Abstract
In the title compound, C12H11FN4S, the thiadiazine ring adopts a twist-boat conformation. The dihedral angle between the triazolothiadiazine system and the benzene ring is 10.54 (9)°. The crystal structure is characterized by C—H⋯N hydrogen bonds. The crystal packing also exhibits π–π interactions, with a centroid–centroid distance of 3.6348 (15) Å.
Related literature
For biological properties of triazolothiadiazines, see: Feng et al. (1992 ▶); Mohan & Anjaneyalu (1987 ▶); Holla et al. (2001 ▶); Walser et al. (1991 ▶); Hirota et al. (1991 ▶); Bradbury & Rivett (1991 ▶); Heindel & Reid (1980 ▶); Heidelberger et al. (1957 ▶). For related structures, see: Andersson & MacGowan (2003 ▶); Novak et al. (2006 ▶).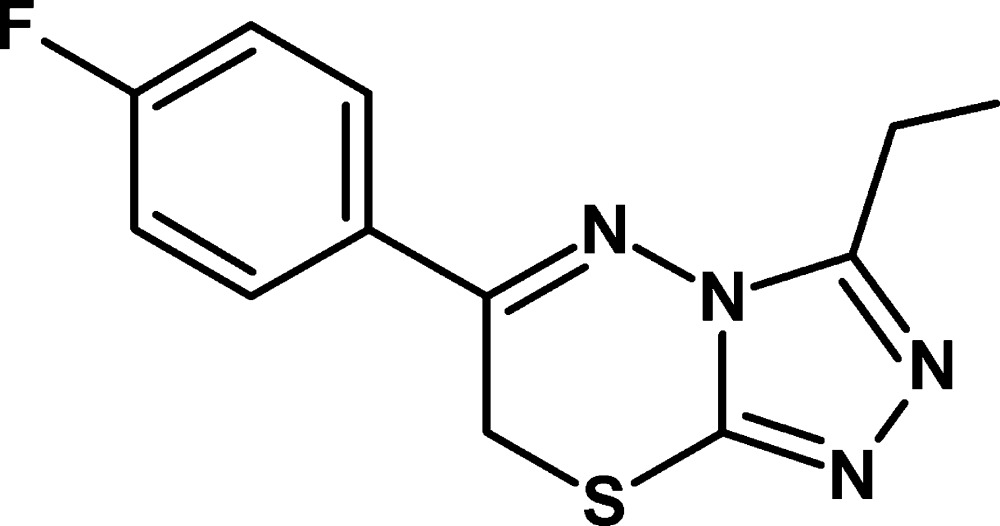
Experimental
Crystal data
C12H11FN4S
M r = 262.31
Monoclinic,

a = 13.322 (3) Å
b = 13.017 (3) Å
c = 7.1912 (16) Å
β = 105.308 (4)°
V = 1202.8 (4) Å3
Z = 4
Mo Kα radiation
μ = 0.27 mm−1
T = 293 K
0.24 × 0.20 × 0.12 mm
Data collection
Bruker SMART CCD area-detector diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2001 ▶) T min = 0.770, T max = 1.000
11097 measured reflections
2119 independent reflections
1828 reflections with I > 2σ(I)
R int = 0.023
Refinement
R[F 2 > 2σ(F 2)] = 0.041
wR(F 2) = 0.106
S = 1.06
2119 reflections
164 parameters
H-atom parameters constrained
Δρmax = 0.22 e Å−3
Δρmin = −0.22 e Å−3
Data collection: SMART (Bruker, 2001 ▶); cell refinement: SAINT (Bruker, 2001 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 (Farrugia, 1997 ▶); software used to prepare material for publication: SHELXL97.
Supplementary Material
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S160053681202185X/bt5922sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S160053681202185X/bt5922Isup2.hkl
Supplementary material file. DOI: 10.1107/S160053681202185X/bt5922Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C1—H1⋯N2i | 0.93 | 2.51 | 3.428 (3) | 172 |
| C8—H8A⋯N1ii | 0.97 | 2.50 | 3.410 (3) | 156 |
| C8—H8B⋯N2i | 0.97 | 2.30 | 3.228 (3) | 160 |
Symmetry codes: (i)  ; (ii)
; (ii)  .
