Abstract
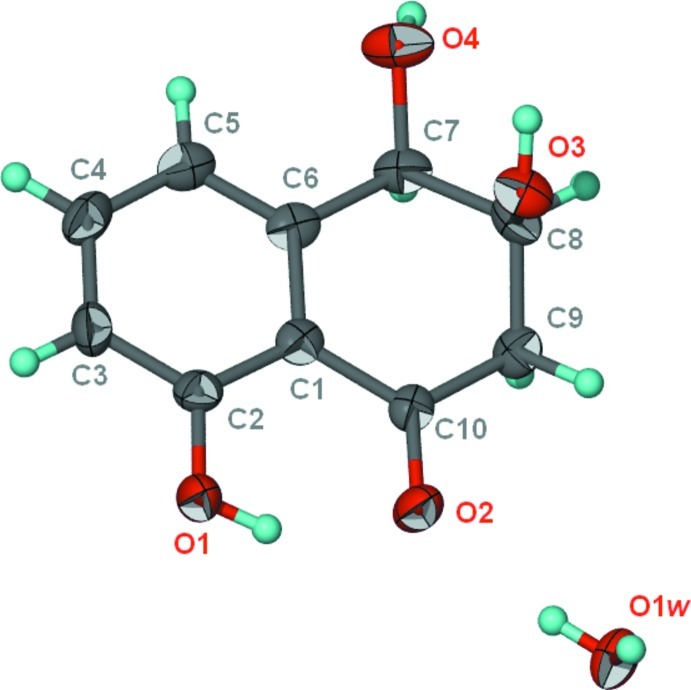
In the title hydrate, C10H10O4·H2O, the six-membered aliphatic ring that is fused to the benzene ring has a sofa shape, with the hydroxy group in the 3-position (that represents the sofa back) of the aliphatic ring occupying a quasi-axial position. The hydroxy group of the aromatic ring is hydrogen-bond donor to the carbonyl O atom; other O—H⋯O hydrogen bonds link the organic molecules and the water molecules into a three-dimensional network.
Related literature
For the isolation of the title compound from other fungi, see: Borgschulte et al. (1991 ▶); Iwasaki et al. (1972 ▶); Trisuwan et al. (2008 ▶). The absolute configuration was assumed from published assignments, see: Trisuwan et al. (2008 ▶).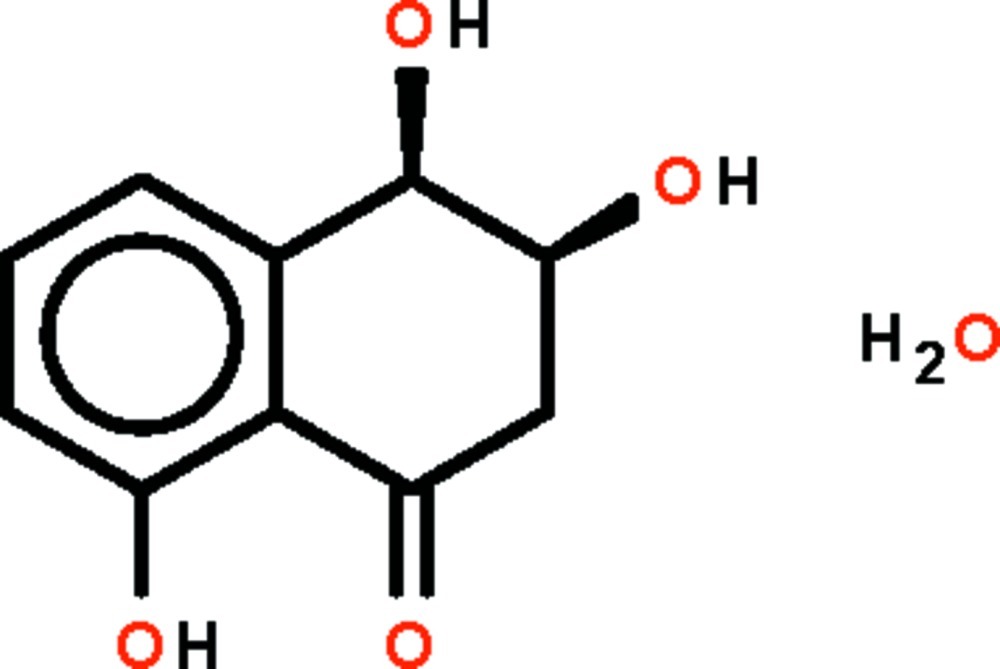
Experimental
Crystal data
C10H10O4·H2O
M r = 212.20
Orthorhombic,
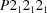
a = 4.6430 (4) Å
b = 14.3904 (11) Å
c = 14.4976 (10) Å
V = 968.65 (13) Å3
Z = 4
Mo Kα radiation
μ = 0.12 mm−1
T = 293 K
0.31 × 0.28 × 0.24 mm
Data collection
Bruker APEX DUO diffractometer
6331 measured reflections
1320 independent reflections
916 reflections with I > 2σ(I)
R int = 0.063
Refinement
R[F 2 > 2σ(F 2)] = 0.039
wR(F 2) = 0.096
S = 0.99
1320 reflections
156 parameters
5 restraints
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.21 e Å−3
Δρmin = −0.21 e Å−3
Data collection: APEX2 (Bruker, 2010 ▶); cell refinement: SAINT (Bruker, 2010 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: X-SEED (Barbour, 2001 ▶); software used to prepare material for publication: publCIF (Westrip, 2010 ▶).
