Abstract
The thermal stability of nucleic acid structures is perturbed under the conditions that mimic the intracellular environment, typically rich in inert components and under osmotic stress. We now describe the thermodynamic stability of DNA oligonucleotide structures in the presence of high background concentrations of neutral cosolutes. Small cosolutes destabilize the basepair structures, and the DNA structures consisting of the same nearest-neighbor composition show similar thermodynamic parameters in the presence of various types of cosolutes. The osmotic stress experiments reveal that water binding to flexible loops, unstable mismatches, and an abasic site upon DNA folding are almost negligible, whereas the binding to stable mismatch pairs is significant. The studies using the basepair-mimic nucleosides and the peptide nucleic acid suggest that the sugar-phosphate backbone and the integrity of the basepair conformation make important contributions to the binding of water molecules to the DNA bases and helical grooves. The study of the DNA hydration provides the basis for understanding and predicting nucleic acid structures in nonaqueous solvent systems.
Introduction
There has been much interest in biomolecular interactions in cosolute-containing solutions from the point of view of understanding biomolecular reactions in the environment rich in macromolecules and small metabolite and osmolyte molecules (1). Water-soluble cosolutes change the solvent properties, including water activity, viscosity, and dielectric constant; in particular, a large polymer cosolute generates the condition of steric crowding between molecules. Aqueous solutions containing a concentrated neutral cosolute have been widely used for the study of influences of the intracellular media on protein-protein interactions and the catalysis of enzymes (reviewed in (1–3)). The cosolute molecules also affect the nucleic acid interactions involving polar purine and pyrimidine bases and negatively charged phosphate groups. The thermal stability of Watson-Crick basepairs is perturbed in the solutions containing a small organic compound or a polymer cosolute, such as an alcohol, dextran, or poly(ethylene glycol) (PEG) (4–11). Our previous study showed that small neutral cosolutes did not influence the basepaired structures of DNA but destabilized the duplex structure of DNA oligonucleotides more than polymer cosolutes did because small cosolutes act as an osmolyte that reduces the solution water activity (12). This observation agrees with the structural aspects that many water molecules bind to the sugar-phosphate backbone and the polar bases exposed to the major and minor grooves of a DNA duplex (13–17). Hydration is an important factor in the helical integrity and folding stability of nucleic acids, but the water binding to basepaired and not basepaired sites and their energy contributions have not yet been fully understood.
The interaction energy for DNA and RNA basepairing has been extensively studied using dilute aqueous solutions containing salts, and the thermodynamic parameters based on the nearest-neighbor interaction model, which are useful to predict the hybridization energy and folding structures, have been reported (18). However, the nearest-neighbor parameters determined using a dilute solution may not be appropriate when cosolutes exist at high concentration. In this study, we investigate the thermodynamic parameters for DNA structure formations in the presence of various types of cosolutes. We demonstrate the DNA hairpins or double-stranded duplexes consisting of the same nearest-neighbor composition that show similar thermodynamic parameters in solutions containing various types of cosolute molecules in an amount comparable to that of intracellular macromolecules. In an attempt to better understand DNA hydrations, we investigate the hydration changes of different DNA structures and those containing nucleic acid analogs upon oligonucleotide folding, and the water binding to basepaired, looped, and mismatched sites are compared. The data provide the source of DNA hydration and the basis for predicting nucleic acid folding structures in nonaqueous solvent systems such as the intracellular condition of a living cell.
Materials and Methods
Materials
High-performance liquid chromatography (HPLC) grade natural DNA oligonucleotides were purchased from Hokkaido System Science (Sapporo, Japan) and Japan Bio Services (Asagiri, Japan). Oligonucleotides containing an ethylene glycol (EG) linker and the basepair analog of N6-(N′-naphthylcarbamoyl)-2′-deoxycytidine (Cnaph) or N6-(N′-phenylcarbamoyl)-2′-deoxyadenosine (Aphe) were synthesized on a controlled-pore glass support column (Glen Research, Sterling, VA) on the basis of the phosphoramidite chemistry using an automated DNA synthesizer (Applied Biosystems, Foster City, CA, 3400 DNA Synthesizer) and purified by HPLC using a C18 reverse phase column (TOSOH, Tokyo, Japan). The peptide nucleic acid (PNA) was synthesized from the Fmoc-protected N-(2-aminoethyl)glycine units (Panagene, Daejeon, Korea) on a solid support of SAL-PEG resin (Watanabe Chemical Industries, Hiroshima, Japan) on the basis of the Fmoc strategy by manual coupling using O-(7-azabenzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium hexafluorophosphate as an activator. The product, capped with an acetyl group at the N-terminus and an amino group at the C-terminus, was purified by HPLC, and the molecular weight was confirmed on the basis of the matrix-assisted laser desorption ionization time-of-flight mass spectra (Bruker, Billerica, MA, Autoflex III). All reagents for preparing buffer solutions were purchased from Wako Pure Chemical Industries (Osaka, Japan) except for Na2EDTA (Dojindo, Kumamoto, Japan), PEG with the molecular weight of 8000 (MP Biomedicals, Solon, OH), dextrans and Ficoll (GE Healthcare, Uppsala, Sweden), and 2-methoxyethanol (MME) (TCI, Tokyo, Japan). These reagents were used without further purification. The solution pH was adjusted after the addition of salts and cosolutes.
The solutions containing a polymer cosolute were prepared using dextran, Ficoll (as a polysaccharide), or PEG. The cosolutes with different molecular weights were examined; dextran with average molecular weights of 7 × 104 (Dex70) and 1 × 104 (Dex10), and PEG with average molecular weights of 8000 (PEG8000) and 200 (PEG200). We also used Ficoll of a highly branched copolymer of sucrose and epichlorohydrin with an average molecular weight of 7 × 104. We also prepared the solutions containing EG, glycerol (Glyc), 1,3-propanediol (PDO), MME, 1,2-dimethoxyethane (DME), methanol (MeOH), ethanol (EtOH), or 1-propanol (PrOH). These small cosolutes as well as the polysaccharides and PEGs contain hydroxyl and/or methoxy groups attached to a saturated carbon atom.
Ultraviolet melting curve
The thermal melting curve of oligonucleotide structures was obtained by monitoring the absorption at 260 nm at a heating rate of 1 or 0.5°C min−1 using the spectrophotometer equipped with a temperature controller (Shimadzu, Kyoto, Japan, UV1800). The cuvette was sealed with an adhesive sheet to prevent evaporation. The melting curve was measured in a buffer consisting of 1 M NaCl, 10 mM Na2HPO4 (pH 7.0), and 1 mM Na2EDTA in the absence and presence of cosolutes, unless otherwise mentioned. For DNA hairpins, we examined the oligonucleotide concentrations of 50 and 2 μM, except for the PEG8000 data that used 10 and 2 μM due to its limited solubility, to verify melting the intramolecular structure. All melting curves of DNA hairpins and double-stranded duplexes followed the two-state transition model describing the helix-coil transition and agreed well with the theoretical curves providing the thermodynamic parameters ΔH°, ΔS°, and ΔG° at 37°C (19,20).