.
Acknowledgments
The authors thank Professor T. N. Guru Row, Solid State and Structural Chemistry Unit, Indian Institute of Science, Bangalore, for the data collection. SJ thanks the University Grants Commission (India), for the award of a Teacher Fellowship under the Faculty Development Programme (UGC-SWRO File No.: FIP/11 th Plan/KAMY006 TF, Dt.: 06/08/2010).
supplementary crystallographic information
Comment
Triazoles fused with six membered ring systems are found to be associated with diverse pharmacological activity. A number of thiadiazines have been shown to exhibit antimicrobial (Feng et al., 1992) and dieretic properties (Mohan et al., 1987) and also serves as photographic couplers (Holla et al., 2001). The analgesic, anti-asthmatic, diuretic, anti-hypertensive, anti-cholinergic, antibacterial, antifungal, anti-inflammatory, hypoglycemic, anti-tubercular and antiviral properties exhibited by various N-bridged heterocycles derived from a variety of 4-amino-5-mercapto-1,2,4-triazoles, have made them an important chemotherapeutic agents (Walser et al., 1991; Hirota et al., 1991; Bradbury et al., 1991). The 1,2,4- triazoles nucleus has recently been incorporated into a wide variety of therapeutically interesting drugs including H1/H2 histamine receptor blockers, cholinesterase active agents, CNS stimulants, anti-anxiety agents and sedatives (Heindel et al., 1980). Further fluorinated heterocycles have been shown to possess wide variety of biocidal activities. Compounds such as fluorouracil and fluoroquinolone have been used as anticancer agents and antibiotics (Heidelberger et al., 1957; Andersson et al., 2003; Novak et al., 2006).
The asymmetric unit of 3-ethyl-6-(4-fluorophenyl)-7H-[1,2,4]triazolo[3,4-b] [1,3,4]thiadiazine is shown in Fig. 1. The triazolo-thiadiazine ring system is not planar. The dihedral angle between the triazolo-thiadiazine ring system (S1/N1–N4/C7–C10) and the benzene ring (C1–C6) is 10.54 (9)°.
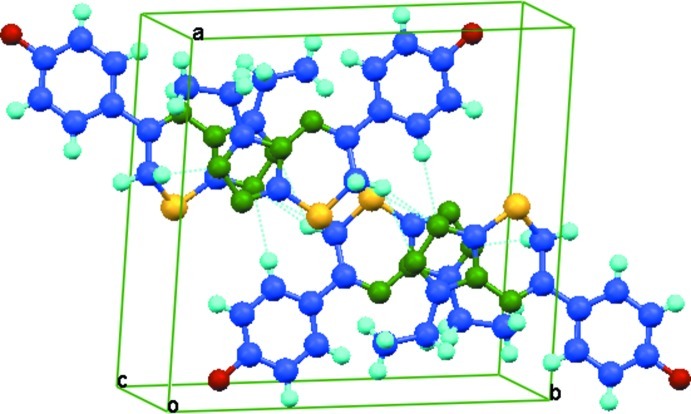
In the crystal structure (Fig. 2), the molecules are connected via intermolecular C1—H1···N2, C8—H8A···N1 and C8—H8B···N2 hydrogen bonds (Table 1). Furthermore, the crystal structure features a π-π interaction, with a centroid-centroid Cg1 (C9/C10/N1–N3) distance of 3.5728 (16) Å.
Experimental
A mixture of triazole (1) (0.01 mol) and p-fluorophenacyl bromide (0.01 mol) in ethanol (25 ml) was heated under reflux for 1–2 hrs. The reaction mixture was cooled to room temparature and neutralized with sodium acetate (5%). The precipitated triazolothiadiazines were collected by filtration, washed with water and recrystallized from ethanol. Yield 82%; m.p.455 K.
Refinement
All H atoms were positioned geometrically, with C—H = 0.93 Å for aromatic H, C—H = 0.97 Å for methylene H and C—H = 0.96 Å for methyl H, and refined using a riding model with Uiso(H) = 1.5Ueq(C) for methyl H and Uiso(H) = 1.2Ueq(C) for all other H.
