Abstract
Diatoms are considered the most successful and widespread group of photosynthetic eukaryotes. Their contribution to primary production is remarkably significant to the earth's ecosystems. Diatoms are composed of two orders: Centrales and Pennales. Thus far, viruses infecting centric diatom species have been isolated and characterized; however, viruses infecting pennates have not been reported. Here, we describe the first isolations and preliminary characterizations of two distinct pennate diatom viruses, AglaRNAV (31 nm in diameter, accumulates in the host cytoplasm) and TnitDNAV (35 nm in diameter, accumulates in the host nuclei) infecting Asterionellopsis glacialis and Thalassionema nitzschioides, respectively. Their genomes contain a single-stranded RNA of approximately 9.5 kb, and a closed, circular single-stranded DNA of approximately 5.5 kb harboring a partially double-stranded region, respectively. Further analysis of these viruses may elucidate many aspects of diatom host–virus relationships.
Keywords: diatom, virus, pennate
Introduction
Diatoms constitute one of the major lineages of photosynthetic eukaryotes on earth, and there are probably more than approximately 105 species (Mann and Droop, 1996). Their contribution to global biogeochemical cycling is significant (Smetacek, 1999), and accounts for a large part of marine primary production, up to 35–75% in the oceans (Nelson et al., 1995). On the basis of morphological features, diatom species are composed of two groups: centric and pennate. Complete genome sequences have recently been reported for members of these groups (Armbrust et al., 2004; Bowler et al., 2008), which revealed many aspects of diatom evolutionary history, ecology, metabolism and influence on biogeochemical cycling (Bowler et al., 2010).
Marine virus studies during the last 2 decades have revealed the significance of viruses to host population dynamics (Suttle, 2005). Diatom viruses are also considered to be important for the understanding of diatom life cycles and the impact of diatoms on biogeochemical cycling. So far, isolations of several viruses infecting centric diatoms have been reported (Tomaru et al., 2011a). However, viruses have not been isolated from pennate species, constituting major components of marine environments (Kooistra et al., 2007). Here, we report preliminary characterizations of viruses infecting the pennate diatoms Asterionellopsis glacialis (Castracane) Round and Thalassionema nitzschioides (Grunow) Mereschkowsky.
Materials, methods, results and discussion
The algal strains A. glacialis IT09-K25 (Figure 1a) and T. nitzschioides AR-TN01 (Figure 1b) were used for the experiments (Supplementary Table 1). Algal cultures were grown in modified SWM-3 medium (Itoh and Imai, 1987) at 15 °C under a 12/12 h light–dark cycle with a light intensity of 110–150 μmol photons per m2 s−1.
Figure 1.
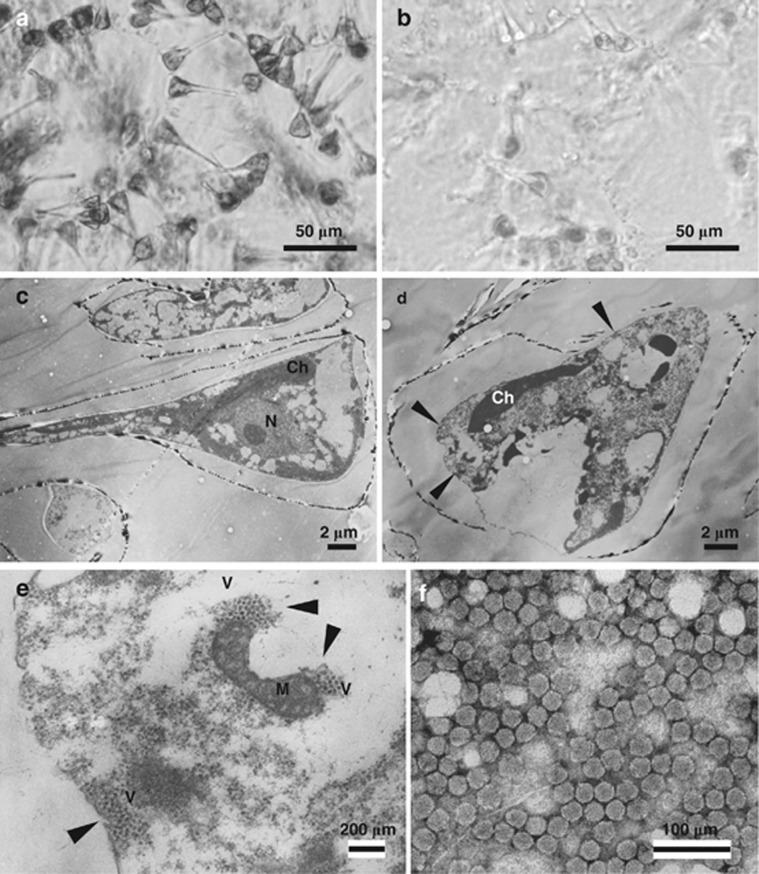
Asterionellopsis glacialis IT09-K25 isolated from surface water in Hiroshima Bay, Japan. (a) Optical micrograph of intact cells. (b) Cultures inoculated with AglaRNAV at 2 d.p.i.. Transmission electron micrographs of ultra-thin sections of A. glacialis and negatively stained AglaRNAV particles. (c) Healthy cell. (d and e) Cells infected with AglaRNAV at 2 d.p.i. (d) Degraded host cytoplasm and chloroplast. (e) Higher magnification of randomly aggregated VLPs in the host cytoplasm in panel d. Arrows denote the accumulation of VLPs. (f) Negatively stained AglaRNAV particles in purified culture lysate. N, nucleus; Ch, chloroplast; M, mitochondrion; V, virus-like particles.