Determination of the number of water bindings upon DNA folding
The thermodynamic parameters for oligonucleotide folding were obtained from fitting the ultraviolet melting curve, and also the Tm−1 versus log(Ct) plot for the double-stranded duplex sequences (19,20). The water activity aw was determined by using the vapor phase osmometry (Wescor, Logan, UT, 5520XR) or the freezing point depression osmometry (KNAUER, Berlin, Germany, Halbmikro Typ Dig.L). We consider the equilibrium for DNA folding from a single strand accompanied by water release (12). The number of water molecules released upon a DNA structure formation Δnw was obtained using Eq. 1,
| (1) |
where R is the gas constant and T is the absolute temperature, by assuming a fixed number regardless of the amount of cosolutes. Because the plot of ΔG° versus ln aw presented in this study was found to be linear with a correlation coefficient r2 >0.97, Δnw and the error value were calculated from the slope of the linear plot weighted by the errors in ΔG°.
Results
Nearest-neighbor interaction of the DNA hairpins in cosolute-containing solutions
The thermal stability of oligonucleotide basepairs in a dilute solution can be accounted for by the nearest-neighbor model, which enables predictions of the interaction energy for hybridization and folding of nucleic acids (21,22). To verify the validity of the interaction model in aqueous mixture solutions, we prepared DNA hairpin sets of the 16-mer (1a and 1b) and 18-mer sequences (2a and 2b) consisting of the same nearest-neighbor composition (Fig. 1 A). Each oligonucleotide set forms the hexaloop of C or T with a hairpin stem of the same dinucleotide frequency and basepairs at both stem ends. The 16-mer hairpins form three G/C and two A/T basepairs, and the 18-mers form two G/C and four A/T basepairs.
Figure 1.
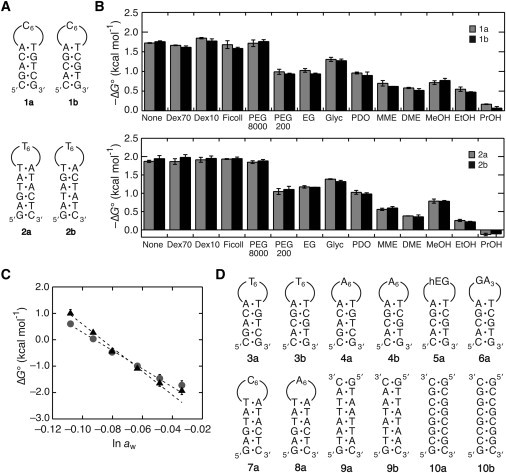
(A) DNA hairpin sets of 1a and 1b, and 2a and 2b, consisting of the same nearest-neighbor composition and the loop sequence. (B) Comparison of the values of ΔG° of the DNA hairpins 1a and 1b (upper) and 2a and 2b (lower) having the same nearest-neighbor composition in the absence and presence of cosolutes at 20 wt%. The ΔG° was calculated at 37°C, and the error value was determined from the data obtained using different DNA concentrations. (C) Plots of the hairpin stability ΔG° of 1a (circles) and 2a (triangles) versus the logarithm of the solution water activity changed by adding PEG200. (D) DNA sequences and the abbreviations used in Table 1 that are not given in the panel A. The sequence of 5a forms the hairpin structure with a hexa(ethylene glycol) loop represented by hEG.
The free energy change ΔG° at 37°C for DNA hairpin formations in 1 M NaCl was measured in the absence and presence of a cosolute at 20 wt%, because the nearest-neighbor model is broadly used for predicting the interaction energy of oligonucleotide structures at the high salt concentration (18). The thermal melting curves obtained with 50 and 2 μM DNA fitted well to the two-state transition model, and their ΔG° values were nearly identical indicating the intramolecular melting. Without a cosolute, the DNA hairpins having the same nearest-neighbor composition showed similar folding energies of –1.72 and –1.76 kcal mol−1 (1 kcal = 4.18 kJ) for 1a and 1b, respectively, and –1.87 and –1.94 kcal mol−1 for 2a and 2b, respectively, agreed with the nearest-neighbor interaction model. Although the polymer cosolutes of Dex70, Dex10, Ficoll, and PEG8000 less significantly changed the hairpin stability, small cosolutes decreased the stability by 0.4–2.0 kcal mol−1 depending on the type of molecule (Fig. 1 B). Importantly, there was a similarity in the stabilities of each DNA set in all the cosolute-containing solutions. Additionally, the enthalpy and entropy changes were also similar in each hairpin set (Tables S1 and S2 in the Supporting Material). These observations indicate that the DNA hairpins consisting of the same nearest-neighbor composition show similar thermodynamic parameters even when a large number of cosolutes exist.
Hydration to the loop nucleotides
The data in Fig. 1 B indicate that the small cosolutes exert a much greater influence than the polymer cosolutes. The cosolutes are highly soluble in water and hydrate more than polymer cosolutes, thereby substantially lowering the activity of water in aqueous mixtures by acting as an osmolyte (23,24). Our previous study showed that the water activity as well as the chemical structure of a cosolute was important for determining the thermodynamic stability of DNA duplexes (12). PEG is one of the most convenient cosolutes for hydration studies (details are given in the Discussion section). We then evaluated the hydration changes upon DNA folding by using PEG with a molecular weight of 200 in the following studies.
According to the thermal melting curve data, 1a and 2a in the absence of PEG showed similar Tm values of around 54.5°C. Their hairpin stabilities gradually decreased as the PEG amount increased, and the Tm of 2a became less than that of 1a at 50 wt% PEG (27.0 and 30.0°C, respectively, as shown in Fig. S1 A). The free energy change ΔG° at 37°C for the hairpin formations of 1a and 2a linearly increased with decreasing the logarithm of the water activity aw, as shown in Fig. 1 C. The plots of ΔG° versus ln aw for 1a and 2a are almost linear with negative slopes and correlation coefficients r2 of 0.989 and 0.972, respectively, indicating that the number of water molecules that bind upon the hairpin formation is greater than the number of water molecules that dissociate, e.g., from the hydrated DNA bases. The –Δnw for 1a was distinctly different from that for 2a, 56 ± 2 and 73 ± 3, respectively. The values of ΔG° for each hairpin set of 1a and 1b or 2a and 2b were similar to each other for all the examined PEG amounts (data not shown), thus giving similar –Δnw numbers for each DNA set. The plots of the data obtained with different salts (KCl and CsCl, instead of NaCl) overlay (Fig. S1 B). Remarkably, although Fig. 1 B shows the different hairpin stabilities obtained with the same amount of cosolutes, the data points plotted against the logarithm of the water activity for different cosolutes (PEG8000, DME, and PrOH, instead of PEG200, but with different amounts) almost overlay (Fig. S1 C). The data points obtained with binary mixture solutions of these cosolute molecules also fall along the same line (Fig. S1 D). There is an argument that PEG preferentially interacts with the DNA surface, and the effect of PEG is relevant to the amount of DNA surface exposed during DNA melting and to the amount of PEG surface exposed to the solvent (11). The Δnw numbers determined from the slope of the plot may not represent the true hydration number if PEG associates with the DNA through hydrophobic interaction. However, the number still provides, at least qualitatively, an estimate of the trends in hydration changes upon DNA folding (see the Discussion section) (25,26).