Figures
Fig. 1.
The molecular structure of the title compound. Displacement ellipsoids are drawn at the 50% probability level.
Fig. 2.
The packing of molecules in the title structure.
Crystal data
| C12H11FN4S | F(000) = 544 |
| Mr = 262.31 | Dx = 1.449 Mg m−3 |
| Monoclinic, P21/c | Melting point: 455 K |
| Hall symbol: -P 2ybc | Mo Kα radiation, λ = 0.71073 Å |
| a = 13.322 (3) Å | Cell parameters from 2119 reflections |
| b = 13.017 (3) Å | θ = 2.2–25.0° |
| c = 7.1912 (16) Å | µ = 0.27 mm−1 |
| β = 105.308 (4)° | T = 293 K |
| V = 1202.8 (4) Å3 | Plate, colourless |
| Z = 4 | 0.24 × 0.20 × 0.12 mm |
Data collection
| Bruker SMART CCD area-detector diffractometer | 2119 independent reflections |
| Radiation source: fine-focus sealed tube | 1828 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.023 |
| ω and φ scans | θmax = 25.0°, θmin = 2.2° |
| Absorption correction: multi-scan (SADABS; Bruker, 2001) | h = −15→15 |
| Tmin = 0.770, Tmax = 1.000 | k = −15→15 |
| 11097 measured reflections | l = −8→8 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.041 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.106 | H-atom parameters constrained |
| S = 1.06 | w = 1/[σ2(Fo2) + (0.0487P)2 + 0.4632P] where P = (Fo2 + 2Fc2)/3 |
| 2119 reflections | (Δ/σ)max < 0.001 |
| 164 parameters | Δρmax = 0.22 e Å−3 |
| 0 restraints | Δρmin = −0.22 e Å−3 |
Special details
| Geometry. All s.u.'s (except the s.u. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell s.u.'s are taken into account individually in the estimation of s.u.'s in distances, angles and torsion angles; correlations between s.u.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell s.u.'s is used for estimating s.u.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| S1 | 0.50854 (4) | 0.90687 (4) | 0.19250 (9) | 0.0542 (2) | |
| F1 | 0.05432 (12) | 1.31988 (11) | −0.1252 (2) | 0.0845 (5) | |
| N1 | 0.37247 (14) | 0.64191 (13) | 0.1417 (3) | 0.0511 (5) | |
| N2 | 0.46299 (14) | 0.70215 (13) | 0.1828 (3) | 0.0550 (5) | |
| N3 | 0.32718 (12) | 0.80332 (11) | 0.1211 (2) | 0.0397 (4) | |
| N4 | 0.26399 (12) | 0.88834 (12) | 0.0605 (2) | 0.0403 (4) | |
| C1 | 0.28158 (17) | 1.16575 (16) | 0.0713 (3) | 0.0534 (6) | |
| H1 | 0.3526 | 1.1746 | 0.1256 | 0.064* | |
| C2 | 0.21920 (19) | 1.25088 (18) | 0.0115 (4) | 0.0626 (6) | |
| H2 | 0.2476 | 1.3166 | 0.0253 | 0.075* | |
| C3 | 0.11625 (18) | 1.23664 (17) | −0.0673 (4) | 0.0566 (6) | |