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536812022623/xu5544sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812022623/xu5544Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536812022623/xu5544Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1—H1⋯O2 | 0.84 (1) | 1.87 (3) | 2.590 (3) | 143 (3) |
| O1—H1⋯O1wi | 0.84 (1) | 2.27 (3) | 2.829 (3) | 124 (3) |
| O3—H2⋯O1ii | 0.84 (1) | 2.15 (3) | 2.924 (3) | 153 (5) |
| O4—H3⋯O1wiii | 0.85 (1) | 1.82 (1) | 2.657 (3) | 170 (4) |
| O1w—H4⋯O2 | 0.84 (1) | 1.98 (1) | 2.805 (3) | 167 (4) |
| O1w—H5⋯O4iv | 0.85 (1) | 1.88 (1) | 2.726 (3) | 177 (3) |
Symmetry codes: (i) 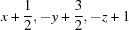 ; (ii)
; (ii) 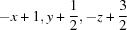 ; (iii)
; (iii) 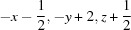 ; (iv)
; (iv) 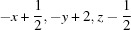 .
.
Acknowledgments
We thank Université Mohammed V-Agdal, and the Ministry of Higher Education of Malaysia (grant No. UM.C/HIR/MOHE/SC/12) for supporting this study
supplementary crystallographic information
Comment
3,4-Dihydro-3,4,8-trihydroxy-1[2H]-naphthalenone is a secondary metabolite produced by several endophytic fungi,e.g., Hypoxylon mammatum (Borgschulte et al., 1991), Nigrospora sp. (Trisuwan et al., 2008) and Pyrichularia orayzae (Iwasaki et al., 1972). The compound was isolated from Embellisia eureka in this study; the compound was found to crystallize as a monohydrate (Scheme I). In the hydrate, C10H10O4.H2O, the six-membered aliphatic ring that is fused to the benzene ring has a soft shape. The C-3 atom represents the sofa back. The hydroxy group of the aliphatic ring occupies a quasi-axial position (Fig. 1). The hydroxy group of the aromatic ring is hydrogen-bond donor to the carbonyl O atom; other O—H···O hydrogen bonds link the organic molecule and water molecule to form a 3D network (Table 1).
Experimental
Fungal extraction
The fungal strain, Embellisia eureka, was identified by PCR. About 250 ml of ethyl acetate was added into each culture material of the fungus in an Erlenmeyer flask. The ethyl acetate phase was then concentrated under reduced pressure. The residue was diluted in 90% aqueous methanol and further extracted with n-hexane to remove fatty acids and other non-polar constituents. The remaining 90% methanol phase was evaporated under reduced pressure to yield 3.0 g of crude product.
Isolation protocol of 3,4-dihydro-3,4,8-trihydroxy-1[2H]-naphthalenone
The 90% methanol extract was submitted to vacuum liquid chromatography on a column packed with silica as the stationary phase,. The resulting fraction was submitted two successive fractionations on a Sephadex column packed with Sephadex LH-20 as stationary phase. The mobile phase was the 100% methanol. This gave 113.4 mg of a material that was purified by using the semi-preparative HPLC to give 7.0 mg of the pure compound. Crystals were obtained by slow evaporation of a methanol: water (9:1) solution of the compound.
Refinement
The aromatic and methylene H-atoms were placed in calculated positions (C–H 0.93–0.97 Å) and were included in the refinement in the riding model approximation, with U(H) set to 1.2U(C). The hydroxy and water H-atoms were located in a difference Fourier map, and were refined with a distance restraint of 0.84±0.01 Å; their temperature factors were refined.
The (0 1 1) reflection was omitted owing to bad disagreement.
