Expanded repeats of r(CGG) in the 5′-untranslated region of the fragile X mental retardation protein mRNA cause fragile X and fragile X-associated tremor/ataxia syndromes. Expanded repeats fold into an RNA hairpin with repeating 5′-CGG/3′-GGC motifs. Herein, we report a structure of a model RNA duplex with three copies of the 5′-CGG/3′-GGC motif (PDB ID: 3JS2), refined to 1.36 Å. All three GG internal loops have N1-carbonyl, N7-amino pairs and are closed by standard Watson–Crick CG pairs. The results expand the available structures of triplet repeating transcripts and provide information to help understand how these RNAs bind small-molecule and protein ligands.
Although RNA is an important target for small molecules, most functionally important RNAs have not been exploited in targeting endeavors.1 This is thought to be due generally to the lack of a fundamental understanding of RNA motifs that specifically bind small molecules and the small molecules that specifically bind RNA motifs. One important class of RNAs that can be exploited as a drug target by small molecules is that of triplet-repeating transcripts. A variety of diseases, including fragile X syndrome (FXS, r(CGG)), Friedreich’s ataxia (r(CGG)), the spinocerebellar ataxias (r(CAG) or r(CUG)), and myotonic dystrophy (r(CUG)) are caused by triplet-repeating RNAs.2 These expanded repeating transcripts fold into higher-order hairpin structures with regularly repeating 1×1 nucleotide internal loops separated by two GC base pairs, as determined by chemical probing of RNA structure (Figure 1A).3
Figure 1.
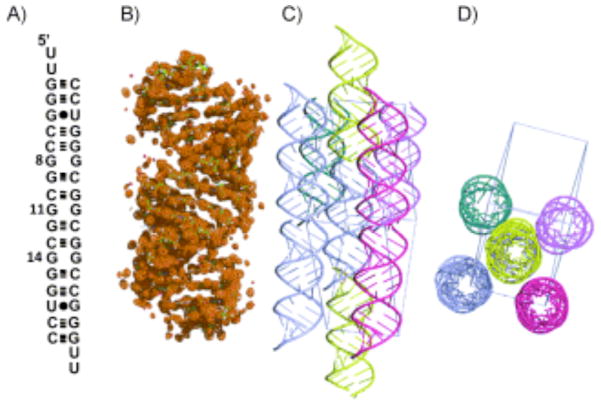
The secondary structure, refined structure, and crystal packing of the RNA construct. A) The secondary structure of the oligonucleotide r(CGG) repeat duplex model that allowed crystal growth. B) The global structure of the RNA including the electron density map at 1.03σ. C) Side and D) top views of the crystal packing that was observed in the unit cell within 5 Å.
Two different general mechanisms for how triplet-repeating transcripts cause disease have been established. In the first mechanism, which has been best established for Huntington’s disease, a repeating transcript of r(CAG) is present in an mRNA coding region. When that transcript is translated, polyQ proteins are synthesized and cause toxicity.4 In a second mechanism, which has been best established for myotonic dystrophy type 1 (DM1), the expanded repeat is present in a non-coding region in an mRNA, such as a 3′-untranslated region (UTR). The transcribed repeat sequesters RNA-binding proteins such as muscleblind-like protein 1 (MBNL1), and this controls pre-mRNA splicing.5 Sequestration of MBNL1 by expanded r(CUG) repeats causes both translational defects of the mRNA with the expanded repeat and pre-mRNA splicing defects.5
Recent studies have shown that the pathology of fragile X-associated tremor/ataxia syndrome (FXTAS) can be due to the latter disease mechanism and FXS is due to a well-established translational-defect mechanism.6 FXTAS is caused when 55–200 r(CGG) repeats are present in the 5′-UTR of the fragile × mental retardation 1 mRNA (FMR1).
A Drosophila model of FXTAS was developed by heterologous expression of 90 r(CGG) repeats that were not translated.7 The repeats alone caused a neurodegenerative phenotype associated with FXTAS. In patient-derived FXTAS cell lines, r(CGG) repeats form inclusion complexes that contain, for example, the Src-associated, 68 kDa (Sam68) protein in mitosis, heterogeneous nuclear ribonucleoprotein G (hnRNP-G) protein, and MBNL1.8 These studies also showed that r(CGG) repeats first bind a protein, which has yet to be determined, that first recruits Sam68, which further recruits hnRNP-G and MBNL1 to the r(CGG) repeats. Furthermore, pre-mRNA splicing of Sam68-controlled transcripts is affected in FXTAS-patient-derived cell lines.8
These mechanistic studies suggest a therapeutic strategy towards developing a treatment for FXTAS in which a small-molecule ligand binds expanded r(CGG) repeats and inhibits protein binding, thereby freeing the protein to perform its normal physiological roles. Such strategies have already been implemented for DM1. For example, oligonucleotides9 and a small molecule, pentamidine,10 target expanded r(CUG) repeats and correct DM1-associated splicing defects in mouse models. General strategies for targeting triplet-repeating transcripts with small molecules have been developed and are centered on a modular-assembly strategy.11
In order to develop an atomic understanding of the structure of r(CGG) repeats, we have refined diffraction data on a model RNA duplex that contains three copies of the 5′-CGG/3′-GGC motif present in FXS and FXTAS hairpin mRNA to 1.36 Å. The secondary structure, overall three-dimensional structure, and the crystal packing are shown in Figure 1.
