Abstract
The human gastrointestinal tract (GI tract) harbors a complex community of microbes. The microbiota composition varies between different locations in the GI tract, but most studies focus on the fecal microbiota, and that inhabiting the colonic mucosa. Consequently, little is known about the microbiota at other parts of the GI tract, which is especially true for the small intestine because of its limited accessibility. Here we deduce an ecological model of the microbiota composition and function in the small intestine, using complementing culture-independent approaches. Phylogenetic microarray analyses demonstrated that microbiota compositions that are typically found in effluent samples from ileostomists (subjects without a colon) can also be encountered in the small intestine of healthy individuals. Phylogenetic mapping of small intestinal metagenome of three different ileostomy effluent samples from a single individual indicated that Streptococcus sp., Escherichia coli, Clostridium sp. and high G+C organisms are most abundant in the small intestine. The compositions of these populations fluctuated in time and correlated to the short-chain fatty acids profiles that were determined in parallel. Comparative functional analysis with fecal metagenomes identified functions that are overrepresented in the small intestine, including simple carbohydrate transport phosphotransferase systems (PTS), central metabolism and biotin production. Moreover, metatranscriptome analysis supported high level in-situ expression of PTS and carbohydrate metabolic genes, especially those belonging to Streptococcus sp. Overall, our findings suggest that rapid uptake and fermentation of available carbohydrates contribute to maintaining the microbiota in the human small intestine.
Keywords: ecological model, function, microbiota, phylogeny, small intestine
Introduction
The human gastrointestinal tract (GI tract) contains several compartments with distinct anatomy and function, and is of utmost importance in supplying the body with energy and essential nutrients by converting and absorbing food components. The GI tract is colonized with approximately 1000 microbial species, commonly called the microbiota, and may harbor more than nine million unique genes (Zoetendal et al., 2008; Yang et al., 2009). Culture-independent approaches such as ribosomal RNA (rRNA) targeting technologies have revealed that the microbiota is stable in time in healthy adults, and host- and location specific (Rajilić-Stojanović et al., 2007; Zoetendal et al., 2008). Random metagenomic analysis of fecal microbiota suggested that it complements human physiology with a range of essential functions that were canonically encountered in different adults, and that a potential link between microbiota-derived gene pool and health and disease exists (Gill et al., 2006; Kurokawa et al., 2007; Turnbaugh et al., 2009; Qin et al., 2010). Integrated approaches that combine multiple ‘omics' approaches permit moving from such ‘parts list' approaches to a deeper understanding of the ecosystem functioning (Raes et al., 2007; Raes and Bork, 2008; Verberkmoes et al., 2009).
A major drawback of the use of fecal samples to determine the intestinal microbial composition is the fact that fecal microbiota represents only the end of the colon, leaving other parts of the GI tract, particularly the small intestine, unexplored. The small intestine is a harsh environment for microbial life because of the short transit time and excretion of digestive enzymes and bile (Johnson, 2006), thereby requiring different survival strategies of microbes compared with those residing in the colon. The few existing studies on the small intestine have generally employed invasive sampling procedures or material from sudden death victims (Wang et al., 2003, 2005; Hayashi et al., 2005; Ahmed et al., 2007; Willing et al., 2010), empeding the study of the dynamics of microbiota over time or as a function of diet. Ileostomists provide a powerful alternative to this problem (Gorbach et al., 1967). Ileostomists underwent surgery to remove the complete colon because of a clinical condition, such as colon cancer or inflammatory bowel diseases, and have the terminal part of the ileum connected to a stoma allowing non-invasive, repeated sampling of the ileostomy effluent. In general, ileostomists that recover within months following the operation, can be considered healthy and enjoy an active life. A recent study suggested that oxygen might penetrate the small intestine via the stoma, resulting in increased facultative anaerobe populations (Hartman et al., 2009), but other studies have clearly established the significant abundance of typical strict anaerobes in effluent samples (Booijink et al., 2010).
Here we compare the microbiota composition in samples obtained from different small intestinal positions (jejunum, ileum and terminal ileum) in healthy subjects to those obtained from ileostomists. In addition, metagenomic and metatranscriptomic analyses, complemented by fermentation end-product profiling of multiple ileostomy effluent samples provided insight in the microbiota metabolic potential, its dynamics and the activity of the microbiota in the small intestine. From these data, we deduce a model that exemplifies that fast uptake and conversion of carbohydrates contributes to maintaining the microbiota in the human small intestine.
Materials and methods
Ethics statement
The study was approved by the University Hospital Maastricht Ethical Committee, and conducted in full accordance with the principles of the ‘Declaration of Helsinki' (52nd WMA General Assembly, Edinburgh, Scotland, October 2000). All volunteers were informed about the study orally and in writing, and signed a written informed consent before participation.
Sample collection
For phylogenetic profiling ileostomy effluent, small intestinal content and feces was freshly collected from five healthy ileostomists (three male, two female; 60.2±7.1 years; (Booijink et al., 2010); individuals A–E) and four healthy subjects (four male; 24±4.5 years; individuals F–I), and two healthy subjects (one male, one female; 56.6±3.5 years; individuals J–K). All ileostomists had an intact small intestine with the exception of the terminal few centimeters of the ileum, which were removed during surgical removal of the colon. For metagenome and metatranscriptome analyses, ileostomy effluent samples were collected from a healthy, 74-year-old male subject who has been carrying an ileostoma for 20 years. Apart from absence of the colon, the subject had no known abnormalities of the digestive system and had not been subjected to any nutrition intervention trial, specific diet or antibiotic treatment for at least 1 year before sampling. To generate maximum coverage of the population dynamics of the small intestine microbiota (4), samples were taken at day 1 (morning and afternoon; 1M and 1A, respectively), day 7 (afternoon; 7A), and compared with a morning sample that was obtained 1 year earlier (A; Supplementary Table S1). Ileostomy effluent samples were freshly collected in an unused ileostomy appliance and frozen immediately in dry ice for DNA isolation or quenched in methanol–HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) buffer (±1:4), homogenized and stored on dry ice as described previously for RNA isolation (Pieterse et al., 2006). Small intestinal fluid samples were obtained after a 10-h fasting period from the healthy volunteers without a history of GI complaints, using an intraluminal naso-ileal catheter that was placed nasogastrically with the catheter tip positioned in the ileum. These samples included a jejunal sample and an ileum sample from subject G, a single ileum sample from subjects F and H, and two terminal ileum samples from subject I. Catheter positioning was performed as described previously (Troost et al., 2008), under short-interval fluoroscopic control. No sedatives were given to the volunteers, and the bowel was not prepared before sampling. The sampling location was determined by assessing the distance from the mouth to the catheter tip. In this way, sample locations can be specified as jejunum, ileum and terminal ileum, but not beyond this classification. Samples were collected at Maastricht University Medical Centre and frozen immediately in liquid nitrogen, stored at −80 °C until analysis. Similarly, fecal samples were immediately frozen after defecation and stored at −80 °C until analysis.