The absolute configuration was assumed from published assignments (Trisuwan et al., 2008); 892 Friedel pairs were merged.
Figures
Fig. 1.
Thermal ellipsoid plot (Barbour, 2001) of C10H10O4.H2O at the 70% probability level. Hydrogen atoms are drawn as spheres of arbitrary radius.
Crystal data
| C10H10O4·H2O | F(000) = 448 |
| Mr = 212.20 | Dx = 1.455 Mg m−3 |
| Orthorhombic, P212121 | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: P 2ac 2ab | Cell parameters from 1057 reflections |
| a = 4.6430 (4) Å | θ = 2.8–21.8° |
| b = 14.3904 (11) Å | µ = 0.12 mm−1 |
| c = 14.4976 (10) Å | T = 293 K |
| V = 968.65 (13) Å3 | Prism, brown |
| Z = 4 | 0.31 × 0.28 × 0.24 mm |
Data collection
| Bruker APEX DUO diffractometer | 916 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.063 |
| Graphite monochromator | θmax = 27.5°, θmin = 2.8° |
| ω scans | h = −6→5 |
| 6331 measured reflections | k = −18→17 |
| 1320 independent reflections | l = −18→18 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.039 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.096 | H atoms treated by a mixture of independent and constrained refinement |
| S = 0.99 | w = 1/[σ2(Fo2) + (0.0507P)2] where P = (Fo2 + 2Fc2)/3 |
| 1320 reflections | (Δ/σ)max = 0.001 |
| 156 parameters | Δρmax = 0.21 e Å−3 |
| 5 restraints | Δρmin = −0.21 e Å−3 |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.5051 (5) | 0.75199 (14) | 0.74392 (14) | 0.0280 (6) | |
| O2 | 0.1740 (5) | 0.83227 (14) | 0.62424 (12) | 0.0281 (5) | |
| O3 | 0.1933 (5) | 1.08380 (14) | 0.69719 (15) | 0.0262 (5) | |
| O4 | −0.0651 (5) | 1.10191 (16) | 0.87414 (16) | 0.0318 (5) | |
| O1w | 0.0565 (5) | 0.82560 (15) | 0.43458 (14) | 0.0268 (5) | |
| C1 | 0.1862 (7) | 0.8812 (2) | 0.77911 (18) | 0.0198 (6) | |
| C2 | 0.3877 (7) | 0.81217 (18) | 0.80514 (18) | 0.0226 (7) | |
| C3 | 0.4788 (7) | 0.8053 (2) | 0.89602 (18) | 0.0270 (7) | |
| H3A | 0.6084 | 0.7592 | 0.9134 | 0.032* | |
| C4 | 0.3753 (7) | 0.8673 (2) | 0.96024 (19) | 0.0278 (8) | |
| H4A | 0.4351 | 0.8622 | 1.0213 | 0.033* | |
| C5 | 0.1843 (7) | 0.9371 (2) | 0.9362 (2) | 0.0247 (7) | |
| H5A | 0.1202 | 0.9787 | 0.9809 | 0.030* | |
| C6 | 0.0881 (7) | 0.94556 (19) | 0.8461 (2) | 0.0211 (6) | |
| C7 | −0.1173 (7) | 1.02214 (19) | 0.8181 (2) | 0.0244 (7) | |
| H7 | −0.3147 | 1.0008 | 0.8293 | 0.029* | |
| C8 | −0.0888 (7) | 1.0480 (2) | 0.7169 (2) | 0.0235 (7) | |
| H8 | −0.2352 | 1.0942 | 0.7004 | 0.028* | |
| C9 | −0.1250 (6) | 0.96220 (19) | 0.6583 (2) | 0.0236 (7) | |
| H9A | −0.3192 | 0.9385 | 0.6658 | 0.028* | |
| H9B | −0.0993 | 0.9786 | 0.5940 | 0.028* | |
| C10 | 0.0858 (6) | 0.88749 (19) | 0.68341 (18) | 0.0193 (6) | |
| H1 | 0.439 (8) | 0.761 (2) | 0.6908 (13) | 0.049 (12)* | |