| C4 | 0.07182 (18) | 1.14193 (19) | −0.0911 (4) | 0.0625 (6) | |
| H4 | 0.0008 | 1.1346 | −0.1464 | 0.075* | |
| C5 | 0.13374 (16) | 1.05684 (17) | −0.0319 (3) | 0.0536 (6) | |
| H5 | 0.1042 | 0.9916 | −0.0479 | 0.064* | |
| C6 | 0.24016 (15) | 1.06767 (15) | 0.0515 (3) | 0.0407 (5) | |
| C7 | 0.30545 (14) | 0.97569 (14) | 0.1164 (3) | 0.0378 (4) | |
| C8 | 0.41148 (15) | 0.98739 (15) | 0.2543 (3) | 0.0446 (5) | |
| H8A | 0.4073 | 0.9702 | 0.3833 | 0.054* | |
| H8B | 0.4330 | 1.0586 | 0.2552 | 0.054* | |
| C9 | 0.43346 (15) | 0.79735 (15) | 0.1685 (3) | 0.0436 (5) | |
| C10 | 0.29301 (15) | 0.70301 (14) | 0.1038 (3) | 0.0423 (5) | |
| C11 | 0.18178 (17) | 0.67302 (16) | 0.0591 (4) | 0.0547 (6) | |
| H11A | 0.1431 | 0.7122 | −0.0514 | 0.066* | |
| H11B | 0.1548 | 0.6907 | 0.1677 | 0.066* | |
| C12 | 0.1639 (2) | 0.56042 (18) | 0.0163 (4) | 0.0671 (7) | |
| H12A | 0.1819 | 0.5441 | −0.1011 | 0.101* | |
| H12B | 0.0920 | 0.5443 | 0.0024 | 0.101* | |
| H12C | 0.2066 | 0.5210 | 0.1204 | 0.101* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S1 | 0.0360 (3) | 0.0450 (3) | 0.0800 (4) | −0.0006 (2) | 0.0126 (3) | 0.0070 (3) |
| F1 | 0.0731 (10) | 0.0583 (9) | 0.1191 (13) | 0.0317 (8) | 0.0204 (9) | 0.0218 (8) |
| N1 | 0.0518 (11) | 0.0354 (9) | 0.0640 (12) | 0.0038 (8) | 0.0113 (9) | −0.0002 (8) |
| N2 | 0.0464 (10) | 0.0411 (10) | 0.0745 (13) | 0.0081 (8) | 0.0108 (9) | 0.0033 (9) |
| N3 | 0.0371 (8) | 0.0311 (8) | 0.0479 (9) | 0.0031 (7) | 0.0060 (7) | 0.0004 (7) |
| N4 | 0.0357 (9) | 0.0329 (9) | 0.0489 (10) | 0.0036 (7) | 0.0053 (7) | −0.0006 (7) |
| C1 | 0.0437 (12) | 0.0385 (12) | 0.0739 (15) | 0.0028 (9) | 0.0084 (10) | 0.0023 (10) |
| C2 | 0.0622 (15) | 0.0361 (12) | 0.0880 (18) | 0.0041 (11) | 0.0175 (13) | 0.0021 (11) |
| C3 | 0.0581 (14) | 0.0450 (13) | 0.0682 (15) | 0.0194 (11) | 0.0194 (11) | 0.0112 (11) |
| C4 | 0.0413 (12) | 0.0615 (15) | 0.0780 (16) | 0.0107 (11) | 0.0039 (11) | 0.0074 (13) |
| C5 | 0.0426 (12) | 0.0425 (12) | 0.0702 (15) | 0.0008 (9) | 0.0049 (10) | 0.0036 (10) |
| C6 | 0.0404 (11) | 0.0367 (10) | 0.0443 (11) | 0.0030 (8) | 0.0099 (9) | 0.0008 (8) |
| C7 | 0.0364 (10) | 0.0350 (10) | 0.0414 (10) | −0.0004 (8) | 0.0091 (8) | 0.0005 (8) |
| C8 | 0.0394 (11) | 0.0332 (10) | 0.0550 (12) | −0.0019 (8) | 0.0013 (9) | −0.0002 (9) |
| C9 | 0.0383 (10) | 0.0400 (11) | 0.0511 (12) | 0.0050 (9) | 0.0091 (9) | 0.0035 (9) |
| C10 | 0.0483 (11) | 0.0312 (10) | 0.0446 (11) | 0.0006 (9) | 0.0071 (9) | −0.0015 (8) |
