyloidogenesis, and the properties of the energy landscape governing structure formation were reconstructed. A gradual transition of the energy landscape between folding and amyloid formation was observed. In the early steps of both folding and misfolding, the protein searches through a hierarchically structured energy landscape to form a molten globule in a few seconds. Depending on the conditions, this intermediate either folds to the native state in a few minutes, or forms amyloid fibers in several days. As conditions are changed from folding to misfolding, the barrier separating the molten globule and native states increases, although the barrier to the amyloid does not change. In the meantime, the native state also becomes more unstable and the amyloid more stable. We conclude that the lower region of the energy landscape determines the final protein structure.
Introduction
Understanding the sequence-specific partitioning of polypeptides between protein folding and aggregation is of great physiological and pathological importance (1). Roughly two dozen human diseases have been linked to the formation of ordered insoluble aggregates called amyloids (2). The illnesses linked to amyloidogenesis include some of the most debilitating conditions, like transmissible spongiform encephalopathy, Alzheimer's, and Parkinson's diseases (3,4).
Proteins that were found to form amyloids in vivo have very diverse sequences (5), and many proteins not related to diseases can form amyloid fibrils in vitro (6–8). All amyloid fibrils show a similar cross β-spine, which is stabilized by sequence-independent hydrogen bonding within the polypeptide backbone (2,9). The role of sequence specific interactions in the formation of amyloid structures has also been shown (9–11), but it is generally believed that amyloid structure formation is a universal property of proteins (2,7,12,13).
Damaschun et al. (8) have demonstrated that yeast phosphoglycerate kinase (PGK) is able to form amyloid fibrils in vitro. Short fibrillar structures were observed few days after initiating amyloid formation. The β-sheet content of the protein in this state was already identical to that adopted in the mature amyloid. Further growth of the fibrillar structures occurs by the association of the protofibrils to form mature amyloids. Fibril growth did not reach saturation even after two weeks. The presence of amyloids was proved by electron microscopy and x-ray scattering. Analyzing the misfolding and amyloid formation of PGK, Modler et al. (14) developed an amyloid growth model based on colloid coagulation theory.
PGK is a 415-residue-large monomeric protein. The native state has two roughly equal-sized domains, linked by a helical hinge. It has a combined total of 20 α-helices, 19 β-strands, and three turns. The structure of the domains is similar, consisting of a core of a six-stranded parallel β-sheet that is surrounded by a series of helices. In addition, the C-terminal domain has three shorter β-strands. It has been used in many unfolding and refolding studies, and different denatured states of PGK have been well characterized (15–19). A hierarchical energy landscape model has been developed to predict the folding kinetics of PGK from milliseconds to the complete folding (20).
Whether the protein chooses the folding or misfolding pathway can be a matter of life and death. Understanding the mechanisms of transition between native and amyloid states and revealing the underlying physicochemical processes is of pivotal importance. The possibility of reversing different steps (21–23) or the whole process (24) of amyloid formation has been shown. Several recent articles also address the relationship between the folding and amyloid formation pathways (25–27), but a lot of research has to be done to understand how the fate of proteins is determined.
Here, using PGK as the model protein, we tune buffer conditions between folding and misfolding and record the kinetics of the structural changes from 10 ms to 20 days. The method allowed us to identify the features of the energy landscape that decide the fate of the protein: folding or amyloidogenesis.
Materials and Methods
Expression, purification, and characterization of PGK
A histidine-tagged version of yeast (Saccharomyces cerevisiae) phosphoglycerate kinase (UniProtKB/Swiss-Prot accession number P00560) was expressed and prepared as described earlier (28). Enzyme activity of the purified PGK was verified using established methods (24,29). Samples were concentrated to 40 μM, dialyzed in 50 mM Tris, 1 mM EDTA, pH 7.0 buffer, aliquoted, and stored at −80°C.