| H2 | 0.228 (12) | 1.1339 (18) | 0.725 (3) | 0.103 (19)* | |
| H3 | −0.228 (4) | 1.118 (3) | 0.895 (2) | 0.061 (13)* | |
| H4 | 0.071 (10) | 0.821 (2) | 0.4923 (8) | 0.060 (13)* | |
| H5 | 0.212 (4) | 0.848 (2) | 0.414 (2) | 0.039 (11)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0361 (14) | 0.0245 (12) | 0.0236 (11) | 0.0091 (10) | −0.0056 (11) | −0.0015 (10) |
| O2 | 0.0319 (12) | 0.0308 (11) | 0.0215 (10) | 0.0076 (11) | −0.0046 (10) | −0.0056 (9) |
| O3 | 0.0207 (11) | 0.0228 (12) | 0.0351 (12) | −0.0052 (10) | 0.0051 (10) | −0.0006 (10) |
| O4 | 0.0180 (11) | 0.0354 (13) | 0.0420 (13) | 0.0034 (11) | 0.0007 (11) | −0.0182 (11) |
| O1w | 0.0195 (12) | 0.0378 (13) | 0.0231 (11) | −0.0054 (11) | −0.0032 (10) | 0.0044 (10) |
| C1 | 0.0176 (15) | 0.0199 (14) | 0.0220 (14) | −0.0065 (12) | 0.0019 (12) | 0.0013 (12) |
| C2 | 0.0261 (17) | 0.0187 (15) | 0.0229 (14) | −0.0045 (13) | −0.0005 (13) | −0.0019 (12) |
| C3 | 0.034 (2) | 0.0230 (16) | 0.0243 (15) | −0.0021 (14) | −0.0067 (14) | 0.0071 (14) |
| C4 | 0.034 (2) | 0.0321 (17) | 0.0175 (14) | −0.0094 (15) | −0.0003 (14) | 0.0024 (13) |
| C5 | 0.0224 (16) | 0.0282 (17) | 0.0235 (15) | −0.0068 (14) | 0.0066 (14) | −0.0030 (13) |
| C6 | 0.0153 (14) | 0.0246 (15) | 0.0234 (15) | −0.0070 (13) | 0.0052 (13) | −0.0015 (12) |
| C7 | 0.0173 (16) | 0.0263 (16) | 0.0298 (16) | −0.0011 (13) | 0.0007 (14) | −0.0069 (14) |
| C8 | 0.0157 (14) | 0.0209 (15) | 0.0340 (16) | 0.0029 (13) | 0.0003 (13) | 0.0015 (13) |
| C9 | 0.0158 (16) | 0.0299 (16) | 0.0252 (16) | 0.0005 (13) | −0.0031 (13) | −0.0006 (13) |
| C10 | 0.0155 (14) | 0.0215 (14) | 0.0211 (13) | −0.0048 (13) | −0.0004 (13) | 0.0016 (12) |
Geometric parameters (Å, º)
| O1—C2 | 1.355 (3) | C3—H3A | 0.9300 |
| O1—H1 | 0.840 (10) | C4—C5 | 1.384 (4) |
| O2—C10 | 1.239 (3) | C4—H4A | 0.9300 |
| O3—C8 | 1.436 (4) | C5—C6 | 1.386 (4) |
| O3—H2 | 0.841 (10) | C5—H5A | 0.9300 |
| O4—C7 | 1.427 (3) | C6—C7 | 1.513 (4) |
| O4—H3 | 0.846 (10) | C7—C8 | 1.520 (4) |
| O1w—H4 | 0.842 (10) | C7—H7 | 0.9800 |
| O1w—H5 | 0.846 (10) | C8—C9 | 1.508 (4) |
| C1—C6 | 1.417 (4) | C8—H8 | 0.9800 |
| C1—C2 | 1.416 (4) | C9—C10 | 1.498 (4) |
| C1—C10 | 1.466 (4) | C9—H9A | 0.9700 |
| C2—C3 | 1.387 (4) | C9—H9B | 0.9700 |
| C3—C4 | 1.377 (4) | ||
| C2—O1—H1 | 111 (3) | O4—C7—C6 | 109.1 (2) |
| C8—O3—H2 | 113 (4) | O4—C7—C8 | 109.7 (2) |
| C7—O4—H3 | 106 (3) | C6—C7—C8 | 112.5 (3) |
| H4—O1w—H5 | 109 (4) | O4—C7—H7 | 108.5 |
| C6—C1—C2 | 119.2 (2) | C6—C7—H7 | 108.5 |
| C6—C1—C10 | 120.4 (3) | C8—C7—H7 | 108.5 |
| C2—C1—C10 | 120.3 (2) | O3—C8—C9 | 106.4 (2) |
| O1—C2—C3 | 117.0 (3) | O3—C8—C7 | 111.0 (2) |
| O1—C2—C1 | 122.7 (2) | C9—C8—C7 | 109.5 (2) |
| C3—C2—C1 | 120.3 (3) | O3—C8—H8 | 109.9 |
| C4—C3—C2 | 119.3 (3) | C9—C8—H8 | 109.9 |