Surface waters from Ariake Sound, Japan (February 2010) and sediment samples (0–1 cm depth) from Ago Bay, Japan (March 2009) were used for isolating viruses infecting T. nitzschioides and A. glacialis, respectively. The water samples were filtered through 0.2-μm filters (Advantec, Tokyo, Japan) to remove microorganisms and the majority of bacteria. The filtrates were preserved at −20 °C until the following isolation procedures. The sediment samples were collected using an Ekman-Birge bottom sampler (Rigo, Tokyo, Japan). Viruses in the sediments were extracted by the method described by Tomaru et al. (2011a).
Aliquots (0.5 mL) of the treated samples were inoculated into 1.0 ml of exponentially growing host cultures and incubated at 15 °C using the light conditions described above. Algal cultures inoculated with SWM-3 served as controls. Cell condition and signs of lysis in each algal culture were monitored by optical microscopy (Tomaru et al., 2011a). The lysed cultures were filtered through a 0.1-μm polycarbonate membrane filter (Whatman, Kent, UK) to exclude bacterial effects, and their lytic activity was reassessed. We assumed the pathogens were viruses and tentatively named the viruses infecting A. glacialis and T. nitzschioides as AglaRNAV and TnitDNAV, respectively.
AglaRNAV was cloned through two extinction dilution cycles (Suttle, 1993). Cloning of TnitDNAV was unsuccessful because the host culture did not grow well in the microplates used for the cloning procedures and we, therefore, used non-cloned TnitDNAV in this study. Transmission electron microscopy observations for ultra-thin sections, negatively stained virions and viral genome analysis were performed using the method described by Tomaru et al. (2009, 2011a).
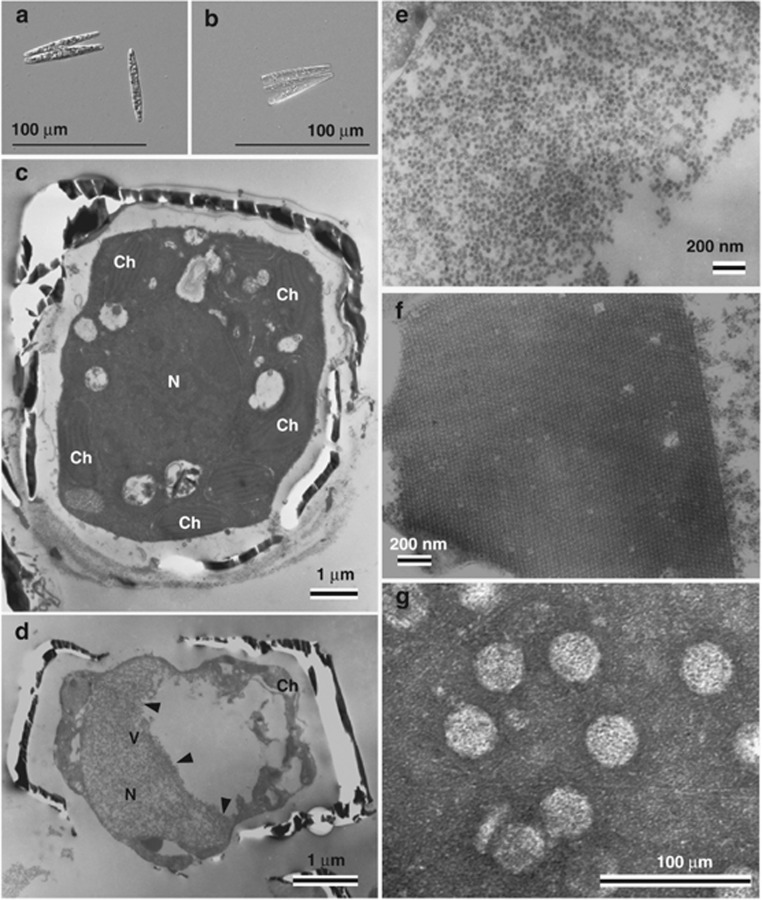
The occurrence of host cell lysis was usually observed 2 and 10 days post inoculation (d.p.i.) in A. glacialis and T. nitzschioides, respectively (Figures 1b and 2b). Thin sections of healthy host cells showed the cytoplasmic organization and frustules that are typical of the diatoms (Figures 1c and 2c). In contrast, electron micrographs of thin-sectioned A. glacialis cells at 2 d.p.i. revealed randomly assembled virus-like particles (VLP) in the host cytoplasm (Figures 1d and e). In T. nitzschioides cells at 8 d.p.i., the presence of randomly assembled or para-crystalline-arrayed VLP was observed in the host nuclei (Figures 2d, e, and f). VLPs were not found in healthy cells of either host species (Figures 1c and 2c). The diameters of VLPs in A. glacialis and T. nitzschioides were 31 (±2) nm (n=40) and 34 (±1) nm (n=40), respectively (Figures 1e and 2f).
Figure 2.
