Abstract
Yeasts are a diverse seemingly ubiquitous group of eukaryotic microbes, and many are naturally associated with fruits. Humans have harnessed yeasts since the dawn of civilisation to make wine, and thus it is surprising that we know little of the distribution of yeast communities naturally associated with fruits. Previous reports of yeast community diversity have been descriptive only. Here we present, we believe, the first robust test for the geographic delineation of yeast communities. Humans have relatively recently employed Saccharomyces cerevisiae as a model research organism, and have long harnessed its ancient adaption to ferment even in the presence of oxygen. However, as far as we are aware, there has not been a rigorous test for the presence of regional differences in natural S. cerevisiae populations before. We combined these community- and population-level questions and surveyed replicate vineyards and corresponding spontaneous ferments from different regions on New Zealand's (NZ's) North Island and analysed the resulting data with community ecology and population genetic tests. We show that there are distinct regional delineations of yeast communities, but the picture for S. cerevisiae is more complex: there is evidence for region-specific sub-populations but there are also reasonable levels of gene flow among these regions in NZ. We believe this is the first demonstration of regional delineations of yeast populations and communities worldwide.
Keywords: biogeography, community ecology, yeast, population structure, Saccharomyces cerevisiae
Introduction
Characterising the distribution of organisms in space and time, and describing the processes that govern these patterns, is central to an integrated description of the natural world (Cox and Moore, 1993). Although the biogeography of plants and animals is reasonably well described (Gaston, 2000), the biogeography of microbes is not (Martiny et al., 2006). Technical advances mean the task of describing microbial diversity is accelerating (Hugenholtz et al., 1998; Green et al., 2004), and not unsurprisingly the emerging picture is more complicated than simply accepting or rejecting the classic Bass–Becking idea that dispersal is not limiting for microbes and thus everything is everywhere but the environment selects (Whitaker et al., 2003; Finlay and Fenchel, 2004; Green et al., 2004; Martiny et al., 2006; Nemergut et al., 2011). Most studies to date have focused on bacterial communities (Martiny et al., 2006), but some notable work has been conducted on the biogeography of aquatic protists (Finlay and Fenchel, 2004), soil fungi (Green et al., 2004), and cacti- and flower-associated yeasts (Lachance et al., 2000, 2001, 2008). However, there are fewer studies that have examined the biogeography of eukaryotic microbes in other niches.
Since the Age of Enlightenment, wine, like many other subjects of human passion, has been investigated from a scientific perspective, and this research has been intertwined with scientific progress generally (Chambers and Pretorius, 2010). Humans have been making wine since at least the dawn of civilisation (McGovern et al., 2004; Chambers and Pretorius, 2010), but it was only within the last 150 years that Pasteur proved that microbes are the agents responsible for fermentation (Pretorius, 2000). Previous work has shown that a number of different yeast (which here we define as single-celled fungi) species are naturally present on grapes and in the resulting juice (Ribereau-Gayon et al., 2006; Ciani et al., 2010), and this includes surveys that span different regions (Schuller et al., 2005). Although rigorous from a technical perspective, the shortcoming of most of these studies is that they are simply descriptive. We require robust ecological approaches in this field, with appropriate replication and the implementation of statistics to test hypotheses; only recently have a number of studies begun to take this approach (for example, Goddard, 2008; Cadez et al., 2010; Goddard et al., 2010; Cordero-Bueso et al., 2011).
Once in the winery, as a spontaneous (as opposed to artificially inoculated) ferment of grape juice proceeds, the species diversity collapses and Saccharomyces species, which are initially rare, take over and complete the ferment (Goddard, 2008; Ciani et al., 2010). Although the species diversity falls, the diversity of different S. cerevisiae genotypes may remain high (Cubillos et al., 2009; Goddard et al., 2010). Saccharomyces ferment even in the presence of oxygen (the Crabtree effect), which is energetically wasteful, but this complex trait manipulates fruits to create hot, anaerobic, alcoholic environments that are essentially toxic to other microbes; thus Saccharomyces eventually dominate (Thomson et al., 2005; Goddard, 2008). Whole genome analyses suggest this trait evolved around the same time as fruiting plants some 100 million years ago (Wolfe and Shields, 1997; Piskur et al., 2006). As well as harnessing this trait to make wine, bread and beer, humans have more recently employed S. cerevisiae as a classic research model (Dujon, 1996; Burke et al., 2000; Pretorius, 2000; Chambers and Pretorius, 2010). However, the complementary research to understand the ecology and population biology of S. cerevisiae has comparatively lagged behind (Landry et al., 2006; Goddard, 2008; Replansky et al., 2008; Liti et al., 2009; Schacherer et al., 2009; Goddard et al., 2010). As a result of its model status, there has been work describing the diversity and population structure of S. cerevisiae (Fay and Benavides, 2005; Ezov et al., 2006; Legras et al., 2007; Liti et al., 2009; Schacherer et al., 2009; Goddard et al., 2010), and its sibling-species S. paradoxus (Johnson et al., 2004; Koufopanou et al., 2006; Liti et al., 2009; Zhang et al., 2010b), on intercontinental scales, but very little work has been conducted on the population structure of S. cerevisiae associated with fruits and ferments. As well as differences over 3 years, Schuller and Casal (2007) showed significant differences between S. cerevisiae populations isolated from ferments of fruit from three different vineyards <100 km apart within the same region in Portugal. However, we are not aware of any previous studies that have conducted robust tests for regional-specific populations of S. cerevisiae, which by definition requires population genetic analyses of replicate samples from multiple regions. In sum, there has not yet been a rigorous test of hypotheses regarding the geographic delineations of either fruit-associated yeast communities or wine ferment yeast populations.
New Zealand (NZ) has only been inhabited by humans in the last ∼700 years (King, 2003), and has only had commercially planted vines in the last ∼100 years (Johnson, 1998). We have recently provided evidence that NZ harbours a genetically distinct population of S. cerevisiae using population genetic inference (Goddard et al., 2010), and now we move on to test whether there are regional-specific populations of microbes within distinct wine-growing areas in NZ. Specifically we test two hypotheses: (1) whether there are different communities of yeasts associated with vineyards in different areas; and, (2) whether there are different populations of S. cerevisiae conducting spontaneous ferments in these same areas. We have tackled these questions by taking multiple samples of replicate Chardonnay and Syrah vineyards, and the spontaneous ferments deriving from them, from three well known and distinct wine-producing areas on NZ's North Island and tested the two main hypotheses outlined above using community ecology and population genetic analyses.