| C4—C3—H3A | 120.3 | C7—C8—H8 | 109.9 |
| C2—C3—H3A | 120.3 | C10—C9—C8 | 112.2 (2) |
| C3—C4—C5 | 121.6 (3) | C10—C9—H9A | 109.2 |
| C3—C4—H4A | 119.2 | C8—C9—H9A | 109.2 |
| C5—C4—H4A | 119.2 | C10—C9—H9B | 109.2 |
| C4—C5—C6 | 120.5 (3) | C8—C9—H9B | 109.2 |
| C4—C5—H5A | 119.8 | H9A—C9—H9B | 107.9 |
| C6—C5—H5A | 119.8 | O2—C10—C1 | 120.7 (3) |
| C5—C6—C1 | 119.0 (3) | O2—C10—C9 | 120.5 (2) |
| C5—C6—C7 | 121.3 (3) | C1—C10—C9 | 118.8 (3) |
| C1—C6—C7 | 119.6 (3) | ||
| C6—C1—C2—O1 | −175.8 (3) | C1—C6—C7—O4 | −148.4 (3) |
| C10—C1—C2—O1 | 2.8 (4) | C5—C6—C7—C8 | 153.2 (3) |
| C6—C1—C2—C3 | 2.6 (4) | C1—C6—C7—C8 | −26.4 (4) |
| C10—C1—C2—C3 | −178.9 (3) | O4—C7—C8—O3 | 59.1 (3) |
| O1—C2—C3—C4 | 177.3 (3) | C6—C7—C8—O3 | −62.5 (3) |
| C1—C2—C3—C4 | −1.2 (4) | O4—C7—C8—C9 | 176.3 (2) |
| C2—C3—C4—C5 | −0.6 (5) | C6—C7—C8—C9 | 54.7 (3) |
| C3—C4—C5—C6 | 1.0 (5) | O3—C8—C9—C10 | 63.4 (3) |
| C4—C5—C6—C1 | 0.5 (4) | C7—C8—C9—C10 | −56.7 (3) |
| C4—C5—C6—C7 | −179.1 (3) | C6—C1—C10—O2 | 178.9 (3) |
| C2—C1—C6—C5 | −2.2 (4) | C2—C1—C10—O2 | 0.4 (4) |
| C10—C1—C6—C5 | 179.2 (3) | C6—C1—C10—C9 | −0.8 (4) |
| C2—C1—C6—C7 | 177.4 (3) | C2—C1—C10—C9 | −179.3 (3) |
| C10—C1—C6—C7 | −1.1 (4) | C8—C9—C10—O2 | −149.4 (3) |
| C5—C6—C7—O4 | 31.2 (4) | C8—C9—C10—C1 | 30.4 (3) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1···O2 | 0.84 (1) | 1.87 (3) | 2.590 (3) | 143 (3) |
| O1—H1···O1wi | 0.84 (1) | 2.27 (3) | 2.829 (3) | 124 (3) |
| O3—H2···O1ii | 0.84 (1) | 2.15 (3) | 2.924 (3) | 153 (5) |
| O4—H3···O1wiii | 0.85 (1) | 1.82 (1) | 2.657 (3) | 170 (4) |
| O1w—H4···O2 | 0.84 (1) | 1.98 (1) | 2.805 (3) | 167 (4) |
| O1w—H5···O4iv | 0.85 (1) | 1.88 (1) | 2.726 (3) | 177 (3) |
Symmetry codes: (i) x+1/2, −y+3/2, −z+1; (ii) −x+1, y+1/2, −z+3/2; (iii) −x−1/2, −y+2, z+1/2; (iv) −x+1/2, −y+2, z−1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: XU5544).
References
- Barbour, L. J. (2001). J. Supramol. Chem. 1, 189–191.
- Borgschulte, K., Rebuffat, S., Trowitzsch-Kienast, W., Schomburg, D., Pinon, J. & Bodo, B. (1991). Tetrahedron, 47, 8351–8360.
- Bruker (2010). APEX2 and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Iwasaki, S., Muro, H., Nozoe, S. & Okuda, S. (1972). Tetrahedron Lett. 1, 13–16.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Trisuwan, K., Rukachaisirikul, V., Sukpondma, V., Preedanon, S., Phongpaichit, S., Rungjindamai, N. & Sakayaroj, J. (2008). J. Nat. Prod. 71, 1323–1326. [DOI] [PubMed]
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536812022623/xu5544sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812022623/xu5544Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536812022623/xu5544Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report