Abstract
Screening for circulating tumor cells (CTCs) in blood has been an object of interest for evidence of progressive disease, status of disease activity, recognition of clonal evolution of molecular changes and for possible early diagnosis of cancer. We describe a new method of microchip-based immunomagnetic CTC detection, in which the benefits of both immunomagnetic assay and the microfluidic device are combined. As the blood sample flows through the microchannel closely above arrayed magnets, cancer cells labeled with magnetic nanoparticles are separated from blood flow and deposited at the bottom wall of the glass coverslip, which allows direct observation of captured cells with a fluorescence microscope. A polydimethylsiloxane (PDMS)-based microchannel fixed on a glass coverslip was used to screen blood samples. The thin, flat dimensions of the microchannel, combined with the sharp magnetic field gradient in the vicinity of arrayed magnets with alternate polarities, lead to an effective capture of labeled cells. Comparing to the commercially available CellSearch™ system, less (25%) magnetic particles are required to achieve a comparable capture rate, while the screening speed (at optimal blood flow rate of 10 mL/hour) is more than five times faster than those reported previously with a microchannel-based assay. For the screening experiment, blood drawn from healthy subjects into CellSave™ tubes was spiked with cultured cancer cell lines of COLO205 and SKBR3. The blood was then kept at room temperature for 48 hours before the screening, emulating the actual clinical cases of blood screening. Customized Fe3O4 magnetic nanoparticles (Veridex Ferrofluid™) conjugated to anti-Epithelial cell adhesion molecule (EpCAM) antibodies were introduced into the blood samples to label cancer cells, and the blood was then run through the microchip device to capture the labelled cells. After capture, the cells were stained with fluorescently labelled anti-cytokeratin, DAPI and anti-CD45. Subsequent immunofluorescence images were taken for the captured cells, followed by comprehensive computer aided analysis based on fluorescence intensities and cell morphology. Rare cancer cells (from ~1000 cells down to ~5 cells per mL) with very low tumor cell to blood cell ratios (about 1: 107~109, including red blood cells) were successfully detected. Cancer cell capture rates of 90% and 86% were demonstrated for COLO205 and SKBR3cells, respectively.
Introduction
Detection of Circulating Tumor Cells (CTCs) in patient blood is on the frontier of next generation diagnostic tools for evidence of progressive disease, status of disease activity, recognition of clonal evolution of molecular changes and for early cancer detection [1–7]. CTCs escape from early stages of carcinoma and circulate in blood and may sow the seeds for metastasis. The numbers of CTCs found from blood samples have been shown to have a strong correlation with the survival rates of the patients [7–9]. Early detection of CTCs will offer a viable means towards better cure rates of cancer. Most commonly utilized for early CTC detection are cytometric methods [10], where the most challenging aspect is that the cancer cells are very rare. The number of leukocytes per CTC is thought to be 106–107, which makes an effective separation or enrichment step challenging yet crucial for further diagnosis.
For highly specific separation of tumor cells, it is desirable to introduce Immunoassay-based detection, in which antibodies for tumor-specific markers are utilized to label target cells. Other methods include morphological separation [11, 12], where size or density is utilized to isolate CTCs from leukocytes that are smaller than the CTCs. These criteria leave a large amount of other types of cells that are morphologically similar to CTCs, and fail to capture cancer cells that are as small as leukocytes. They still require an additional screening process such as immunofluorescence.
Immunomagnetic-particle-based assays, in which specific cells are labeled with antibody-functionalized magnetic carriers and separated by a magnetic field, have been utilized in purification or isolation with human blood [13]. It has been also applied to the CTC detection [14–17] and is one of the most successful approaches, including a commercially available detection tool kit. The size of the particles tested in those studies ranged from 100 nm [17] to 4.5 µm [16]. Some advanced methods even allow automated screening of samples, in which robotic approaches are taken to replace unreliable manual steps of handling small amounts of liquid. The main screeni g process of these methods still relies on conventional tools such as centrifuge tubes, whose designs and dimensions are not necessarily optimized for cell separation.
The recent advent of the microfabrication technique allowed introduction of microchannel-based approaches for capture of these rare cells [18–19]. These methods usually utilize complicated three dimensional microstructures, such as arrays of pillars or slots with antibody-functionalized surfaces to enhance the interactions with cells flowing in the microchannels. Although microchannel-based immunoassays offer potentially precise control over the physical parameters of the screening process such as sample volume and flow rate of the screening process, these structures may cause several practical problems. First, cells other than those targeted ones can also easily stick to the complicated structures reducing the specificity of detection. Second, the height of such three dimensional structures is much larger than the focal depth of the microscope, which makes the following cell identification process much more time-consuming.
Microchip-based immunomagnetic assay is a method which combines the main benefits of both an immunomagnetic assay and a microfluidic device. The screening process can be precisely controlled through optimized engineered design of the microfluidic device, while it can utilize the downscaled immunomagnetic techniques inside micro channels for efficient separation of cells bound to magnetic particles. Previous work on the use of magnetic force with microfluidic devices has been reviewed in the literature [20]. In addition, Wang et al. developed a microfabricated sifter for separation of magnetic nanoparticles with an array of micropatterned slots in a magnetically soft membrane [21]. Ahn et al. developed a generic microfluidic system for magnetic bead-based biochemical detection, where components such as microvalves, flow sensors, filters and immunosensors have been developed and characterized [22]. Ingber et al. developed a microfluidic device which removes E. coli bacteria bound to magnetic nanoparticles from flowing solutions containing red blood cells [23]. They also demonstrated a blood cleansing device that removes Candida albicans fungi from flowing contaminated human whole blood with over 80% clearance at a flow rate of 20 mL/h [24]. Furdui et al. developed an integrated silicon microchip for separation of Jurkat cells from reconstituted horse blood samples [25] as well as human blood (about 1:10,000 ratio of Jurkat cells to blood cells) [26]. Immunomagnetic separation of cancer cells in microchannels has also been studied by Zborowski et al. They theoretically discussed the behavior of particles in a flow channel [27] and demonstrated separation of MCF7 breast carcinoma cells from mixtures of human peripheral leukocytes [28]. They also used a similar device for blood screening to evaluate human malaria, exploiting the fact that Plasmodium species parasites produce hemozoin that gives magnetic susceptibility to parasitized cells [29, 30].
Here we demonstrate microchip-based immunomagnetic separation and identification of tumor cells spiked into whole blood. The uniqueness in our study includes the detection of very small number of tumor cells (from ~1000 cells down to ~5 cells per mL) from whole blood samples that are prepared and scree ed in similar conditions (kept for 24–48 hours before screening) as actual clinical testing of patient samples. Tumor cell to blood cell ratio is extremely small (about 1:109, including red blood cells), and immunofluorescence takes an essential role to identify cancer cells which cannot be differentiated using cytomorphology only.