The DNA hairpins forming a hexanucleotide loop of C, T, and A shown in Fig. 1 D are also compared. The G-loop hairpins are excluded here because a long guanine tract tends to form self-aggregated complexes. In the absence of PEG, the T-loop hairpins were 0.7–0.8 kcal mol−1 more stable than the C-loop and A-loop hairpins with the same stem sequence (Table 1), and the DNA hairpins having the same nearest-neighbor composition showed quite similar free energy values to each other both in the presence and absence of PEG. Table 1 compares the –Δnw numbers for the hairpins 3a–8a having a different loop sequence, including the hexa(ethylene glycol) (hEG) loop and a GA3 tetraloop. The numbers for each DNA set consisting of an identical nearest-neighbor composition and the loop sequence, including 3a and 3b forming the T6 loop with a 5-basepair stem and 4a and 4b forming the A6 loop with a 5-basepair stem, are similar to each other. It is clear that both the 16-mer and 18-mer hairpins exhibit a slightly higher –Δnw number for the T-loop hairpin than those for the C-loop and A-loop hairpins, indicating a slightly higher water-binding capacity in the T loop than in the other loops.
Table 1.
Values of the ΔG° without PEG and the Δnw for the formations of DNA structures
| DNA sequence | Stem length (bp) | Loop sequence | –ΔG° (kcal mol−1)∗ | –Δnw |
|---|---|---|---|---|
| 1a | 5 | C6 | 1.72 | 56 ± 2 |
| 1b | 5 | C6 | 1.76 | 59 ± 3 |
| 2a | 6 | T6 | 1.87 | 73 ± 3 |
| 2b | 6 | T6 | 1.90 | 70 ± 3 |
| 3a | 5 | T6 | 2.48 | 68 ± 3 |
| 3b | 5 | T6 | 2.43 | 70 ± 3 |
| 4a | 5 | A6 | 1.63 | 50 ± 2 |
| 4b | 5 | A6 | 1.68 | 51 ± 2 |
| 5a | 5 | hEG† | 2.72 | 50 ± 4 |
| 6a | 5 | GA3 | 3.23 | 67 ± 4 |
| 7a | 6 | C6 | 1.28 | 58 ± 2 |
| 8a | 6 | A6 | 1.21 | 56 ± 2 |
| 9a | 8 | No loop | 5.26 | 53 ± 10 |
| 9b | 8 | No loop | 4.98 | 49 ± 13 |
| 10a | 8 | No loop | 12.3 | 102 ± 12 |
| 10b | 8 | No loop | 11.7 | 90 ± 6 |
ΔG° was calculated at 37°C and the error value for ΔG° are smaller than 6%.
hEG indicates the hexa(ethylene glycol) linker.
Hydration to the matched and mismatched sites
Fig. 2 A compares the PEG-dependency of the 9-mer basepairs with and without a loop. The DNA hairpins forming a tetraloop of C and a hEG loop showed similar stabilities in the examined PEG amounts and similar –Δnw of ∼86. This number is equivalent, within an experimental uncertainty, to that obtained for the double-stranded 9-mer DNA without the loop (–Δnw = 85 ± 3). The data indicate less water binding to the C-loop and the hEG linker upon the formation of the hairpin loop. We also derive the same conclusion from the data in Table 1 showing the similar –Δnw numbers for the C6 loop (1b), the A6 loop (4b), and the hEG loop (5a). In addition, the number was unchanged when the buffer solutions containing the magnesium ion and cobalt hexamine ion instead of the sodium ion were used (Fig. 2 A), suggesting a similar degree of DNA hydration regardless of the ionic charge of metal ions.
Figure 2.
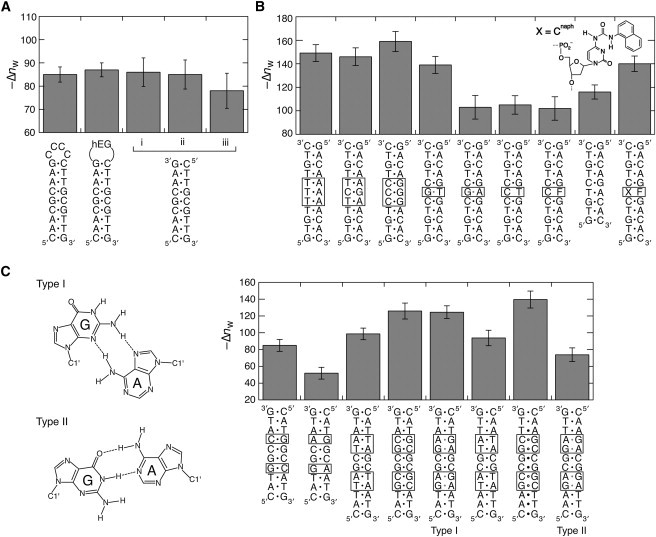
(A) Comparison of the Δnw numbers for the DNA structure formations consisting of the same basepairs. The data for the double-stranded duplex obtained in i), 1 M NaCl, ii), 10 mM MgCl2, and iii), 1 mM [Co(NH3)6]Cl3 are also compared. For the experiments using MgCl2 and [Co(NH3)6]Cl3, the buffer solutions containing 25 mM HEPES (pH 7.0) and 0.1 mM Na2EDTA were used. (B) The Δnw numbers for the 11-mer duplexes having different basepairs in the middle and the 9-mer duplex without trinucleotide interactions in the middle of the 11-mer sequence. F and X indicate the tetrahydrofuran abasic site and Cnaph, respectively. (C) Two types of conformations of the tandem G/A mismatch formed in a DNA sequence 5′-pyrimidine-GA-purine-3′ (type I) and 5′-purine-GA-pyrimidine-3′ (type II). The Δnw numbers of the DNA duplexes forming the type I and type II conformations and those forming corresponding fully matched duplexes or the single G/A mismatch sites are compared.
To address the influence of the basepair sequence, two sets of nonself-complementary sequences having the same but biased nearest-neighbor composition were investigated. The sequences of 9a and 9b are AT-rich and 10a and 10b are GC-rich, forming fully matched basepairs of the same sequence alignment of purine and pyrimidine nucleotides (Fig. 1 D). As shown in Table 1 (Tables S3 and S4), the thermodynamic parameters and the –Δnw were similar for each DNA set, and the basepair substitutions from A/T to G/C increased the duplex stability (by 7 kcal mol−1 in the absence of a cosolute) and the number of water bindings (by ∼45).