The RNA duplex construct was designed to contain 5′-UU dangling ends and a duplex region that surround the 5′-CGG/3′-GGC motifs. In this structure, the 5′-UU dangling ends form a two-hydrogen bonded pair with 5′-UU dangling ends from another helix to create a pseudo-infinite helix.
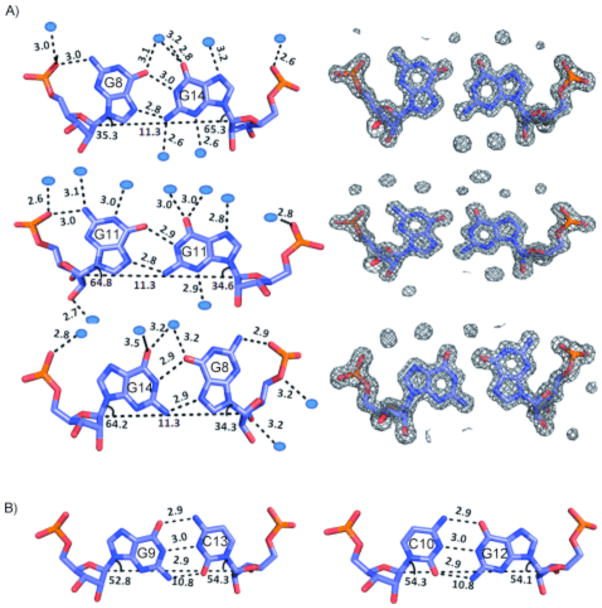
In the refined structure, all three 1×1 nucleotide GG internal loops form well-defined pairs that have the same structure (Figure 2A). In each 1×1 nucleotide GG internal loop one of the G’s is syn and the other is anti. The GG pairs each have three hydrogen bonds two of which are between the Hoogsteen and the Watson–Crick faces (N1-carbonyl, N7-amino pair), and the third is between the syn-G and a 5′-nonbridging pro-R(p) phosphate oxygen (Figure 2A). At the resolution of the structure refined in this report, one can also observe a series of water molecules bound to the GG pairs. In general, bound water molecules are seen to interact with both the major- and minor-groove functional groups in these pairs. The GG pairs fit well into the helix and do not disrupt any of the loop-closing GC pairs, which have typical geometries and standard hydrogen-bonded distances (Figure 2B). This type of GG pairing has been previously observed in single 1×1 nucleotide GG internal loops in the ribosome12 and an NMR structure of an RNA duplex.13 In addition, structures of d(CGG) repeats have shown that the DNA bases are positions as a syn-G/anti-G pairing.14
Figure 2.
The structures of the GG and CG pairs present in the crystal structure. A) The refined structures of the 1×1 nucleotide GG internal loops and the electron density that was refined; the electron density map is shown at 1.53σ. Each type of G–G pair is solvated by several water molecules. B) Refined structures of two loop-closing GC pairs; all GC pairs that close loops have the same standard Watson–Crick geometry.
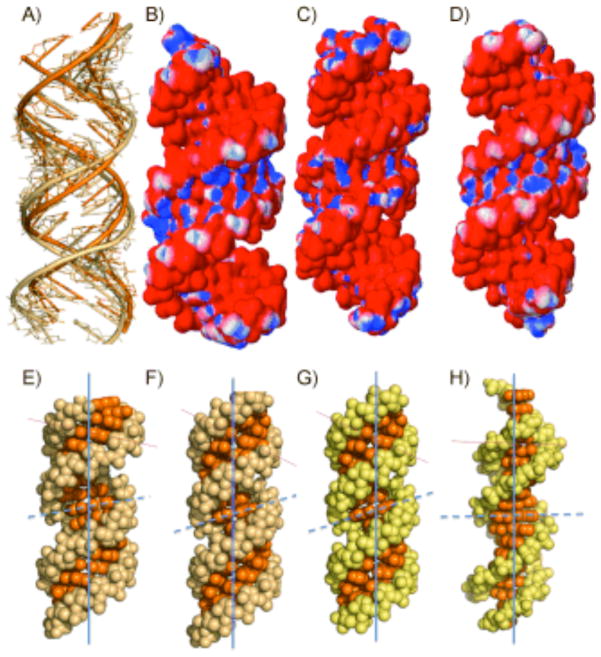
An overlay of the backbone of the r(CGG) structure and a structure in which the 1×1 nucleotide GG internal loops are replaced by GC pairs is shown in Figure 3A. There are differences in the helical geometry of the r(CGG) structure and duplex RNA. In Figure 3B–D the electrostatic distribution of partial charge are shown. The minor groove in the r(CGG) structure has a higher density of partial positive charge than fully canonically paired RNA duplexes in which the 1×1 nucleotide GG internal loop is replaced with either GC or AU pairs.