| C11 | 0.0498 (13) | 0.0416 (12) | 0.0682 (14) | −0.0055 (10) | 0.0075 (11) | −0.0017 (10) |
| C12 | 0.0722 (17) | 0.0486 (13) | 0.0808 (17) | −0.0161 (12) | 0.0210 (14) | −0.0076 (12) |
Geometric parameters (Å, º)
| S1—C9 | 1.724 (2) | C4—C5 | 1.379 (3) |
| S1—C8 | 1.809 (2) | C4—H4 | 0.9300 |
| F1—C3 | 1.359 (2) | C5—C6 | 1.393 (3) |
| N1—C10 | 1.294 (2) | C5—H5 | 0.9300 |
| N1—N2 | 1.403 (2) | C6—C7 | 1.481 (3) |
| N2—C9 | 1.296 (2) | C7—C8 | 1.504 (3) |
| N3—C9 | 1.368 (3) | C8—H8A | 0.9700 |
| N3—C10 | 1.377 (2) | C8—H8B | 0.9700 |
| N3—N4 | 1.389 (2) | C10—C11 | 1.483 (3) |
| N4—C7 | 1.282 (2) | C11—C12 | 1.504 (3) |
| C1—C6 | 1.383 (3) | C11—H11A | 0.9700 |
| C1—C2 | 1.384 (3) | C11—H11B | 0.9700 |
| C1—H1 | 0.9300 | C12—H12A | 0.9600 |
| C2—C3 | 1.351 (3) | C12—H12B | 0.9600 |
| C2—H2 | 0.9300 | C12—H12C | 0.9600 |
| C3—C4 | 1.359 (3) | ||
| C9—S1—C8 | 94.03 (9) | N4—C7—C8 | 123.28 (17) |
| C10—N1—N2 | 108.10 (16) | C6—C7—C8 | 119.81 (16) |
| C9—N2—N1 | 106.96 (17) | C7—C8—S1 | 112.80 (14) |
| C9—N3—C10 | 105.32 (16) | C7—C8—H8A | 109.0 |
| C9—N3—N4 | 128.67 (16) | S1—C8—H8A | 109.0 |
| C10—N3—N4 | 124.63 (15) | C7—C8—H8B | 109.0 |
| C7—N4—N3 | 115.66 (15) | S1—C8—H8B | 109.0 |
| C6—C1—C2 | 121.1 (2) | H8A—C8—H8B | 107.8 |
| C6—C1—H1 | 119.4 | N2—C9—N3 | 110.27 (17) |
| C2—C1—H1 | 119.4 | N2—C9—S1 | 128.82 (16) |
| C3—C2—C1 | 118.7 (2) | N3—C9—S1 | 120.86 (14) |
| C3—C2—H2 | 120.6 | N1—C10—N3 | 109.33 (17) |
| C1—C2—H2 | 120.6 | N1—C10—C11 | 126.78 (18) |
| C2—C3—C4 | 122.5 (2) | N3—C10—C11 | 123.81 (17) |
| C2—C3—F1 | 119.1 (2) | C10—C11—C12 | 113.32 (19) |
| C4—C3—F1 | 118.4 (2) | C10—C11—H11A | 108.9 |
| C3—C4—C5 | 119.0 (2) | C12—C11—H11A | 108.9 |
| C3—C4—H4 | 120.5 | C10—C11—H11B | 108.9 |
| C5—C4—H4 | 120.5 | C12—C11—H11B | 108.9 |
| C4—C5—C6 | 120.6 (2) | H11A—C11—H11B | 107.7 |
| C4—C5—H5 | 119.7 | C11—C12—H12A | 109.5 |
| C6—C5—H5 | 119.7 | C11—C12—H12B | 109.5 |
| C1—C6—C5 | 118.08 (19) | H12A—C12—H12B | 109.5 |
| C1—C6—C7 | 121.91 (18) | C11—C12—H12C | 109.5 |
| C5—C6—C7 | 120.01 (18) | H12A—C12—H12C | 109.5 |
| N4—C7—C6 | 116.73 (16) | H12B—C12—H12C | 109.5 |
| C10—N1—N2—C9 | 0.3 (2) | N4—C7—C8—S1 | 46.9 (2) |
| C9—N3—N4—C7 | −26.4 (3) | C6—C7—C8—S1 | −138.27 (16) |
| C10—N3—N4—C7 | 168.98 (18) | C9—S1—C8—C7 | −49.59 (16) |
| C6—C1—C2—C3 | −0.1 (4) | N1—N2—C9—N3 | 0.5 (2) |
| C1—C2—C3—C4 | −0.4 (4) | N1—N2—C9—S1 | −177.06 (16) |
| C1—C2—C3—F1 | 179.6 (2) | C10—N3—C9—N2 | −1.0 (2) |