Phylogenetic profiling
DNA was extracted from 1.0 ml of ileostomy effluent samples and 0.25 ml from ileal content as described before (Zoetendal et al., 2006b), using the Stool DNA Isolation Kit (Qiagen, Leiden, The Netherlands) and subsequently quantified by a spectrophotometer (Nanodrop ND-1000 spectrophotometer, NanoDrop Technologies, Wilmington, DE, USA). For phylogenetic analysis of the samples, the human intestinal tract chip (HITChip), a phylogenetic microarray containing more than 4800 oligonucleotides based on SSU rRNA gene sequences that target over 1100 intestinal bacterial phylotypes, was used (Rajilić-Stojanović et al., 2009). In short, the SSU rRNA gene was amplified from 10-ng DNA with the T7prom-Bact-27-for and Uni-1492-rev primers followed by in vitro transcription and labeling with Cy3 and Cy5, respectively. The Cy3/Cy5-labeled target mixes were fragmented and hybridized on the arrays at 62.5 °C for 16 h in a rotation oven (Agilent Technologies, Amstelveen, The Netherlands). After washing and drying, the slides were scanned. Data was extracted from the microarray images using the Agilent Feature Extraction software, versions 7.5–9.1 (http://www.agilent.com) and subsequently, the microarray data normalization, and further analyzed using a set of R-based scripts (http://r-project.org) in combination with a custom-designed relational database that runs under the MySQL database management system (http://www.mysql.com;Rajilić-Stojanović et al., 2009). Hierarchical clustering of probe profiles was carried out using the Euclidian distance and Ward's minimum variance method. These small intestinal microbiota composition profiles generated for the samples obtained from healthy subjects were compared with the ileostoma effluent profiles that were already present in the database (Booijink et al., 2010). Multivariate statistical software Canoco 4.5 for windows (Leps and Smilauer, 2003;Biometrix, Plant Research International, Wageningen, The Netherlands) was used to perform the principle component analysis on log-transformed HITChip probe signal-intensity profiles.
High molecular weight DNA isolation from ileostoma effluent samples
From the ileostoma effluent samples, 30–40 g were used for high molecular weight DNA isolation as described by Berridge et al. (1998) with modifications. Effluent samples were resuspended as portions of 10 g in 20 ml 1 × phosphate-buffered saline containing 10–20 glassbeads of 2 mm and vortexed for 3 min. After centrifugation at 700 g for 1 min to remove large-sized debris, the supernatant was centrifuged at 3000 g for 10 min. The pellet was resuspended in 5 ml 1 × phosphate-buffered saline with 2% w/v polyvinylpyrrolidone (PVP), incubated at room temperature for 5 min and centrifuged at 3000 g for 10 min. After washing with 1 × phosphate-buffered saline, the pellet was resuspended in 3 ml lysis buffer (50 mM glucose, 50 mM Tris-HCl (pH 8.0), 100 mM EDTA, 2% w/v PVP, 150 mM NaCl, 0.2 mg ml−1 RNAse A (Sigma-Aldrich, Zwijndrecht, The Netherlands), 0.02 mg ml−1 mutanolysin (Sigma-Aldrich) and 1 mg ml−1 lysozyme (Sigma-Aldrich)), and incubated at 37 °C for 2 h. Following addition of 360 μl 10% w/v SDS and 160 μl of a 10 mg ml−1 Proteinase K (Sigma-Aldrich) solution, the samples were incubated at 45 °C for 2 h. After addition of 600 μl 5 M NaCl and 480 μl CTAB/NaCl solution (10% Cetyl trimethylammonium bromide in 0.7 M NaCl), samples were incubated at 65 °C for 10 min. An equal volume of phenol/chloroform/isoamylalcohol (25:24:1) was added to the sample, followed by gentle mixing of the sample and subsequent centrifugation at 8500 g at 4 °C for 20 min. The aqueous layer was removed and extracted with phenol/chloroform/isoamylalcohol (25:24:1) and chloroform/isoamylalcohol (24:1), respectively, as described before (Sambrook et al., 1989). After removal of the aqueous layer, 0.7 volume of isopropanol was added. Samples were mixed gently by inverting, and incubated at 4 °C for 2 h. Samples were centrifuged at 9000 g for 15 min, washed with 70% EtOH and dissolved in TE (10 mM Tris-HCL (pH 8.0), 1 mM EDTA) by overnight incubation without agitation at 4 °C. DNA quality and quantity were evaluated by electrophoresis on 0.8% agarose gels; samples with appropriate amounts of large-sized, high-quality DNA were shipped to GATC Biotech (Konstanz, Germany) for fosmid library construction.