Kinetic measurements
Structure formation of samples containing 40 μM PGK unfolded in pH 2.0 HCl solution was initiated by an 11-fold mixing with a buffer containing 10 mM Na-citrate, 10 mM Na-phosphate, and 200 mM NaCl using stopped-flow (Applied Photophysics, Leatherhead, UK) or manual mixing. The pH of the diluting buffer was varied between pH 2.0 and 7.0 in 0.5 pH steps, and structure formation of PGK was detected. All kinetic values were recorded at 4°C to avoid unspecific aggregation. Conformational changes were followed by time-resolved tryptophan fluorescence measurements (excited at 295 nm with 5-nm bandwidth and detected between 305 and 400 nm). One-hundred individual mixing experiments were averaged.
Characterization of PGK amyloid
The presence of mature amyloid structures in the sample was checked by electron microscopy (11) and Thioflavin T fluorescence using established methods (24,30).
The fluorescence emission spectrum of freshly prepared 20 μM Thioflavin T solution (in 100 mM Na-phosphate, 100 mM NaCl, pH 8.5) was measured before and after the addition of protein to a final concentration of 4 μM. Spectra were recorded on a FluoroLog-3 fluorometer (Horiba Jobin-Yvon, Edison, NJ) between 455 nm and 530 nm (λexc = 450 nm) with 1-nm bandwidth. No fluorescence change was observed when PGK in the native or unfolded state was added. On the other hand, the fluorescence of the dye increased roughly fivefold upon addition of mature amyloid fibers.
Amyloid fibrils grown in vitro were visualized in a. 7100 transmission electron microscope (Hitachi, Tokyo, Japan) after negative staining. The sample was diluted to 1 μM protein concentration and a 50 μL droplet of it applied to a Formvar-coated copper grid. An equal volume of 2% uranyl-acetate solubilized in 50% methanol was added to stain the sample for 15 min. After this time, the excess solution was removed and the sample let dry for 15 min. No fibrils were detected in samples containing the protein in the native or the unfolded state.
Results
Yeast phosphoglycerate kinase (PGK) was unfolded by dialysis against pH 2 solution with low ionic strength. Structure formation of the unfolded protein was initiated by rapid mixing in buffers containing 200 mM NaCl, and followed from the millisecond to the week timescale. Solvent conditions were gradually changed from favoring amyloid formation to native folding by tuning the pH of the buffer between pH 2.0 and pH 7.0.
Early kinetics of structure formation of the acid unfolded PGK was recorded after stopped-flow mixing into various buffers with different values of pH. The pH of the solution after mixing was varied in 0.5 pH unit steps from pH 2 to pH 7. The time course of tryptophan fluorescence was monitored from 10 ms to 50 s. One-hundred individual traces were averaged, and the fluorescence was normalized to the fluorescence intensity observed at 10 ms. All curves indicate the formation of a hyperfluorescent state with a kinetics that is stretched over several orders of magnitude in time. Fig. 1 shows the kinetics recorded at pH 2.0, 4.5, and 7.0.
Figure 1.
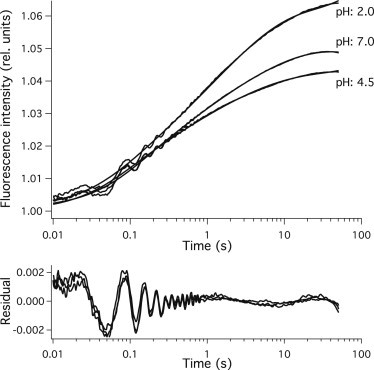
Early steps of structure formation of PGK detected by tryptophan fluorescence. Structure formation of acid unfolded PGK was initiated by stopped flow dilution and followed by tryptophan florescence measurements. Data were fitted with a hierarchic landscape model. The result of the fit is also shown. Residuals obtained as the difference between the measured and fitted curves demonstrate the goodness of fit. Buffer conditions after mixing: 3.6 μM PGK, 9.1 mM Na-citrate, 9.1 mM Na-phosphate, and 182 mM NaCl.
Earlier results demonstrated that at neutral pH, the first steps toward native structure formation of PGK proceed on a hierarchically structured energy landscape (20). Here we used this hierarchic finite level model to fit the early kinetics of PGK structure formation at all pH values.
