Controlled intracellular delivery of genetic material for vaccine or other therapeutic applications in vivo remains a major challenge for non-viral delivery systems.[1,2] As an alternative to cationic polymers and lipids commonly used to form nano-scale complexes for in vitro plasmid transfection,[3–6] microparticles made from biodegradable polymers such as poly(lactide-co-glycolide) (PLGA) and acid-sensitive poly(ortho esters) (POEs) and hydrogels have been pursued as in vivo plasmid DNA carriers.[6–9] Due to their size (typically 1–10 μm), these particles passively target phagocytic antigen presenting cells (APCs) of the immune system for DNA vaccine applications.[8] In addition, these particles may alleviate the shortcomings of cationic polymers and lipids with regard to in vivo targeting, toxicity, and stability.[3] Despite their promise, microparticulate delivery systems explored to date often suffer from uncontrolled initial burst release of upwards of 50% of the encapsulated plasmid, independent of any built-in triggered-release mechanism, making it difficult to achieve rapid yet controlled release in response to a specific stimulus.[10] Furthermore, there have been few reports to date of attempts to extend the application of these systems to target non-phagocytic cells, which are present in much greater quantity throughout the body. Herein we report a tunable and modular microparticle system for plasmid delivery that, we hypothesized, would overcome these problems and simultaneously allow the systematic study of the dependence of transfection efficiency on various formulation parameters including degradation kinetics, use of cationic blend polymers, and surface functionalization for the transfection of non-phagocytic cells.
We recently developed an acid-sensitive delivery system based on acetal-modified dextran (Ac-DEX, Figure 1a),[11] a hydrophobic polymer that can be readily processed into microparticles, which may address the delivery issues described above. Ac-DEX is unique in its ability to provide a tunable degradation rate that is readily controlled by varying the ratio of faster-hydrolyzing acyclic acetals to slower-hydrolyzing cyclic acetals on the polymer backbone.[12] Particle degradation rates can be tuned over the course of minutes to days under endosomal conditions (pH 5). Upon hydrolytic degradation, Ac-DEX reverts back to FDA-approved dextran without the generation of acidic byproducts, which could damage the encapsulated plasmid[13,14] or lead to inflammation,[15,16] as observed with some other commonly used polymer systems.
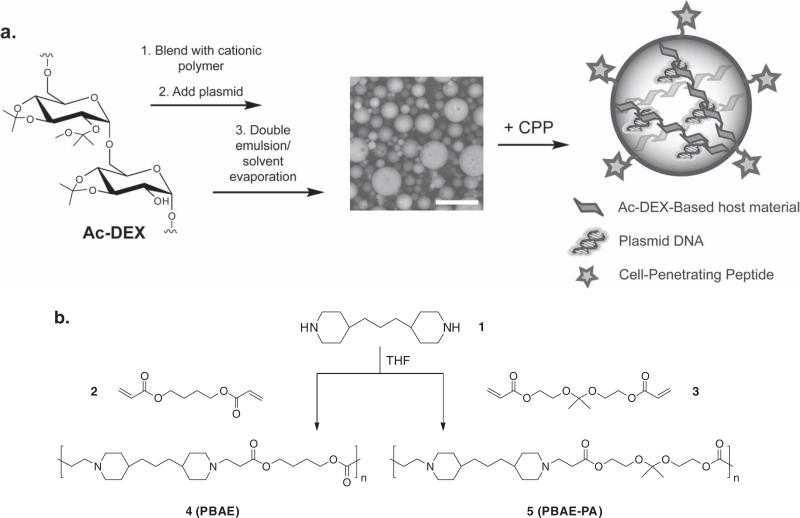
Figure 1.
a) Scheme for blending Ac-DEX and cationic polymers to synthesize plasmid-loaded microparticles, and subsequent functionalization with a nona-arginine cell-penetrating peptide (CPP). In the center, an SEM image of the dry particles is shown (scale bar = 10 μm). b) Synthesis of cationic poly(β-amino ester) 4 (PBAE) and polyacetal analog 5 (PBAE-PA) used in Ac-DEX particle formulations.