Experimental setup
Design of microchip-based immunomagnetic assay
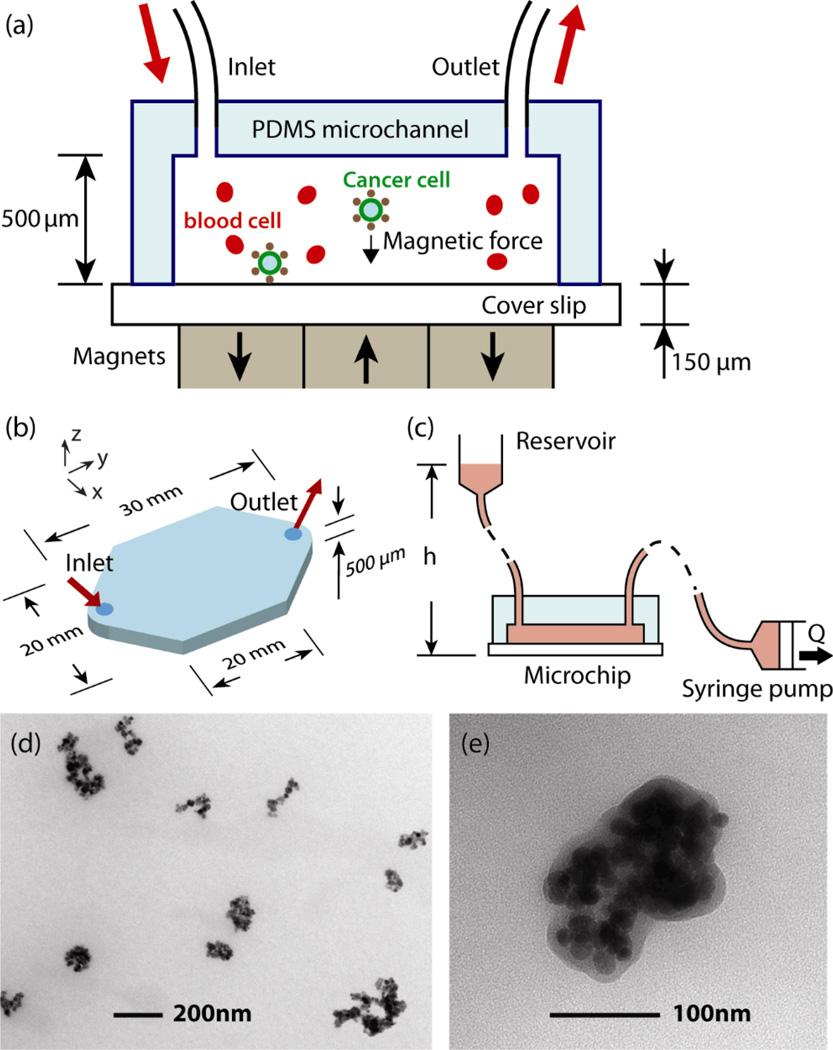
Figure 1 shows an illustration of the CTCs capture system. The blood sample, which has been spiked with cultured carcinoma cells, is combined with magnetic nanoparticles that are functionalized with an antibody to the surface of epithelial cells (anti-Epithelial cell adhesion molecule, EpCAM). The cancer cells in the blood actively bind to the particles and are collected by permanent magnets as the blood is pumped through the microchip (Fig. 1(a)). The microchannel is made by a standard molding technique using PDMS (Sylgard184, Dow Corning, Midland, MI, 10:1 prepolymer to curing agent). UV-Patterned SU-8 photoresist (MicroChem, Newton, MA) on a silicon wafer was used as the master. After removal from the master, the PDMS was manually cut to size and bonded on a 150 µm-thick glass bottom substrate, which serves as a sample slide for the captured tumor cells. Dimensions of the microchannel are shown in Figure 1(b). Figure 1(c) shows the schematic of the pneumatic flow system. To deliver samples to the cancer cell capture device, a pneumatic flow system was constructed using a standard syringe pump. The pump is attached through tubing to the outlet of the microchip to draw the sample with a 14.5 mm-diameter syringe. The reservoir, which is held 100 mm higher than the microchip, is connected to the inlet. The reservoir is open to the atmosphere so the inside pressure of the microchannel is given by ρgh, where ρ, g and h are the density of blood, acceleration of gravity and the height of the sample level in the reservoir, respectively. In this way, the pressure can be kept as low as possible and the risk of possible leakage is minimized. Assuming ρ = 1.05 g/mL, the pressure is calculated to be 0.01 atm. This low pressure configuration allows the use of reversible bonding techniques between the PDMS channel and the bottom glass slide, which is detached from the channel after screening and serves as a regular microscope slide for observation after cell screening. Figure 1(d) and (e) show transmission electron microscopy (TEM) micrographs of the Fe3O4 nanoparticles (Ferrofluid™, Veridex, LLC) used in the experiment. The typical sizes of the particles are in the order of 100 nm.
Fig. 1.
Microchip design for immunomagnetic detection of cancer cell. (a) Schematic showing the principle of operation. CTCs in blood are labelled with EpCAM functionalized Fe3O4 magnetic nanoparticles, and captured by the magnetic field as the blood flows through the microchannel. (b) Dimensions of the microchannel. (c) Schematic of the pneumatic flow system. The flow rate is regulated by the syringe pump from 2.5–10 mL/hour, which draws the blood rather than pushing it to minimize the inside pressure of the chamber. (d–e) TEM images of Fe3O4 magnetic nanoparticles.