The model considers the interconversion of the unfolded (U), intermediate (I), and folded (F) states; amyloid (A) formation is neglected because it is much slower. The measured fluorescence intensity (Fluo(t)) can be calculated from the amount of the different states and their fluorescence efficiency (ΦU, ΦI, ΦF):
Conversion of the unfolded into the intermediate state occurs on a hierarchically structured energy landscape. A multitude of traps that can have N different depths give rise to a large number of relaxation pathways described by N different relaxation times. The amount U(t) of unfolded protein at time t after initiating structure formation can be calculated as
Here U(0) is the amount of unfolded protein at time t = 0. Because structure formation was initiated from the acid unfolded state, U(0) is the total amount of protein in our experiments. The parameters ak are the amplitudes and τk the time constants of the Debye contributions, which correspond to different levels of the energy landscape structure. For a landscape that consists of a multitude of hierarchically structured kinetic barriers, these components are not independent, but their amplitudes and relaxation times obey a simple scaling law:
The fastest relaxation pathway has a time constant of τ1. Every deeper level has λ-times longer relaxation time than the level before it. This early structure formation shows a stretched kinetics that lacks a characteristic time. The value τ1 thus is not a characteristic time of the process, but rather it is a parameter describing the entire hierarchic landscape. The ak/τk ratio shows the fraction of the protein molecules that follows the pathway with the characteristic time τk, thus the sum of the ak/τk values is 1. This relationship and the scaling property of the landscape reduce the number of the free fit parameters to the constants τ1, α, and λ regardless of N.
The above model fitted well the initial steps of the structure formation at all pH values (Fig. 1). The fluorescence intensity of the intermediate structure showed small variations as the pH was changed. The number N = 5 of the possible trap sizes, the parameter α/λ = 1.0 ± 0.15 and λ = 8.1 ± 0.9, did not change significantly as pH was altered. The shortest relaxation time (τ1) was the only parameter of the model that showed significant pH dependence.
Fig. 2 shows the pH dependence of τ1, the shortest relaxation time. The kinetics is fastest at pH 3.5, and it speeds up at neutral pH roughly five times compared to this.
Figure 2.
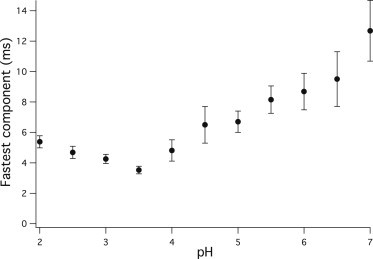
pH dependence of the shortest relaxation time τ1 obtained from the hierarchic finite level model fit of the fast kinetic measurements (see Fig. 1).
Structural changes of PGK from 1 min to 6 h after mixing were followed by tryptophan fluorescence intensity changes. Measurements were carried out between pH 2.0 and 7.0 in 0.5 pH unit steps. Fluorescence intensities were normalized to the fluorescence of the hyperfluorescent peak. Fig. 3 shows the kinetic traces recorded at pH 5.0, 6.0, and 7.0. The observed kinetics indicated a strong pH dependence of the structure formation on this timescale. At neutral pH the tryptophan fluorescence decreased, indicating the formation of native structure, in accordance with earlier results (31). As pH was decreased, the fluorescence intensity change became smaller and slower. Only transition between the intermediate (I) and folded (F) species can be observed on this timescale because the disappearance of the unfolded (U) state is faster and appearance of the amyloid aggregate (A) is slower. The folding kinetics could be fitted well with a single exponential at pH values between 4.5 and 7.0. The characteristic time (τI-F) of the reaction was determined from the fit.
Figure 3.
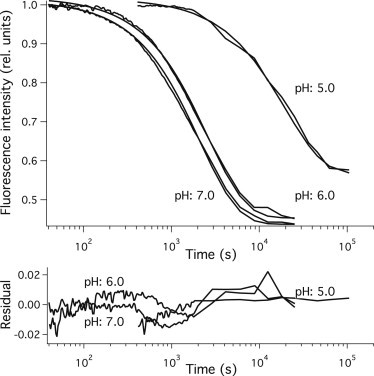
Structural changes of PGK detected by tryptophan fluorescence. Structure formation of acid unfolded PGK recorded by tryptophan fluorescence after a quick dilution by mixing. Monoexponential fit of the observed kinetics is also indicated. Residuals obtained as the difference between the measured and fitted curves are also shown. Buffer conditions after mixing: 3.6 μM PGK, 9.1 mM Na-citrate, 9.1 mM Na-phosphate, and 182 mM NaCl.