Fosmid library construction
DNA isolated from each of the four samples was cloned into the pCC1Fos vector using the protocol and kit provided by Epicentre Biotechnologies (Madison, WI, USA). In short, DNA was not additionally fragmented, as the isolation process fragmented the DNA sufficiently to be cloned directly. Two size ranges were used for cloning. DNA of 40 and 30 kb was isolated from (0.35%) low-melting-point agarose, extracted using agarase and purified by ethanol precipitation. The DNA fragments were ligated to the clone-ready pCC1Fos vector, and packaged and transformed according to the standard protocol provided by the manufacturer. Plasmids were isolated from pools of all transformants per library according to standard procedures performed at the GATC Biotech, and used for phylogenetic comparison with their original samples.
cDNA library construction
Total RNA was isolated from a morning sample according to the method described previously (Zoetendal et al., 2006a). The cDNA library was constructed at the GATC Biotech. Briefly, 10 μg total RNA was treated with Terminator 5′-Phosphate-Dependent Exonuclease (Epicentre Biotechnologies) to digest tRNAs and rRNAs. The RNA was subsequently precipitated with 2.5 M LiCl at 4 °C overnight and resuspended in TE. An additional rRNA removal step was performed using the MicrobeExpress Kit from Ambion (Austin, TX, USA). The enriched mRNA was reverse transcribed and cloned using the SMART cDNA Library Construction Kit (Clontech-Takara Bio Europe, Saint-Germain-en-Laye, France). The resulting double-stranded cDNA was cloned into a bBSII SK+ vector with a modified multiple cloning site containing two Sfi I sites at positions 716 and 740, which allowed the directional cloning of the cDNAs. This resulted in clones of which 3609 of the 10 000 were mRNA derived.
Sanger sequencing
For Sanger end-read sequencing, clones were picked and vector DNA was isolated using the alkaline extraction method (Birnboim, 1983). The vector DNA was sequenced using BigDye Chemistry (Applied Biosystems, Foster City, CA, USA) and reactions were subsequently analyzed on the ABI 3730xl.
Before obtaining all the end reads, the quality of the sublibraries were determined by initially sequencing the end reads of 192 randomly selected clones (stored in 96-well plate). These were blasted against genomes and plasmids from a selection of 50 non-redundant microbes (46 Bacteria, 4 Archaea, see Supplementary Table S2). This indicated that samples A, 1A and 1M harbored a diverse community consisting of a variety of phyla. Sample 7A was excluded for further sequencing as 75% of the sequences originated from E. coli and related bacteria, which suggests it was contaminated with DNA from the cloning host. This was comfirmed by comparative phylogenetic profiling of the original sample and the pooled fosmid DNA (Supplementary Figure S1), which demonstrated the congruency between libraries and original samples, except for sample 7A.
GS-FLX sequencing
Fosmids from the different libraries were pooled in an equimolar mixture. DNA was prepared for sequencing using the Amplicon A kit (Hoffmann-La Roche AG, Basel, Switzerland) and sequenced on the GS-FLX (Hoffmann-La Roche AG) using the manufacturer's protocols. The sequencing data were trimmed to remove the pCC1Fos vector and assembled using the GS De Novo Assembler (Hoffmann-La Roche AG).
Computational and statistical analyses
Sequence analysis was performed using the SMASH pipeline (Arumugam et al., 2010). In short, sequence reads were filtered for the cloning vector pCC1Fos using the BLASTN (using parameters ‘M=5 N=−11 Q=22 R=11 V=10 E=1e-10' and a 90% similarity threshold). The resulting reads were filtered for possible phage sequences using BLASTN against ACLAME viral genes v0.3 (E-value > 1e-10, 90% similarity threshold, >90% of the read must be aligned; (Leplae et al., 2004)). The remaining reads were then assembled using Forge assembler (using ‘—metagenomic' for metagenome assembly; (Diguistini et al., 2009)). Genes were predicted using GeneMark v2.6c with a heuristic model, based on the GC content (Besemer and Borodovsky, 2005). Functional annotation was performed as in Kunin et al. (2008) and Gianoulis et al. (2009). In short, genes were mapped to EggNOG orthologous groups (Jensen et al., 2008), and Kyoto Encyclopedia of Genes and Genomes (KEGG) modules and pathways (Kanehisa et al., 2008). Protein sequences were searched against the EggNOG and KEGG databases using BLASTP (Altschul et al., 1997). A functional entity (Orthologous Group (OG), module, pathway) was called present when a hit matched any of its components (bitscore>=60). All results described were manually scrutinized to reduce artifactual assignments. The functional entity frequency for each sample was calculated by summing the total number of instances of that entity in a particular sample, and then normalized by total number of assignments for all entities in that sample to compensate for sample-coverage differences. For all analyses, entities for which the summed count over all samples constituted less than or equal to 0.01% of the total count were removed to avoid artifacts. Proteins from selected pathways were mapped to nodes of the tree of life (Ciccarelli et al., 2006) using an in-house perl script based upon the last common ancestor approach (Huson et al., 2007). Input data were BLASTp results of the proteins against the STRING 7 database (Jensen et al., 2008). Only hits above 60 bits and whose scores lied within 10% of the best score were considered. Samples were compared with fecal metagenomes from Gill et al. (2006) and Kurokawa et al. (2007). For statistical comparison, the two most similar ileal samples were pooled and functional group counts were compared with the pooled non-infant fecal samples using Fisher's exact test with Benjamini–Hochberg FDR correction for multiple testing. Finally, highlighted case studies were manually scrutinized to exclude any artifacts. Sequence data are available at http://systemsbiology.vub.ac.be/suppl_data/ileum/.
pH and metabolite measurements
Approximately 3 g of the directly frozen ileostomy effluent was defrosted on ice and subsequently centrifuged at 9000 g for 5 min at 4 °C. Afterwards, the supernatant was collected for measuring the pH and the concentration of short-chain fatty acids, lactate and alcohols by high-performance liquid chromatography as described previously (Starrenburg and Hugenholtz, 1991).