Materials and methods
Vineyard samples
We sampled 24 distinct vineyards in triplicate, for a total of 72 samples, across three wine-producing regions of NZ's North Island on an approximate N—S cline spanning ∼350 km: West Auckland, Waiheke Island and Hawke's Bay. While Waiheke Island is only ∼40 km away from the West Auckland region, it is an island in the Hauraki Gulf with a distinct microclimate. The mean distance between vineyards within regions is ∼5 km. Five to six Chardonnay vineyards were sampled within each region and these comprised the main experimental regime to test our hypotheses. We also sampled seven Syrah vineyards across Waiheke Island and Hawke's Bay (there is not enough Syrah planted in West Auckland to have included this region). All samples were taken approximately 1 week before harvest around mid-March in 2010: three whole bunches of fruit that appeared rot-free were selected randomly from each of the 24 vineyards and removed aseptically into a sterile plastic bag. The samples were transported back to the laboratory on ice, where the fruit was crushed in situ and the contents of the bag mixed thoroughly, and the juice was then immediately recovered into a sterile beaker. Serial dilutions were plated onto yeast extract, peptone, glucose agar (1% yeast extract, 2% peptone and 2% glucose) with chloramphenicol (200 μg ml–1) to prevent bacterial growth and incubated at room temperature for 2–3 days to allow single cells to form colonies; 96 of these colonies were then selected randomly from each sample and stored at −80 °C.
Ferment samples
Commercial spontaneous ferments of fruit deriving from the same vineyards as we initially targeted were also sampled around three quarters of the way through ferment (when ∼75% of sugar had been consumed). In all, 50 ml samples were transported to the laboratory on ice where they were serially diluted onto Yeast Extract, Peptone, Glucose with chloramphenicol. In total, 96 colonies were again recovered randomly from the resulting plates.
Molecular methods
In total, 2688 colonies were collected from both vineyard and ferment samples. To discriminate between these, the DNA from each purified colony was extracted using the Chelex method (see Serjeant et al., 2008). The genomic internal transcribed spacer (ITS) region was amplified and subjected to restriction fragment length polymorphism (RFLP) analyses, which is a good method to differentiate between species (Esteve-Zarzoso et al., 1999), and we collated the ITS RFLP patterns into cohorts (see Supplementary Material). To assess the confidence between, and identity of, the different cohorts, we determined the sequence of the diagnostic divergent domains 1 and 2 of the 26S nuclear ribosomal gene (D1/D2 26S) for one to three examples from each cohort (some cohorts only had a single representative; see Supplementary Material). The D1/D2 26S region is commonly used to discriminate between yeast species, and empirical analyses suggests that isolates with <98% homology in this region are likely different species (Kurtzman and Robnett, 2003). We reconstructed a bootstrap consensus tree to examine 26S sequence correlation with ITS-RFLP cohorts using Geneious (Drummond et al., 2011). We also compared the 26S sequences with deposits for typical examples for known yeast species in the National Centre for Biotechnology Information database, allowing us to ascribe species names with some degree of confidence. The allele lengths of isolates identified as S. cerevisiae from ferment samples were analysed at 10 unlinked variable microsatellite repeat loci as described by Richards et al. (2009). The particular combinations of alleles at these loci allow ∼109 strains to be differentiated, but more importantly the relatedness between individuals and specific sub-populations to be inferred (Richards et al., 2009; Goddard et al., 2010).
Data analyses
Differences in community composition between regions were tested with an analysis of similarity (ANOSIM) on square root transformed species incidences with PAST, which is freely available (Hammer et al., 2001). An ANOSIM test makes no assumption about the normality of data and this multivariate test is classically used in community ecology and reports the probability of observing the differences in community composition between regions by chance using permutations of a rank Bray–Curtis similarity matrix to create null distributions (Clarke, 1993). The observed rank abundance difference (R) between regions is reported, and this value ranges from −1 to +1: values of R around zero indicate that there are no differences between regions and values above zero indicate that communities differ between regions. We generated null distributions that recalculated R under a framework that assumed no difference between regions by randomising the labels associated with samples across the entire data set a total of one million independent times.
Routine population genetic analyses of the S. cerevisiae genotypes, including elucidating matching genotypes and the calculation of genetic distance and various F statistics, were conducted using ‘Genetic Analyses in Excel' (Peakall and Smouse, 2006). Outcrossing rates were estimated using Mathematica 7 (code available on request). Allelic richness was estimated using HP-rare (Kalinowski, 2005). Tests for population structure, and assignments to likely sub-populations, employed Bayesian inference as implemented in InStruct (Gao et al., 2007) using a model that incorporated admixture. Five chains of two million Markov Chain Monte Carlo iterations with a burn-in of 100 000 were run for K=1 to 20.
Results
We tested the null hypothesis of no difference in vine- and wine-associated yeasts in different regions of NZ with two approaches: (1) the communities associated with vineyards; and, (2) populations of S. cerevisiae conducting spontaneous ferments.
Community comparisons—vineyard samples
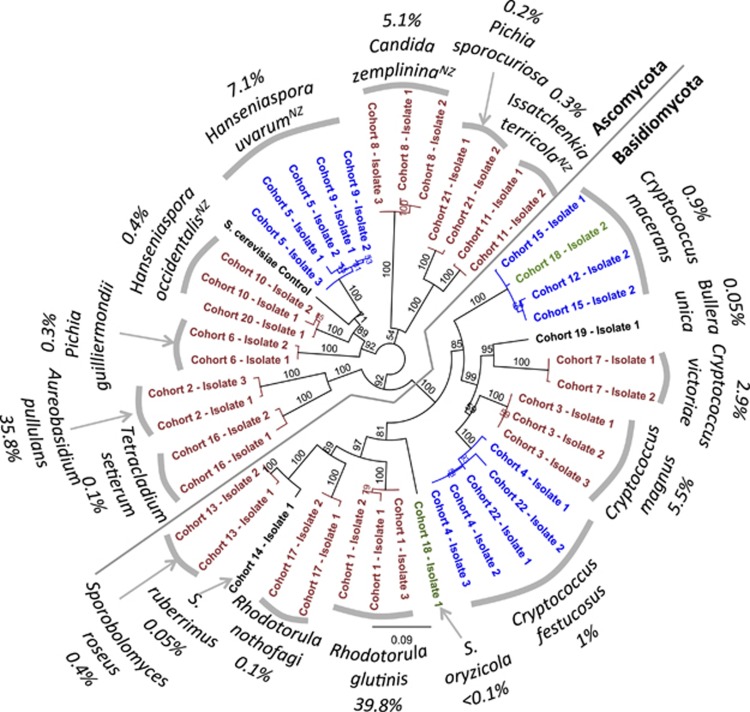
In total, 22 distinct ITS-RFLP cohorts were delineated among the 2304 isolates deriving from 24 vineyards from West Auckland, Waiheke Island and Hawke's Bay (see Figure 1, and Supplementary Material). The 26S sequence data allowed species assignment based on homology, and the assignment of random examples within ITS-RFLP cohorts were all to the same species with 100% bootstrap support apart from one cohort, which contained matches to both Sporobolomyces oryzicola and Cryptococcus macerans (#18; see Figure 1). Isolates with 26S sequences matching to Cryptococcus festucosus and C. macerans derived from cohorts with different RFLP patterns (#4 and #22, and #12, #15 and #18, respectively; see Figure 1) indicating reasonable levels of within species diversity at the ITS region: the respective mean identities at the 26S region of 96% and 90% for these two groupings is below the 98% commonly considered the homology boundary between yeast species (Kurtzman and Robnett, 2003). Thus, these cohorts were considered separately in further analyses. Two other cohorts converged on the same species (#5 and #9 to Hanseniaspora uvarum, and #10 and 20 to H. occidentalis), but as the average 26S homology for these isolates was >99%, these were comfortably considered the same species and thus the cohorts were combined in further analyses. The 20 different final groupings, and their homology to reference sequences at the D1/D2 26S ribosomal DNA region, are shown in Figure 1 and have the National Centre for Biotechnology Information accession numbers JN377440–JN377484 (and see Supplementary Material).
