Abstract
In the title compound, C24H25N5O4, the stereogenic C atom bonded to three N atoms and one C atom has an S configuration and its directly bonded neighbour has an R configuration. An intramolecular N—H⋯O hydrogen bond supports the near coplanarity of the two C3N2-five-membered rings [dihedral angle = 5.64 (10)°]. In the crystal, molecules are linked by N—H⋯N hydrogen bonds, forming a C(8) chain propagating in [001]. The chains are connected by C—H⋯O interactions, generating a three-dimensional network. The previous study [Nagel et al. (1974 ▶). Chem. Commun. pp. 1021–1022] did not establish the absolute structure and no atomic coordinates were published or deposited.
Related literature
For the previous structure, see: Nagel et al. (1974 ▶). For background to oxaline and its properties, see: Steyn (1970 ▶); Koizumi et al. (2004 ▶). For puckering parameters, see: Cremer & Pople (1975 ▶).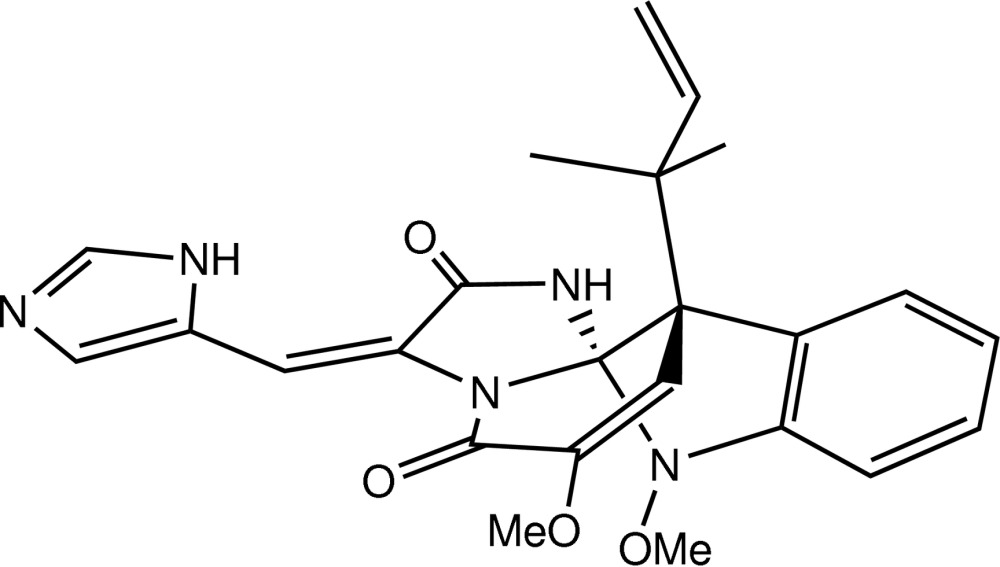
Experimental
Crystal data
C24H25N5O4
M r = 447.49
Orthorhombic,
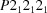
a = 10.7897 (2) Å
b = 13.2457 (3) Å
c = 15.6436 (4) Å
V = 2235.74 (9) Å3
Z = 4
Cu Kα radiation
μ = 0.76 mm−1
T = 100 K
0.60 × 0.20 × 0.12 mm
Data collection
Bruker APEXII CCD diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 2003 ▶) T min = 0.658, T max = 0.914
11202 measured reflections
3786 independent reflections
3766 reflections with I > 2σ(I)
R int = 0.034
Refinement
R[F 2 > 2σ(F 2)] = 0.035
wR(F 2) = 0.089
S = 1.08
3786 reflections
379 parameters
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.64 e Å−3
Δρmin = −0.26 e Å−3
Absolute structure: Flack (1983 ▶), 1403 Friedel pairs
Flack parameter: −0.05 (18)
Data collection: APEX2 (Bruker, 2004 ▶); cell refinement: SAINT (Bruker, 2004 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: XP in SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: WinGX (Farrugia, 1999 ▶).
Supplementary Material
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536812019423/hb6738sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812019423/hb6738Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N17—H17⋯O13 | 0.89 (2) | 1.85 (2) | 2.6562 (19) | 151 (2) |
| N14—H14⋯N19i | 0.83 (2) | 1.97 (2) | 2.798 (2) | 175 (2) |
| C4—H4⋯O9ii | 0.97 (2) | 2.54 (2) | 3.154 (2) | 121.5 (16) |
| C20—H20⋯O13iii | 1.00 (2) | 2.54 (2) | 3.353 (2) | 137.9 (16) |
| C24—H24C⋯O13iv | 0.98 (3) | 2.47 (3) | 3.382 (2) | 154 (2) |
Symmetry codes: (i) 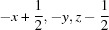 ; (ii)
; (ii) 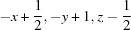 ; (iii)
; (iii) 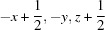 ; (iv)
; (iv) 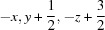 .
.