Thalassionema nitzschioides AR-TN01 isolated from surface water in Ariake Sound, Japan. (a) Optical micrograph of intact cells. (b) Cultures inoculated with TnitDNAV at 15 d.p.i.. Transmission electron micrographs of ultra-thin sections of T. nitzschioides and negatively stained TnitDNAV particles. (c) Healthy cell. (d, e and f) Cells infected with TnitDNAV at 8 d.p.i. (d) Degraded host cytoplasm and chloroplast. (e) Higher magnification of randomly aggregated VLPs in the host nucleus in panel d. (f) Para-crystalline array aggregation of VLPs in an infected host nucleus. (g) Negatively stained TnitDNAV particles in purified culture lysate. Arrows denote the accumulation of VLPs. N, nucleus; Ch, chloroplast; V, virus-like particles.
VLPs were also observed in culture lysates by negative staining transmission electron microscopy. VLPs in the lysate of A. glacialis and T. nitzschioides were 31 (±1) nm (n=47) and 36 (±2) nm (n=40) in diameter, respectively (Figures 1f and 2g). They appeared similar to the VLP observed in their respective host cytoplasm and/or nuclei (Figures 1e and 2e). The particle diameters of these viruses were within a range of those of the centric diatom viruses, 22–38 nm (Tomaru et al., 2009; Tomaru et al., 2011a).
The basic genome characters of the two viruses were examined by preliminary tests. Denaturing gel electrophoresis showed that the nucleic acids extracted from AglaRNAV were 9.5 kb, 4.0 kb and 2.5 kb (Supplementary Figure 1). These bands were sensitive to RNase A, both under high- and low-salt concentrations, but not to DNase I (Supplementary Figure 1), suggesting that AglaRNAV genome constituted a single-stranded RNA (ssRNA) molecule of approximately 9.5 kb. The lower bands were considered to be components of the viral genome, but not revealed in this study. Similar results were reported on ssRNA centric diatom virus CtenRNAV (Shirai et al., 2008).
In a gel electrophoresis of closed circular single-stranded DNA (ssDNA) diatom viral genome harboring a complementary sequence fragment, two larger bands with intact genome and single lower band with S1 nuclease treatment are usually observed at 5.0–6.0 kb and 0.8–1.0 kb, respectively (for example, Tomaru et al. (2011a)). The intact TnitDNAV genome exhibited two bands of nucleic acids of approximately 5.5 and 5.0 kb, and double-stranded DNA of 0.6 kbp remained undigested after S1 nuclease treatment (Supplementary Figure 2), which were considerably similar to the features of the ssDNA diatom viral nucleic acids. The above results suggested that the genome of TnitDNAV consists of a closed circular ssDNA molecule of approximately 5.5 kb, which contains a complementary fragment of ca 0.6 kb.
Previously reported centric diatom ssRNA and ssDNA viruses accumulate in their host cytoplasm and nucleus, respectively (Tomaru et al., 2009; Tomaru et al., 2011b), which agrees to the present observations. On the basis of the above results, we concluded that the particles in the host cells and in the lysates were viruses.