Figure 1.
Correlation between ITS-RFLP patterns and D1/D2 26s RNA sequence data. Neighbour-joining consensus tree of 1000 bootstrap replicates reconstructed from an alignment of the D1/D2 26s RNA region for 45 isolates associated with Chardonnay and Syrah fruit (see Supplementary Material). Random isolates from each of the 22 ITS-RFLP cohorts were selected. Isolates from the same cohort whose 26S sequences match to the same type species and are placed together with 100% support are shown in red. Isolates from different ITS-RFLP cohorts whose 26S sequences match to the same species in GenBank and are placed together with 100% support are shown in blue. The two examples from cohort 18 that yielded different 26s RNA sequences are in green. The >98% homology of our query sequences to type-strain deposits in GenBank allow species names to be ascribed to well-supported groups and the abundance of each group among the 2688 isolates is shown. Those species previously reported as associated with juice/wines in NZ are denoted with a superscript.
Approximately half of the species identified belong to the Basidiomycota, and the remainder to the Ascomycota. Rhodotorula glutinis and Aureobasidium pullulans combined made up 75.6% of isolates, so are by far the most common species. Following these two species, there is a tail of rarer species, as is common in ecological sampling (Pedros-Alio, 2007). The most abundant budding yeasts are Hanseniaspora uvarum (7.1%) followed by Candida zemplinina (5.1%)—these species are commonly reported associated with wine grapes and ferments from other winemaking countries (Cadez et al., 2010; Ciani et al., 2010). Only four of the species found here have previously been reported as associated with grape juice in NZ (Goddard, 2008; Zhang et al., 2010a; see Figure 1).
Our preliminary analyses showed that the communities associated with Chardonnay and Syrah differed significantly (R=0.27; P=0.013; ANOSIM) regardless of region. The subject of test here although is whether communities differ between regions, and the primary and balanced data set gathered to test this derived from Chardonnay: Table 1 shows the regional distributions. West Auckland and Hawke's Bay both have Rhodotorula glutinis as the dominant species, while Aureobasidium pullulans is dominant on Waiheke Island. The number of species also differs between regions with West Auckland containing the lowest species diversity and Waiheke Island the greatest. The Shannon and Simpson indices measure diversity and ‘evenness' and these metrics suggest that the West Auckland community is the most skewed toward a dominant species. ANOSIMs report positive R-values for the three possible pair-wise region comparisons, suggesting that each region indeed harbours different communities. Comparison of the observed R-values to null distributions show the communities associated with Chardonnay differ significantly between regions with Waiheke Island having the most distinct community. The West Auckland and Hawke's Bay communities differed moderately (R=0.213; P=0.017). Both comparisons of West Auckland and Hawke's Bay with Waiheke Island produced greater R-values, indicating the communities in these areas differed greatly (R=0.66 and 0.71, respectively; both P=0.002).
Table 1. Species identities, based on D1/D2 26S region sequence homology to deposits described as ‘type strains' in NCBI, and abundances based on ITS RFLP, of isolates from ripe Chardonnay fruit by region.
| Species | West Auckland | Hawkes Bay | Waiheke Island |
|---|---|---|---|
| Rhodotorula glutinis | 67.0% | 61.7% | 13.8% |
| Aureobasidium pullulans | 26.2% | 22.1% | 64.2% |
| Hanseniaspora uvarum | 0% | 5.7% | 17% |
| Cryptococcus magnus | 6.6% | 7.0% | 0.7% |
| Candida zemplinina | 0% | 0.9% | 1.8% |
| Cryptococcus festucosus #1 | 0% | 2.7% | 0% |
| Cryptococcus macerans #1 | 0% | 0% | 1.1% |
| Pichia sporocuriosa | 0% | 0% | 0.9% |
| Cryptococcus festucosus #2 | 0.2% | 0% | 0.5% |
| Number of species | 4 | 6 | 8 |
| Number of individuals | 564 | 561 | 441 |
| Shannon index | 0.8092 | 1.119 | 1.084 |
| Simpson index | 0.4776 | 0.5619 | 0.5395 |
Abbreviations: ITS, internal transcribed spacer; RFLP, restriction fragment length polymorphism.
S. cerevisiae population structure—ferment samples
We ascertained the microsatellite profiles of 353 individual S. cerevisiae isolates from 16 Chardonnay and five Syrah ferments across the three regions. A total of 274 different genotypes were revealed, indicating a highly diverse population of S. cerevisiae. Of the 216 alleles we identified, 125 were novel compared with a previous NZ population examined from Auckland ferment and environmental samples (Goddard et al., 2010); however, all the genotypes discovered here were distinct. We compared these genotypes with our database of 79 commercial wine strains commonly used in NZ (Richards et al., 2009), and only one of the 82 isolates deriving from the Hawke's Bay Chardonnay ferments matched. Six genotypes differed at just one allele from a commercial strain, but the remaining genotypes differed at many alleles; thus, these data further support the concept that a diverse natural population of S. cerevisiae resides in NZ (Goddard et al., 2010). A total of 303 different genotypes were used in further analyses in order to accurately reflect genotype presence–absence across ferments (some genotypes were present in more than one ferment); only a single representative of each genotype from each ferment was included in analyses to remove the effect of within-ferment clonal expansion of genotypes.