Figure 3.
Comparison of the refined r(CGG) structure to canonically paired duplexes. A) Overlay of the backbone of the r(CGG) structure (beige) and a model in which the 1×1 nucleotide GG internal loops are replaced with GC pairs (orange). Panels B–D show electrostatic charge distributions of B) the r(CGG) structure and structures in which the 1×1 nucleotide GG internal loops in the CGG construct were replaced with C) AU and D) GC pairs. Panels E–H show ball-and-stick models of a variety nucleic acid helical forms: E) r(CGG) structure (A′-form RNA); constructs in which the 1×1 nucleotide GG internal loops in the r(CGG) construct are replaced with F) GC and G) AU pairs (A form); H) a DNA duplex in which the 1×1 nucleotide GG internal loops are replaced with GC pairs (B form). Vertical lines show the helical axis, and horizontal lines show the inclination axes.
The global helical architecture of the RNA was analyzed with the 3DNA software package.15 The C1′ = C1′ distances are, on average, 11.3 Å for each GG pair; for the GC pairs, the C1′ = C1′ distances are 10.8 Å (Figure 2B). Thus, introduction of the purine–purine pair increases the C1′ = C1′ distance somewhat; however, the syn geometry of one of the G’s allows the pairs to fit into the helix. There is local unwinding of the helix at syn-guanines (Supporting Information). Further analysis of the structure shows that it has several features in common with the A′ form of RNA owing to global widening of the major groove and base-pair inclinations near the 1×1 nucleotide GG internal loops (Figure 3E–H). A direct comparison of the r(CGG) structure (A′ form) to base-paired A-form RNA helices in which the 1×1 nucleotide GG internal loops are replaced by GC or AU pairs and a B-form DNA helix are shown in Figure 3E–H. A′-form RNA structures have been observed previously in both NMR16 and a crystal structure17 of RNA helices with internal loops and bulges.
Structures of model triplet repeating transcripts of r(CUG)18 and r(CAG)19 have been previously reported. For the r(CUG) repeats in DM1, different conformations of the UU loop have been observed in several constructs. These include structures with one or two hydrogen-bonded 1×1 nucleotide UU internal loops. In the structure of r(CAG), the AA pair has one hydrogen bond between C2 = H2 and N1; both A’s are in the anti conformation. In addition, the 1×1 nucleotide AA internal loops have a sulfate anion bound near the major groove.18
The r(CGG) structure has some notable differences compared to the other triplet repeating transcripts reported to date. For example, the 1×1 nucleotide GG internal loops have one anti- and one syn-G. In addition, no ions are found within 6.5 Å of the 1×1 nucleotide GG internal loops in this structure. The lack of ion density is not surprising, as in the r(CAG) structure, contacts to the sulfate are formed with the exocyclic amine of two unpaired A’s in the 1×1 nucleotide internal loop. In the r(CGG) structure, these exocyclic amines are tied up in GG pairs and are thus not free to form similar contacts with anions. These data point to differences in the ligand-binding capacity of r(CAG) versus r(CGG) repeats.
There are several features of this refined structure that could provide unique binding sites for protein or small-molecule ligands over canonically paired RNA. First, the A′-form geometry of the RNA around the GG pairs opens up the major groove and makes it more accessible for binding proteins or small molecules (Figure 3E–H).16–17 Second, the electrostatic differences between paired and r(CGG)-repeat RNAs show that r(CGG) repeats have a larger density of partial positive charge in the minor groove (Figure 3B–D). Third, the positioning of hydrogen-bond acceptors and donors in the major and minor grooves are different in the r(CGG) repeats from those in duplex RNA (Figure 2). Subsequent structural investigations of small molecules binding to r(CGG) repeats can help unravel features in this RNA that drive specific recognition.
Supplementary Material
Acknowledgments
This work was funded by the National Institutes of Health (3R01M079235-02S1 and 1R01GM079235-01A2 to M.D.D.) and by The Scripps Research Institute. M.D.D. is a Dreyfus New Faculty Awardee, a Dreyfus Teacher-Scholar, and a Research Corporation Cottrell Scholar.