Acknowledgments
This work was supported by grants from the NSFC (No. 21172204), the Special Fund for Marine Scientific Research in the Public Interest of China (No. 2010418022–3) and the PCSIRT (No. IRT0944).
supplementary crystallographic information
Comment
Oxaline is a member of a class of biologically active indole alkaloids, characterized by a unique indoline spiroaminal framework and substitution of a 1,1-dimethylallyl ("reverse-prenyl") group at the benzylic ring junction. Oxaline was originally isolated from the culture broth of Penicillium oxalicum HK14–01 containing several unique structural features, including the N-OMe group, the unusual coupling of tryptophan and histidine, a single carbon atom bearing three nitrogen functionalities and a reversed prenyl group (Steyn, 1970). Besides, oxaline was found to inhibit tubulin polymerization in Jarkat cells, resulting in cell cycle arrest at the M phase (Koizumi et al., 2004). The X-ray structure of oxaline on Mo—Kα data was determined without definite absolute configuration (Nagel, et al., 1974). We isolated oxaline as part of our ongoing studies on characterizing bioactive metabolites from marine-derived halotolerant fungi. And the crystal structure on Cu—Kα and the absolute configuration are reported here.
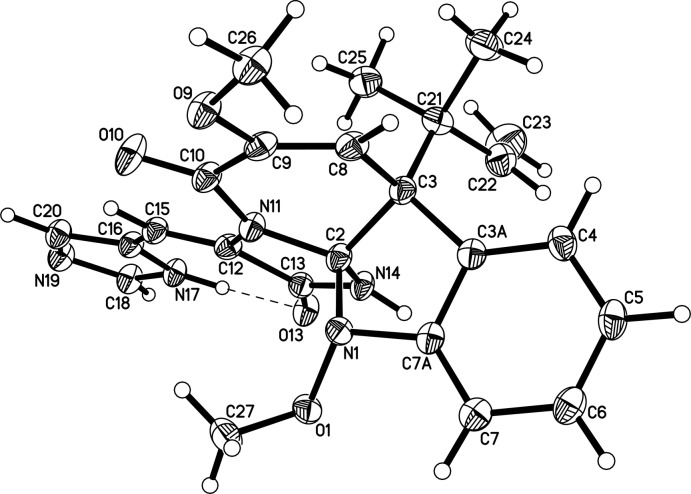
The title compound I contains a four fused rings structure as illustrated in Fig. 1. Two chiral atoms of C2 and C3 have the absolute configurations of S and R, respectively. Atom of N1 is S but it can invert in solution. Atom O1 in I (S-) has a short intra-contact of O1···N14 [2.7018 (19) Å]. While in R- one, the short contact are 2.882(C3), 2.581(C8), 2.390(C9), 2.662(C10) and 2.675 Å(N11), which indicates a unfavorable configuration. Both bonds of C8═C9 and C12═C15 are E but cis conformation. The five-membered ring of N1—C2—C3—C3A—C7A adopts envelope conformation with the puckering parameters (Cremer and Pople, 1975) of Q[0.3968 (17) Å] and φ[34.8 (2)°]. The six-membered ring of C2—C3—C8—C9—C10—N11 has the puckering parameters of Q = 0.4342 (17) Å, θ = 69.0 (2)° and φ = 76.8 (2)°, which implies a conformation among boat, twist-boat and half-chair.
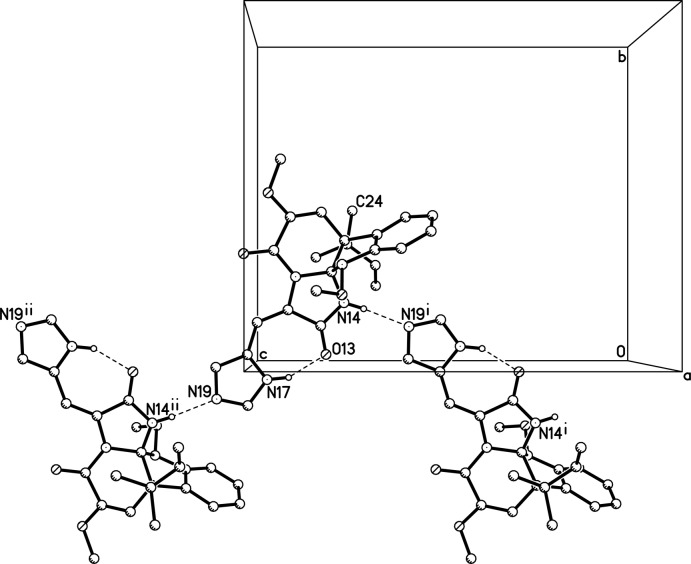
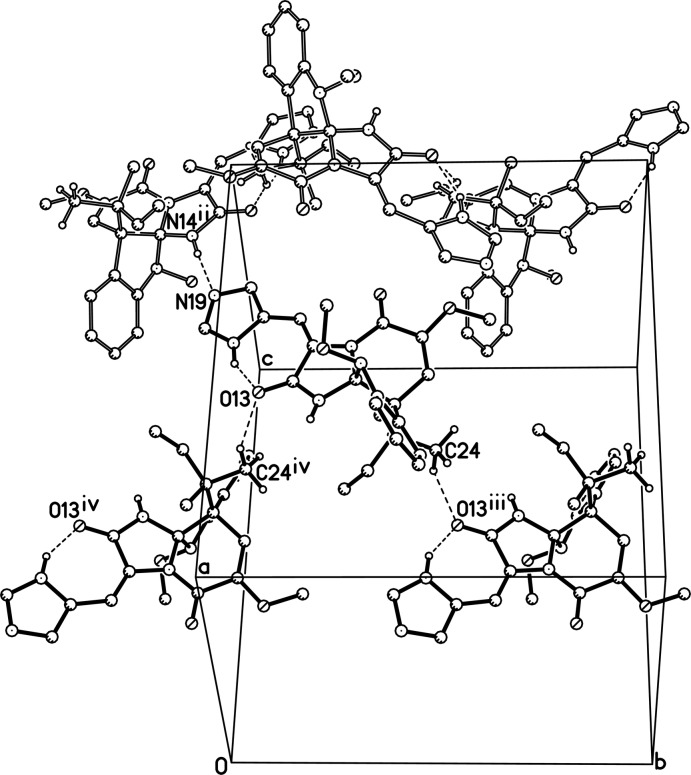
In the crystal, there are a one-dimensional classical hydrogen bonding chain parallel to the c axis (Fig. 2, Table 2) and a non-classical one along the b axis. These two kinds of chains together weave a three-dimensional supramolecular structure (Fig. 3).