When populations are broken down by region and variety, the greatest pairwise Fst is between West Auckland Chardonnay and Waiheke Chardonnay (0.12), and the least between Hawke's Bay Chardonnay and Syrah (0.02); the pairwise Fst between genotypes when simply grouped by variety (Chardonnay vs Syrah) is only 0.025 indicating very little signal for population delineation by variety, regardless of region. However, the mean pairwise Fst is more than doubled to 0.055 when genotypes are instead partitioned by region of isolation, indicating moderate genetic differentiation. Together, this suggests geographic origin accounts for a greater portion of signal for population differentiation than variety in these data (Hartl and Clark, 1997). When the data are partitioned by region, then the largest pairwise difference is between Waiheke Island and West Auckland populations (Fst=0.084), which are only ∼40 km apart, followed by West Auckland—Hawke's Bay (Fst=0.056) and Waiheke Island—Hawke's Bay populations (Fst=0.025), which are both ∼350 km apart, suggesting isolation by distance per se is not a major descriptor of these data. Although some of the genotypes were found in multiple ferments, these were mostly from ferments within the same region and ∼94% of genotypes are region specific (see Table 2).
Table 2. Population genetic summary for spontaneous NZ Chardonnay and Syrah ferment S. cerevisiae genotypes from different regions.
| Region (n) | Mean allelic richnessa | Fis b | Outcrossing ratec | Region-specific genotypes (%)d |
|---|---|---|---|---|
| West Auckland (82) | 7±1.5 | 0.73±0.19 | 0.12–0.16–0.20 | 100 |
| Waiheke Island (96) | 7.6±2.6 | 0.61±0.20 | 0.08–0.11–0.14 | 90 |
| Hawkes Bay (124) | 9.9±3.3 | 0.67±0.12 | 0.12–0.15–0.19 | 92 |
Abbreviations: n, the number of genotypes; NZ, New Zealand.
Mean allelic richness was calculated for each locus independently by rarefaction using HP-rare and the mean and s.d. across all loci are reported.
The mean inbreeding coefficients across all ten loci (±s.d.); a value of 1 indicates a completely inbred population.
Average proportion of the mating of spores from independent tetrads per meiotic generation surrounded by 2 log likelihood support limits, which are equivalent to the 95% confidence intervals, calculated following Johnson et al. (2004).
Percentage of genotypes found in that region only.
All 10 loci are significantly out of Hardy–Weinberg equilibrium (P<0.001), and this correlates with previous reports from natural S. cerevisiae populations from South America, Europe and NZ (Schuller et al., 2005; Cubillos et al., 2009; Goddard et al., 2010), and is expected given the life history of S. cerevisiae predisposes it to inbreed (Knop, 2006). However, that these populations have inbreeding coefficients (Fis) well below 1 (mean 0.67, Table 2) indicates that they are sexually outcrossed (that is, engage in mating between spores from independent parents) to some extent. The inbreeding system that S. cerevisiae displays means it is not appropriate to use Wright's inbreeding coefficient as classically calculated to estimate the rates of inbreeding vs outcrossing: this must be modified as matings between the four related spores from the same tetrad lead to a reduction in heterozygosity of one-third per generation, and not one-half as is the case for fusion of gametes from independent meiosis (Falconer and Mackay, 1996; Goddard et al., 2001). Following Johnson et al. (2004), we estimated the most likely outcrossing rate with the more appropriate model (Table 2), and this averages 14% outcrossed matings per meiotic generation among all the populations, and strongly suggests that a reasonable extent of genetic mixing is occurring within this population.
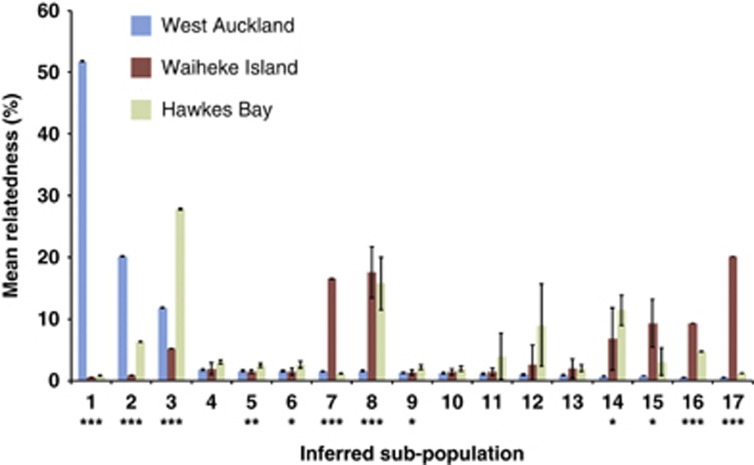
The fact that most genotypes were recovered from multiple different ferments within but not between regions (only 10 isolates overlapped between Waiheke and Hawke's Bay) lends support to the idea that there are region-specific sub-populations, but it does not prove it. For example, in outcrossed metazoans populations each individual has a unique genotype (barring identical twins), and thus each genotype is by definition unique to an area: this does not prove that there are region-specific populations. It is more appropriate to test hypotheses concerning the degree of relatedness of individuals within and between regions. If there were no average difference in relatedness between S. cerevisiae among different regions, then this population would appear homogenous with no sub-structure: genotypes from different regions would be equally related. However, if different regions contain distinct sub-populations of more closely related individuals then the population would appear structured. The usual methods to infer population structure are sensitive to inbreeding (Falush et al., 2003; Huelsenbeck and Andolfatto, 2007) and thus we employed a Bayesian approach, which accounts for both admixture (we have shown this population is outcrossed to some extent) and S. cerevisiae's tendency for inbreeding, to infer whether population structure exists, and then estimate the relatedness of genotypes among different sub-populations (Gao et al., 2007). Importantly, this approach does not use a priori information on the origin of genotypes to detect structure. These analyses unequivocally allow us to reject the null hypothesis of a homogeneous population (P<0.0001): the presence of 17 sub-populations, each containing closely related individuals, is inferred. Multiple Markov Chain Monte Carlo chains (5 runs) converged on identical inferences for the number of sub-populations.