Magnetic field distribution
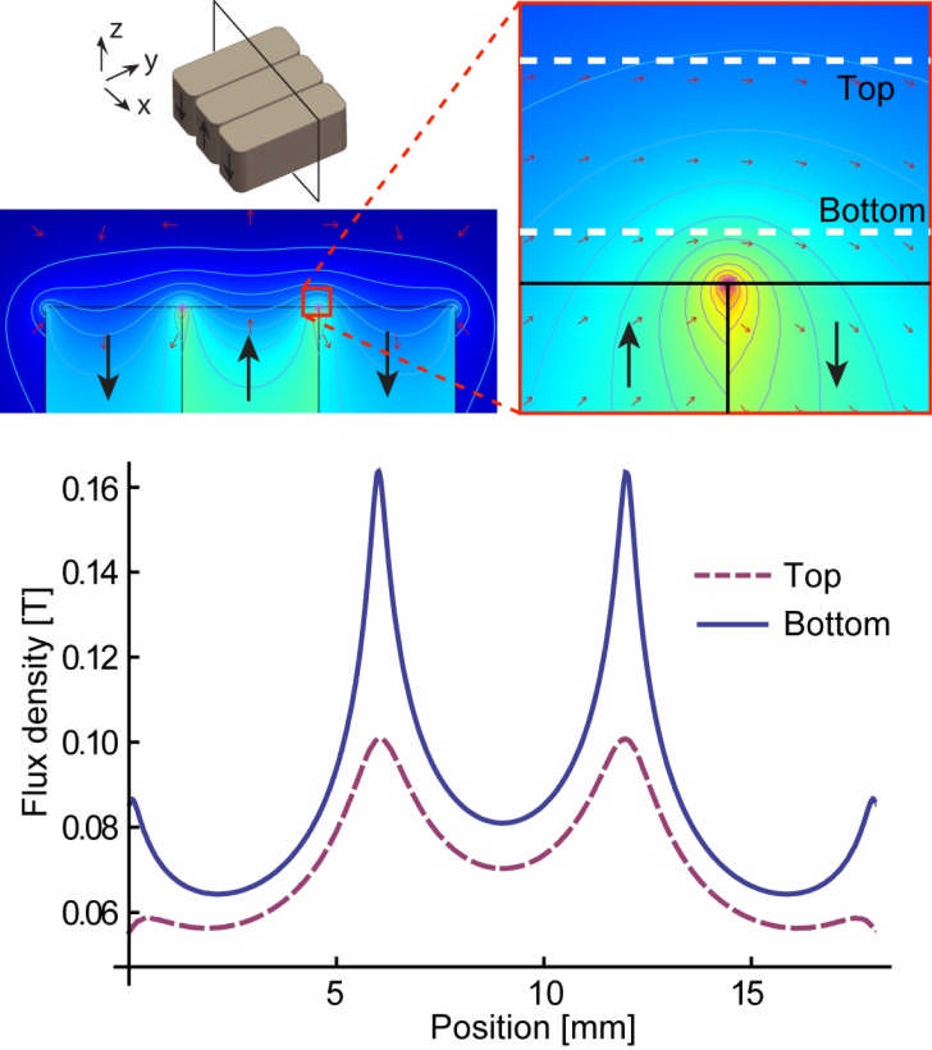
Figure 2 shows a simulated distribution of magnetic flux density made by an array of three magnets. For efficient attraction of magnetic nanoparticles, which essentially act as small dipoles, magnetic field gradient rather than absolute field strength is required. A strong but uniform magnetic field rotates the dipole orientation but does not attract it. Alternately arranged magnets with opposite polarities next to each other make a large gradient. We used three NdFeB block magnets with maximum energy product of 42 MGOe (grade N42). A gauss meter was first used to measure the magnetic field induced by one magnet, and the obtained intensity value was used to further calculate the result with three magnets. The microchannel is located on top of the array. The flux densities at the bottom (150 µm from the magnet) and the top (650 µm from the magnet) of the channel are also shown in the Figure 2. A strong gradient is created to attract nanopa ticles between adjacent magnets.
Fig. 2.
Simulated magnetic flux density. The Contour plot is for the magnetic potentials. Magnetic flux densities at the top and bottom of the microchannel are plotted. A large magnetic field gradient is induced by the alternating arrangement of the three magnets.
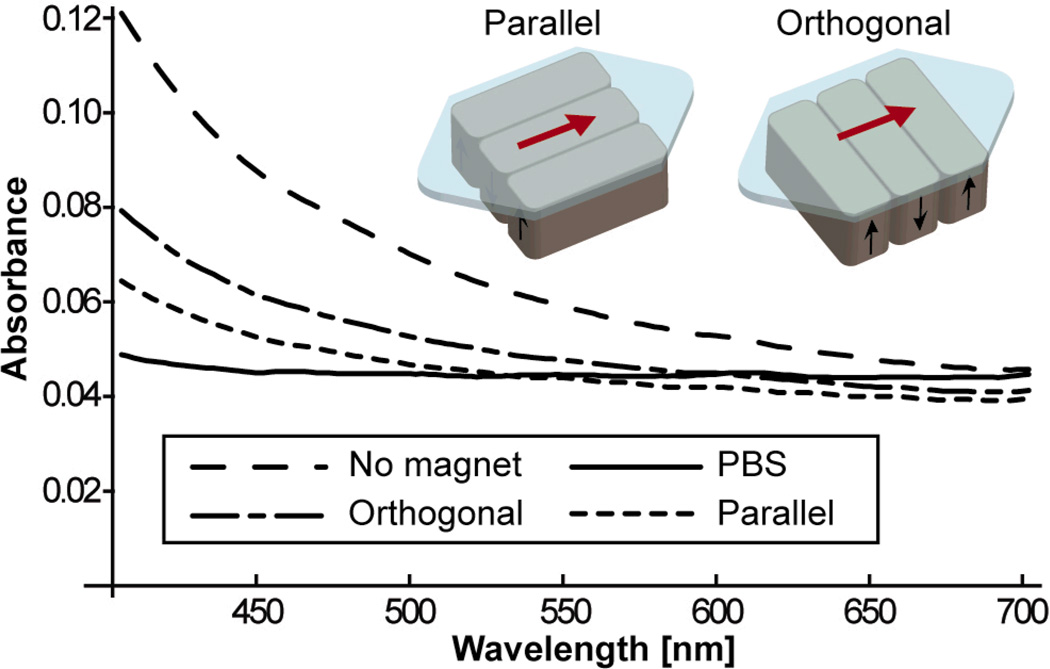
To assess the efficacy of the magnetic field, control experiments with Ferrofluid nanoparticles suspended in a buffer solution were performed with different magnet orientations. Optical densities of the nanoparticle solution were used to obtain quantitative evaluation of the capture efficiency of magnetic particles. We tested two types of magnet orientations. Figure 3 is the result of the measurements with three magnets placed in orthogonal and parallel arrangement with regard to the flow direction. With lower flow rates such as 5 mL/hour, almost all of the particles are captured, and it is difficult to assess the capture efficiency. Therefore the flow rate was deliberately set to be 40 mL/hour for the measurements, which is much higher than those used in experiments involving cancer cells. As shown in the magnetic flux density plot in figure 2, the dominant areas for attracting nanoparticles are the lines between two adjacent magnets. The parallel arrangement gives particles longer exposure to the magnetic field gradient induced by those areas. All the following experiments were made with the three magnets in the parallel arrangement.
Fig. 3.
Control screening experiment with only nanoparticles in buffer solution in the flow channel. Optical transmission is measured for orthogonal and parallel arrangements of magnets. The parallel arrangement showed the better capture efficiency.
Experimental
Sample preparation
Blood samples from a healthy person were spiked with control carcinoma cell lines and screened to calculate the capture rates. Vacutainer™ tubes (BD, Franklin Lakes, NJ) containing ethylenediaminetetraacetic acid as anticoagulant (EDTA tubes) and CellSave™ tubes (Veridex, LLC, NJ) were used to draw blood. We mainly used a human cancer cell line COLO205 (a type of colon cancer), which is reported to have less EpCAM markers [31] than commonly used MCF7 [28] (a type of breast cancer), and provide good simulation of clinical screening. First, cultured cells were harvested, centrifuged, and resuspended in buffer solution. Cells were counted with a hemacytometer and diluted in phosphate buffered saline (PBS) to prepare a solution with approximately 2,000 cells/mL. Then 100 µL was added to each 2.5 mL aliquot of blood to prepare a sample spiked with 200 cells. The same amount of solution was also dispensed on each of two glass slides as counting controls. The number of cells actually spiked into the blood was determined by using the average of the two control slides as 100%, and then the percent recovery was calculated.. Normal blood samples which are not spiked with cancer cells are prepared along with spiked ones, and all the following procedures were performed in parallel.