Results and discussion
Small intestine versus ileostomy
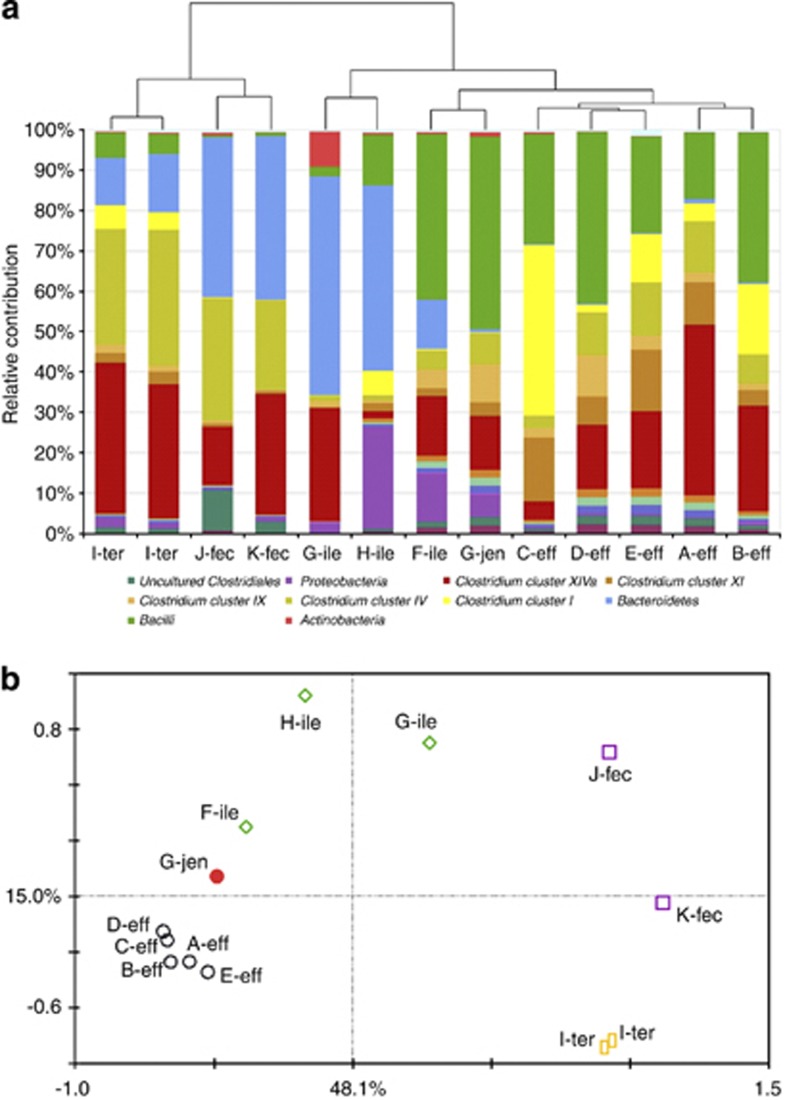
To address the comparability of the ileostoma effluent and small intestinal lumen, we used the GI tract-specific phylogenetic microarray HITChip (Rajilić-Stojanović et al., 2009) to compare the microbiota composition of ileostoma effluent with fecal samples and samples from the small intestine of healthy individuals that were obtained by an extended oral catheter that was placed by peristalsis (Troost et al., 2008). HITChip analysis of small intestine samples from four healthy subjects revealed a wide diversity at phylum level, depending on the subject and sampling location within the subject (Figure 1a). Bacteroidetes, Clostridium cluster XIVa and Proteobacteria were among the dominant groups in the ileum of subjects G and H, and terminal ileum of subject I. Notably, the jejunal sample (subject G) and one ileum sample (subject F) were dominated by Bacilli (Streptococcus sp.), Clostridium clusters IX (Veillonella sp.) and XIVa (several genera), and several Gamma Proteobacteria, thereby displaying higher similarity with microbiota composition profiles obtained from ileostoma effluent samples (Booijink et al., 2010), which was supported by cluster analysis (Figure 1a). Principle component analysis analysis suggested that ileostomy effluent cluster closely to the jejunal sample, whereas the terminal ileum samples were more similar to feces as explained by 48% of the data along the first principle component (Figure 1b). The ileum samples were positioned in between ileostomy effluent and feces and display large subject-specific differences. These results indicated that the microbiota composition that is encountered in ileostomy effluent can also be detected in the small intestine of healthy subjects. However, the limited number of samples from healthy subjects and the impact of subject-specificity on the overall variation between samples hamper a more detailed comparison between ileostomy effluent and small intestinal lumen samples.
Figure 1.
(a) Relative contribution of different microbial groups that are present in samples derived from the small intestine and feces of four and two healthy individuals, respectively, and those from five healthy ileostomists. The taxonomic classification is performed as described previously (Rajilić-Stojanović et al., 2009). The tree represents the Euclidian clustering of the HITChip probe profiles. A–K encode the subjects; eff, jen, ile, ter, fec encode ileostomy effluent, jenunum, ileum, terminal ileum, and feces, respectively. Microbial groups that are 2.5% of the total community in at least one of the samples are represented in the legend. (b) Principle component analysis of the microbiotas based on the HITChip probe signal profiles. Sample identities are identical as in Figure 1a. The two first components and their respective percentage of variation they explain are indicated.
Microbiome of small intestine is less complex than that of the colon
To obtain insight into the genetic potential and population dynamics within the small intestinal microbiota, a metagenomic library was constructed from the ileostoma effluent from a healthy individual who had the stoma for more than 20 years and did not require any stoma-related medication. As the ileostomy effluent microbiota composition fluctuates over time (Booijink et al., 2010), the metagenome library was constructed on four different morning and afternoon effluent samples. The entire library encompassed 25 344 fosmid clones with an average insert size of more than 25 000 bp, and thus, containing in total more than 700 Mb (Supplementary Table S1).