As the optimal inferred number of sub-populations is 17, this means that population sub-division is not as simple as a direct correlation with region (or variety), where only 2–3 sub-populations would be inferred; thus the pattern of population sub-structure is more complicated. There might be any number of reasons why these genotypes are structured into sub-populations other than it being driven by geographic origin, and so having established that population structure exists, the next question is whether any of these sub-populations correlate with geographic location or not. If region-specific populations existed, it would mean that genotypes from the same area would be more closely related to one another than to genotypes from other regions. Is there any evidence for this? The null expectation, if geographic origin has no bearing on population sub-structure, is that isolates from different regions would be equally related within the various sub-populations. We analysed relatedness (proportion of shared ancestry) between genotypes within each sub-population from the different regions and analysis of variances (on arc-sined transformed ancestry proportions) show that while some sub-populations contain genotypes that are equally related between regions, relatedness among other sub-populations significantly differs between regions. The probability of isolates from each region being equally related, and their mean relatedness among the 17 inferred sub-populations are shown in Figure 2. It is clear that the isolates are not assigned to sub-populations randomly with respect to region. The between run variance for population assignment and relatedness in each subpopulation for each genotype was generally very small, with the average s.d. for relatedness being just ∼1% a few sub-populations have a larger variance, suggesting a less certain relatedness for these genotypes. There are clear examples of region-specific sub-populations. For example, 55% of the West Auckland genotypes are more closely related to one another than to any genotypes from other regions (P<0.001 by an honestly significant difference test) and these are all classified into sub-population 1. Similarly sub-populations 7 and 17 are made up of closely related genotypes that all come from Waiheke and are more closely related to one another than genotypes from other regions (P<0.001 by a honestly significant difference test). The case is less clear-cut for Hawke's Bay genotypes: isolates that have significantly greater relatedness within sub-population 3 all come from the Hawke's Bay, but there is also a reasonable number of isolates from the other regions in this group, but these have lower relatedness. The interpretation of these data is that groups of highly related genotypes exist, and some of these are strongly described by the region of isolation, but other groups of closely related genotypes comprise strains from different regions. Sixty-three percent of genotypes have at least 90% of the their ancestry inferred to be in one sub-population only, and thus approximately one-third of genotypes have ancestry among multiple sub-populations and accordingly many of these genotypes are hybrids (chimeras) with parents (or grandparents and so on) from different sub-populations. This inference correlates with the earlier suggestion of reasonable levels of outcrossed matings and thus genetic mixing.
Figure 2.
The average relatedness of 303 S. cerevisiae genotypes isolated from 21 spontaneous ferments among the 17 inferred sub-populations, partitioned by region. The proportion of ancestry of each genotype was inferred from five Markov Chain Monte Carlo chains with K=17 using InStruct; the grand mean for ancestry of genotypes for each region in each sub-population was calculated from the average of these five runs and is plotted, along with the s.d. Blue bars: genotypes from West Auckland; red bars: Waiheke Island; green bars: Hawkes Bay. The sum of any one isolate's ancestry across all sub-populations is one. Deviation in relatedness between regions within each sub-population was tested with a one-way analysis of variance, and those sub-populations with a significant difference in relatedness between regions are indicated (*P<0.01; **P<0.001; ***P<0.0001).
Discussion
To our knowledge, there have been no previous reports of the diversity of microbes directly associated with fruit in NZ, and the list of genera, including those that are dominant, appears similar to reports from vineyard microbial communities in Europe (Cadez et al., 2010) and Australia (Prakitchaiwattana et al., 2004) with the exceptions of Bullera (interestingly first isolated in NZ from Melicytus macrophyllus (Hamamoto and Nakase, 1996)) and Tetracladium. All the vineyards sampled are commercially managed, which includes the application of anti-fungal sprays, which does not seem to have a significant effect on yeast communities that contribute to ferments at least (Cadez et al., 2010). Close to 300 different genotypes of S. cerevisiae were recovered from the spontaneous ferments; while our sampling effort was large, it seems that we have only scratched the surface of the diversity of S. cerevisiae in NZ, and this is in line with a large S. cerevisiae diversity recovered from two spontaneous Chardonnay ferments in previous years (Goddard et al., 2010).
This study's principal aim was to test for geographic differences in microbial communities and populations. The community composition tests we conducted allow us to categorically reject the null hypothesis of no difference in microbial communities on ripe Chardonnay fruit between West Auckland, Waiheke Island and Hawke's Bay. We are not aware of any previous study to objectively show regional community differences of microbes associated with fruit. We are aware of just one previous study attempting to test for regional geographic delineations of S. cerevisiae populations: this found no clear correlation between 64 S. cerevisiae from South America and Europe based on the clustering of RAPD profiles, but this conclusion was based on observation only and was not objectively tested (Martinez et al., 2007). Population genetic analyses here show that different regions of NZ contain different genetically distinct and highly related sub-populations of S. cerevisiae driving spontaneous ferments of Chardonnay and Syrah. However, some genotypes are equally related to genotypes isolated from alternate regions, and this strongly suggests the movement of strains. There are also many hybrid genotypes with ancestry in more than one sub-population. This pattern is similar to the global population structure of S. cerevisiae (Liti et al., 2009), as well as other populations from NZ (Goddard et al., 2010). A helpful analogy to interpret this population pattern might be with a human example: within any one region, there are individuals, which belong to families that have resided in that region for a considerable period of time and thus many individuals in that area will be highly related (share ancestors); however, other individuals will have migrated into to the region in the recent past, and share little or no ancestry with, and thus have low relatedness to, the indigenous residents. New migrants may mate with the local population to produce offspring that will thus have relatedness spread among different ancestral groups from different regions. Together, these data and tests allow us to accept the general alternate hypotheses that there are region-specific communities and populations of microbes associated with vines and wines, at least on the North Island of NZ: we believe this is the first demonstration of regional differences worldwide.
There are very few reported observations of changes in vine and wine communities and populations over time (Beltran et al., 2002) and our data clearly do not allow us to make any statements about such changes. One obvious negative manifestation of microbes associated with fruit is the disease or rot some cause. If grapes are infected by organisms such as Botrytis cinerea (grey and noble rot) or Erysiphe necator (powdery mildew), then the damage caused will allow other microbial populations (including yeasts) to expand. Thus, we were careful to randomly select fruit that was visually rot-free to minimise this influence on our inference of differences in microbial communities between regions. Healthy fruit was selected at random from replicate vineyards within each region, and all samples processed identically: with the implementation of the ANOSIM we feel this provides a comparable and fair estimate of the differences in communities associated with vines among these regions of NZ. One other potential criticism of this work is that it is culture based. There are no previous reports of direct nucleic acid based estimates for microbial communities associated with vines and wines (but we are pursuing this approach). Although we may have missed some proportion of these communities, because this would have been a systematic bias (the same isolation procedure was used throughout), the replicate estimates from regions allow us to make statistically meaningful comparisons of communities and thus our conclusions are valid.
The population genetic data suggest that there is a degree of migration of S. cerevisiae between regions within NZ. This may occur by any number of vectors, but humans, insects and birds seem likely candidates, and the two former have been previously implicated (Goddard et al., 2010). The observation that a diverse natural population of S. cerevisiae contributed to these spontaneous ferments is another piece of evidence again showing that spontaneous ferments are not driven by ‘escaped' commercial isolates (Blanco et al., 2011), even in large commercial wineries that also run parallel inoculated ferments (as was the case here), but driven by a diversity of natural isolates.