Fig. 4 shows the pH dependence of the time constant of folding to the native state from the intermediate conformation (Fig. 4 A) and the fluorescence change associated with it (Fig. 4 B). No native structure formation was observed below pH 4.5.
Figure 4.
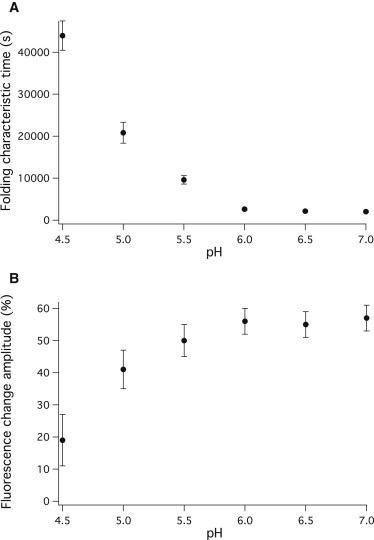
pH dependence of the time constant and amplitude of the fit shown in Fig. 3. (A) pH dependence of the characteristic time obtained by a monoexponential fit of the kinetic traces shown in Fig. 3. (B) pH dependence of the amplitude of the monoexponential fit shown in Fig. 3.
Fig. 5 represents the formation of amyloid structures monitored by Thioflavin T fluorescence from 1 min to 20 days. The gradual increase of the fluorescence intensity observed for the samples kept at low pH indicates amyloid accumulation. The observed kinetics do not change significantly between pH 2.0 and 4.5. Above pH 4.5, however, Thioflavin T fluorescence did not indicate amyloid formation.
Figure 5.
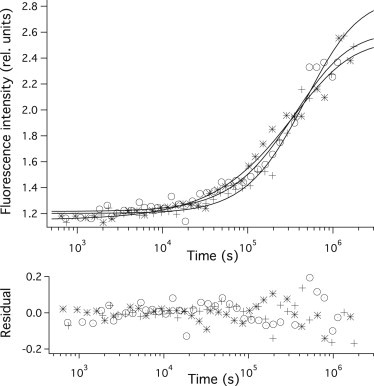
Amyloid formation of PGK detected by Thioflavin T fluorescence. Structure formation of acid unfolded PGK was initiated by mixing to pH 2.0 (circles), 3.0 (crosses), or 4.0 (stars) final acidity. (Continuous lines) Fit to the data with the Smoluchowski. The kinetics data were fitted with a coagulation model using size-independent rate constants. The results of the model fit are also indicated. Residuals obtained as the difference between the measured and fitted curves are also shown. Buffer conditions after mixing: 3.6 μM PGK, 9.1 mM Na-citrate, 9.1 mM Na-phosphate, and 182 mM NaCl.
Modler et al. (14) have shown that a good description of PGK amyloid growth can be given by the Smoluchowski coagulation theory, using size-independent association rate-constants. We used this model to fit the observed amyloid formation kinetics. The model gave a good fit to our measurements at all pH values.
If the sample contains only monomers at time t = 0, the concentrations ni of the oligomers containing i associated protein molecules can be calculated at any time t (14):
Here tc denotes the coagulation time, a constant characteristic to the process. Because there are only monomers present at t = 0, n1(0) is the protein concentration.
It is known that Thioflavin T binds to fibrillar amyloid structures, but not to monomeric protein. We assumed that the dye binds to the forming PGK aggregate when the oligomer becomes bigger than m monomeric units. The fraction fm of the protein molecules that are in oligomers smaller than or equal to m monomers is
By simplifying the equation analytically, the following formula was obtained:
The measured Thioflavin T fluorescence intensity can be calculated as
Here Φ0 is the fluorescence of the dye in the absence of amyloid, and ΦA is the Thioflavin T fluorescence if all the protein is in amyloid fibers.
The tc characteristic time of coagulation was extracted from the model fit of the kinetics. This parameter does not show a significant variation with pH.