We created a small library of Ac-DEX-based microparticles to determine the carrier characteristics needed for efficient gene delivery in phagocytic and non-phagocytic cell lines. Fast-hydrolyzing (f-Ac-DEX) and slow-hydrolyzing (s-Ac-DEX) Ac-DEX polymer variants were synthesized by altering the reaction time for converting the hydroxyl groups of the native dextran to pendant acetals (see Supporting Information for experimental details).[12] For reference, the degradation half-lives of particles prepared from the two polymers at pH 5 and 37 °C were approximately 0.3 h for f-Ac-DEX and 18 h for s-Ac-DEX particles. Particles made from both polymers were relatively stable at pH 7.4 (vide infra).
We next prepared a tertiary-amine-containing poly(β-amino ester) polymer (PBAE, polymer 4, Figure 1b)[17] commonly used in small quantities to enhance transfection in PLGA-based formulations,[18,19] presumably through an endosomal proton-sponge effect.[20,21] The protonated form of polymer 4 hydro-lyzes relatively slowly at pH 5, with a half-life of about 8 h. [17] We hypothesized that more closely matching the hydrolysis rate of this polymer to that of f-Ac-DEX particles might increase the availability of plasmid for transfection, as has been suggested for cationic polymer systems.[22] Thus, a polyacetal analog of polymer 4 (PBAE-PA, polymer 5 , Figure 1b) was synthesized with a dimethyl acetal monomer expected to hydrolyze at a rate similar to that of f-Ac-DEX.
Ac-DEX particles encapsulating a luciferase reporter plasmid were made from blends of Ac-DEX and either PBAE or PBAE-PA using a double emulsion technique (Figure 1a). Interestingly, particles did not aggregate during processing, a common problem with plasmid-loaded PLGA-based microparticles.[19] As a result, precautions such as working up particles in the cold and adding saccharide-based cryoprotectants to the formulation were not required for Ac-DEX particles.[21,23] Quantitative incorporation of PBAE or PBAE-PA blend polymers into particles was confirmed by NMR (see Supporting Information). When visualized by scanning electron microscopy (SEM)(Figure 1a) and confocal microscopy (Figure S1 in the Supporting Information), particles were found to be spherical in shape and porous. All particles had average diameters in the range of 3–6 μm (Figure S2). Plasmid loading efficiency ranged from 10% to 100% depending on the formulation (Figure S3, 100% efficiency ≈ 10 μg plasmid per mg of particles). Improved efficiencies were seen upon the addition of PBAE or PBAE-PA to the system, likely due to the partial cationic nature of these polymers. Electrophoretic analysis of extracts from particles revealed that encapsulated plasmid retained active conformations (super-coiled and relaxed) following particle fabrication, with no major differences observed among the formulations (Figure S4).[24] The surface charge of unblended particles (from zeta-potential measurements) was highly negative due to the encapsulated DNA (Figure 2a and Table S1). Adding PBAE or PBAE-PA to the particle formulation increased the surface charge with increasing blend polymer content (making it less negative), again owing to the cationic nature of these polymers. Particles containing the largest amounts of cationic polymer (30 wt%) had nearly neutral surface potentials.