We did not have enough time or resources to go on to test ideas as to why there are differences between regions, but we will look to test some of the main ideas around this in the future. One possibility suggests that dispersal is limiting between these regions, but this seems not to be supported by our data, at least for S. cerevisiae: maybe the Bass–Becking idea is correct, and it is local adaptation that determines the differences? Climate is one of the more obvious factors that might explain why certain species are more prevalent in different areas; other factors include the soil and substrate nutrient composition and availability. It is likely that a combination of factors will be responsible, but it will be of interest to ascertain the relative contributions of each.
Finally, it is widely accepted that the particular climatic and geographic/soil conditions within an area interact with different grape varieties differently and serve to make distinctive wines, and this underlies the concept of ‘terroir' or a wine's sense of place (Goode, 2006). Previous data suggest that many of the S. cerevisiae in spontaneous NZ ferments derive from the local environment (Goddard et al., 2010), but it is also possible that some populations may reside ‘in the cellar'. Either way, we provide some evidence for regional delineations of natural S. cerevisiae populations and the use of region-specific yeasts to make spontaneously fermented wine is precisely in keeping with the terroir concept as these wines more faithfully reflect their sense of place. There is also strong scientific evidence to show that different yeast species and strains act on grape precursors differently to produce different types and amounts of metabolites (Romano et al., 2003; Swiegers and Pretorius, 2005, 2007; Howell et al., 2006; Anfang et al., 2009) and that these may have significant effects on the flavour and aroma of the resulting wine (Lambrechts and Pretorius, 2000; Cordente et al., 2007; Ciani et al., 2010; King et al., 2010). Thus, the logical extension of this is that if different regions harbour different communities and populations, as we have shown, that these may, in part, contribute to the distinctiveness of wines deriving from that area: that is, there is a microbial aspect to terroir. However, the extent to which different microbes contribute to the uniqueness of wine from different regions is not yet proven and the stability of these communities and populations over time remains to be tested. Finally, it is well known that fungi and plants have intimate associations (rhizomes being the obvious example). Another aspect of a microbial influence on terroir worth considering is the degree to which microbes may manipulate vine and fruit development. It is possible that the different species in different areas modulate vine development differently such that different types or amounts of compounds, including precursors that yeasts act on during the ferment, are present and thus translate into altered sensory properties of wine. This also remains to be tested.
Acknowledgments
We thank Michael Brajkovich MW at Kumeu River Wines, Pernod-Ricard NZ (especially Tony Robb), Waiheke Winegrowers (Obsidian, Mudbrick, Kennedy Point), Trinity Hill, and Mission Estate for their support and involvement with this project. We also thank the anonymous reviewers for comments on drafts of this paper. This work was directly supported by a Faculty Research grant to MG, but is part of a larger research program supported by New Zealand Winegrowers and the Ministry for Science and Innovation.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Anfang N, Brajkovich M, Goddard MR. Co-fermentation with Pichia kluyveri increases varietal thiol concentrations in Sauvignon Blanc. Aust J Grape Wine R. 2009;15:1–8. [Google Scholar]
- Beltran G, Torija MJ, Novo M, Ferrer N, Poblet M, Guillamon JM, et al. Analysis of yeast populations during alcoholic fermentation: a six year follow-up study. Syst Appl Microbiol. 2002;25:287–293. doi: 10.1078/0723-2020-00097. [DOI] [PubMed] [Google Scholar]
- Blanco P, Orriols I, Losada A. Survival of commercial yeasts in the winery environment and their prevalence during spontaneous fermentations. J Ind Microbiol Biot. 2011;38:235–239. doi: 10.1007/s10295-010-0818-2. [DOI] [PubMed] [Google Scholar]
- Burke D, Dawson D, Stearns T. Methods in Yeast Genetics. A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor Press: Cold Spring Harbor; 2000. [Google Scholar]
- Cadez N, Zupan J, Raspor P. The effect of fungicides on yeast communities associated with grape berries. FEMS Yeast Res. 2010;10:619–630. doi: 10.1111/j.1567-1364.2010.00635.x. [DOI] [PubMed] [Google Scholar]
- Chambers PJ, Pretorius IS. Fermenting knowledge: the history of winemaking, science and yeast research. EMBO Rep. 2010;11:914–920. doi: 10.1038/embor.2010.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciani M, Comitini F, Mannazzu I, Domizio P. Controlled mixed culture fermentation: a new perspective on the use of non-Saccharomyces yeasts in winemaking. FEMS Yeast Res. 2010;10:123–133. doi: 10.1111/j.1567-1364.2009.00579.x. [DOI] [PubMed] [Google Scholar]
- Clarke KR. Non-parametric multivariate analyses of changes in community structure. Aust J Ecol. 1993;18:117–143. [Google Scholar]
- Cordente AG, Swiegers JH, Hegardt FG, Pretorius IS. Modulating aroma compounds during wine fermentation by manipulating carnitine acetyltransferases in Saccharomyces cerevisiae. FEMS Microbiol Lett. 2007;267:159–166. doi: 10.1111/j.1574-6968.2006.00548.x. [DOI] [PubMed] [Google Scholar]
- Cordero-Bueso G, Arroyo T, Serrano A, Tello J, Aporta I, Velez MD, et al. Influence of the farming system and vine variety on yeast communities associated with grape berries. Intl J Food Micro. 2011;145:132–139. doi: 10.1016/j.ijfoodmicro.2010.11.040. [DOI] [PubMed] [Google Scholar]
- Cox CB, Moore PD. Biogeography, an Ecological and Evolutionary Approach. Blackwell: London; 1993. [Google Scholar]
- Cubillos FA, Vasquez C, Faugeron S, Ganga A, Martinez C. Self-fertilization is the main sexual reproduction mechanism in native wine yeast populations. FEMS Microbiol Ecol. 2009;67:162–170. doi: 10.1111/j.1574-6941.2008.00600.x. [DOI] [PubMed] [Google Scholar]
- Drummond A, Ashton B, Cheung M, Heled J, Kearse M, Moir R, et al. 2011Geneious v5.4Available from http://www.geneious.com/ .