Fig. 2 B shows the influences of basepair substitutions in the 11-mer DNA duplexes containing continuous A/T or C/G basepairs in the middle. There were changes in –Δnw from 146 to 159 when the T/A basepairs were substituted by C/G basepairs. On the other hand, single G/A and C/T mismatches and a tetrahydrofuran abasic site (F) in the 11-mer duplex, which lowered the structural stability by 4.9, 5.8, and 5.0 kcal mol−1, respectively, showed the –Δnw in the range from 102 to 105, which was similar to that of the 9-mer duplex lacking trinucleotide interactions in the middle of the 11-mer duplex (–Δnw = 116 ± 6). In contrast, the G/T mismatch, forming two hydrogen bonds known as the Wobble pair, showed the –Δnw of 139 ± 7 that was comparable to those of the fully matched 11-mer duplexes.
We also examined the tandem G/A mismatches adopting different basepair conformations through hydrogen bonds depending on the flanking sequence. Fig. 2 C shows the type I and type II conformations formed in two adjacent mismatched guanine and adenine bases in the 5′-pyrimidine-GA-purine-3′ and 5′-purine-GA-pyrimidine-3′ sequences, respectively (27). We used the self-complementary sequences forming two tandem G/A mismatch sites that adopt the type I or type II conformation. The DNA sequences exhibited different circular dichroism spectra (Fig. S2), consistent with the adoption of different mismatch geometries. The 5′-pyrimidine-GA-purine-3′ sequence formed a much more stable duplex than the 5′-purine-GA-pyrimidine-3′ sequence (–12.4 kcal mol−1 and –9.30 kcal mol−1, respectively). Although the introduction of single G/A mismatches in the duplex significantly decreased the hydration number, the tandem G/A mismatches did not. Intriguingly, in comparison with the corresponding fully matched duplex, the number of water bindings obviously decreased for the type II conformation, but not for the type I conformation that gave the –Δnw similar to that of the DNA duplex replacing the G/A mismatches by G/C base pairs.
Influence on the nucleic acid analogs
Fig. 2 B includes the data of the DNA duplex containing the deoxycytidine derivative Cnaph that covalently tethers a stacking moiety of the naphthyl group as a simple structural analog of the DNA purine base. Our previous study revealed that the Cnaph/F pair allowed the intercalation of the naphthyl group into a DNA duplex without substantial perturbation of the overall double helical structure (28,29). Although the abasic site introduced in the sequence significantly decreased the duplex stability and the –Δnw, incorporation of Cnaph opposite the abasic site restored the values. We also obtained similar results when a deoxyadenosine derivative Aphe, tethering the phenyl group as a nonpolar analog of the pyrimidine base (29,30), was tested (Fig. S3). These data suggest that the polar bases are not absolutely essential for the water binding upon the duplex formations.
The PNA is a nucleic acid analog that possesses the DNA base moiety with the uncharged peptide backbone of N-(2-aminoethyl)glycine units (Fig. 3 A). Complementary PNA strands hybridize with one another through Watson-Crick basepairing and form a stable double helical structure (31). PNA strands also form the hybridization complex with a complementary DNA, and the thermal stability of the short hybrid duplexes follows the nearest-neighbor model (32). We measured the stability of a PNA duplex formed by the self-complementary sequence of NATGCGCATC. In our previous study, the stability of the DNA duplex having the same nucleotide sequence 5′ATGCGCAT3′ showed a large dependence on the changes in water activity by cosolutes (12). In contrast, the PNA duplex stability was unchanged by PEG and other cosolutes, as shown in Fig. S4. The nonself-complementary sequences were also tested to compare the four types of duplexes formed by PNA and DNA, i.e., the PNA duplex, PNA/DNA, and DNA/PNA hybrid duplexes, and DNA duplex, shown in Fig. 3 B. The PNA duplex was the most stable and showed a markedly different dependence of water activity from the other duplexes. The –Δnw for the PNA duplex formation was nearly 0, as observed for the self-complementary sequence. Remarkably, the hybrid duplexes showed a moderate dependence and their –Δnw numbers were almost half that for the DNA duplex (Fig. 3 C). These data suggest that the sugar-phosphate backbone of each DNA strand also plays a crucial role in the water binding.
Figure 3.
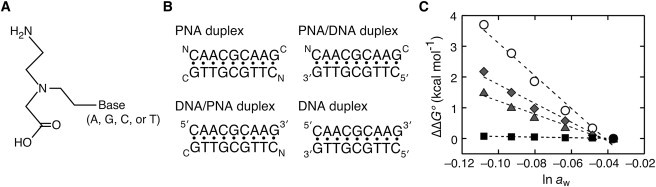
(A) Structure of the PNA monomer unit with the N-(2-aminoethyl)glycine backbone. (B) Four types of duplexes formed by the nonself-complementary PNA and DNA strands. (C) Changes in ΔG° (ΔΔG°) of the four types of duplexes by adding PEG; PNA duplex (squares), PNA/DNA duplex (diamonds), DNA/PNA duplex (triangles), and DNA duplex (circles).
Discussion
Cosolute effects on the nearest-neighbor interaction of DNA
We examined the DNA oligonucleotide structures forming the G/C basepair at both sequence ends to avoid fraying of the helix termini that could underestimate the hydration number. In all cases, the basepair stabilities ΔG° at 37°C were, to a greater or lesser extent, decreased by the presence of polymer and small cosolutes. The destabilizations are attributed to several reasons: small aliphatic alcohols provide the solution environment favorable for solvating hydrophobic groups and thus stabilizing an unstructured state of DNA. The presence of a cosolute also changes the thermodynamic activity of solute molecules and solution properties, particularly a polymer cosolute generates macromolecular crowding, restricting the dynamic behavior of molecules and the volume available to oligonucleotides (2). However, as indicated in Fig. 1 B, the hairpin stability was less affected by the polymer cosolutes. The stability was adversely affected by small cosolutes that provide an osmotic stress greater than polymer cosolutes of the same weight quantities. The formation of DNA basepairs brings the sugar-phosphate backbone close to each other and establishes the hydrogen-bonding network of water surrounding DNA strands. Thus, the basepair stability (Fig. 1 B) is highly sensitive to the amount of osmotically active water, and the water activity of solutions used for the experiments shown in Fig. 1 B ranges from 0.97 to 0.87. The cosolute effect is attributed to its chemical structure as well as the water activity; the hairpin stability decreased as the number of hydroxyl groups decreased (e.g., among glycerol, 1,3-propanediol, and PrOH or between EG and EtOH) and as the number of alkyl or methyl groups increased (e.g., among methanol, EtOH, and PrOH or among EG, MME, and DME) when the same amount of cosolutes is compared. The number of hydroxyl groups and carbon groups seems to influence the degree of the hairpin destabilization, possibly by direct interactions with an oligonucleotide (12,33–37) and by indirect repulsive interactions that disrupts the hydrogen-bonding network of water molecules where the balance between the hydroxyl oxygens and alkyl carbon of the alcohol molecules are reported to be significant on the extent of exclusion from a DNA surface (38,39). We can speculate that the intracellular condition, in which small hydrophilic molecules are abundant and the water activity has been altered, strongly influences the interaction energy and folding structures of nucleic acids. The nearest-neighbor model is widely used for the predictions of hybridization energy and folding structures of oligonucleotides in dilute aqueous solutions (18), and the nearest-neighbor parameters obtained with 1 M NaCl can be extended for the predictions under low salt conditions by taking into account the correction term for the salt concentration (40). Similarly, the parameters could be used for the predictions in nonaqueous solvent systems by taking into account the hydration term, which is to be addressed in the future work.