Experimental
The halotolerant fugal strain Penicillium chrysogenum HK14–01, was isolated from the sediments collected in the Yellow River Delta, Dongying, Shandong, China. The working strain was cultured under static conditions at 298 K for 35 days in two hundred 1L conical flasks containing the liquid medium (300 ml/flask) composed of glucose (10 g/L), peptone (5 g/L), yeast extract (3 g/L), malt extract (1.5 g/L), marinum salt (100 g/L). The fermented whole broth (60 L) was filtered through cheese cloth to separate into supernatant and mycelia. The mycelia was extracted three times with acetone. The acetone solution was concentrated under reduced pressure to afford an aqueous solution. The acetone solution was extracted three times with ethyl acetate to give an ethyl acetate solution which was concentrated under reduced pressure to give a crude extract (39 g). The crude extract, which was subjected to chromatography over silica gel column using a stepwise gradient elution of CH2Cl2/petroleum ether(50–100%,V/V) and CH2Cl2/MeOH (0–100%,V/V),to yield twelve fractions (Fr.1-Fr.12). Fr.9, was fractionated on a C-18 ODS column using a step gradient elution of MeOH/H2O (60–100%,V/V) and was separated into 6 subfractions (Fr.9.1-Fr.9.6). Fr.9.3 was applied on Sephadex LH-20 using CH2Cl2/MeOH (1:1) to yield the title compound (145.0 mg). Colourless blocks were obtained by slow evaporation of petroleum ether/acetone (1:1) solution at 298 K.
Refinement
H atoms on C23 and C25 were placed in calculated positions, with C—H distances of 0.95 (C23) and 0.98 Å (C25), and were included in the final cycles of refinement in a riding model, with Uiso(H) values equal to 1.2Ueq(C23) or 1.5Ueq(C25). All other H atoms were located in a difference Fourier map and included in structure-factor calculations with free refinement. The highest difference peak is 0.83Å from atom H25C.
Figures
Fig. 1.
The molecular structure of (I), with displacement ellipsoids shown at the 50% probability level. Dashed lines indicates a intramolecular hydrogen bond.
Fig. 2.
A one-dimensional classical hydrogen-bonding chain along the c axis. [Symmetry code: (i) 1/2 - x, -y, z - 1/2; (ii) 1/2 - x, -y, z + 1/2]
Fig. 3.
A view of a three-dimensional hydrogen-bonding networks assembled by the classical chains above and the nonclassical ones parallel to the b axis. [Symmetry code: (ii) 1/2 - x, -y, z + 1/2; (iii) -x, y + 1/2, 3/2 - z; (iv) -x, y - 1/2, 3/2 - z]
Crystal data
| C24H25N5O4 | F(000) = 944 |
| Mr = 447.49 | Dx = 1.329 Mg m−3 |
| Orthorhombic, P212121 | Cu Kα radiation, λ = 1.54178 Å |
| Hall symbol: P 2ac 2ab | Cell parameters from 9936 reflections |
| a = 10.7897 (2) Å | θ = 2.8–69.0° |
| b = 13.2457 (3) Å | µ = 0.76 mm−1 |
| c = 15.6436 (4) Å | T = 100 K |
| V = 2235.74 (9) Å3 | Block, colourless |
| Z = 4 | 0.60 × 0.20 × 0.12 mm |
Data collection
| Bruker APEXII CCD diffractometer | 3786 independent reflections |
| Radiation source: fine-focus sealed tube | 3766 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.034 |
| φ and ω scans | θmax = 69.4°, θmin = 4.4° |
| Absorption correction: multi-scan (SADABS; Sheldrick, 2003) | h = −13→12 |
| Tmin = 0.658, Tmax = 0.914 | k = −15→15 |