- Dujon B. The yeast genome project: what did we learn. Trends Genet. 1996;12:263–270. doi: 10.1016/0168-9525(96)10027-5. [DOI] [PubMed] [Google Scholar]
- Esteve-Zarzoso B, Belloch C, Uruburu F, Querol A. Identification of yeasts by RFLP analysis of the 5.8S rRNA gene and the two ribosomal internal transcribed spacers. Int J Syst Bacteriol. 1999;49:329–337. doi: 10.1099/00207713-49-1-329. [DOI] [PubMed] [Google Scholar]
- Ezov TK, Boger-Nadjar E, Frenkel Z, Katsperovski I, Kemeny S, Nevo E, et al. Molecular-genetic biodiversity in a natural population of the yeast Saccharomyces cerevisiae from ‘Evolution canyon': microsatellite polymorphism, ploidy and controversial sexual status. Genetics. 2006;174:1455–1468. doi: 10.1534/genetics.106.062745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconer DS, Mackay TFC.1996Introduction to Quantitative Genetics4th edn.Pearson, Prentice Hall: New Jersey [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay JC, Benavides JA. Evidence for domesticated and wild populations of Sacchoromyces cerevisiae. PLOS Genetics. 2005;1:66–71. doi: 10.1371/journal.pgen.0010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay BJ, Fenchel T. Cosmopolitan metapopulations of free-living microbial eukaryotes. Protist. 2004;155:237–244. doi: 10.1078/143446104774199619. [DOI] [PubMed] [Google Scholar]
- Gao H, Williamson S, Bustamante CD. A Markov chain Monte Carlo approach for joint inference of population structure and inbreeding rates from multilocus genotype data. Genetics. 2007;176:1635–1651. doi: 10.1534/genetics.107.072371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaston KJ. Global patterns in biodiversity. Nature. 2000;405:220–227. doi: 10.1038/35012228. [DOI] [PubMed] [Google Scholar]
- Goddard MR. Quantifying the complexities of Saccharomyces cerevisiae′s ecosystem engineering via fermentation. Ecology. 2008;89:2077–2082. doi: 10.1890/07-2060.1. [DOI] [PubMed] [Google Scholar]
- Goddard MR, Anfang N, Tang R, Gardner RC, Jun C. A distinct population of Saccharomyces cerevisiae in New Zealand: evidence for local dispersal by insects and human-aided global dispersal in oak barrels. Environ Microbiol. 2010;12:63–73. doi: 10.1111/j.1462-2920.2009.02035.x. [DOI] [PubMed] [Google Scholar]
- Goddard MR, Greig D, Burt A. Outcrossed sex allows a selfish gene to invade yeast populations. Proc R Soc Lond B-Biol Sci. 2001;268:2537–2542. doi: 10.1098/rspb.2001.1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode J. The Science of Wine. University of California Press: Berkeley; 2006. [Google Scholar]
- Green JL, Holmes AJ, Westoby M, Oliver I, Briscoe D, Dangerfield M, et al. Spatial scaling of microbial eukaryote diversity. Nature. 2004;432:747–750. doi: 10.1038/nature03034. [DOI] [PubMed] [Google Scholar]
- Hamamoto M, Nakase T. Ballistosporous yeasts found on the surface of plant materials collected in New Zealand—the genera Bensingtonia and Bullera with descriptions of five new species. Antonie Van Leeuwenhoek Int J Gen Mol Microbiol. 1996;69:279–291. doi: 10.1007/BF00399617. [DOI] [PubMed] [Google Scholar]
- Hammer O, Harper DAT, Ryan PD.2001PAST: paleontological statistics software package for education and data analysis Palaeontologia Electronica 4art. 49online only http://palaeo-electronica.org/2001_1/past/issue1_01.htm ). [Google Scholar]
- Hartl DL, Clark AG.1997Principles of Population Genetics3rd edn.Sinauer: Sunderland [Google Scholar]
- Howell KS, Cozzolino D, Bartowsky EJ, Fleet GH, Henschke PA. Metabolic profiling as a tool for revealing Saccharomyces interactions during wine fermentation. FEMS Yeast Res. 2006;6:91–101. doi: 10.1111/j.1567-1364.2005.00010.x. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Andolfatto P. Inference of population structure under a Dirichlet process model. Genetics. 2007;175:1787–1802. doi: 10.1534/genetics.106.061317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugenholtz P, Goebel BM, Pace NR. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol. 1998;180:4765–4774. doi: 10.1128/jb.180.18.4765-4774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson H. The Story of Wine. Simon and Schuster: London; 1998. [Google Scholar]
- Johnson LJ, Koufopanou V, Goddard MR, Hetherington R, Schafer SM, Burt A. Population genetics of the wild yeast Saccharomyces paradoxus. Genetics. 2004;166:43–52. doi: 10.1534/genetics.166.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinowski ST. HP-RARE 1.0: a computer program for performing rarefaction on measures of allelic richness. Mol Ecol Notes. 2005;5:187–189. [Google Scholar]
- King ES, Kievit RL, Curtin C, Swiegers JH, Pretorius IS, Bastian SEP, et al. The effect of multiple yeasts co-inoculations on Sauvignon Blanc wine aroma composition, sensory properties and consumer preference. Food Chem. 2010;122:618–626. [Google Scholar]
- King M. History of New Zealand. Penguin: Auckland; 2003. [Google Scholar]
- Knop M. Evolution of the hemiascomycete yeasts: on life styles and the importance of inbreeding. Bioessays. 2006;28:696–708. doi: 10.1002/bies.20435. [DOI] [PubMed] [Google Scholar]
- Koufopanou V, Hughes J, Bell G, Burt A. The spatial scale of genetic differentiation in a model organism: the wild yeast Saccharomyces paradoxus. Philos T R Soc B. 2006;361:1941–1946. doi: 10.1098/rstb.2006.1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtzman CP, Robnett CJ. Phylogenetic relationships among yeasts of the ‘Saccharomyces complex' determined from multigene sequence analyses. FEMS Yeast Res. 2003;3:417–432. doi: 10.1016/S1567-1356(03)00012-6. [DOI] [PubMed] [Google Scholar]
- Lachance MA, Lawrie D, Dobson J, Piggott J. Biogeography and population structure of the Neotropical endemic yeast species Metschnikowia lochheadii. Antonie Van Leeuwenhoek Int J Gen Mol Microbiol. 2008;94:403–414. doi: 10.1007/s10482-008-9258-7. [DOI] [PubMed] [Google Scholar]
- Lachance MA, Starmer WT, Bowles JM, Phaff HJ, Rosa CA. Ribosomal DNA, species structure, and biogeography of the cactophilic yeast Clavispora opuntiae. Can J Microbiol. 2000;46:195–210. doi: 10.1139/w99-130. [DOI] [PubMed] [Google Scholar]