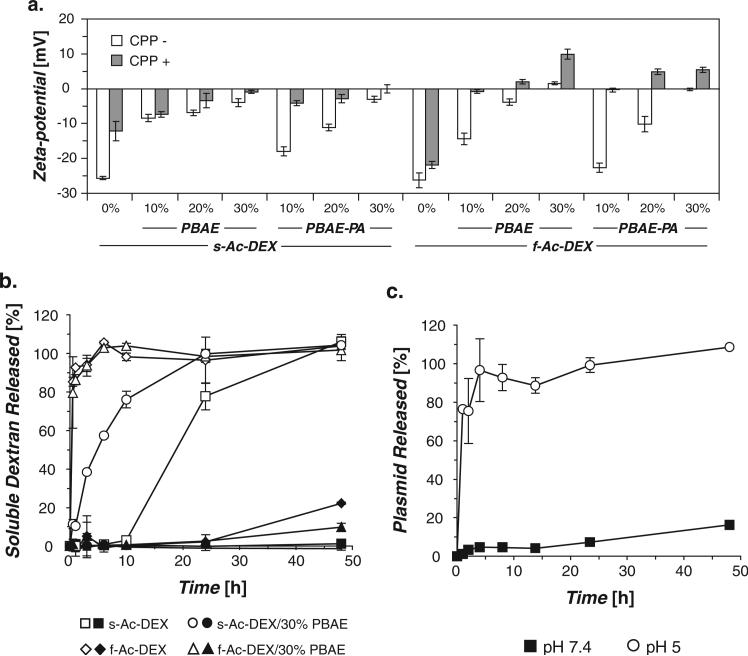
Figure 2.
a) Particle zeta-potentials measured pre- (−) and post-modification (+) with a nona-arginine CPP. b) Degradation of particles at pH 7.4 (closed symbols) and pH 5 (open symbols), determined by analysis of released soluble dextran. Data for 30% PBAE-PA blends were nearly identical to their 30% PBAE blend counterparts and were omitted for clarity (see Figure S5a in the Supporting Information). c) Release of plasmid from f-Ac-DEX particles incubated at pH 7.4 or pH 5. Plasmid release profiles for additional particle formulations can be found in Figure S5b in the Supporting Information.
To expand the application of this system beyond its use with phagocytic APCs, particles were functionalized with a nonaarginine cell-penetrating peptide (CPP) to enable transfection of non-phagocytic cells.[25] The CPP used in this study was functionalized at its N-terminus with an alkoxyamine moiety, which we have previously shown can be used to stably modify the terminal reducing end of dextran chains via oxime bonds. [26] No significant changes in particle size were observed following the ligation step, indicating minimal particle degradation under the reaction conditions (Table S1). Surface modification was corroborated by zeta-potential measurements that showed a general increase in surface charge (less negative or more positive) following the reaction (Figure 2a and Table S1).
Ac-DEX particles are designed to degrade at a rate related to the acetal composition of their base polymer. To determine if the added PBAE and PBAE-PA blend polymers affected this degradation rate, representative formulations were incubated at 37 °C at either pH 5 or pH 7.4 and monitored for the release of soluble dextran. In general, we found that particle degradation was not hindered by the presence of either blend polymer (Figures 2b and S5a). Particles made from f-Ac-DEX degraded too quickly at pH 5 to detect any significant difference in dextran release rate associated with the blend polymers. However, we did observe a marked degradation rate increase for s-Ac-DEX-blended particles at pH 5, likely due to further protonation and associated solubilization of the blend polymers at this pH.[21] We monitored the release of plasmid under similar conditions as described above to determine if this release occurred in a controlled manner. Minimal burst release was observed at extracellular pH (7.4) (Figures 2c and S5b). Typically, 10% or less plasmid was released in the first 24 h at pH 7.4, as compared to bursts of up to 50% seen in other systems in the same time frame.[19,27] We observed no effect of CPP modification on plasmid release rate (Figure S6).
For in vitro testing, RAW 264.7 macrophages and HeLa cells were used as model phagocytic and non-phagocytic systems, respectively. We first confirmed that Ac-DEX particles were capable of being taken up by these cell lines using confocal microscopy. Following incubation in the presence of fluorescently-tagged particles (CPP- for macrophages, CPP+ for HeLa cells), imaging revealed that particles were indeed restricted to the cells (Figures S7 and S8). In the case of HeLa cells, CPP was necessary for particle uptake (Figure S8).