After initial quality control (see Supplementary Materials), three libraries representing the morning (1M) and afternoon (1A) of the same day and a morning sample taken 1 year earlier (A) were subjected to end sequencing (Sanger) and GS-FLX random sequencing (Supplementary Table S1), generating a total of 178 Mb of sequence information. 146 Mb of sequence information could be assembled into contigs that collectively encompass 63 Mb, with the largest contig being 78 kb, and leaving only 13% of the sequence-reads unassembled. This assembled proportion of sequences is considerably higher than previously observed with fecal metagenomes (Gill et al., 2006; Kurokawa et al., 2007), indicating a lower species diversity in the small intestine as compared with the colon, which is in line with the previous observations based on 16S rRNA genes (Hartman et al., 2009; Booijink et al., 2010). More than 170 000 genes could be assigned in the assembled contigs, of which 16% was complete.
Microbiome of the small intestine consists of various microbial phyla
Phylogenetic positioning of sequences suggested that they originated from a wide variety of phylotypes, with Clostridium sp., Streptococcus sp. and coliforms as dominant phylogenetic groups (Supplementary Figure S1), which is consistent with the 16S rRNA-based analyses. In addition, an unexpectedly large fraction of high G+C Gram positives was observed, which may be due to the commonly encountered underestimation of these microbial groups by regular 16S rRNA gene amplification (Hayashi et al., 2004). Moreover, a fraction of sequences mapped at low phylogetic resolution, indicating the presence of several uncultured species from which close cultured relatives have not been characterized by full genome sequencing.
Comparative phylogenetic profiling of the individual sublibraries revealed that all groups encountered in the overall analysis were present in each of the sublibraries, but displayed inequal distributions in the different sublibraries (Supplementary Figure S2). The phylogenetic distribution of sequences derived from samples taken on the same day (1A and 1M) were similar to each other, but differed from the sample that was taken a year earlier (A). Notably, Streptococcus sp. and high G+C Gram positives were overrepresented in the 1A and 1M libraries, whereas Clostridium sp. and coliforms were more prominently represented in the A library, which was confirmed by the HITChip analysis of the fosmid libraries (Supplementary Figure S1). These findings support previous observations that underline the fluctuation of the microbiota composition in ileostomy effluent over time (Booijink et al., 2010), which contrasts the relatively stable composition in the colon (Zoetendal et al., 2008). Moreover, it demonstrates that the metagenome of a single sample provides only a snapshot of an ecosystem and may underestimate the consequences of population dynamics.
Small intestinal microbiome reflects a community that can quickly respond to varying carbohydrate availability
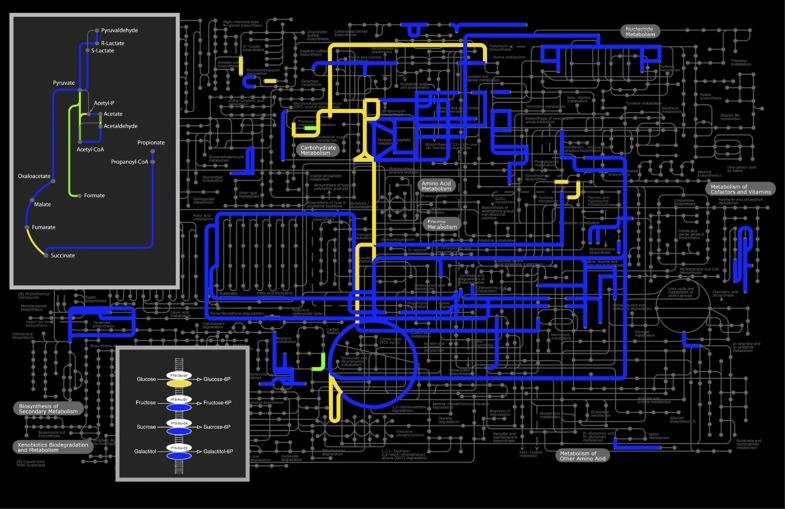
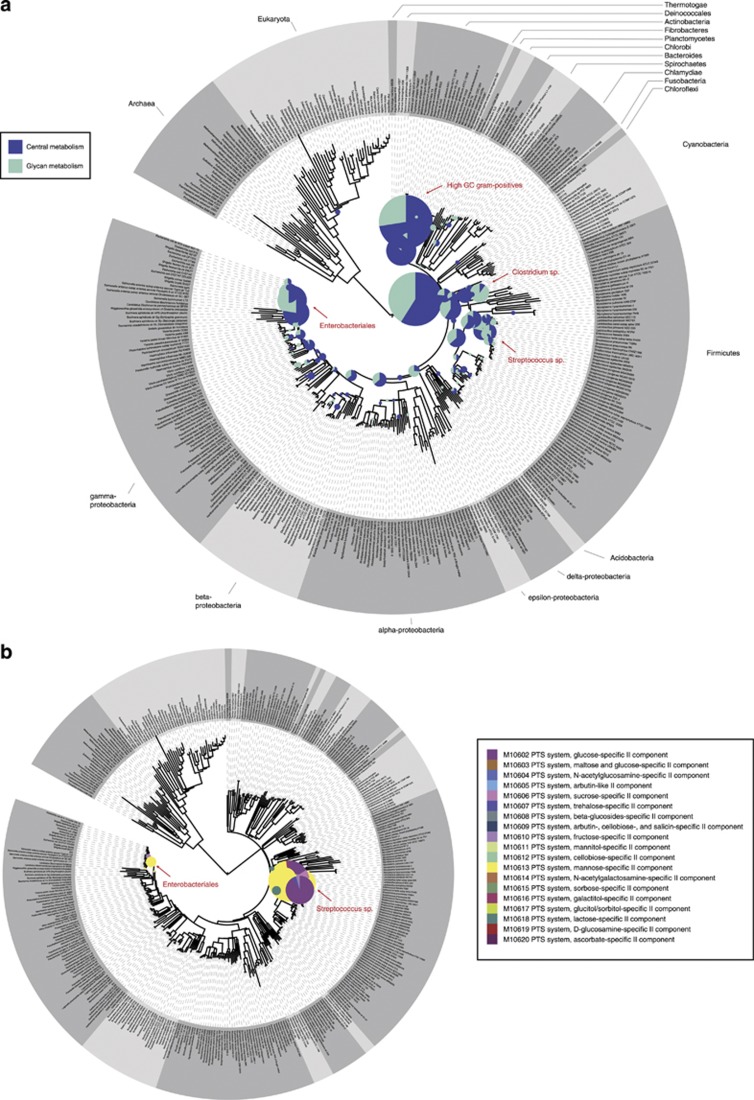
Comparative metagenomics indicated that several pathways and functions related to carbohydrate uptake and metabolism were highly enriched in the collective small intestinal metagenome compared with those of fecal metagenomes (Figure 2; Supplementary Table S3 and S4). In contrast, membrane proteins, enzymes related to metal binding and proteins with unknown functions were enriched in the fecal metagenomes. Focusing on the KEGG modules, several sugar phosphotransferase systems (PTS), enzymes related to central metabolism (such as pentose phosphate pathway) and fermentation pathways (such as lactate and propionate fermentation), and amino acid metabolism, were highly enriched in the small intestine (Figure 2). As amino acids are regarded as fuel for the human small intestine (Wu, 1998), our findings suggest that the availability of amino acids could be limiting for bacteria in the small intestine, stimulating their de novo synthesis. The phylogenetic distribution of the genes related to central metabolic pathways was scattered across a variety of bacterial phyla (Figure 3a), indicating that microbes in the small intestine harbor an extensive repertoire of genes involved in import and utilization of a variety of sugars. Remarkably, genes related to synthesis