- Lachance MA, Starmer WT, Rosa CA, Bowles JM, Barker JSF, Janzen DH. Biogeography of the yeasts of ephemeral flowers and their insects. FEMS Yeast Res. 2001;1:1–8. doi: 10.1111/j.1567-1364.2001.tb00007.x. [DOI] [PubMed] [Google Scholar]
- Lambrechts MG, Pretorius IS. Yeast and its importance to wine aroma—a review. S Afr J Enol Vitic. 2000;21:97–129. [Google Scholar]
- Landry CR, Townsend JP, Hartl DL, Cavalieri D. Ecological and evolutionary genomics of Saccharomyces cerevisiae. Mol Ecol. 2006;15:575–591. doi: 10.1111/j.1365-294X.2006.02778.x. [DOI] [PubMed] [Google Scholar]
- Legras JL, Merdinoglu D, Cornuet JM, Karst F. Bread, beer and wine: Saccharomyces cerevisiae diversity reflects human history. Mol Ecol. 2007;16:2091–2102. doi: 10.1111/j.1365-294X.2007.03266.x. [DOI] [PubMed] [Google Scholar]
- Liti G, Carter DM, Moses AM, Warringer J, Parts L, James SA, et al. Population genomics of domestic and wild yeasts. Nature. 2009;458:337–341. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez C, Cosgaya P, Vasquez C, Gac S, Ganga A. High degree of correlation between molecular polymorphism and geographic origin of wine yeast strains. J Appl Microbiol. 2007;103:2185–2195. doi: 10.1111/j.1365-2672.2007.03493.x. [DOI] [PubMed] [Google Scholar]
- Martiny JBH, Bohannan BJM, Brown JH, Colwell RK, Fuhrman JA, Green JL, et al. Microbial biogeography: putting microorganisms on the map. Nat Rev Microbiol. 2006;4:102–112. doi: 10.1038/nrmicro1341. [DOI] [PubMed] [Google Scholar]
- McGovern PE, Zhang J, Tang J, Zhang Z, Hall GR, Moreau RA, et al. Fermented beverages of pre- and proto-historic China. Proc Natl Acad Sci USA. 2004;101:17593–17598. doi: 10.1073/pnas.0407921102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemergut DR, Costello EK, Hamady M, Lozupone C, Jiang L, Schmidt SK, et al. Global patterns in the biogeography of bacterial taxa. Environ Microbiol. 2011;13:135–144. doi: 10.1111/j.1462-2920.2010.02315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peakall R, Smouse PE. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes. 2006;6:288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedros-Alio C. Dipping into the rare biosphere. Science. 2007;315:192–193. doi: 10.1126/science.1135933. [DOI] [PubMed] [Google Scholar]
- Piskur J, Rozpedowska E, Polakova S, Merico A, Compagno C. How did Saccharomyces evolve to become a good brewer. Trends Genet. 2006;22:183–186. doi: 10.1016/j.tig.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Prakitchaiwattana CJ, Fleet GH, Heard GM. Application and evaluation of denaturing gradient gel electrophoresis to analyse the yeast ecology of wine grapes. FEMS Yeast Res. 2004;4:865–877. doi: 10.1016/j.femsyr.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Pretorius IS. Tailoring wine yeast for the new millennium: novel approaches to the ancient art of winemaking. Yeast. 2000;16:675–729. doi: 10.1002/1097-0061(20000615)16:8<675::AID-YEA585>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Replansky T, Koufopanou V, Greig D, Bell G. Saccharomyces sensu stricto as a model system for evolution and ecology. Trends Ecol Evol. 2008;23:494–501. doi: 10.1016/j.tree.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Ribereau-Gayon P, Dobourdieu D, Doneche B, Lonvaud A.2006Handbook of EnologyVol. 1 The Microbiology of Wine and Vinifications.John Wiley & Sons, Ltd: Chichetser, UK [Google Scholar]
- Richards KD, Goddard MR, Gardner RC. A database of microsatellite genotypes for Saccharomyces cerevisiae. Antonie Van Leeuwenhoek Int J Gen Mol Microbiol. 2009;96:355–359. doi: 10.1007/s10482-009-9346-3. [DOI] [PubMed] [Google Scholar]
- Romano P, Fiore C, Paraggio M, Caruso M, Capece A. Function of yeast species and strains in wine flavour. Intl J Food Micro. 2003;86:169–180. doi: 10.1016/s0168-1605(03)00290-3. [DOI] [PubMed] [Google Scholar]
- Schacherer J, Shapiro JA, Ruderfer DM, Kruglyak L. Comprehensive polymorphism survey elucidates population structure of Saccharomyces cerevisiae. Nature. 2009;458:342–345. doi: 10.1038/nature07670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller D, Alves H, Dequin S, Casal M. Ecological survey of Saccharomyces cerevisiae strains from vineyards in the Vinho Verde Region of Portugal. FEMS Microbiol Ecol. 2005;51:167–177. doi: 10.1016/j.femsec.2004.08.003. [DOI] [PubMed] [Google Scholar]
- Schuller D, Casal M. The genetic structure of fermentative vineyard-associated Saccharomyces cerevisiae populations revealed by microsatellite analysis. Antonie Van Leeuwenhoek Int J Gen Mol Microbiol. 2007;91:137–150. doi: 10.1007/s10482-006-9104-8. [DOI] [PubMed] [Google Scholar]
- Serjeant K, Tang R, Anfang N, Beggs JR, Goddard MR. Yeasts associated with the New Zealand Nothofagus honeydew system. New Zeal J Ecol. 2008;32:209–213. [Google Scholar]
- Swiegers JH, Pretorius IS. Yeast modulation of wine flavor. Adv Appl Microbiol. 2005;57:131–175. doi: 10.1016/S0065-2164(05)57005-9. [DOI] [PubMed] [Google Scholar]
- Swiegers JH, Pretorius IS. Modulation of volatile sulfur compounds by wine yeast. Appl Microbiol Biotechnol. 2007;74:954–960. doi: 10.1007/s00253-006-0828-1. [DOI] [PubMed] [Google Scholar]
- Thomson JM, Gaucher EA, Burgan MF, De Kee DW, Li T, Aris JP, et al. Resurrecting ancestral alcohol dehydrogenases from yeast. Nat Genet. 2005;37:630–635. doi: 10.1038/ng1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitaker RJ, Grogan DW, Taylor JW. Geographic barriers isolate endemic populations of hyperthermophilic archaea. Science. 2003;301:976–978. doi: 10.1126/science.1086909. [DOI] [PubMed] [Google Scholar]
- Wolfe KH, Shields DC. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- Zhang HY, Lee SA, Bradbury JE, Warren RN, Seth H, Hooks D, et al. Yeasts isolated from New Zealand vineyards and wineries. Aust J Grape Wine R. 2010a;16:491–496. [Google Scholar]
- Zhang HY, Skelton A, Gardner RC, Goddard MR. Saccharomyces paradoxus and Saccharomyces cerevisiae reside on oak trees in New Zealand: evidence for migration from Europe and interspecies hybrids. FEMS Yeast Res. 2010b;10:941–947. doi: 10.1111/j.1567-1364.2010.00681.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.