Osmotic stress experiments using PEG
An osmotic stress experiment, which uses a cosolute as an osmolyte, has been employed to study DNA hydrations (24). PEG has a high solubility in water and a relatively low vapor pressure, which are great advantages for the experiment in which oligonucleotides are thermally melted. In our previous study, we found a linear correlation between DNA duplex stability and the logarithm of the water activity of solutions containing PEG with molecular weights ranging from 200 to 8000 (12). The values of –ΔnW determined from the slope of the linear plot may not represent the true hydration number if cosolutes directly interact with the DNA. There are reports regarding the effects of cosolute molecules that are not excluded from the vicinity of DNA, and different slopes are formed depending on the type of cosolute selected; however, different cosolutes give similar trends in hydration changes (8,10,41,42). Our previous study proposed that cosolute molecules with two or more hydroxyl groups in the vicinal position caused the number of bound water molecules to be underestimated (12). This can be because of direct interactions with an oligonucleotide through multiple hydrogen bonding, or destructive overlap of the hydration spheres of hydroxyl groups in close proximity as proposed for glycerol and monosaccharides (38). Note that PEG has two hydroxyl groups separated by several methylene groups, which would be unsuitable for establishing stable hydrogen bonds with DNA, although hydroxyl groups of PEG could still interact with certain bases because of using a large amount. It is also proposed that PEG may interact with exposed DNA bases because of their hydrophobic nature (11). However, we show the data that the direct interaction of PEG with DNA bases might contribute very little, as the following: 1), PEG essentially does not change the stability of PNA duplexes possessing a DNA base moiety. 2), The basepair-mimic nucleosides having a nonpolar aromatic group, which provides a hydrophobic binding site for PEG, and show destabilizations that are comparable to or smaller than those of the natural duplexes of the same length. 3), In disagreement with the model of PEG access to the DNA surface, the degree of PEG-promoted destabilizations is distinctly different among oligonucleotides of the same length. 4), The hydrophobicity of PEG substantially increases when the temperature is raised. However, the DNA basepairs with and without a loop provide similar Δnw numbers (Fig. 2 A), although there is a difference of 40°C in their melting temperatures. 5), The cosolutes without hydroxyl groups in the vicinal position (PEG200, PEG8000, DME, and PrOH) and their binary mixture solutions showed the same water activity plots, consistent with the water activity being the major determinant of the DNA stability.
Hydration changes on the paired and unpaired sites in DNA
The osmotic stress experiments reveal that the DNA hairpin formation is the water uptake reaction in which water molecules bind both to basepaired and looped nucleotides and that more water molecules bind to the T-loop hairpin than to the C-loop and A-loop hairpins. It is reported that the tetraloop of T in a DNA hairpin has a distinct connectivity with the adjacent basepair of a stem duplex and provides more interaction energy than other loops (43). Because the hairpin with the T6 loop is obviously more stable than the other hairpins by 0.7–1.1 kcal mol−1 in –ΔG° (Table 1), there are supposedly interactions involving the loop nucleotides that accompany the binding of water molecules. In agreement with the idea, when the GA3 tetraloop, known as an unusually stable hairpin loop motif caused by substantial interactions within the loop nucleotides (44), was employed, a higher water-binding number –Δnw of 67 ± 4 was obtained (Table 1). On the other hand, less water binding to the hEG loop, which is highly flexible and has no substantial interaction in the loop, is shown in Table 1 and Fig. 2 A. It is thus probable that water molecules preferentially bind to structurally ordered DNA sites. Whether the number of water bindings is directly relevant to the thermodynamic stability of DNA structures (Fig. S5 A) remains unclear, which requires further study.
If the hydration change around the loop nucleotides is ignored, the number of water bindings to Watson-Crick basepairs obtained in this study ranges from 46 to 157. These data give a correlation between the number of water bindings and the base pair length (Fig. S5 B). On average, ∼6–14 water molecules are calculated to associate with every single basepair formation. We find moderate increases in the –Δnw when A/T basepairs are substituted by G/C pairs (Table 1 and Fig. 2, B and C). More water bindings to G/C basepairs than to A/T basepairs have also been reported in other studies using short oligonucleotides of biased sequences (15,42). If the data of the biased DNA sequences of continuous A/T or G/C pairs longer than four are excluded, the –Δnw data given previously provide the average number of water binding of 9–14. Accordingly, there is a sequence dependence on the –Δnw number for strongly biased DNA sequences but this becomes weakly significant for random sequences. The correlations may be used to derive the hydration term for the prediction in the presence of cosolutes.
Although we cannot exclude the possibility that certain mismatch bases interact favorably with PEG or water, it is more likely that the water binding to mismatch sites significantly differs depending on the conformation of mismatch pairs. Although the G/T mismatch forming interbase hydrogen bonds only slightly changed the –Δnw compared to the numbers for the Watson-Crick basepairs, the unpaired sites of G/A and C/T mismatches and an abasic site significantly changed the number (Fig. 2 B). The data suggest substantial water binding to the G/T mismatch site due to a minimum local disruption of the double helical structure but less binding to the structurally unstable G/A and C/T mismatches and the abasic site. It is probable that the formation of stable basepairs is absolutely important for the water binding and that the unpaired sites disrupt water binding but only locally.
The idea is verified from the data for tandem G/A mismatches shown in Fig. 2 C. The type I conformation is more stable than the type II conformation, attributed to their different stacking geometries with adjacent bases (45). The type I mismatch forms edge-to-edge hydrogen bonds involving the nitrogen atoms and the amino groups of guanine and adenine. Although the interaction geometry of the mismatched bases is different from the Watson-Crick basepairs, the type I conformation fits well into the standard duplex structure with their backbone angles deviated from the normal helicity of DNA (45). The duplex forming the type I conformation show the –Δnw as high as that of the duplex replacing the G/A mismatches by G/C basepairs despite their basepair geometries being different. On the other hand, the type II mismatch adopts the face-to-face geometry analogous to the Watson-Crick basepair, but markedly widens the minor groove and increases the interphosphate distance resulting from a greater distance between the C1′ atoms compared to that of Watson-Crick basepairs (1.25 nm instead of 1.05 nm) (46). We find that the type II mismatch sites decrease the hydration number. Thus, the sugar-phosphate backbone conformation is also important for the water binding.