Processing of blood samples
Samples of blood we prepared in CellSave™ tubes were screened 24–48 hours after spiking, simulating the actual screening situation of patient blood. The CellSave™ tube contains fixative agent so samples can be sent from distant locations. As a reference, we also performed testing with blood prepared in standard EDTA tubes. The EDTA tube we used here (Vacutainer™ tube) is for general purpose blood collection and does not contain preservative or fixative agents. The samples were screened just after spiking to minimize possible antigen deactivation. Before screening, the blood was processed as follows: First, 3.5 mL of dilution buffer solution (Veridex, LLC) is added to the blood and centrifuged at 800G for 10 min. Supernatant containing plasma as well as the buffer solution is removed and the buffer solution is added again to make a total of 3.5 mL of sample. These steps replace blood plasma with the dilution buffer. Viscosities measured for blood samples in an EDTA tube and a CellSave tube after the dilution step are shown in Table 1. A cone plate viscometer (DV-I+, Brookefield, Middleboro, MA) was used for the measurement. The EDT and CellSave samples were drawn from a same subject at the same time. The viscosities of the EDTA samples were measured just after drawing, and the CellSave sample was processed 24 hours after drawing as used in the screening. The CellSave tubes contain cell fixative reagents and tend to make the blood sample more viscous than the EDTA samples. Secondly, a suspension of anti-Epithelial cell adhesion molecule (EpCAM) functionalized Fe3O4 nanoparticles (Ferrofluid™, Veridex, LLC) along with Capture Enhancement Reagent (Veridex, LLC) are added to the blood. The screening process starts 15 minutes after those reagents are added. We tested different amounts of nanoparticle suspension, namely 75 µL and 18.8 µL, added to the above processed samples, which contain 2.5 mL of blood, while 75 µL is the standard amount used in the CellSearch™ system.
Table 1.
Viscosities [cP = 10−3 kg m−1 s−1] of blood samples for different shear rates from 2/s to 200/s.
| Tube | Shear rate | ||
|---|---|---|---|
| 2 /s | 20 /s | 200 /s | |
| EDTA | 6.1 cP | 3.3 cP | 2.3 cP |
| CellSave | 7.2 cP | 3.9 cP | 3.0 cP |
Screening process
Before the blood sample is introduced into the reservoir, the microchannel is filled with PBS to eject air bubbles. The spiked solutions were then driven pneumatically at a continuous rate. We tested flow rates of 2.5 mL/hour to 10 mL/hour. After the blood screening, PBS is introduced and flowed continuously until the red blood cells are not visible in the microchannel. This process removes unwanted blood cells from the bottom glass coverslip. The same rate as the blood flow is used for the flushing step, where typically 4 mL of PBS is added. After flushing, 1 mL of ice-cold acetone at the same rate is introduced to the channel to fix cancer cells on to the glass coverslip. After being disassembled and dried completely, the bottom glass coverslip was stored at 5 °C until staining.
Cell identification
The cell is fluorescently stained with anti-cytokeratin (CK, protein found in epithelial tissue, positive test, mouse anti-cytokeratin, pan-FITC, Sigma-Aldrich, St. Louis, MO), anti-CD45 (found on leukocytes, negative test, AlexaFluor 568, Invitrogen, Carlsbad, CA, bound to mouse anti-human clone 9.1 made in Dr. Jonathan Uhr’s lab at University of Texas Southwestern Medical Center), and DAPI (staines DNA found in cell nucleus, positive test, Vectashield Mounting Medium with DAPI, Vector Laboratories, Inc, Burlingame, CA). Samples from normal blood, which are not spiked are also stained side by side to perform blind observation, where the observer was not informed of which sample was spiked.
Analysis of device functionality
Device model
We build a computer based analytical program to assess the device functionality. The program traces trajectory of a particle in the microchannel. The capture rate for a particle suspension can be estimated by computing calculations for particles uniformly entering the device.
A simplified model of the microdevice is defined with the following assumptions:
The magnetic field is uniform in y direction
The flow is laminar and uniform in x and y direction.
The velocity profile in z direction follows a simple parabola defined by the flow rate Q.
Two important forces acting on a particle are the drag force Fd and the magnetic force Fm, which are given in the following way [27,32].
| (1) |
| (2) |
where η the medium viscosity, R is the particle radius, Δv is the particle relative velocity to the medium, Δχ is effective magnetic volumetric susceptibility, μ0 = 4π × 10−7 T · m · A−1 is the magnetic permeability of vacuum, B is the magnetic field intensity which is shown in figure 2.
When we assume a quasi-static motion of particles, the two forces equal each other, namely
| (3) |
Using (1), (2) and V = (4 / 3)πR3 for (3), the relative velocity Δv is given as:
| (4) |
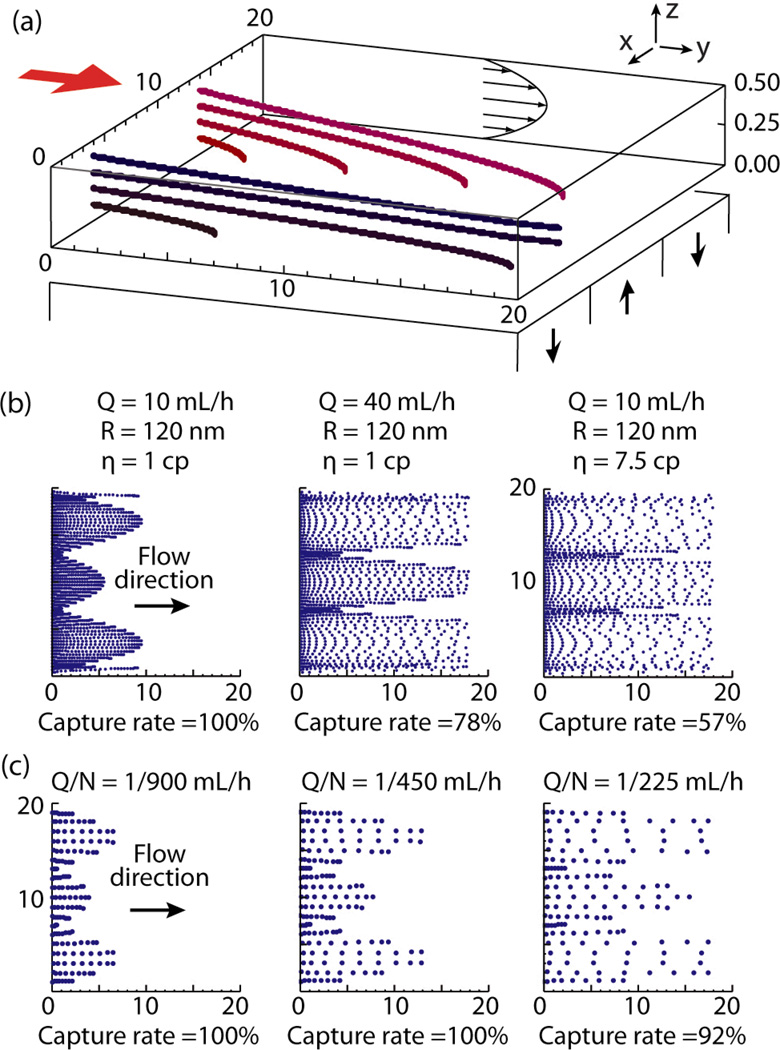
Figure 4(a) shows an example of trajectories of particles with different initial positions. Some particles are trapped by the magnetic force while the other escape through the micro channel. Parameters used for calculation are: Q = 10 mL/hour, R = 50 nm and η = 10−3 kg · m−1 · s−1.