| 11202 measured reflections | l = −16→18 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.035 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.089 | w = 1/[σ2(Fo2) + (0.0539P)2 + 0.608P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.08 | (Δ/σ)max < 0.001 |
| 3786 reflections | Δρmax = 0.64 e Å−3 |
| 379 parameters | Δρmin = −0.26 e Å−3 |
| 0 restraints | Absolute structure: Flack (1983), 1403 Friedel pairs |
| Primary atom site location: structure-invariant direct methods | Flack parameter: −0.05 (18) |
Special details
| Geometry. All s.u.'s (except the s.u. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell s.u.'s are taken into account individually in the estimation of s.u.'s in distances, angles and torsion angles; correlations between s.u.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell s.u.'s is used for estimating s.u.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C2 | 0.32699 (15) | 0.27923 (12) | 0.80027 (10) | 0.0186 (3) | |
| C3 | 0.26044 (15) | 0.37581 (12) | 0.76650 (10) | 0.0195 (3) | |
| C3A | 0.33279 (15) | 0.39156 (12) | 0.68341 (11) | 0.0194 (3) | |
| C4 | 0.30763 (17) | 0.44534 (13) | 0.60975 (11) | 0.0229 (4) | |
| C5 | 0.39516 (18) | 0.44605 (14) | 0.54394 (11) | 0.0253 (4) | |
| C6 | 0.50732 (17) | 0.39574 (14) | 0.55266 (12) | 0.0246 (4) | |
| C7 | 0.53572 (16) | 0.34468 (13) | 0.62767 (11) | 0.0220 (4) | |
| C7A | 0.44724 (15) | 0.34255 (12) | 0.69104 (10) | 0.0182 (3) | |
| C8 | 0.29907 (16) | 0.46075 (12) | 0.82645 (11) | 0.0211 (4) | |
| C9 | 0.34100 (16) | 0.44460 (12) | 0.90560 (11) | 0.0210 (3) | |
| C10 | 0.33551 (16) | 0.34357 (13) | 0.94788 (11) | 0.0214 (3) | |
| C12 | 0.28689 (15) | 0.16359 (13) | 0.91257 (11) | 0.0190 (3) | |
| C13 | 0.26417 (14) | 0.11484 (12) | 0.82818 (10) | 0.0187 (3) | |
| C15 | 0.27783 (15) | 0.12521 (13) | 0.99230 (11) | 0.0200 (3) | |
| C16 | 0.24075 (15) | 0.02576 (13) | 1.01914 (10) | 0.0204 (3) | |
| C18 | 0.18204 (17) | −0.13354 (13) | 1.01993 (11) | 0.0233 (4) | |
| C20 | 0.24315 (17) | −0.01288 (13) | 1.10109 (11) | 0.0231 (3) | |
| C21 | 0.11422 (16) | 0.36829 (14) | 0.75912 (12) | 0.0245 (4) | |
| C22 | 0.07620 (17) | 0.30327 (15) | 0.68323 (13) | 0.0292 (4) | |
| C23 | −0.0179 (2) | 0.23927 (19) | 0.68053 (16) | 0.0467 (6) | |
| H23A | −0.0694 | 0.2306 | 0.7293 | 0.056* | |
| H23B | −0.0341 | 0.2020 | 0.6299 | 0.056* | |
| C24 | 0.06073 (19) | 0.47547 (16) | 0.74467 (14) | 0.0319 (4) | |
| C25 | 0.05692 (16) | 0.32850 (15) | 0.84208 (12) | 0.0281 (4) | |
| H25A | −0.0330 | 0.3387 | 0.8407 | 0.042* | |
| H25B | 0.0921 | 0.3650 | 0.8908 | 0.042* | |
| H25C | 0.0750 | 0.2563 | 0.8478 | 0.042* | |
| C26 | 0.38241 (19) | 0.61629 (14) | 0.92987 (12) | 0.0261 (4) | |
| C27 | 0.62470 (18) | 0.21991 (16) | 0.83731 (13) | 0.0304 (4) | |
| N1 | 0.45699 (13) | 0.29884 (10) | 0.77360 (9) | 0.0185 (3) | |
| N11 | 0.32034 (13) | 0.26396 (10) | 0.89240 (9) | 0.0194 (3) | |
| N14 | 0.28079 (13) | 0.18525 (10) | 0.76808 (9) | 0.0192 (3) | |
| N17 | 0.19945 (13) | −0.05261 (11) | 0.96885 (9) | 0.0211 (3) | |
| N19 | 0.20655 (14) | −0.11236 (11) | 1.10046 (9) | 0.0243 (3) | |
| O1 | 0.52932 (11) | 0.20898 (8) | 0.77451 (8) | 0.0220 (3) | |
| O9 | 0.38366 (12) | 0.51505 (9) | 0.96104 (8) | 0.0242 (3) | |