The investigations using crystallography, molecular dynamics simulations, and other methods have revealed that the double helical grooves of DNA are surrounded by a hydrogen-bonded water network. Around 20 water molecules are found as the primary hydration shell per nucleotide pair, some of which bridge the polar atoms of DNA bases and the sugar-phosphate backbone belonging to the same or adjacent nucleotide (13–16,47). It is reported that the hydration patterns seen in the crystal structures of nucleic acid duplexes qualitatively agree with the hydration data obtained from osmotic stress measurements (48,49). We find that the number of water bindings energetically coupled to DNA folding is within the scope of the nearest-neighbor interaction model. Therefore, the long-range network of water molecules beyond the nearest-neighbor model would not significantly contribute to the thermodynamics for DNA folding.
Formations of nucleic acid structures are directed by basepair formations accompanying the bindings of metal ions as well as water molecules. Metal ions associate with a nucleic acid duplex predominantly through Coulomb interactions that screen the electrostatic repulsion among phosphate groups (33), and the metal ions occupy some of the hydration sites in a DNA duplex (15). The data included in Fig. 2 A and Fig. S1 B indicate the similar PEG-dependence on the basepair stability regardless of the salt used. Because the ionic radius of these metal ions is different, there are considerable differences in the charge interaction energy and the hydration energy among the ions. Therefore, the charge density, specific metal ion coordination, and ion exclusion in the media of decreased water activity have a small contribution to the PEG effect. Notably, [Co(NH3)6]3+ is an exchange-inert metal complex and has the size and coordination geometry similar to hydrated Mg2+. It is likely that the nonspecific outer sphere binding distant from the DNA surface is predominant and dehydrations of metal ions and the phosphate groups are insignificant.
Hydration study using the nucleic acid analogs
The comparison with data of the basepair-mimic nucleosides reveals the role of polar DNA bases. Our previous study demonstrated that Cnaph or Aphe inserted opposite to the abasic site in a DNA duplex adopted a basepair-like conformation by intercalating the nonpolar group without perturbing the integrity of the double helical structure (28,29). If the hydrogen-bonding network of water, which is mediated by interactions with polar DNA bases, is disrupted by the presence of the nucleic acid analogs, Δnw should be largely changed. However, the DNA duplexes containing the Cnaph/F and Aphe/F pairs show the Δnw numbers comparable to those of the fully matched duplexes (Fig. 2 B and Fig. S3). This observation suggests that the integrity of the double helical structure is important for the water binding.
The PNA data reveal the role of the backbone in water binding. The PNA backbone is composed of N-(2-aminoethyl)-glycine units linked by peptide bonds, and the oxygen and nitrogen atoms of the peptide bond as well as the DNA base moieties are capable of hydration. Nevertheless, the PNA duplexes formed by the self- and nonself-complementary sequences showed no dependence of water activity on their stabilities (Fig. 3 C and Fig. S4). The difference in the hydration degrees between the duplexes of DNA and PNA has been studied using organic solvents (50,51). We find by employing cosolutes that the numbers of water molecules bound to the hybrid duplexes formed by PNA and DNA, which consists of one sugar-phosphate backbone strand, are as high as half the number for the DNA duplex. According to the crystal structure data, a PNA duplex forms a slightly different double helical structure compared to a DNA duplex (31,52,53). The polar groups in the purine and pyrimidine bases of a PNA duplex are exposed to the double helical grooves, but the degrees of water binding to the bases and the backbone appear to be relatively small. Moreover, the hydrogen-bonding network surrounding PNA strands appears to be less organized, however the crystal structure gives only qualitative information on the water binding relevant to the duplex stability. In contrast, the sugar-phosphate backbone and the basepairs of a DNA duplex are well hydrated, and the phosphate anionic oxygen atoms, phosphodiester oxygen atoms, and the furanose O4′ atoms participate in the binding of water molecules that directly or indirectly interact with DNA bases (15,54–56). We propose that the sugar-phosphate backbone is indispensable for the groove hydration and acts as a scaffold for the water binding to DNA base pairs.
Conclusions
In this study, we show that the DNA oligonucleotide structures consisting of the same nearest-neighbor composition show similar thermodynamic parameters in the solutions mixed with cosolutes in an amount comparable to that of intracellular molecules. Because the hydration number determines the degree of destabilization of DNA basepairs in decreased water activity, the hydration study based on the nearest-neighbor model provides the basis for the prediction of nucleic acid structures in nonaqueous solvent systems. The osmotic pressure experiments using PEG show the differences in the hydration numbers between the folded and unfolded states of DNA. We find that there is no significant water binding to less-ordered sites, such as a flexible loop and unpaired mismatch pairs, where the hydration degree is similar to that of the isolated bases in a single-strand state, whereas the stable mismatches forming stable hydrogen bonds show the hydration numbers as high as those of the Watson-Crick basepairs. It is probable that water molecules preferentially bind to structurally ordered DNA sites at which a stable hydrogen-bonding network of water can be established. The studies using DNA mismatches and the nucleic acid analogs suggest that the conformations of the sugar-phosphate backbone and the integrity of basepair stacking are important for the binding of water molecules to the DNA bases and helical grooves. The cosolute study provides valuable insights to explore the experimental conditions optimized for biochemical assays and to study the thermodynamics for nucleic acid folding for their applications in molecular biology and medicine.
Acknowledgments
The basepair-mimic nucleosides Cnaph and Aphe were provided from Dr. Masayuki Fujii at Kinki University, Japan. We also thank Junpei Ueno, Yoko Funaji, Minako Nakajima, Yoko Hasegawa, and Yuichi Kitagawa for technical assistance.
This work was supported in part by Grants-in-Aid for Scientific Research, the “Core Research” Project (2009-2014) and the “Academic Frontier” Project (2004-2009) from the Ministry of Education, Culture, Sports, Science and Technology, Japan, and the Hirao Taro Foundation of the Konan University Association for Academic Research.
Contributor Information
Shu-ichi Nakano, Email: shuichi@center.konan-u.ac.jp.
Naoki Sugimoto, Email: sugimoto@konan-u.ac.jp.