Fig. 4.
Computer based analysis of the device functionality. (a) Example of particle trajectories. Q = 10 mL/hour, R = 50 nm and η = 1 cP. (b) Ferrofluid particle captured on slides. Left two panels are for buffer solution (η = 1 cP), Panel on the right is for blood (η = 7.5 cP). (c) Cancer cell captured on slides. Flow rate Q and number of Ferrofluid particles per cell N defines the distribution of the cells.
Ferrofluid particles
When calculating the motion of Ferrofluid particles, equation (4) should not be simply applied, because the magnetic interaction between particles takes an important part and particles assemble themselves into larger aggregations [31,34]. However, it still helps to estimate the particle behavior by using particle radius R as a variable. Here we consider the control experiments with Ferrofluid nanoparticles suspended in a buffer solution as discussed in Figure 3. Brownian motion can be neglected under a large magnetic field [34]. The susceptibility ΔχF is estimated from the magnetization curve measured for Fe3O4 particles in literature [33]:
| (5) |
Water viscosity is used as the medium viscosity:
| (6) |
One can see from (4), the capture rate is a monotonically increasing function of R. Testing with different R values showed that the condition of R = 120 nm yields capture rates of 78% at the flow rates of Q = 40 mL/hour. The result seems to match the optical density measurement described in Figure 3. This suggests that the particles are aggregated to form larger particles since most of the particles in the TEM images in Figure 1 (d) and (e) seem to be smaller than R = 120 nm. Examples of particle distributions for different flow rates are shown in the left two panels of Figure 4(b). The right panel in Figure 4(b) is a result with blood as the medium. Flow rates of Q = 10 mL/hour and the medium viscosity of ηB = 7.5 × 10−3 kg · m−1 · s−1 were used for the calculation (see (10)).
Cancer cell capture rate
The effective magnetic volumetric susceptibility of a cancer cell ΔχC is given as:
| (7) |
where N is the number of Ferrofluid particles attached to the cell, RF and RC are radii of the Ferrofluid particle and the cancer cell, respectively, and ΔχF is the volumetric susceptibility of the Ferrofluid particle.
From the TEM images in Figure 1(d)(e) and the observed average cell diameter, RF and RC are defined as:
| (8) |
| (9) |
Blood viscosity after dilution is taken from table 1 at a shear rate of 2/s, considering the shear rate to be in the order of Δv/R with the cell radius of 7.5 µm and the approximated relative velocity Δv of ~10 µm/s.
| (10) |
The value of N has to be decided to match the experimental result. In this simplified model, the ratio of the flow rate Q to N defines the distribution of captured cells. Cell distributions and the capture rates were calculated for different Q/N values in Figure 4(c). Experimental result will be compared and discussed with the simulations.
Experimental results
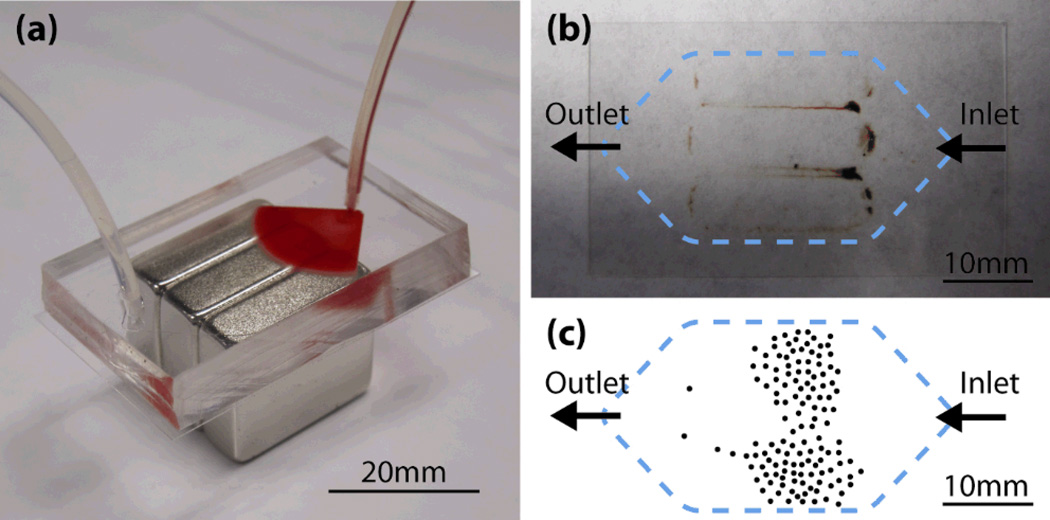
Figure 5 shows the results of blood screening experiments. Figure 5(a) shows a photograph of the microchannel in which a blood sample is being introduced. In the actual experiments, the microchannel and magnets are placed in a mechanical holder that clamps the components together. Figure 5(b) is a photograph of a glass coverslip detached from the microchannel after screening. Nanoparticle aggregations can be seen around the edges of the magnets. Large aggregations of particles are observed at the areas between magnets, where the strongest magnetic density was shown in the calculation of Figure 2. The result is also predicted by the numerical calculation in Figure 4(b) right. Figure 5(c) is an example of a manually drawn sketch showing the locations of the COLO205 cells found on the slide. Unlike the nanoparticles, which are easily affected by the magnetic field rather than the flow, cancer cells are more evenly distributed, with more cells found near the inlet.
Fig. 5.
Blood screening experiment. (a) Photograph of the experimental setup. Blood sample is being introduced in the microchannel. (b) Bottom glass slide removed from the channel after screening. (c) Example of COLO205 cell distribution on the glass slide. The picture is a trace of a manually drawn sketch, approximating the coordinates of the cells.