| O10 | 0.33924 (13) | 0.33375 (10) | 1.02538 (8) | 0.0297 (3) | |
| O13 | 0.23426 (11) | 0.02613 (9) | 0.81455 (7) | 0.0227 (3) | |
| H4 | 0.230 (2) | 0.4816 (18) | 0.6029 (14) | 0.036 (6)* | |
| H5 | 0.3779 (19) | 0.4836 (16) | 0.4900 (13) | 0.023 (5)* | |
| H6 | 0.5638 (17) | 0.3927 (14) | 0.5057 (12) | 0.015 (4)* | |
| H7 | 0.613 (2) | 0.3112 (16) | 0.6374 (13) | 0.026 (5)* | |
| H8 | 0.2979 (17) | 0.5235 (15) | 0.8010 (12) | 0.015 (4)* | |
| H14 | 0.284 (2) | 0.1668 (17) | 0.7172 (15) | 0.027 (5)* | |
| H15 | 0.3009 (18) | 0.1647 (16) | 1.0401 (13) | 0.021 (5)* | |
| H17 | 0.198 (2) | −0.0459 (17) | 0.9125 (15) | 0.029 (5)* | |
| H18 | 0.1494 (17) | −0.1977 (14) | 0.9977 (12) | 0.016 (4)* | |
| H20 | 0.269 (2) | 0.0184 (16) | 1.1564 (14) | 0.028 (5)* | |
| H22 | 0.118 (3) | 0.318 (2) | 0.6324 (17) | 0.050 (7)* | |
| H24A | 0.095 (2) | 0.5062 (18) | 0.6938 (15) | 0.037 (6)* | |
| H24B | 0.080 (2) | 0.522 (2) | 0.7904 (16) | 0.042 (7)* | |
| H24C | −0.028 (3) | 0.4672 (19) | 0.7331 (16) | 0.045 (7)* | |
| H26A | 0.4374 (17) | 0.6236 (14) | 0.8794 (12) | 0.015 (4)* | |
| H26B | 0.412 (2) | 0.6584 (18) | 0.9782 (16) | 0.040 (6)* | |
| H26C | 0.294 (2) | 0.6392 (17) | 0.9142 (14) | 0.029 (5)* | |
| H27A | 0.676 (2) | 0.2766 (19) | 0.8265 (15) | 0.035 (6)* | |
| H27B | 0.668 (2) | 0.1612 (18) | 0.8349 (15) | 0.033 (6)* | |
| H27C | 0.590 (2) | 0.2315 (17) | 0.8929 (14) | 0.028 (5)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C2 | 0.0247 (8) | 0.0158 (7) | 0.0153 (8) | −0.0008 (6) | −0.0003 (6) | −0.0016 (6) |
| C3 | 0.0253 (8) | 0.0165 (7) | 0.0166 (8) | 0.0022 (6) | 0.0003 (6) | −0.0005 (6) |
| C3A | 0.0255 (8) | 0.0138 (7) | 0.0189 (8) | −0.0011 (7) | −0.0012 (7) | −0.0018 (6) |
| C4 | 0.0291 (9) | 0.0183 (8) | 0.0213 (8) | 0.0007 (7) | −0.0043 (7) | 0.0012 (7) |
| C5 | 0.0352 (9) | 0.0207 (9) | 0.0199 (8) | −0.0039 (7) | −0.0023 (7) | 0.0046 (7) |
| C6 | 0.0305 (9) | 0.0223 (9) | 0.0210 (8) | −0.0057 (7) | 0.0028 (7) | 0.0005 (7) |
| C7 | 0.0258 (8) | 0.0180 (8) | 0.0223 (8) | −0.0022 (7) | 0.0005 (7) | −0.0026 (7) |
| C7A | 0.0259 (8) | 0.0120 (7) | 0.0167 (7) | −0.0028 (6) | −0.0016 (6) | −0.0026 (6) |
| C8 | 0.0273 (8) | 0.0132 (8) | 0.0229 (9) | 0.0008 (6) | 0.0041 (7) | −0.0003 (7) |
| C9 | 0.0258 (8) | 0.0165 (8) | 0.0206 (8) | −0.0009 (7) | 0.0045 (7) | −0.0040 (7) |
| C10 | 0.0286 (8) | 0.0174 (8) | 0.0181 (8) | −0.0010 (7) | 0.0014 (7) | −0.0034 (6) |
| C12 | 0.0215 (7) | 0.0157 (7) | 0.0198 (8) | 0.0002 (6) | 0.0017 (6) | −0.0014 (7) |
| C13 | 0.0209 (7) | 0.0170 (8) | 0.0181 (8) | 0.0009 (6) | −0.0005 (6) | 0.0001 (6) |
| C15 | 0.0252 (8) | 0.0173 (8) | 0.0176 (8) | 0.0007 (7) | 0.0001 (6) | −0.0023 (6) |
| C16 | 0.0241 (8) | 0.0191 (8) | 0.0179 (8) | −0.0003 (7) | 0.0006 (6) | −0.0021 (7) |
| C18 | 0.0310 (8) | 0.0176 (8) | 0.0213 (8) | −0.0049 (7) | 0.0007 (7) | 0.0020 (7) |
| C20 | 0.0306 (8) | 0.0199 (8) | 0.0190 (8) | −0.0018 (7) | 0.0005 (7) | 0.0003 (7) |
| C21 | 0.0238 (8) | 0.0233 (9) | 0.0264 (9) | 0.0050 (7) | −0.0002 (7) | −0.0004 (7) |
| C22 | 0.0275 (9) | 0.0345 (10) | 0.0257 (9) | 0.0009 (8) | −0.0031 (8) | −0.0011 (8) |
| C23 | 0.0451 (12) | 0.0505 (13) | 0.0446 (13) | −0.0061 (11) | 0.0018 (11) | −0.0054 (11) |
| C24 | 0.0290 (10) | 0.0293 (10) | 0.0374 (11) | 0.0085 (8) | 0.0022 (8) | 0.0036 (10) |