of cofactors, such as cobalamin and biotin, were strongly enriched (Figure 2; Supplementary Table S3 and S4). The biotin synthesis genes were phylogenetically mostly linked to Proteobacteria and higher ancestral origins, but were also associated with Firmicutes and Bacteroidetes (Supplementary Figure S3). As cobalamin and biotin are important cofactors in central metabolic reactions, it is not surprising that their biosynthetic pathways are overrepresented in different phylogenetic levels as well. Notably, biotin absorption by epithelia takes place in the intestine, suggesting that the residing microbes may significantly contribute to the human biotin supply (Said, 2009).
Figure 2.
KEGG modules plotted at the metabolic pathways. Pathways that are overrepresented in small intestine versus colon are indicated in blue; pathways that are overrepresented in metatranscriptome versus metagenome are indicated in yellow; overrepresented pathways in metatranscriptome and small intestine are indicated in green. The fermentation pathways and PTS are highlighted in boxes.
Figure 3.
Phylogenetic representation of the reads. The size of the bullets represents the relative abundance of the reads per phylogenetic position. Highly abundant phylogenetic groups are indicated. (a) Representation of GS-FLX reads that showed hits to genes involved in central (purple) and glycan (green) metabolism. (b) Representation of the cDNA reads that showed hits to PTS. The colors indicate the different types of PTS.
Remarkably, Cluster of Orthologous Groups (COG) analysis also indicated enrichment of several (pro-)phages (Supplementary Table S4), which is intriguing in the light of the presence of phage-related sequences in the minimal gut metagenome (Qin et al., 2010), in combination with the recently proposed role of phages in sustaining microbial diversity via the ‘killing-the-winner' principle (Rodriguez-Valera et al., 2009). However, these suggestions appear to be contradicted by a recent report, which concludes that large intestinal phages could not be correlated to a diversity-sustaining role (Reyes et al., 2010). Consequently, the importance of phages in sustaining microbial diversity within the small intestinal microbiota remains speculative.
pH and metabolite profiles confirm the genetic potential of the microbiota
To determine if the genetic potential within the metagenomes is reflected in their activity, pH and metabolites were determined in the respective effluent samples. The pH in the morning sample (1M) was more than 1 unit lower than the afternoon sample (1A), which reflects the high nutrient availability fluctuations and cognate microbial activity in the small intestine (Table 1). Considerable concentrations of acetate, lactate and butyrate, and sometimes formate, were detected (Table 1), confirming the activity of fermenting bacteria in the small intestine. Concentrations of acetate and butyrate in the small intestine are similar to those observed in fecal samples, whereas propionate concentrations are 3–5 times lower (Schwiertz et al., 2010). The high concentrations of lactate and butyrate are in agreement with the presence of Streptococcus sp. and Clostridium cluster XIVa sp., respectively. Genes assigned to the butyrate fermentation pathway were frequently detected in the small intestinal metagenome, but were not enriched compared with colon metagenomes, which agrees with the dominance of butyrate producers in the latter niche (Pryde et al., 2002). Notably, the lactate concentration in sample A was much lower than samples 1A and 1M, whereas acetate levels were higher. This is in agreement with the low abundance of metagenome sequences associated with Streptococcus and the relative abundance of E. coli and Clostridium in sample A (Supplementary Figures S1 and S2), confirming high population and functional dynamics in this ecosystem.
Table 1. pH and metabolites (mM) in ileostomy effluent.
| Sample | A | 1M | 1A |
|---|---|---|---|
| pH | 6.3 | 5.6 | 6.8 |
| Citric acid | 0.6 | 1.1 | 8.2 |
| Pyruvate | n.a. | n.a. | n.a. |
| Succinate | 1.0 | 2.5 | 0.8 |
| Lactate | 2.5 | 11.0 | 11.6 |
| Formate | n.a. | 26.0 | 7.8 |
| Acetate | 98.2 | 53.8 | 41.7 |
| Propionate | 3.4 | 2.7 | 3.9 |
| Butyrate | 17.4 | 17.5 | 6.3 |
| Acetoin | n.a. | n.a. | n.a. |
| Ethanol | n.a. | n.a. | n.a. |
Abbreviations: A, afternoon; M, morning; n.a., not applicable.