Discussion
Structure formation initiated from the acid unfolded state of PGK was tuned from folding to amyloid formation by adjusting pH from 7.0 to 2.0. Earlier results found that at neutral pH, PGK folded in the enzymatically active structure (15,32), whereas amyloid formation was observed at pH 2 (11,33). Conformational changes were followed at 4°C in the 10 ms to 20 days time range by tryptophan and Thioflavin T fluorescence. Depending on the pH, either folding to the native state or amyloid formation occurred in the samples. Amyloid fibers were visualized after 20 days by electron microscopy.
Analyzing kinetics recorded at various pH indicates significant accumulation of only four states: unfolded, intermediate, native, and amyloid. In such a simple scenario, the molten globule intermediate is off pathway to the amyloid formation only if the amyloid is formed directly from the unfolded state. Earlier results, however, proved that interaction between the two folding domains of PGK directs the tertiary structure formation during amyloid growth (11). This indicates that amyloid formation happens from a state in which the domain structure of the protein is already formed, i.e., the molten globule intermediate and not the unfolded state. We also showed earlier that folding to the native state happens through the molten globule intermediate (20).
The early steps of structure formation of PGK followed stretched kinetics in the entire studied pH range (Fig. 1). Stretched kinetics was observed earlier in the folding of PGK at neutral pH (31,34), and explained by a hierarchic landscape structure with a finite number of possible relaxation pathways (20). Opposite to other landscape models, this model was developed using no a priori assumptions, giving the strongest evidence that time is important for determining the existence of complex folding landscape structures. Our landscape can be described using three parameters, one of which is a time constant. This time constant, however, is not a characteristic time of an exponential.
Changing this time constant affects the entire kinetics through the scaling relationship of the model, and a very sensitive fit of this parameter is possible. The measurements show a vibration artifact often observed in stopped-flow data. The largest modulation amplitude of the vibration artifacts, however, is 3–5% of the amplitude of the stretched fluorescence increase, and the modulation decreases quickly with time. Because the fit is done to the whole process extending through three-and-a-half orders of magnitude in time (10 ms to 50 s), the artifact does not affect the fit with the hierarchic landscape model. The model fitted well to our observation at all pH values.
We found that that the parameters of the upper part of the energy landscape, which direct the early steps of structure formation, were pH-independent. The number N = 5 of the possible relaxation times, the parameter α/λ = 1.0 ± 0.15, and the parameter λ = 8.1 ± 0.9, did not change significantly as pH was altered. This means that the structure of the upper region of the energy landscape, which governs the pathway and mechanism of the early conformational changes, is the same for folding and for amyloid formation. The number N and the scaling factor λ of the relaxation times were identical to those found earlier for the refolding from the GuHCl unfolded state at neutral pH and room temperature (20). This indicates that the refolding landscapes were essentially the same in the two experiments. The α/λ ratio of the model is the scaling factor for the number of molecules that follow different relaxation pathways. The difference between folding from acid denatured state at 4°C (α/λ = 1.0 ± 0.15) and refolding from GuHCl unfolded state at room temperature (α/λ = 0.75 ± 0.17) comes from the different distribution of the unfolded ensemble in the two experiments.
The shortest relaxation time (τ1) was the only parameter of the model that showed significant pH dependence (Fig. 2). This pH dependence reflects the effect of acid denaturation on the upper regions of the energy landscape. As pH is decreased from neutral, early structure formation of PGK speeds up showing a flattening of the upper regions of the landscape. It was found that the quick relaxation from the unfolded to the intermediate state is fastest at pH 3.5. Decreasing the pH further results in a slowing of the early structural changes.
Based on the above analysis of the early steps of structure formation of PGK, we conclude that the folding and the misfolding preceding amyloid formation follow the same path up to the formation of a hyperfluorescent intermediate. Earlier measurements done at neutral pH and room temperature showed that this intermediate has molten globule structure (20).
From the fast kinetic measurements, we conclude that the upper region of the energy landscape does not influence the final structure of PGK. The fate of the protein is determined by the lower part of the energy landscape. To explore this landscape region, structure formation from the intermediate state was followed by tryptophan fluorescence measurements from 1 min to 1 day (Fig. 3). This part of the kinetics followed a monoexponential time dependence indicating a simple cross-barrier reaction. Folding to the native enzyme structure was observed at neutral pH in accordance with earlier studies (20). Decreasing sample temperature to 4°C from room temperature, where earlier experiments were carried out, slowed PGK folding down roughly eightfold (20).