| C25 | 0.0258 (8) | 0.0300 (9) | 0.0286 (9) | 0.0006 (7) | 0.0018 (7) | 0.0002 (8) |
| C26 | 0.0373 (10) | 0.0160 (9) | 0.0250 (9) | −0.0044 (8) | 0.0029 (8) | −0.0023 (7) |
| C27 | 0.0309 (9) | 0.0292 (10) | 0.0310 (11) | 0.0080 (8) | −0.0100 (8) | −0.0010 (8) |
| N1 | 0.0244 (7) | 0.0121 (6) | 0.0189 (7) | 0.0027 (5) | 0.0004 (6) | 0.0024 (5) |
| N11 | 0.0274 (7) | 0.0149 (7) | 0.0161 (7) | −0.0006 (5) | 0.0004 (5) | −0.0001 (5) |
| N14 | 0.0280 (7) | 0.0152 (7) | 0.0145 (7) | −0.0004 (5) | −0.0012 (5) | −0.0016 (5) |
| N17 | 0.0273 (7) | 0.0195 (7) | 0.0164 (7) | −0.0026 (6) | 0.0004 (6) | 0.0009 (6) |
| N19 | 0.0329 (7) | 0.0188 (7) | 0.0211 (7) | −0.0022 (6) | 0.0019 (6) | 0.0027 (6) |
| O1 | 0.0284 (6) | 0.0152 (6) | 0.0222 (6) | 0.0062 (5) | −0.0023 (5) | −0.0006 (5) |
| O9 | 0.0369 (6) | 0.0156 (6) | 0.0202 (6) | −0.0040 (5) | 0.0016 (5) | −0.0021 (5) |
| O10 | 0.0526 (8) | 0.0206 (6) | 0.0159 (6) | −0.0076 (6) | 0.0019 (6) | −0.0022 (5) |
| O13 | 0.0340 (6) | 0.0157 (6) | 0.0184 (6) | −0.0052 (5) | −0.0009 (5) | −0.0011 (5) |
Geometric parameters (Å, º)
| O1—N1 | 1.4233 (17) | C12—C13 | 1.490 (2) |
| O1—C27 | 1.430 (2) | C12—C15 | 1.350 (2) |
| O9—C9 | 1.354 (2) | C15—C16 | 1.439 (2) |
| O9—C26 | 1.427 (2) | C16—C20 | 1.381 (2) |
| O10—C10 | 1.220 (2) | C21—C22 | 1.523 (3) |
| O13—C13 | 1.237 (2) | C21—C24 | 1.549 (3) |
| N1—C2 | 1.486 (2) | C21—C25 | 1.531 (2) |
| N1—C7A | 1.419 (2) | C22—C23 | 1.324 (3) |
| N11—C2 | 1.457 (2) | C4—H4 | 0.97 (3) |
| N11—C10 | 1.375 (2) | C5—H5 | 1.00 (2) |
| N11—C12 | 1.413 (2) | C6—H6 | 0.955 (19) |
| N14—C2 | 1.432 (2) | C7—H7 | 0.95 (2) |
| N14—C13 | 1.336 (2) | C8—H8 | 0.92 (2) |
| N17—C16 | 1.377 (2) | C15—H15 | 0.95 (2) |
| N17—C18 | 1.350 (2) | C18—H18 | 0.984 (19) |
| N19—C18 | 1.317 (2) | C20—H20 | 1.00 (2) |
| N19—C20 | 1.376 (2) | C22—H22 | 0.93 (3) |
| N14—H14 | 0.83 (2) | C23—H23A | 0.9500 |
| N17—H17 | 0.89 (2) | C23—H23B | 0.9500 |
| C2—C3 | 1.559 (2) | C24—H24A | 0.97 (2) |
| C3—C3A | 1.531 (2) | C24—H24B | 0.97 (3) |
| C3—C8 | 1.523 (2) | C24—H24C | 0.98 (3) |
| C3—C21 | 1.585 (2) | C25—H25A | 0.9800 |
| C3A—C4 | 1.382 (2) | C25—H25B | 0.9800 |
| C3A—C7A | 1.400 (2) | C25—H25C | 0.9800 |
| C4—C5 | 1.397 (3) | C26—H26A | 0.993 (19) |
| C5—C6 | 1.388 (3) | C26—H26B | 0.99 (2) |
| C6—C7 | 1.389 (3) | C26—H26C | 1.03 (2) |
| C7—C7A | 1.377 (2) | C27—H27A | 0.95 (2) |
| C8—C9 | 1.336 (2) | C27—H27B | 0.91 (2) |
| C9—C10 | 1.494 (2) | C27—H27C | 0.96 (2) |
| N1—O1—C27 | 108.47 (12) | N11—C12—C13 | 104.59 (13) |
| C9—O9—C26 | 115.18 (13) | N11—C12—C15 | 125.37 (16) |
| O1—N1—C2 | 111.62 (12) | C13—C12—C15 | 130.04 (15) |
| O1—N1—C7A | 113.01 (12) | O13—C13—N14 | 125.21 (15) |
| C2—N1—C7A | 104.88 (12) | O13—C13—C12 | 127.39 (15) |
| C2—N11—C10 | 120.79 (14) | N14—C13—C12 | 107.39 (13) |
| C2—N11—C12 | 111.34 (13) | C12—C15—C16 | 129.35 (16) |
| C10—N11—C12 | 127.64 (14) | C12—C15—H15 | 120.2 (13) |
| C2—N14—C13 | 113.94 (14) | C16—C15—H15 | 110.4 (13) |
| C2—N14—H14 | 125.3 (16) | N17—C16—C15 | 127.83 (15) |
| C13—N14—H14 | 118.2 (16) | N17—C16—C20 | 104.91 (14) |
| C16—N17—C18 | 107.78 (14) | C15—C16—C20 | 127.23 (16) |
| C16—N17—H17 | 120.0 (15) | N17—C18—N19 | 111.65 (15) |