PTS and carbohydrate metabolism are highly expressed in small intestine
As the metagenome provides only insight into the genetic potential of the ecosystem, total RNA was isolated from an ileostomy effluent sample and enriched for mRNA by selective capture methodology, followed by metatranscriptome analysis. Phylogenetic profiling of the mRNA-derived sequences indicated that the majority of these transcripts originated from Streptococcus sp. and coliforms (Supplementary Figure S4). Relative to the metagenome, the fast-growing facultative anaerobic organisms were prominently represented at the transcription level, whereas transcripts from the strict anaerobes were virtually absent from this library. As intestinal facultative anaerobes are, in general, opportunists with an r-strategy, that is they increase their proportion in the total community under favorable conditions, we speculate that they adapt fast to the fluctuating conditions in the small intestine and hence may contain higher mRNA levels when compared with strict anaerobes. The metatranscriptome was strongly enriched for PTS and other carbohydrate transport systems, as well as energy- and central metabolic, and amino acid conversion pathways as compared with the metagenome, which is indicative for fast adaptation to nutrient availability (Figure 3b; Supplementary Table S5 and S6). Phylogenetic profiling of the PTS transcripts indicated that a majority was expressed by streptococci, suggesting that these bacteria are the main utilizers of the available carbohydrates in the small intestinal lumen (Figure 3b). Although genes related to formate and acetate production were found to be overrepresented in the metatranscriptome compared with the metagenome, transcripts of other Short Chain Fatty Acids (SCFAs) and lactate fermentation pathways were also frequently detected. Notably, carbohydrate utilization and central metabolism functions were also overrepresented in fecal communities, but appeared to be more dedicated to the degradation of complex carbohydrates rather than simple sugars (Klaassens et al., 2009; Turnbaugh et al., 2010). Moreover, shotgun metaproteomics revealed the enrichment of translation, energy production and carbohydrate metabolism in feces (Verberkmoes et al., 2009). Neither of these studies revealed a high enrichment of sugar PTS transport modules, which contrast to our observations of the small intestine microbiota. These findings indicate that rapid internalization and conversion of the available (simple) carbohydrates to support growth is a prominent strategy for microbial proliferation and maintenance in the small intestine.
Ecological model of the small intestinal microbiota
In agreement with previous findings (Finegold et al., 1970, 1983; Booijink et al., 2010), our data indicate that the microbiota of the small intestine consists of facultative and strict anaerobes. However, we observed a large proportion of anaerobes from the Clostridium cluster XIVa in our effluent samples, such as Coprococcus eutactus and relatives, which was detected up to 6% of the total community. Therefore, our data do not confirm a recent study that concluded oxygen exposure to have a major impact at the end of the terminal ileum in ileostomy patients (Hartman et al., 2009). Although we cannot fully exclude a potential role of oxygen, we consider it more likely that the absence of a colonic reflux explains our observations. We generically profiled the microbiota of non-medicated subjects that had an ileostoma for many years, whereas Hartman et al. (2009) studied selected microbial targets by quantitative PCR in patients that recently underwent surgery, which might explain the difference.
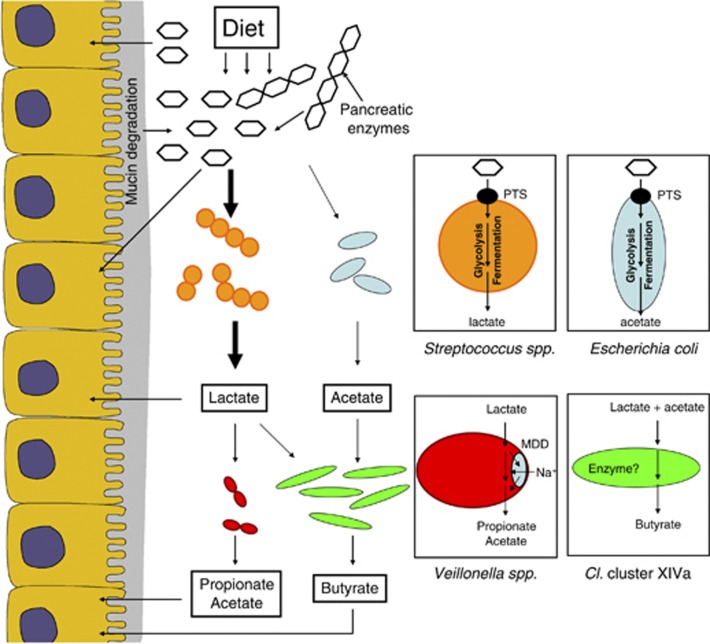
In this study, we generated a combination of molecular ecological data sets to describe the small intestine, using an ecosystems biology approach (Raes and Bork, 2008). On the basis of all data, we hypothesize that microbial communities in the human small intestine are dependent on the capacities of microbes to rapidly import and ferment the available carbohydrates, which are combined into a metabolic cooperative network to support maintenance of various microbial groups (Figure 4). This model is in agreement with the dominance and high-level activity of the small intestinal streptococci, which are renowned fast-growing facultative anaerobes that can ferment relatively simple carbohydrates at a high rate to produce mainly lactate (Holt, 1994). To successfully utilize such carbohydrates in the small intestine, these microbes have to compete with the host for many of these substrates. Consequently, PTS and other transport systems, as well as the downstream conversion pathways (e.g., glycolysis, pentose phosphate pathway), need to be efficient and highly expressed, which was clearly reflected by the metatranscriptome sequences. Although PTS systems of E. coli and relatives were also found in the metatranscriptome, their abundance was less compared with those of the Streptococcus sp., suggesting that the latter bacteria most effectively fulfill the selective demands of the small intestinal habitat. This is in line with previous studies that describe the dominant abundance of Streptococcus sp. in the human small intestine (Hayashi et al., 2005; Wang et al., 2005; Booijink, 2009; Booijink et al., 2010). As the host absorbs most simple carbohydrates in the proximal small intestine, it can be anticipated that part of the simple carbohydrates that are available in more distal small intestine regions derive from food polysaccharides that are converted by enzymes excreted by the host, or from microbial conversion of complex carbohydrates, such as mucin or dietary carbohydrates that are not converted by the host's digestive enzymes. The fermentation products from these facultative anaerobes will be available to the host epithelial cells as additional energy source, but can also serve as substrate for other microbes, such as Veillonella sp., members of the Clostridium cluster XIVa group that are able to convert lactate and acetate into propionate and butyrate, respectively (Ng and Hamilton, 1971; Pryde et al., 2002; Duncan et al., 2004).