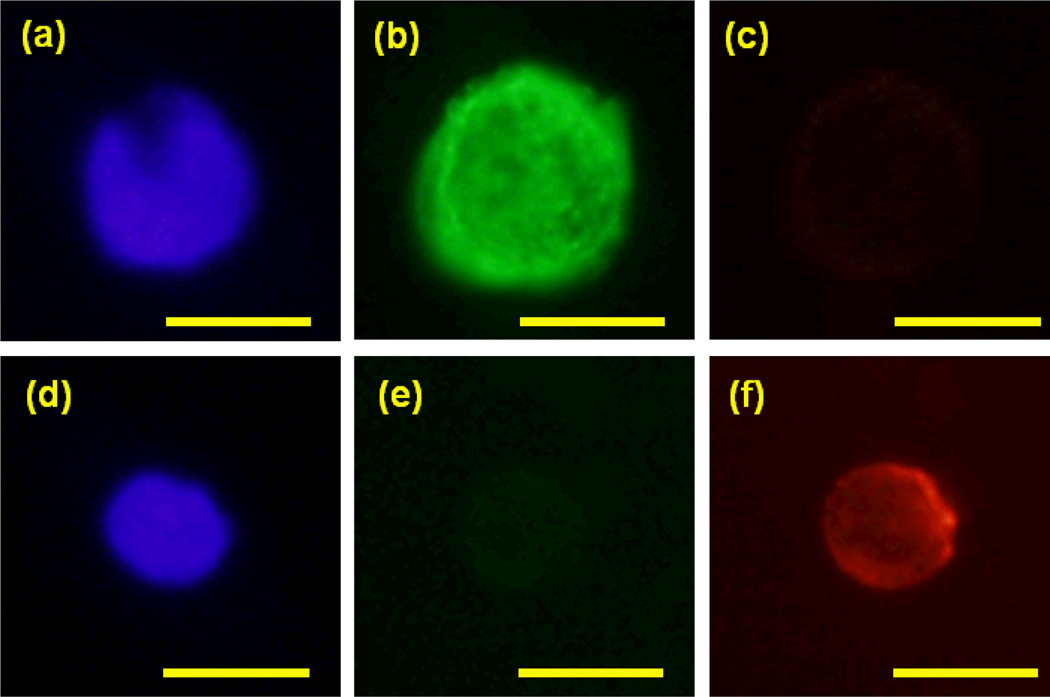
Figure 6 shows fluorescence images of a captured COLO205 cell and a leukocyte using a color CCD camera (Olympus DP71) and separate filter cubes for red (excitation peak wavelength (Ex) 535 nm, bandwidth (BW) 50 nm, dichroic mirror (DM) 565 nm long-pass (LP) and emission peak wavelength (EM) 610 nm, BW 75 nm), green (Ex 480 nm BW 40 nm, DM 505 nm LP, Em 535 nm BW 50 nm) and blue(Ex 350 nm BW 50 nm, DM 400 nm LP, Em 460 nm BW 50 nm). A trained observer identifies cancer cells fro other cells that still remain on the slide after the flushing step. Cancer cells are (a) DAPI positive, (b) CK positive, and (c) CD45 negative, while leukocytes are (d) DAPI positive, (e) CK negative, and (f) CD45 positive.
Fig. 6.
Example of (a) DAPI(blue), (b) CK (green) and (c) CD45 (red) fluorescence images of captured COLO205 cell. (d) DAPI, (e) CK and (f) CD45 images of a white blood cell, which shows clear CK negative and CD45 positive images, is also shown. Bars = 10 µm.
Computer-aided cell identification
It is reported that different observers can make different decisions in tumor cell enumeration process [7]. To implement objective identification, numerical analysis of the size and the fluorescence values of cells were performed. Fluorescence intensity was accessed by Weber contrast defined as:
| (11) |
In which I is the mean intensity of a cell, Ib is the background intensity. An idealized diameter was calculated from the cell area of a binary image calculated from the Cytokeratin image. Table 2 shows the mean values and the standard deviations of the three fluorescent contrast values and the idealized diameters for 61 cells indentified as cancer cells by a trained observer. Based on the values, we made a normalized evaluation function in the following formula.
| (12) |
where, DAPI, CK, CD45 and Dia. are the contrast values and the diameter of a single cell, . are the mean contrast values and diameter of the 61 cells, and σDAPI, σCK, σCD45 and σDia. are the standard deviations of the contrast values and the diameters of the same 61 cells, respectively. We found 60 of the 61 cancer cells satisfied E < 10, 55 satisfied E < 8, and 50 satisfied E < 5, while none of the cells identified as non-cancer cells satisfied the same criteria. The largest E found among cancer cells is 11.9, which is the only cancer cell with the E more than 10. This was caused by a large diameter (21.9 µm, contribution to E : 9.6). Other larger E values tend to be caused by a large diameter or an intense CK value (largest contribution from CK: 7.5), but E values still fell within 10. On the other hand, the two smallest E values found in non-cancer cells were 10.7 and 13.6, which were exceptionally small. For other non-cancer cells, even though we just calculated E values for those that are somehow categorized as “confusing” cells, values are mostly more than 30 or even more than 100. E = 10 seems to be a safe value as an elimination factor. The result ensures its agreement with the subjective cell identification process made by the trained observer, as well as the feasibility of a future computer-based automated cell identification process.
Table 2.
Mean and standard deviations of fluorescence intensities as contrast values and idealized diameters of captured COLO205 cells.
| DAPI | CK | CD45 | Dia. [µm] | |
|---|---|---|---|---|
| Mean | 6.6 | 9.5 | 0.6 | 15.2 |
| Std. Dev. | 2.0 | 1.7 | 0.3 | 2.2 |
Capture rates
Capture rates are calculated in the following way:
| (13) |
As described above, two control slides are prepared from the same cell suspension and at the same time as the blood sample is spiked. In typical cases, cancer cells found in EDTA were relatively intact and easy to identify. Similar capture rates were found regardless of how long (0–48 hours) the samples were kept following spiking. This result shows promise for future clinical applications. With slower flow rates, many of the cells tend to be trapped with aggregations of nanoparticles on the glass coverslip. Sometimes weakly CK+ cells are found behind a particle aggregation, but do not satisfy either the observer’s or the computer’s criteria to be counted as a cancer cell, resulting in relatively low capture rates. Such a “over-aggregated” situation can be also seen in the simulation result in Figure 4(c) left. Note that for the simulation, the capture rate remains 100% simply because no observation process is needed. With a smaller amount of added particle suspension (7.5 µL per 1 mL of pre-processed blood) and faster flow ra e, cells are spread in larger areas and easier to observe, which made the optimal condition for the higher average capture rates of 90%. Considering the obtained capture rates and the sketch in Figure 5(c), we assume the Q/N value discussed for calculation in Figure 4(c) to be:
| (14) |
Since the flow rate was Q = 10 mL/hour, the number of Ferrofluid particles per cancer cell is estimated to be:
| (15) |
The results are summarized in Table 3. Results made with SKBR3 cells are also shown in the table. We have also tested PC3 (prostate cancer) cells and a comparable result with capture rate of 88% has been demonstrated.
Table 3.