Footnotes
Note in added proof
A structure of r(CGG) repeats that is similar to the structure here was recently reported in Nucleic Acids Research, 10.1093/nar/gkr368.
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/cbic.201100337.
References
- 1.Thomas JR, Hergenrother PJ. Chem Rev. 2008;108:1171–1224. doi: 10.1021/cr0681546. [DOI] [PubMed] [Google Scholar]
- 2.Caskey CT, Pizzuti A, Fu YH, Fenwick RG, Jr, Nelson DL, Kuhl DP. Science. 1992;256:784–789. doi: 10.1126/science.1589758. [DOI] [PubMed] [Google Scholar]
- 3.Sobczak K, Michlewski G, de Mezer M, Kierzek E, Krol J, Olejniczak M, Kierzek R, Krzyzosiak WJ. J Biol Chem. 2010;285:12755–12764. doi: 10.1074/jbc.M109.078790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.MacDonald ME, Gusella JF. Curr Opin Neurobiol. 1996;6:638–643. doi: 10.1016/s0959-4388(96)80097-3. [DOI] [PubMed] [Google Scholar]
- 5.Wheeler TM. Neurotherapeutics. 2008;5:592–600. doi: 10.1016/j.nurt.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jin P, Alisch RS, Warren ST. Nat Cell Biol. 2004;6:1048–1053. doi: 10.1038/ncb1104-1048. [DOI] [PubMed] [Google Scholar]
- 7.Jin P, Zarnescu DC, Zhang F, Pearson CE, Lucchesi JC, Moses K, Warren ST. Neuron. 2003;39:739–747. doi: 10.1016/s0896-6273(03)00533-6. [DOI] [PubMed] [Google Scholar]
- 8.Sellier C, Rau F, Liu Y, Tassone F, Hukema RK, Gattoni R, Schneider A, Richard S, Willemsen R, Elliott DJ, Hagerman PJ, Charlet-Berguerand N. EMBO J. 2010;29:1248–1261. doi: 10.1038/emboj.2010.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wheeler TM, Sobczak K, Lueck JD, Osborne RJ, Lin X, Dirksen RT, Thornton CA. Science. 2009;325:336–339. doi: 10.1126/science.1173110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Warf MB, Nakamori M, Matthys CM, Thornton CA, Berglund JA. Proc Natl Acad Sci USA. 2009;106:18551–18556. doi: 10.1073/pnas.0903234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pushechnikov A, Lee MM, Childs-Disney JL, Sobczak K, French JM, Thornton CA, Disney MD. J Am Chem Soc. 2009;131:9767–9779. doi: 10.1021/ja9020149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee MM, Pushechnikov A, Disney MD. ACS Chem Biol. 2009;4:345–355. doi: 10.1021/cb900025w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagaswamy U, Voss N, Zhang Z, Fox GE. Nucleic Acids Res. 2000;28:375–376. doi: 10.1093/nar/28.1.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burkard ME, Turner DH. Biochemistry. 2000;39:11748–11762. doi: 10.1021/bi000720i. [DOI] [PubMed] [Google Scholar]
- 14.Mariappan SV, Catasti P, Chen X, Ratliff R, Moyzis RK, Bradbury EM, Gupta G. Nucleic Acids Res. 1996;24:784–792. doi: 10.1093/nar/24.4.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mitas M, Yu A, Dill J, Haworth IS. Biochemistry. 1995;34:12803–12811. doi: 10.1021/bi00039a041. [DOI] [PubMed] [Google Scholar]
- 15.Lu XJ, Olson WK. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bullock SL, Ringel I, Ish-Horowicz D, Lukavsky PJ. Nat Struct Mol Biol. 2010;17:703–709. doi: 10.1038/nsmb.1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tanaka Y, Fujii S, Hiroaki H, Sakata T, Tanaka T, Uesugi S, Tomita K, Kyogoku Y. Nucleic Acids Res. 1999;27:949–955. doi: 10.1093/nar/27.4.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mooers BH, Logue JS, Berglund JA. Proc Natl Acad Sci USA. 2005;102:16626–16631. doi: 10.1073/pnas.0505873102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiliszek A, Kierzek R, Krzyzosiak WJ, Rypniewski W. Nucleic Acids Res. 2009;37:4149–4156. doi: 10.1093/nar/gkp350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parkesh R, Disney MD, Fountain M. Biochemistry. 2011;50:599–601. doi: 10.1021/bi101896j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kiliszek A, Kierzek R, Krzyzosiak WJ, Rypniewski W. Nucleic Acids Res. 2010;38:8370–8376. doi: 10.1093/nar/gkq700. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.