Supporting Material
References
- 1.Luby-Phelps K. Cytoarchitecture and physical properties of cytoplasm: volume, viscosity, diffusion, intracellular surface area. Int. Rev. Cytol. 2000;192:189–221. doi: 10.1016/s0074-7696(08)60527-6. [DOI] [PubMed] [Google Scholar]
- 2.Minton A.P. Molecular crowding: analysis of effects of high concentrations of inert cosolutes on biochemical equilibria and rates in terms of volume exclusion. Methods Enzymol. 1998;295:127–149. doi: 10.1016/s0076-6879(98)95038-8. [DOI] [PubMed] [Google Scholar]
- 3.Zhou H.X., Rivas G., Minton A.P. Macromolecular crowding and confinement: biochemical, biophysical, and potential physiological consequences. Annu Rev Biophys. 2008;37:375–397. doi: 10.1146/annurev.biophys.37.032807.125817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Albergo D.D., Turner D.H. Solvent effects on thermodynamics of double-helix formation in (dG-dC)3. Biochemistry. 1981;20:1413–1418. doi: 10.1021/bi00509a002. [DOI] [PubMed] [Google Scholar]
- 5.Hickey D.R., Turner D.H. Solvent effects on the stability of A7U7p. Biochemistry. 1985;24:2086–2094. doi: 10.1021/bi00329a042. [DOI] [PubMed] [Google Scholar]
- 6.Woolley P., Wills P.R. Excluded-volume effect of inert macromolecules on the melting of nucleic acids. Biophys. Chem. 1985;22:89–94. doi: 10.1016/0301-4622(85)80029-6. [DOI] [PubMed] [Google Scholar]
- 7.Spink C.H., Chaires J.B. Selective stabilization of triplex DNA by poly(ethylene glycols) J. Am. Chem. Soc. 1995;117:12887–12888. [Google Scholar]
- 8.Spink C.H., Chaires J.B. Effects of hydration, ion release, and excluded volume on the melting of triplex and duplex DNA. Biochemistry. 1999;38:496–508. doi: 10.1021/bi9820154. [DOI] [PubMed] [Google Scholar]
- 9.Goobes R., Kahana N., Minsky A. Metabolic buffering exerted by macromolecular crowding on DNA-DNA interactions: origin and physiological significance. Biochemistry. 2003;42:2431–2440. doi: 10.1021/bi026775x. [DOI] [PubMed] [Google Scholar]
- 10.Spink C.H., Garbett N., Chaires J.B. Enthalpies of DNA melting in the presence of osmolytes. Biophys. Chem. 2007;126:176–185. doi: 10.1016/j.bpc.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 11.Knowles D.B., LaCroix A.S., Record M.T., Jr. Separation of preferential interaction and excluded volume effects on DNA duplex and hairpin stability. Proc. Natl. Acad. Sci. USA. 2011;108:12699–12704. doi: 10.1073/pnas.1103382108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakano S., Karimata H., Sugimoto N. The effect of molecular crowding with nucleotide length and cosolute structure on DNA duplex stability. J. Am. Chem. Soc. 2004;126:14330–14331. doi: 10.1021/ja0463029. [DOI] [PubMed] [Google Scholar]
- 13.Brandes R., Vold R.R., Kearns D.R. Effects of hydration on purine motion in solid DNA. Biochemistry. 1986;25:7744–7751. doi: 10.1021/bi00371a069. [DOI] [PubMed] [Google Scholar]
- 14.Feig M., Pettitt B.M. A molecular simulation picture of DNA hydration around A- and B-DNA. Biopolymers. 1998;48:199–209. doi: 10.1002/(SICI)1097-0282(1998)48:4<199::AID-BIP2>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 15.Auffinger P., Westhof E. Water and ion binding around r(UpA)12 and d(TpA)12 oligomers—comparison with RNA and DNA (CpG)12 duplexes. J. Mol. Biol. 2001;305:1057–1072. doi: 10.1006/jmbi.2000.4360. [DOI] [PubMed] [Google Scholar]
- 16.Arai S., Chatake T., Niimura N. Complicated water orientations in the minor groove of the B-DNA decamer d(CCATTAATGG)2 observed by neutron diffraction measurements. Nucleic Acids Res. 2005;33:3017–3024. doi: 10.1093/nar/gki616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ganguly M., Wang F., Gold B. A study of 7-deaza-2′-deoxyguanosine 2′-deoxycytidine base pairing in DNA. Nucleic Acids Res. 2007;35:6181–6195. doi: 10.1093/nar/gkm670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turner D.H. Conformational changes. In: Bloomfield V.A., Crothers D.M., Tinoco I. Jr., editors. Nucleic Acids: Structures, Properties and Functions. University Science Books Press; Sausalito, CA: 2000. pp. 259–334. [Google Scholar]
- 19.Marky L.A., Breslauer K.J. Calculating thermodynamic data for transitions of any molecularity from equilibrium melting curves. Biopolymers. 1987;26:1601–1620. doi: 10.1002/bip.360260911. [DOI] [PubMed] [Google Scholar]
- 20.Puglisi J.D., Tinoco I., Jr. Absorbance melting curves of RNA. Methods Enzymol. 1989;180:304–325. doi: 10.1016/0076-6879(89)80108-9. [DOI] [PubMed] [Google Scholar]
- 21.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kraeva R.I., Krastev D.B., Stoynov S.S. Stability of mRNA/DNA and DNA/DNA duplexes affects mRNA transcription. PLoS ONE. 2007;2:e290. doi: 10.1371/journal.pone.0000290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blandamer M.J., Engberts J.B., Reis J.C. Activity of water in aqueous systems; a frequently neglected property. Chem. Soc. Rev. 2005;34:440–458. doi: 10.1039/b400473f. [DOI] [PubMed] [Google Scholar]
- 24.Rozners E. Determination of nucleic acid hydration using osmotic stress. Curr. Protoc. Nucleic Acid Chem. 2010;Chapter 7 doi: 10.1002/0471142700.nc0714s43. Unit 7 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Timasheff S.N. In disperse solution, “osmotic stress” is a restricted case of preferential interactions. Proc. Natl. Acad. Sci. USA. 1998;95:7363–7367. doi: 10.1073/pnas.95.13.7363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parsegian V.A., Rand R.P., Rau D.C. Osmotic stress, crowding, preferential hydration, and binding: A comparison of perspectives. Proc. Natl. Acad. Sci. USA. 2000;97:3987–3992. doi: 10.1073/pnas.97.8.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y., Zon G., Wilson W.D. Thermodynamics of DNA duplexes with adjacent G.A mismatches. Biochemistry. 1991;30:7566–7572. doi: 10.1021/bi00244a028. [DOI] [PubMed] [Google Scholar]
- 28.Nakano S., Oka H., Sugimoto N. Dynamics and energetics of the base flipping conformation studied with base pair-mimic nucleosides. Biochemistry. 2009;48:11304–11311. doi: 10.1021/bi901496q. [DOI] [PubMed] [Google Scholar]
- 29.Nakano S., Oka H., Sugimoto N. Stacking interaction in the middle and at the end of a DNA helix studied with non-natural nucleotides. Mol. Biosyst. 2010;6:2023–2029. doi: 10.1039/c0mb00002g. [DOI] [PubMed] [Google Scholar]