Next, macrophages and HeLa cells were incubated with particles equivalent to 50 ng of plasmid per well for 36 h and analyzed for the expression of luciferase. As shown in Figure 3, particles not blended with either PBAE or PBAE-PA exhibited minimal transfection. However, adding as little as 10 wt% PBAE to the particle formulation resulted in a clear increase in transfection in macrophages (Figure 3a ). However, this change in formulation alone was insufficient to transfect HeLa cells (Figure S9). Consistent with an earlier proof-of-concept experiment using slowly-degrading Ac-DEX particles,[26] functionalization of the particle formulations with a surface-bound ligand was necessary to transfect non-phagocytic cells (Figures S9 and 3b). To address the possibility of electrostatic adsorption of the CPP onto particles with negative surface charge, we incubated identical particles with a non-reactive control CPP. Particles functionalized with alkoxyamine-terminated CPP, showed a 4- to 7-fold increase in transfection over identical particles treated with an analogous, but non-reactive, acrylamide-terminated CPP (Figure S10).
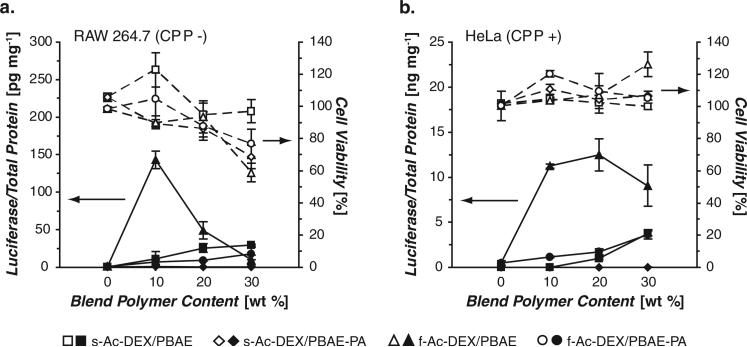
Figure 3.
In vitro transfection of a) RAW 264.7 macrophages and b) HeLa cells with plasmid-loaded microparticles (closed symbols), combined with results from a concurrently performed cytotoxicity assay (open symbols). Particles were either not-modified (−) or modifi ed (+) with a nona-arginine CPP.
In both cell lines, a significant enhancement in transfection was observed using f-Ac-DEX particles relative to particles made with s-Ac-DEX. One formulation in particular, f-Ac-DEX/10% PBAE, was approximately 10-fold and 100-fold more effective than the best performing s-Ac-DEX particles at the same blend polymer content, for macrophages and HeLa cells, respectively. For reference, this optimal formulation was within 15-fold (for macrophages) or 24-fold (for HeLa cells) of the transfection levels observed with a commercially available cationic-lipid-based reagent used exclusively for in vitro transfection (Lipofectamine 2000, data not shown). Particles prepared with s-Ac-DEX showed low, albeit increasing transfection with higher blend content, possibly due to the increasing cationic nature of the particles, but also possibly due to increased particle degradation rates (Figure 2b ).
High PBAE/PBAE-PA blend content resulted in particle-associated cytotoxicity in macrophages, particularly with f-Ac-DEX-blended particles, which may explain the decrease in transfection observed at blend contents above 10 wt%. In contrast, similar cytotoxicity was not observed in HeLa cells, where high transfection levels were observed at up to 30 wt% PBAE content. Interestingly, f-Ac-DEX particles blended with PBAE-PA exhibited much lower transfection efficiency compared to particles blended with the more hydrolytically stable PBAE polymer. This behavior suggests that although fast carrier degradation is generally beneficial, the formation of relatively stable PBAE/plasmid complexes during particle degradation, and retention of complex integrity on the time scale needed for plasmid trafficking to the nucleus, may be required for effective transfection in microparticle systems.