Implication
Considering the potential similarities between pennate and centric diatom virus features, diatom host–virus relationships may have been established in the early evolutionary stage of the diatoms. A recent study showed evidence for a horizontal gene transfer event from non-retroviral RNA viruses into the genomes of distantly related plant lineages (Chiba et al., 2011), suggesting that lateral gene transfer could occur through viral infections. The understanding of pennate and centric diatom viruses may elucidate their hosts' phylogenetic relationships and evolution. The genome sequences and the precise features of the viruses described in this study are currently under analysis, and will be reported in the near future.
Acknowledgments
This study was partly supported by Grants-in-Aid for Young Scientists (A) (22688016) from the Ministry of Education, Science and Culture of Japan.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Armbrust EV, Berges JA, Bowler C, Green BR, Martinez D, Putnam NH, et al. The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science. 2004;306:79–86. doi: 10.1126/science.1101156. [DOI] [PubMed] [Google Scholar]
- Bowler C, Allen AE, Badger JH, Grimwood J, Jabbari K, Kuo A, et al. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature. 2008;456:239–244. doi: 10.1038/nature07410. [DOI] [PubMed] [Google Scholar]
- Bowler C, Vardi A, Allen AE. Oceanographic and biogeochemical insights from diatom genomes. Annu Rev Mar Sci. 2010;2:333–365. doi: 10.1146/annurev-marine-120308-081051. [DOI] [PubMed] [Google Scholar]
- Chiba S, Kondo H, Tani A, Saisho D, Sakamoto W, Kanematsu S, et al. Widespread endogenization of genome sequences of non-retroviral RNA viruses into plant genomes. PLoS pathogens. 2011;7:e1002146. doi: 10.1371/journal.ppat.1002146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh K, Imai I.1987Rafido so (Raphidophyceae)In: Japan Fisheries Resource Council Association (ed).A Guide for Studies of Red Tide Organisms Shuwa: Tokyo; 122–130. [Google Scholar]
- Kooistra WHCF, Gersonde R, Medlin LK, Mann DG.2007The origin and evolution of the diatoms: Their adaptation to a planktonic existenceIn: Falkowski PG, Knoll A (eds). Evolution of Primary Producers in the Sea Elsevier: San Diego; 207–249. [Google Scholar]
- Mann DG, Droop SJM. 3. Biodiversity, biogeography and conservation of diatoms. Hydrobiologia. 1996;336:19–32. [Google Scholar]
- Nelson DM, Treguer P, Brzezinski MA, Leynaert A, Queguiner B. Production and dissolution of biogenic silica in the ocean: Revised global estimates, comparison with regional data and relationship to biogenic sedimentation. Global Biogeochem Cycle. 1995;9:359–372. [Google Scholar]
- Shirai Y, Tomaru Y, Takao Y, Suzuki H, Nagumo T, Nagasaki K. Isolation and characterization of a single-stranded RNA virus infecting the marine planktonic diatom Chaetoceros tenuissimus Meunier. Appl Environ Microbiol. 2008;74:4022–4027. doi: 10.1128/AEM.00509-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smetacek V. Diatoms and the ocean carbon cycle. Protist. 1999;150:25–32. doi: 10.1016/S1434-4610(99)70006-4. [DOI] [PubMed] [Google Scholar]
- Suttle CA.1993Enumeration and isolation of virusesIn: Kemp PF, Sherr E, Cole JJ (eds).Handbook of methods in aquatic microbial ecology Lewis Publishers: Boca Raton; 121–137. [Google Scholar]
- Suttle CA. Viruses in the sea. Nature. 2005;437:356–361. doi: 10.1038/nature04160. [DOI] [PubMed] [Google Scholar]
- Tomaru Y, Takao Y, Suzuki H, Nagumo T, Nagasaki K. Isolation and characterization of a single-stranded RNA virus Infecting the bloom forming diatom Chaetoceros socialis. Appl Environ Microbiol. 2009;75:2375–2381. doi: 10.1128/AEM.02580-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaru Y, Shirai Y, Toyoda K, Nagasaki K. Isolation and characterisation of a single-stranded DNA Virus infecting the marine planktonic diatom Chaetoceros tenuissimus Meunier. Aquat Microb Ecol. 2011a;64:175–184. doi: 10.1128/AEM.00509-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaru Y, Takao Y, Suzuki H, Nagumo T, Koike K, Nagasaki K. Isolation and characterization of a single-stranded DNA virus infecting Chaetoceros lorenzianus Grunow. Appl Environ Microbiol. 2011b;77:5285–5293. doi: 10.1128/AEM.00202-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.