| C2—C3—C4—C5 | 0.3 (4) | N4—N3—C9—N2 | −167.93 (18) |
| F1—C3—C4—C5 | −179.7 (2) | C10—N3—C9—S1 | 176.72 (15) |
| C3—C4—C5—C6 | 0.2 (4) | N4—N3—C9—S1 | 9.8 (3) |
| C2—C1—C6—C5 | 0.6 (3) | C8—S1—C9—N2 | −156.4 (2) |
| C2—C1—C6—C7 | −179.4 (2) | C8—S1—C9—N3 | 26.27 (18) |
| C4—C5—C6—C1 | −0.6 (3) | N2—N1—C10—N3 | −1.0 (2) |
| C4—C5—C6—C7 | 179.3 (2) | N2—N1—C10—C11 | −177.8 (2) |
| N3—N4—C7—C6 | 179.22 (16) | C9—N3—C10—N1 | 1.2 (2) |
| N3—N4—C7—C8 | −5.8 (3) | N4—N3—C10—N1 | 168.80 (17) |
| C1—C6—C7—N4 | −167.62 (19) | C9—N3—C10—C11 | 178.14 (19) |
| C5—C6—C7—N4 | 12.5 (3) | N4—N3—C10—C11 | −14.3 (3) |
| C1—C6—C7—C8 | 17.2 (3) | N1—C10—C11—C12 | −13.0 (3) |
| C5—C6—C7—C8 | −162.73 (19) | N3—C10—C11—C12 | 170.6 (2) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C1—H1···N2i | 0.93 | 2.51 | 3.428 (3) | 172 |
| C8—H8A···N1ii | 0.97 | 2.50 | 3.410 (3) | 156 |
| C8—H8B···N2i | 0.97 | 2.30 | 3.228 (3) | 160 |
Symmetry codes: (i) −x+1, y+1/2, −z+1/2; (ii) x, −y+3/2, z+1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: BT5922).
References
- Andersson, M. I. & MacGowan, A. P. (2003). J. Antimicrob. Chemother. 51, 1–11. [DOI] [PubMed]
- Bradbury, R. H. & Rivett, J. E. (1991). J. Med. Chem. 34, 151–157. [DOI] [PubMed]
- Bruker (2001). SMART, SAINT and SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Farrugia, L. J. (1997). J. Appl. Cryst. 30, 565.
- Feng, X. M., Chen, R. & Yang, W. D. (1992). Chem. J. Chin. Univ. 13, 187–194.
- Heidelberger, C., Chaudhuri, N. K., Danneberg, P., Mooren, D., Greisbach, L., Duschinsky, R., Scnnitzer, R. J., Pleaven, E. & Scheiner, J. (1957). Nature (London), 179, 663–666. [DOI] [PubMed]
- Heindel, N. D. & Reid, J. R. (1980). J. Heterocycl. Chem 17, 1087–1088.
- Hirota, T., Sasaki, K., Yamamoto, H. & Nakayama, T. (1991). J. Heterocycl. Chem 28, 257–261.
- Holla, B. S., Akberali, P. M. & Shivananda, M. K. (2001). Il Farmaco, 56, 919–927. [DOI] [PubMed]
- Mohan, J. & Anjaneyalu, G. S. R. (1987). Pol. J. Chem. 61, 547–551.
- Novak, M. J., Baum, J. C. & Buhrow, J. A. (2006). Olson. Surf. Sci 600, L269–L273.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Walser, A., Flynn, T. & Mason, C. (1991). J. Heterocycl. Chem 28, 1121–1125.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S160053681202185X/bt5922sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S160053681202185X/bt5922Isup2.hkl
Supplementary material file. DOI: 10.1107/S160053681202185X/bt5922Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report