As the pH was decreased, the characteristic time obtained from the fit increased and the amplitude of the change decreased (Fig. 4). This indicates that the native structure is formed in a decreasing fraction of the molecules and more slowly as pH is decreased from the neutral. As Fig. 4 B indicates, the midpoint pH where half of the molecules are in folded, half in intermediate state in equilibrium, is roughly pH 4.7. Practically no native structure formation was observed at pH 4.0 and below it. These results suggest that the native state is destabilized compared to the intermediate state as the pH is decreased. At approximately pH 4.7 the two states become equally stable. Lowering the pH further, the intermediate becomes more stable compared to the folded conformation, and no native PGK is formed. Lengthening of the characteristic time shows the increase of the barrier separating the intermediate from the native state.
We detected amyloid formation from 1 min to 20 days using Thioflavin T fluorescence (Fig. 5). The fluorescence of the dye did not change above pH 4.5, but its intensity increased 2–3 times during the experiment at pH <4.5. This indicates that amyloid fibers are formed only below pH 4.5.
Modler et al. (14) found that the amyloid formation of PGK at pH 2.0 can be described using the Smoluchowski coagulation theory and size-independent association rate-constants. The Thioflavin T fluorescence kinetics could be well fitted with the model at all pH values (Fig. 5). For several proteins it has been found that the mechanism of amyloid formation depends on the buffer conditions, e.g., pH (7). Our results indicate that in the case of PGK, the amyloid growth mechanism is the same throughout the studied pH range. The characteristic coagulation time tc does not show significant pH dependence, indicating that the barrier between the molten globule and amyloid states is not sensitive to pH. The lack of large change in the amyloid growth kinetics as a function of pH makes kinetic control of partitioning between amyloid and native structures unlikely.
The only rationale for kinetically controlled amyloid formation would be the appearance of a disproportionately high kinetic barrier between the intermediate and native structures as pH is decreased whereas the folded structure remains the thermodynamically most stable state on the energy landscape. The fact that the native structure of PGK unfolds quickly under acidic conditions contradicts the above rationale. The fact that no amyloid structure is accumulated above pH 4.5 even after long waiting times also indicates that the fate of the protein is not kinetically determined. Lowering the pH makes the amyloid fibers more stable than the intermediate, allowing the growth of amyloid fibers.
Conclusions
Early steps of the folding and the amyloid formation of PGK follow the same path. The protein searches through a hierarchically structured energy landscape to form a molten globule intermediate (I).
Here the pathways leading toward the native and the amyloid structures diverge. Fig. 6 illustrates the changes in the energy landscape of PGK as conditions are gradually tuned from folding to misfolding. Above pH 4.5, PGK folds to the native state (F), which is thermodynamically the most stable conformation.
Figure 6.
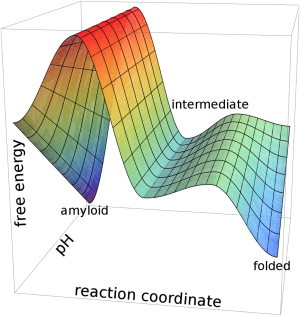
Simple scheme of the pH dependence of the energy landscape governing the folding and amyloid formation of PGK.
As pH is decreased, the native state becomes gradually less stable and less accessible from the molten globule, whereas the amyloid state becomes more stable. The kinetic barrier separating the intermediate from the amyloid does not depend on pH. Below pH 4.5, the amyloid structure represents a global minimum of the energy landscape, and PGK forms amyloid fibers.
Acknowledgments
This work was funded by the Hungarian Scientific Research Fund, grant OTKA K-77730; the János Bolyai Research Scholarship of the Hungarian Academy of Sciences, grant BO/00468/09; and the Social Renewal Operational Programme, grant TÁMOP-4.2.1/B-09/1/KMR-2010-0001. The research leading to these results has received funding from the European Union's Seventh Framework Programme (FP7/2007-2013) under grant agreement HEALTH-F2-2011-278850 (INMiND).