Capture rates of spiked blood experiment
| Cell | Tube | Ferrofluid /blood [µL/mL] |
Flow rate [mL/hour] |
Ctrl 1 | Ctrl 2 | Ctrl average (A) |
Cells found (C) |
Capture rate (C/A) |
Average capture rate |
|---|---|---|---|---|---|---|---|---|---|
| COLO-205 | EDTA | 30 | 2.5 | 75 | 46 | 60.5 | 61 | 108% | 79% |
| EDTA | 30 | 2.5 | 115 | 114 | 114.5 | 92 | 80% | ||
| EDTA | 30 | 2.5 | 139 | 130 | 134.5 | 92 | 68% | ||
| CellSave | 30 | 2.5 | 75 | 46 | 60.5 | 40 | 66% | 65% | |
| CellSave | 30 | 2.5 | 139 | 130 | 134.5 | 70 | 52% | ||
| CellSave | 30 | 2.5 | 48 | 53 | 50.5 | 49 | 96% | ||
| CellSave | 7.5 | 2.5 | 109 | 126 | 117.5 | 125 | 106% | 53% | |
| CellSave | 7.5 | 2.5 | 180 | 193 | 186.5 | 83 | 28% | ||
| CellSave | 7.5 | 2.5 | 180 | 193 | 186.5 | 53 | 44% | ||
| CellSave | 7.5 | 10 | 252 | 275 | 263.5 | 239 | 91% | 90% | |
| CellSave | 7.5 | 10 | 252 | 275 | 263.5 | 257 | 98% | ||
| CellSave | 7.5 | 10 | 252 | 275 | 263.5 | 267 | 101% | ||
| CellSave | 7.5 | 10 | 252 | 275 | 263.5 | 228 | 87% | ||
| CellSave | 7.5 | 10 | 252 | 275 | 263.5 | 240 | 91% | ||
| CellSave | 7.5 | 10 | 252 | 275 | 263.5 | 203 | 77% | ||
| SKBR3 | CellSave | 7.5 | 10 | 711 | 927 | 819.0 | 713 | 87% | 86% |
| CellSave | 7.5 | 10 | 711 | 927 | 819.0 | 704 | 86% |
Side-by-side blind observation, where one spiked and one non-spiked samples were prepared and the observer is not informed of which sample is spiked, was performed for all the spiking experiments shown in Table 3. The observer did not find a false cancer cell from non-spiked samples, and all the cancer cells were counted from the spiked samples..
We also performed screening experiments with different numbers of COLO205 cells spiked in the samples ranging from ~10 to ~1500 in 2.5 mL blood samples. CellSave tubes were used, 7.5 µL Ferrofluid suspension is added per 1 mL blood, the flow rate is 10 mL/hour. The result is shown in Table 4. Comparable results were obtained for cases with both the smaller (~10) and larger numbers (>1000) of cells.
Table 4.
Capture rates for different numbers of spiked cells.
| Ctrl1 | 14 | 52 | 81 | 164 | 968 | 1477 |
| Ctrl2 | 12 | 45 | 84 | 150 | 893 | 1496 |
| Ctrl average | 13.0 | 48.5 | 82.5 | 157.0 | 930.5 | 1486.5 |
| Cells found | 7 | 37 | 58 | 160 | 625 | 1129 |
| Capture rate | 54% | 76% | 70% | 102% | 62% | 76% |
In order to assess the number of cancer cells that are non-selectively trapped in the device, a blood sample of 2.5 mL was spiked with approximately 100,000 cells and screened normally but without the permanent magnets. No cancer cell was captured by the experiment, which proves the high selectivity of our device.
Conclusion
We have developed a microchip-based CTC capture system, which utilizes immunomagnetic nanoparticles to selectively label tumor cells spiked into blood samples. A strong magnetic field gradient is created in the microfluidic channel, resulting in a high capture rate with a fast flow rate of 10 mL/hour. The amount of nanoparticle suspension added was also reduced to 25% of the amount used in the commercially available CellSearch system. Rare cancer cells (from ~1000 cells down to ~5 cells per mL) with a very low tumor cell to blood cell ratio (about 1:109, including red blood cells) were successfully detected. With the optimal conditions, average capture rates of 90% and 86% were demonstrated for samples with COLO205 and SKBR3 cells, respectively. Numerical analysis of the immunofluorescence observation compares favorably with the subjective cell identification process made by the trained observer. Such microchip-based CTC detection in patient blood enables new diagnostic tools to record s atus of disease activity as well as clonal evolution of molecular changes for early cancer detection.
Acknowledgements
We would like to thank Drs Konstantin V. Sokolov and Keith P. Johnston of the University of Texas at Austin for their fruitful discussions on nanoparticle imaging and functionization and Dr. Hirofumi Tanaka of the University of Texas at Austin for his help in measuring blood viscosities. We are grateful for National Nanotechnology Infrastructure Network (NNIN) Undergraduate Researcher Mr. Fraser. R. Downing from University of Colorado at Boulder for the help in creating the MATLAB program for cell identification. We are grateful for the support from National Institute of Health (NIH) National Cancer Institute (NCI) Cancer Diagnosis Program under the grant 1R01CA139070.
Footnotes
Electronic Supplementary Information (ESI) available: [details of any supplementary information available should be included here]. See DOI: 10.1039/b000000x/
Contributor Information
Kazunori Hoshino, Email: hoshino@mail.utexas.edu.
Xiaojing Zhang, Email: John.Zhang@engr.utexas.edu.