| C18—N17—H17 | 131.7 (15) | N17—C18—H18 | 121.8 (11) |
| C18—N19—C20 | 105.57 (15) | N19—C18—H18 | 126.4 (11) |
| N1—C2—N11 | 110.39 (13) | N19—C20—C16 | 110.06 (15) |
| N1—C2—N14 | 112.43 (13) | N19—C20—H20 | 118.9 (12) |
| N1—C2—C3 | 101.31 (12) | C16—C20—H20 | 131.0 (12) |
| N11—C2—N14 | 102.12 (13) | C3—C21—C22 | 111.13 (14) |
| N11—C2—C3 | 115.23 (13) | C3—C21—C25 | 111.21 (14) |
| N14—C2—C3 | 115.70 (13) | C3—C21—C24 | 108.89 (15) |
| C2—C3—C3A | 99.48 (13) | C22—C21—C24 | 107.71 (15) |
| C2—C3—C8 | 105.75 (13) | C22—C21—C25 | 110.94 (15) |
| C2—C3—C21 | 115.56 (14) | C24—C21—C25 | 106.77 (15) |
| C3A—C3—C8 | 106.43 (13) | C21—C22—C23 | 126.4 (2) |
| C3A—C3—C21 | 117.03 (14) | C21—C22—H22 | 114.8 (17) |
| C8—C3—C21 | 111.33 (14) | C23—C22—H22 | 118.2 (17) |
| C3—C3A—C4 | 132.71 (16) | C22—C23—H23A | 120.0 |
| C3—C3A—C7A | 108.31 (14) | C22—C23—H23B | 120.0 |
| C4—C3A—C7A | 118.92 (16) | H23A—C23—H23B | 120.0 |
| C3A—C4—C5 | 119.03 (17) | C21—C24—H24A | 111.4 (14) |
| C3A—C4—H4 | 121.0 (14) | C21—C24—H24B | 113.5 (15) |
| C5—C4—H4 | 119.9 (14) | H24A—C24—H24B | 104.9 (19) |
| C4—C5—C6 | 120.91 (16) | C21—C24—H24C | 106.7 (15) |
| C4—C5—H5 | 120.0 (12) | H24A—C24—H24C | 105 (2) |
| C6—C5—H5 | 119.0 (12) | H24B—C24—H24C | 115 (2) |
| C5—C6—C7 | 120.61 (17) | C21—C25—H25A | 109.5 |
| C5—C6—H6 | 120.1 (11) | C21—C25—H25B | 109.5 |
| C7—C6—H6 | 119.2 (11) | C21—C25—H25C | 109.5 |
| C6—C7—C7A | 117.74 (16) | H25A—C25—H25B | 109.5 |
| C6—C7—H7 | 123.6 (13) | H25A—C25—H25C | 109.5 |
| C7A—C7—H7 | 118.6 (13) | H25B—C25—H25C | 109.5 |
| N1—C7A—C3A | 109.40 (14) | O9—C26—H26A | 111.0 (11) |
| N1—C7A—C7 | 127.75 (15) | O9—C26—H26B | 105.4 (14) |
| C3A—C7A—C7 | 122.73 (15) | O9—C26—H26C | 111.6 (13) |
| C3—C8—C9 | 123.06 (15) | H26A—C26—H26B | 111.1 (18) |
| C3—C8—H8 | 113.4 (11) | H26A—C26—H26C | 109.5 (16) |
| C9—C8—H8 | 123.3 (12) | H26B—C26—H26C | 108.2 (19) |
| O9—C9—C8 | 126.76 (16) | O1—C27—H27A | 112.0 (14) |
| O9—C9—C10 | 110.32 (14) | O1—C27—H27B | 104.8 (14) |
| C8—C9—C10 | 122.71 (15) | O1—C27—H27C | 111.2 (13) |
| O10—C10—N11 | 123.31 (16) | H27A—C27—H27B | 112.0 (19) |
| O10—C10—C9 | 122.29 (15) | H27A—C27—H27C | 105.0 (19) |
| N11—C10—C9 | 114.34 (14) | H27B—C27—H27C | 112.0 (19) |
| C2—N1—O1—C27 | −116.05 (15) | C12—C15—C16—N17 | −3.6 (3) |
| C7A—N1—O1—C27 | 126.02 (15) | C12—C15—C16—C20 | 173.90 (17) |
| C8—C9—O9—C26 | 0.1 (2) | N17—C16—C20—N19 | 1.06 (19) |
| C10—C9—O9—C26 | −174.67 (15) | C15—C16—C20—N19 | −176.90 (16) |
| C3—C21—C22—C23 | 142.1 (2) | C8—C3—C21—C22 | 165.31 (14) |
| C24—C21—C22—C23 | −98.7 (2) | C3A—C3—C21—C22 | 42.6 (2) |
| C25—C21—C22—C23 | 17.8 (3) | C2—C3—C21—C22 | −74.04 (19) |
| N14—C2—C3—C8 | 164.60 (14) | C8—C3—C21—C25 | −70.57 (19) |
| N11—C2—C3—C8 | 45.63 (18) | C3A—C3—C21—C25 | 166.75 (14) |
| N1—C2—C3—C8 | −73.52 (15) | C2—C3—C21—C25 | 50.1 (2) |
| N14—C2—C3—C3A | −85.22 (16) | C8—C3—C21—C24 | 46.82 (19) |
| N11—C2—C3—C3A | 155.80 (14) | C3A—C3—C21—C24 | −75.86 (19) |
| N1—C2—C3—C3A | 36.66 (15) | C2—C3—C21—C24 | 167.47 (15) |
| N14—C2—C3—C21 | 41.0 (2) | C7—C7A—N1—O1 | −35.1 (2) |
| N11—C2—C3—C21 | −78.00 (18) | C3A—C7A—N1—O1 | 148.75 (13) |
| N1—C2—C3—C21 | 162.85 (14) | C7—C7A—N1—C2 | −156.93 (16) |
| C8—C3—C3A—C4 | −89.6 (2) | C3A—C7A—N1—C2 | 26.94 (16) |