References
- 1.Luheshi L.M., Dobson C.M. Bridging the gap: from protein misfolding to protein misfolding diseases. FEBS Lett. 2009;583:2581–2586. doi: 10.1016/j.febslet.2009.06.030. [DOI] [PubMed] [Google Scholar]
- 2.Jahn T.R., Radford S.E. Folding versus aggregation: polypeptide conformations on competing pathways. Arch. Biochem. Biophys. 2008;469:100–117. doi: 10.1016/j.abb.2007.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ross C.A., Poirier M.A. Protein aggregation and neurodegenerative disease. Nat. Med. 2004;10(Suppl):S10–S17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- 4.Chiti F., Dobson C.M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 5.Dobson C.M. Protein misfolding, evolution and disease. Trends Biochem. Sci. 1999;24:329–332. doi: 10.1016/s0968-0004(99)01445-0. [DOI] [PubMed] [Google Scholar]
- 6.Guijarro J.I., Sunde M., Dobson C.M. Amyloid fibril formation by an SH3 domain. Proc. Natl. Acad. Sci. USA. 1998;95:4224–4228. doi: 10.1073/pnas.95.8.4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiti F., Webster P., Dobson C.M. Designing conditions for in vitro formation of amyloid protofilaments and fibrils. Proc. Natl. Acad. Sci. USA. 1999;96:3590–3594. doi: 10.1073/pnas.96.7.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Damaschun G., Damaschun H., Zirwer D. Proteins can adopt totally different folded conformations. J. Mol. Biol. 1999;291:715–725. doi: 10.1006/jmbi.1999.3009. [DOI] [PubMed] [Google Scholar]
- 9.Sawaya M.R., Sambashivan S., Eisenberg D. Atomic structures of amyloid cross-β spines reveal varied steric zippers. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 10.Nelson R., Sawaya M.R., Eisenberg D. Structure of the cross-β spine of amyloid-like fibrils. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Osváth S., Jäckel M., Fidy J. Domain interactions direct misfolding and amyloid formation of yeast phosphoglycerate kinase. Proteins. 2006;62:909–917. doi: 10.1002/prot.20823. [DOI] [PubMed] [Google Scholar]
- 12.Dobson C.M. Protein folding and misfolding. Nature. 2003;426:884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 13.Wang L., Maji S.K., Riek R. Bacterial inclusion bodies contain amyloid-like structure. PLoS Biol. 2008;6:e195. doi: 10.1371/journal.pbio.0060195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Modler A.J., Gast K., Damaschun G. Assembly of amyloid protofibrils via critical oligomers—a novel pathway of amyloid formation. J. Mol. Biol. 2003;325:135–148. doi: 10.1016/s0022-2836(02)01175-0. [DOI] [PubMed] [Google Scholar]
- 15.Chardot T., Mitraki A., Yon J.M. The effect of phosphate on the unfolding-refolding of phosphoglycerate kinase induced by guanidine hydrochloride. FEBS Lett. 1988;228:65–68. doi: 10.1016/0014-5793(88)80586-6. [DOI] [PubMed] [Google Scholar]
- 16.Griko Y., Venyaminov S., Privalov P.L. Heat and cold denaturation of phosphoglycerate kinase (interaction of domains) FEBS Lett. 1989;244:276–278. doi: 10.1016/0014-5793(89)80544-7. [DOI] [PubMed] [Google Scholar]
- 17.Lillo M.P., Beechem J.M., Mas M.T. Design and characterization of a multisite fluorescence energy-transfer system for protein folding studies: a steady-state and time-resolved study of yeast phosphoglycerate kinase. Biochemistry. 1997;36:11261–11272. doi: 10.1021/bi9707887. [DOI] [PubMed] [Google Scholar]
- 18.Damaschun G., Damaschun H., Zirwer D. Denatured states of yeast phosphoglycerate kinase. Biochemistry (Mosc.) 1998;63:259–275. [PubMed] [Google Scholar]