Finally, we noted that transfection did not require an overall positive surface charge. Particles made from the optimal f-Ac-DEX/10% PBAE blend transfected both macrophages (CPP-particles) and HeLa cells (CPP+ particles) (Figures 3a and b), even though these formulations had negative zeta-potentials (Figure 2a ). This behavior is interesting, as the few examples of microparticles tested for plasmid delivery in non-phagocytic cells thus far have generally had highly positive surface charges due to blending with cationic species such as polyethylenimine.[28] Such systems may result in non-specific transfection of tissues in vivo,[29] which is problematic if targeted gene delivery using a cell-specific peptide ligand is the primary goal. Since Ac-DEX particles can be formulated with minimal cationic character yet still effectively transfect non-phagocytic cells, our system may be an attractive platform for targeted gene delivery when coupled with cell-specific ligands.
In conclusion, Ac-DEX-based microparticles represent a novel class of biocompatible materials for gene delivery, capable of transfecting both phagocytic and non-phagocytic cells. Facile tuning of Ac-DEX polymer degradation rates enabled the rapid release of plasmid under endosomal conditions and significantly enhanced transfection efficiency. We anticipate that these properties, coupled with a modular ability to control surface charge, modification with peptide targeting ligands, incorporation of additional molecules for co-delivery, [30] and minimal blending with cationic polymers (ensuring effective transfection while minimizing cytotoxicity), will enable use of this system in a wide range of gene delivery applications in vivo.
Supplementary Material
Acknowledgements
This work was supported by National Institutes of Health grants (PEN Grant 1 U01 HL080729-01 and R01 EB005824). We would like to acknowledge Ann Fischer and Michelle Yasukawa for help with cell culture, and Dr. Peter Friebe and Dr. Eva Harris for assistance with and the use of their luminescence plate reader.
Footnotes
Experimental Section
Experimental materials and methods can be found in the Supporting Information. The Supporting Information includes methods for the synthesis of all polymers used in this study, particle fabrication and characterization, microscopy studies, transfection and viability assays, Figures S1–S10, and Table S1.
Supporting Information
Supporting Information is available online from Wiley InterScience or from the author.
References
- 1.Glover DJ, Lipps HJ, Jans DA. Nat. Rev. Genet. 2005;6:299. doi: 10.1038/nrg1577. [DOI] [PubMed] [Google Scholar]
- 2.Putnam D. Nat. Mater. 2006;5:439. doi: 10.1038/nmat1645. [DOI] [PubMed] [Google Scholar]
- 3.Pack DW, Hoffman AS, Pun S, Stayton PS. Nat. Rev. Drug Discovery. 2005;4:581. doi: 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]
- 4.Park TG, Jeong JH, Kim SW. Adv. Drug Delivery Rev. 2006;58:467. doi: 10.1016/j.addr.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 5.Li W, Szoka FC. Pharm. Res. 2007;V24:438. doi: 10.1007/s11095-006-9180-5. [DOI] [PubMed] [Google Scholar]
- 6.Luten J, van Nostrum CF, De Smedt SC, Hennink WE. J. Controlled Release. 2008;126:97. doi: 10.1016/j.jconrel.2007.10.028. [DOI] [PubMed] [Google Scholar]
- 7.Panyam J, Labhasetwar V. Adv. Drug Delivery Rev. 2003;55:329. doi: 10.1016/s0169-409x(02)00228-4. [DOI] [PubMed] [Google Scholar]
- 8.Nguyen DN, Green JJ, Chan JM, Langer R, Anderson DG. Adv. Mater. 2008;20:1. [Google Scholar]