Notes and references
- 1.Kaiser J. Cancer’s Circulation Problem. Science. 2010;327:1072–1074. doi: 10.1126/science.327.5969.1072. [DOI] [PubMed] [Google Scholar]
- 2.Paterlini-Brechot P, Linda Benali N. Circulating tumor cells (CTC) detection: Clinical impact and future directions. Cancer Lett. 2007;253:180–204. doi: 10.1016/j.canlet.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 3.Mostert B, Sleijfer S, Foekens JA, Gratama JW. Circulating tumor cells (CTCs): Detection methods and their clinical relevance in breast cancer. Cancer Treat. Rev. 2009;35:463–474. doi: 10.1016/j.ctrv.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 4.Allard WJ, Matera J, Miller MC, Repollet M, Connelly MC, Rao C, Tibbe AG, Uhr JW, Terstappen LW. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin. Cancer Res. 2004;10:6897–6904. doi: 10.1158/1078-0432.CCR-04-0378. [DOI] [PubMed] [Google Scholar]
- 5.Fehm T, Sagalowsky A, Clifford E, Beitsch P, Saboorian H, Euhus D, Meng S, Morrison L, Tucker T, Lane N, Ghadimi BM, Heselmeyer-Haddad K, Ried T, Rao C, Uhr J. Cytogenetic evidence that circulating epithelial cells in patients with carcinoma are malignant. Clin. Cancer Res. 2002;8:2073–2084. [PubMed] [Google Scholar]
- 6.Budd GT, et al. Circulating tumor cells versus imaging–predicting overall survival in metastatic breast cancer. Clin. Cancer Res. 2006;12:6403–6409. doi: 10.1158/1078-0432.CCR-05-1769. [DOI] [PubMed] [Google Scholar]
- 7.Miller MC, Doyle GV, Terstappen LWMM. Significance of Circulating Tumor Cells Detected by the CellSearch System in Patients with Metastatic Breast Colorectal and Prostate Cancer. J. Oncol. 2010;2010:1–8. doi: 10.1155/2010/617421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen S, et al. Relationship of Circulating Tumor Cells to Tumor Response, Progression-Free Survival, and Overall Survival in Patients With Metastatic Colorectal Cancer. J. Clin. Oncol. 2008;26:3213–3221. doi: 10.1200/JCO.2007.15.8923. [DOI] [PubMed] [Google Scholar]
- 9.He W, et al. Quantitation of circulating tumor cells in blood samples from ovarian and prostate cancer patients using tumor-specific fluorescent ligands. Int. J. Cancer. 2008;123:1968–1973. doi: 10.1002/ijc.23717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gross HJ, Verwer B, Houck D, Hoffman RA, Recjtenwald D. Model study detecting breast cancer cells in peripheral blood mononuclear cells at frequencies as low as 10-7. Proc. Natl. Acad. Sci. USA. 1995;92:537–541. doi: 10.1073/pnas.92.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin HK, et al. Portable filter-based microdevice for detection and characterization of Circulating Tumor Cells. Clin. Cancer Res. 2010;16:5011–5018. doi: 10.1158/1078-0432.CCR-10-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tan SJ, Yobas L, Lee GYH, Ong CN, Lim CT. Microdevice for the isolation and enumeration of cancer cells from blood. Biomed. Microdevices. 2009;11:883–892. doi: 10.1007/s10544-009-9305-9. [DOI] [PubMed] [Google Scholar]
- 13.Hansel TT, De Vries IJM, Iff T, Rihs S, Wandzilak M, Betz S, Blaser K, Walker C. An improved immunomagnetic procedure for the isolation of highly purified human blood eosinophils. J. Immunol. Methods. 1991;145:105–110. doi: 10.1016/0022-1759(91)90315-7. [DOI] [PubMed] [Google Scholar]
- 14.Racila E, Euhus D, Weiss AJ, Rao C, McConnell J, Terstappen Leon W. M. M, Uhr JW. Detection and characterization of carcinoma cells in the blood. Proc. Natl. Acad. Sci.USA. 1998:4589–4594. doi: 10.1073/pnas.95.8.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Balasubramanian P, Yang L, Lang JC, Jatana KR, Schuller D, Agrawal A, Zborowski M, Chalmers JJ. Confocal Images of Circulating Tumor Cells Obtained Using a Methodology and Technology That Removes Normal Cells. Molecul. Pharm. 2009;6:1402–1408. doi: 10.1021/mp9000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Talasaz AH, et al. Isolating highly enriched populations of circulating epithelial cells and other rare cells from blood using a magnetic sweeper device. Proc. Natl. Acad. Sci.USA. 2009;106:3970–3975. doi: 10.1073/pnas.0813188106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liberti PA, Rao CG, Terstappen LWMM. Optimization of ferrofluids and protocols for the enrichment of breast tumor cells in blood. J. Magn. Magn., Mater. 2001;225:301–307. [Google Scholar]
- 18.Nagrath S, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stott SL, et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc. Natl. Acad. Sci.USA. 2010;107:1012539. doi: 10.1073/pnas.1012539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pamme N. Magnetism and microfluidics. Lab Chip. 2006;6:24–38. doi: 10.1039/b513005k. [DOI] [PubMed] [Google Scholar]
- 21.Earhart CM, Wilson RJ, White RL, Pourmand N, Wang SX. Microfabricated magnetic sifter for high-throughput and high-gradient magnetic separation. J. Magn. Magn., Mater. 2009;321:1436–1439. doi: 10.1016/j.jmmm.2009.02.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi JW, et al. Development and Characterization of Microfluidic Devices and Systems for Magnetic Bead-Based Biochemical detection. Biomed Microdev. 2001;3:191–200. [Google Scholar]
- 23.Xia N, Hunt TP, Mayers BT, Alsberg E, Whitesides GM, Westervelt RM, Ingber DE. Combined microfluidic-micromagnetic separation of living cells in continuous flow. Biomed Microdev. 2006;8:299–308. doi: 10.1007/s10544-006-0033-0. [DOI] [PubMed] [Google Scholar]
- 24.Yung CW, Fiering J, Muellerb AJ, Ingber DE. Micromagnetic–microfluidic blood cleansing device. Lab Chip. 2009;9:1171–1177. doi: 10.1039/b816986a. [DOI] [PubMed] [Google Scholar]
- 25.Furdui VI, Kariuki JK, Harrison DJ. Microfabricated electrolysis pump system for isolating rare cells in blood. J. Micromech. Microeng. 2003;13:S164. [Google Scholar]
- 26.Furdui VI, Harrison DJ. Immunomagnetic T cell capture from blood for PCR analysis using microfluidic systems. Lab Chip. 2004;4:614. doi: 10.1039/b409366f. [DOI] [PubMed] [Google Scholar]
- 27.Zborowski M, Fuh CB, Green R, Sun L, Chalmers JJ. Analytical Magnetopheresis of Ferritin-labeled Lymphocytes. Anal. Chem. 1995;67:3702–3712. doi: 10.1021/ac00116a014. [DOI] [PubMed] [Google Scholar]
- 28.Fan B, Zborowski M, Moore LR. Detection of rare MCF-7 breast carcinoma cells from mixtures of human peripheral leukocytes by magnetic deposition analysis. Cytometry. 1999;36:294–302. [PubMed] [Google Scholar]
- 29.Karl S, David M, Moore L, Grimberg BT, Michon P, Muellter I, Zborowski M, Zimmerman PA. Enhanced detection of grametocytes by magnetic deposition microscopy predicts higher potential for Palsmodium falciparum transmission. Malar. J. 2008;7:66. doi: 10.1186/1475-2875-7-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moore LR, Fujioka H, Williams PS, Chalmers JJ, Grimberg B, Zimmerman P, Zborowski M. Hemoglobin degradation in malaria-infected erythrocytes determined from live cell magnetophoresis. FASEB J. 2006;20:747–749. doi: 10.1096/fj.05-5122fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rao CG, Chianese D, Doyle GV, Miller MC, Russell T, Sanders RA, Jr, Terstappen LW. Expression of epithelial cell adhesion molecule in carcinoma cells present in blood and primary and metastatic tumors. Int J. Oncol. 2005;27:49–57. [PubMed] [Google Scholar]
- 32.Pankhurst QA, Connolly J, Jones SK, Dobson J. Applications of magnetic nanoparticles in biomedicine. J. Phys. D: Appl. Phys. 2003;36:R167–R181. [Google Scholar]
- 33.Wang AY, Kuo CL, Lin JL, Fu CM, Wang YF. Study of magnetic ferrite nanoparticles labeled with 99mTc-pertechnetate. J Radioanal Nucl Chem. 2010;284:405–413. [Google Scholar]
- 34.Liu D, Maxey MR, Karniadakis GE. Simulations of dynamic self-assembly of paramagnetic microspheres in confined microgeometries. J. Micromech. Microeng. 2005;15:2298–2308. [Google Scholar]