- 19.Dhar A., Samiotakis A., Cheung M.S. Structure, function, and folding of phosphoglycerate kinase are strongly perturbed by macromolecular crowding. Proc. Natl. Acad. Sci. USA. 2010;107:17586–17591. doi: 10.1073/pnas.1006760107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Osváth S., Herényi L., Köhler G. Hierarchic finite level energy landscape model: to describe the refolding kinetics of phosphoglycerate kinase. J. Biol. Chem. 2006;281:24375–24380. doi: 10.1074/jbc.M601915200. [DOI] [PubMed] [Google Scholar]
- 21.Foguel D., Suarez M.C., Silva J.L. Dissociation of amyloid fibrils of α-synuclein and transthyretin by pressure reveals their reversible nature and the formation of water-excluded cavities. Proc. Natl. Acad. Sci. USA. 2003;100:9831–9836. doi: 10.1073/pnas.1734009100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dirix C., Meersman F., Heremans K. High hydrostatic pressure dissociates early aggregates of TTR105-115, but not the mature amyloid fibrils. J. Mol. Biol. 2005;347:903–909. doi: 10.1016/j.jmb.2005.01.073. [DOI] [PubMed] [Google Scholar]
- 23.Kardos J., Micsonai A., Goto Y. Reversible heat-induced dissociation of β2-microglobulin amyloid fibrils. Biochemistry. 2011;50:3211–3220. doi: 10.1021/bi2000017. [DOI] [PubMed] [Google Scholar]
- 24.Agócs G., Solymosi K., Osváth S. Recovery of functional enzyme from amyloid fibrils. FEBS Lett. 2010;584:1139–1142. doi: 10.1016/j.febslet.2010.01.058. [DOI] [PubMed] [Google Scholar]
- 25.Apetri A.C., Maki K., Surewicz W.K. Early intermediate in human prion protein folding as evidenced by ultrarapid mixing experiments. J. Am. Chem. Soc. 2006;128:11673–11678. doi: 10.1021/ja063880b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Apetri A.C., Surewicz K., Surewicz W.K. The effect of disease-associated mutations on the folding pathway of human prion protein. J. Biol. Chem. 2004;279:18008–18014. doi: 10.1074/jbc.M313581200. [DOI] [PubMed] [Google Scholar]
- 27.Chen K.C., Xu M., Roder H. Microsecond unfolding kinetics of sheep prion protein reveals an intermediate that correlates with susceptibility to classical scrapie. Biophys. J. 2011;101:1221–1230. doi: 10.1016/j.bpj.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Osváth S., Gruebele M. Proline can have opposite effects on fast and slow protein folding phases. Biophys. J. 2003;85:1215–1222. doi: 10.1016/S0006-3495(03)74557-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cserpán I., Vas M. Effects of substrates on the heat stability and on the reactivities of thiol groups of 3-phosphoglycerate kinase. Eur. J. Biochem. 1983;131:157–162. doi: 10.1111/j.1432-1033.1983.tb07243.x. [DOI] [PubMed] [Google Scholar]
- 30.LeVine H., 3rd Thioflavine T interaction with synthetic Alzheimer's disease β-amyloid peptides: detection of amyloid aggregation in solution. Protein Sci. 1993;2:404–410. doi: 10.1002/pro.5560020312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Osváth S., Köhler G., Fidy J. Asymmetric effect of domain interactions on the kinetics of folding in yeast phosphoglycerate kinase. Protein Sci. 2005;14:1609–1616. doi: 10.1110/ps.051359905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ptitsyn O.B., Pain R.H., Razgulyaev O.I. Evidence for a molten globule state as a general intermediate in protein folding. FEBS Lett. 1990;262:20–24. doi: 10.1016/0014-5793(90)80143-7. [DOI] [PubMed] [Google Scholar]
- 33.Damaschun G., Damaschun H., Zirwer D. Conversion of yeast phosphoglycerate kinase into amyloid-like structure. Proteins. 2000;39:204–211. doi: 10.1002/(sici)1097-0134(20000515)39:3<204::aid-prot20>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 34.Sabelko J., Ervin J., Gruebele M. Observation of strange kinetics in protein folding. Proc. Natl. Acad. Sci. USA. 1999;96:6031–6036. doi: 10.1073/pnas.96.11.6031. [DOI] [PMC free article] [PubMed] [Google Scholar]