- 9.Goh SL, Murthy N, Xu M, Fréchet JMJ. Bioconjugate Chem. 2004;15:467. doi: 10.1021/bc034159n. [DOI] [PubMed] [Google Scholar]
- 10.Fu K, Harrell R, Zinski K, Um C, Jaklenec A, Frazier J, Lotan N, Burke P, Klibanov AM, Langer R. J. Pharm. Sci. 2003;92:1582. doi: 10.1002/jps.10414. [DOI] [PubMed] [Google Scholar]
- 11.Bachelder EM, Beaudette TT, Broaders KE, Dashe J, Fréchet JMJ. J. Am. Chem. Soc. 2008;130:10494. doi: 10.1021/ja803947s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Broaders KE, Cohen JA, Beaudette TT, Bachelder EM, Fréchet JMJ. Proc. Natl. Acad. Sci. USA. 2009;106:5497. doi: 10.1073/pnas.0901592106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu K, Pack DW, Klibanov AM, Langer R. Pharm. Res. 2000;17:100. doi: 10.1023/a:1007582911958. [DOI] [PubMed] [Google Scholar]
- 14.Ding A, Schwendeman S. Pharm. Res. 2008;25:2041. doi: 10.1007/s11095-008-9594-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim MS, Ahn HH, Shin YN, Cho MH, Khang G, Lee HB. Biomaterials. 2007;28:5137. doi: 10.1016/j.biomaterials.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 16.Sy JC, Seshadri G, Yang SC, Brown M, Oh T, Dikalov S, Murthy N, Davis ME. Nat. Mater. 2008;7:863. doi: 10.1038/nmat2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lynn DM, Langer R. J. Am. Chem. Soc. 2000;122:10761. [Google Scholar]
- 18.Little SR, Lynn DM, Ge Q, Anderson DG, Puram SV, Chen J, Eisen HN, Langer R. Proc. Natl. Acad. Sci. USA. 2004;101:9534. doi: 10.1073/pnas.0403549101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Little SR, Lynn DM, Puram SV, Langer R. J. Controlled Release. 2005;107:449. doi: 10.1016/j.jconrel.2005.04.022. [DOI] [PubMed] [Google Scholar]
- 20.Sonawane ND, Szoka FC, Jr., Verkman AS. J. Biol. Chem. 2003;278:44826. doi: 10.1074/jbc.M308643200. [DOI] [PubMed] [Google Scholar]
- 21.Lynn DM, Amiji MM, Langer R. Angew. Chem. Int. Ed. 2001;40:1707. [PubMed] [Google Scholar]
- 22.Ko IK, Ziady A, Lu S, Kwon YJ. Biomaterials. 2008;29:3872. doi: 10.1016/j.biomaterials.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 23.Abdelwahed W, Degobert G, Stainmesse S, Fessi H. Adv. Drug Delivery Rev. 2006;58:1688. doi: 10.1016/j.addr.2006.09.017. [DOI] [PubMed] [Google Scholar]
- 24.Adami RC, Collard WT, Gupta SA, Kwok KY, Bonadio J, Rice KG. J. Pharm. Sci. 1998;87:678. doi: 10.1021/js9800477. [DOI] [PubMed] [Google Scholar]
- 25.Wender PA, Mitchell DJ, Pattabiraman K, Pelkey ET, Steinman L, Rothbard JB. Proc. Natl. Acad. Sci. USA. 2000;97:13003. doi: 10.1073/pnas.97.24.13003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beaudette TT, Cohen JA, Bachelder EM, Broaders KE, Cohen JL, Engleman EG, Fréchet JMJ. J. Am. Chem. Soc. 2009 doi: 10.1021/ja903984s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nguyen DN, Raghavan SS, Tashima LM, Lin EC, Fredette SJ, Langer RS, Wang C. Biomaterials. 2008;29:2783. doi: 10.1016/j.biomaterials.2008.03.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang X-Q, Intra J, Salem AK. J. Microencapsulation. 2008;25:1. doi: 10.1080/02652040701659347. [DOI] [PubMed] [Google Scholar]
- 29.Walter E, Merkle HP. J. Drug Targeting. 2002;10:11. doi: 10.1080/10611860290007478. [DOI] [PubMed] [Google Scholar]
- 30.Bachelder EM, Beaudette TT, Broaders KE, Fréchet JMJ, Albrecht MT, Mateczun AJ, Ainslie KM, Pesce JT, Keane-Myers AM. Mol. Pharmaceutics. 2010 doi: 10.1021/mp900311x. DOI:10.1021/mp900311x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.