- 30.Nakano S., Uotani Y., Sugimoto N. DNA base flipping by a base pair-mimic nucleoside. Nucleic Acids Res. 2005;33:7111–7119. doi: 10.1093/nar/gki1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wittung P., Nielsen P.E., Nordén B. DNA-like double helix formed by peptide nucleic acid. Nature. 1994;368:561–563. doi: 10.1038/368561a0. [DOI] [PubMed] [Google Scholar]
- 32.Sugimoto N., Satoh N., Nakano S. Stabilization factors affecting duplex formation of peptide nucleic acid with DNA. Biochemistry. 2001;40:8444–8451. doi: 10.1021/bi010480m. [DOI] [PubMed] [Google Scholar]
- 33.Record M.T., Jr., Zhang W., Anderson C.F. Analysis of effects of salts and uncharged solutes on protein and nucleic acid equilibria and processes: a practical guide to recognizing and interpreting polyelectrolyte effects, Hofmeister effects, and osmotic effects of salts. Adv. Protein Chem. 1998;51:281–353. doi: 10.1016/s0065-3233(08)60655-5. [DOI] [PubMed] [Google Scholar]
- 34.Del Vecchio P., Esposito D., Barone G. The effects of polyols on the thermal stability of calf thymus DNA. Int. J. Biol. Macromol. 1999;24:361–369. doi: 10.1016/s0141-8130(99)00058-6. [DOI] [PubMed] [Google Scholar]
- 35.Nordstrom L.J., Clark C.A., Schwinefus J.J. Effect of ethylene glycol, urea, and N-methylated glycines on DNA thermal stability: the role of DNA base pair composition and hydration. Biochemistry. 2006;45:9604–9614. doi: 10.1021/bi052469i. [DOI] [PubMed] [Google Scholar]
- 36.Lambert D., Draper D.E. Effects of osmolytes on RNA secondary and tertiary structure stabilities and RNA-Mg2+ interactions. J. Mol. Biol. 2007;370:993–1005. doi: 10.1016/j.jmb.2007.03.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koumoto K., Ochiai H., Sugimoto N. Structural effect of synthetic zwitterionic cosolutes on the stability of DNA duplexes. Tetrahedron. 2008;64:168–174. [Google Scholar]
- 38.Engberts J.B.F.N., Blandamer M.J. Reactant–solute encounters in aqueous solutions studied by kinetic methods: hydration cosphere overlap and camouflage effects. J. Phys. Org. Chem. 1998;11:841–846. [Google Scholar]
- 39.Stanley C.B., Rau D.C. Preferential hydration of DNA: the magnitude and distance dependence of alcohol and polyol interactions. Biophys. J. 2006;91:912–920. doi: 10.1529/biophysj.106.086579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.SantaLucia J., Jr. A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl. Acad. Sci. USA. 1998;95:1460–1465. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hong J., Capp M.W., Record M.T., Jr. Preferential interactions of glycine betaine and of urea with DNA: implications for DNA hydration and for effects of these solutes on DNA stability. Biochemistry. 2004;43:14744–14758. doi: 10.1021/bi049096q. [DOI] [PubMed] [Google Scholar]
- 42.Rozners E., Moulder J. Hydration of short DNA, RNA and 2′-OMe oligonucleotides determined by osmotic stressing. Nucleic Acids Res. 2004;32:248–254. doi: 10.1093/nar/gkh175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Senior M.M., Jones R.A., Breslauer K.J. Influence of loop residues on the relative stabilities of DNA hairpin structures. Proc. Natl. Acad. Sci. USA. 1988;85:6242–6246. doi: 10.1073/pnas.85.17.6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Antao V.P., Lai S.Y., Tinoco I., Jr. A thermodynamic study of unusually stable RNA and DNA hairpins. Nucleic Acids Res. 1991;19:5901–5905. doi: 10.1093/nar/19.21.5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Greene K.L., Jones R.L., Wilson W.D. Solution structure of a GA mismatch DNA sequence, d(CCATGAATGG)2, determined by 2D NMR and structural refinement methods. Biochemistry. 1994;33:1053–1062. doi: 10.1021/bi00171a003. [DOI] [PubMed] [Google Scholar]
- 46.Privé G.G., Heinemann U., Dickerson R.E. Helix geometry, hydration, and G.A mismatch in a B-DNA decamer. Science. 1987;238:498–504. doi: 10.1126/science.3310237. [DOI] [PubMed] [Google Scholar]
- 47.Chalikian T.V., Sarvazyan A.P., Breslauer K.J. Influence of base composition, base sequence, and duplex structure on DNA hydration: apparent molar volumes and apparent molar adiabatic compressibilities of synthetic and natural DNA duplexes at 25°C. Biochemistry. 1994;33:2394–2401. doi: 10.1021/bi00175a007. [DOI] [PubMed] [Google Scholar]
- 48.Li F., Pallan P.S., Egli M. Crystal structure, stability and in vitro RNAi activity of oligoribonucleotides containing the ribo-difluorotoluyl nucleotide: insights into substrate requirements by the human RISC Ago2 enzyme. Nucleic Acids Res. 2007;35:6424–6438. doi: 10.1093/nar/gkm664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kolarovic A., Schweizer E., Rozners E. Interplay of structure, hydration and thermal stability in formacetal modified oligonucleotides: RNA may tolerate nonionic modifications better than DNA. J. Am. Chem. Soc. 2009;131:14932–14937. doi: 10.1021/ja904926e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sen A., Nielsen P.E. Unique properties of purine/pyrimidine asymmetric PNA.DNA duplexes: differential stabilization of PNA.DNA duplexes by purines in the PNA strand. Biophys. J. 2006;90:1329–1337. doi: 10.1529/biophysj.105.073213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sen A., Nielsen P.E. On the stability of peptide nucleic acid duplexes in the presence of organic solvents. Nucleic Acids Res. 2007;35:3367–3374. doi: 10.1093/nar/gkm210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rasmussen H., Kastrup J.S., Nielsen P.E. Crystal structure of a peptide nucleic acid (PNA) duplex at 1.7 Å resolution. Nat. Struct. Biol. 1997;4:98–101. doi: 10.1038/nsb0297-98. [DOI] [PubMed] [Google Scholar]
- 53.Menchise V., De Simone G., Pedone C. Insights into peptide nucleic acid (PNA) structural features: the crystal structure of a D-lysine-based chiral PNA-DNA duplex. Proc. Natl. Acad. Sci. USA. 2003;100:12021–12026. doi: 10.1073/pnas.2034746100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Westhof E. Hydration of oligonucleotides in crystals. Int. J. Biol. Macromol. 1987;9:186–192. [Google Scholar]
- 55.Schneider B., Berman H.M. Hydration of the DNA bases is local. Biophys. J. 1995;69:2661–2669. doi: 10.1016/S0006-3495(95)80136-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Narayana N., Weiss M.A. Crystallographic analysis of a sex-specific enhancer element: sequence-dependent DNA structure, hydration, and dynamics. J. Mol. Biol. 2009;385:469–490. doi: 10.1016/j.jmb.2008.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