| C2—C3—C3A—C4 | 160.80 (18) | N14—C2—N1—C7A | 84.27 (15) |
| C21—C3—C3A—C4 | 35.6 (3) | N11—C2—N1—C7A | −162.41 (12) |
| C8—C3—C3A—C7A | 87.46 (16) | C3—C2—N1—C7A | −39.85 (15) |
| C2—C3—C3A—C7A | −22.17 (16) | N14—C2—N1—O1 | −38.44 (17) |
| C21—C3—C3A—C7A | −147.36 (14) | N11—C2—N1—O1 | 74.88 (16) |
| C7A—C3A—C4—C5 | 2.4 (2) | C3—C2—N1—O1 | −162.57 (12) |
| C3—C3A—C4—C5 | 179.14 (17) | O10—C10—N11—C12 | 11.2 (3) |
| C3A—C4—C5—C6 | −1.6 (3) | C9—C10—N11—C12 | −165.97 (16) |
| C4—C5—C6—C7 | −0.9 (3) | O10—C10—N11—C2 | −174.82 (17) |
| C5—C6—C7—C7A | 2.5 (3) | C9—C10—N11—C2 | 8.0 (2) |
| C6—C7—C7A—C3A | −1.7 (2) | C15—C12—N11—C10 | −8.3 (3) |
| C6—C7—C7A—N1 | −177.36 (15) | C13—C12—N11—C10 | 171.09 (15) |
| C4—C3A—C7A—C7 | −0.7 (2) | C15—C12—N11—C2 | 177.29 (16) |
| C3—C3A—C7A—C7 | −178.25 (15) | C13—C12—N11—C2 | −3.37 (17) |
| C4—C3A—C7A—N1 | 175.62 (15) | N14—C2—N11—C10 | −168.15 (14) |
| C3—C3A—C7A—N1 | −1.89 (17) | N1—C2—N11—C10 | 72.10 (18) |
| C3A—C3—C8—C9 | −126.85 (17) | C3—C2—N11—C10 | −41.9 (2) |
| C2—C3—C8—C9 | −21.7 (2) | N14—C2—N11—C12 | 6.74 (17) |
| C21—C3—C8—C9 | 104.56 (19) | N1—C2—N11—C12 | −113.01 (14) |
| C3—C8—C9—O9 | 176.00 (16) | C3—C2—N11—C12 | 133.00 (14) |
| C3—C8—C9—C10 | −9.9 (3) | O13—C13—N14—C2 | −174.18 (15) |
| C8—C9—C10—O10 | −158.30 (18) | C12—C13—N14—C2 | 6.41 (18) |
| O9—C9—C10—O10 | 16.7 (2) | N11—C2—N14—C13 | −8.14 (18) |
| C8—C9—C10—N11 | 18.9 (2) | N1—C2—N14—C13 | 110.17 (15) |
| O9—C9—C10—N11 | −166.11 (14) | C3—C2—N14—C13 | −134.10 (15) |
| C15—C12—C13—O13 | −1.8 (3) | N19—C18—N17—C16 | 1.5 (2) |
| N11—C12—C13—O13 | 178.90 (15) | C20—C16—N17—C18 | −1.49 (18) |
| C15—C12—C13—N14 | 177.59 (17) | C15—C16—N17—C18 | 176.46 (16) |
| N11—C12—C13—N14 | −1.72 (17) | N17—C18—N19—C20 | −0.8 (2) |
| N11—C12—C15—C16 | 177.70 (16) | C16—C20—N19—C18 | −0.2 (2) |
| C13—C12—C15—C16 | −1.5 (3) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N17—H17···O13 | 0.89 (2) | 1.85 (2) | 2.6562 (19) | 151 (2) |
| N14—H14···N19i | 0.83 (2) | 1.97 (2) | 2.798 (2) | 175 (2) |
| C4—H4···O9ii | 0.97 (2) | 2.54 (2) | 3.154 (2) | 121.5 (16) |
| C20—H20···O13iii | 1.00 (2) | 2.54 (2) | 3.353 (2) | 137.9 (16) |
| C24—H24C···O13iv | 0.98 (3) | 2.47 (3) | 3.382 (2) | 154 (2) |
Symmetry codes: (i) −x+1/2, −y, z−1/2; (ii) −x+1/2, −y+1, z−1/2; (iii) −x+1/2, −y, z+1/2; (iv) −x, y+1/2, −z+3/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: HB6738).
References
- Bruker (2004). APEX2, SAINT and SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Cremer, D. & Pople, J. A. (1975). J. Am. Chem. Soc. 97, 1354–1358.
- Farrugia, L. J. (1999). J. Appl. Cryst. 32, 837–838.
- Flack, H. D. (1983). Acta Cryst. A39, 876–881.
- Koizumi, Y., Arai, M., Tomoda, H. & Omura, S. (2004). Biochim. Biophys. Acta, 1693, 47–55. [DOI] [PubMed]
- Nagel, D. W., Pachler, K. G. R., Steyn, P. S., Wessels, P. L., Gafner, G. & Kruger, G. J. (1974). Chem. Commun. pp. 1021–1022.
- Sheldrick, G. M. (2003). SADABS University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Steyn, P. S. (1970). Tetrahedron, 26, 51–57. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536812019423/hb6738sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812019423/hb6738Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report