Figure 4.
Small intestinal microbiome model based on the 16S rRNA, metabolite, metagenome and metatranscriptome data.
The relatively high concentration of SCFAs in the effluent samples was found to be similar to those frequently observed in the feces. This might be explained by the high activity of the microbes in the small intestine, which is required for their persistence in this ecosystem that is characterized by short residence times. In addition, this finding may in part explain why people without a colon can in general live a normal life, which by itself is remarkable, and deserves more attention in future studies.
Overall, this is the first study that presents an ecological model of the small intestinal microbiota, based on metagenome, -transcriptome, metabolite and community-profiling data. To facilitate the construction of this ecological model, we focused in this study on several samples from one individual rather than single time points from several individuals to capture population dynamics, which is more prominent in the small intestine compared with the large intestine (Booijink et al., 2010). Our study indicates that the small intestine communities are depending on the capacity for fast import and conversion of relatively simple carbohydrates and rapid adaptation to the overall nutrient availability, which contrasts the specialization towards the efficient degradation of complex carbohydrates that is seen in colonic microbial communities.
Acknowledgments
We are grateful to all volunteers who provided their small intestinal content. We also acknowledge Professor Robert-Jan Brummer (now at Örebro University), and Jeroen Maljaars and Daniel Keszthelyi who assisted during collection of the small intestinal content at Maastricht University Medical Centre. GATC Biotech is thanked for construction and sequencing of the metagenome and metatranscriptome libraries. Dr Peter Schaap of Wageningen University, Professor Roland Siezen of Radboud University Nijmegen and MSc K Cuelenaere of Dalicon BV were involved in setting up some initial bioinformatics analyses and discussing metagenome data analysis.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Ahmed S, Macfarlane GT, Fite A, McBain AJ, Gilbert P, Macfarlane S. Mucosa-associated bacterial diversity in relation to human terminal ileum and colonic biopsy samples. Appl Environ Microbiol. 2007;73:7435–7442. doi: 10.1128/AEM.01143-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumugam M, Harrington ED, Foerstner KU, Raes J, Bork P. SmashCommunity: a metagenomic annotation and analysis tool. Bioinformatics. 2010;26 (23:2977–2978. doi: 10.1093/bioinformatics/btq536. [DOI] [PubMed] [Google Scholar]
- Berridge BR, Fuller JD, de Azavedo J, Low DE, Bercovier H, Frelier PF. Development of specific nested oligonucleotide PCR primers for the Streptococcus iniae 16S-23S ribosomal DNA intergenic spacer. J Clin Microbiol. 1998;36:2778–2781. doi: 10.1128/jcm.36.9.2778-2781.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besemer J, Borodovsky M. GeneMark: web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Res. 2005;33:W451–W454. doi: 10.1093/nar/gki487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim HC. A rapid alkaline extraction method for the isolation of plasmid DNA. Methods Enzymol. 1983;100:243–255. doi: 10.1016/0076-6879(83)00059-2. [DOI] [PubMed] [Google Scholar]
- Booijink CCGM. Analysis of Diversity and Function of the Human Small Intestinal Microbiota. Wageningen University: Wageningen, The Netherlands; 2009. [Google Scholar]
- Booijink CCGM, El-Aidy S, Rajilić-Stojanović M, Heilig HGHJ, Troost FJ, Smidt H, et al. High temporal and inter-individual variation detected in the human ileal microbiota. Environ Microbiol. 2010;12:3213–3227. doi: 10.1111/j.1462-2920.2010.02294.x. [DOI] [PubMed] [Google Scholar]
- Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;312:697. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- Diguistini S, Liao NY, Platt D, Robertson G, Seidel M, Chan SK, et al. De novo genome sequence assembly of a filamentous fungus using Sanger, 454 and Illumina sequence data. Genome Biol. 2009;10:R94. doi: 10.1186/gb-2009-10-9-r94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan SH, Louis P, Flint HJ. Lactate-utilizing bacteria, isolated from human feces, that produce butyrate as a major fermentation product. Appl Environ Microbiol. 2004;70:5810–5817. doi: 10.1128/AEM.70.10.5810-5817.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finegold SM, Sutter VL, Boyle JD, Shimada K. The normal flora of ileostomy and transverse colostomy effluents. J Infect Dis. 1970;122:376–381. doi: 10.1093/infdis/122.5.376. [DOI] [PubMed] [Google Scholar]
- Finegold SM, Sutter VL, Mathisen GE.1983Normal indigenous intestinal floraIn: Hentges DJ (ed).Human Intestinal Microflora in Health and Disease Academic press: New York; 3–31. [Google Scholar]
- Gianoulis TA, Raes J, Patel PV, Bjornson R, Korbel JO, Letunic I, et al. Quantifying environmental adaptation of metabolic pathways in metagenomics. Proc Natl Acad Sci USA. 2009;106:1374–1379. doi: 10.1073/pnas.0808022106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbach SL, Nahas L, Weinstein L, Levitan R, Patterson JF. Studies of intestinal microflora IV. The microflora of ileostomy effluent: a unique microbial ecology. Gastroenterol. 1967;53:874–880. [PubMed] [Google Scholar]
- Hartman AL, Lough DM, Barupal DK, Fiehn O, Fishbein T, Zasloff M, et al. Human gut microbiome adopts an alternative state following small bowel transplantation. Proc Natl Acad Sci USA. 2009;106:17187–17192. doi: 10.1073/pnas.0904847106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi H, Sakamoto M, Benno Y. Evaluation of three different forward primers by terminal restriction fragment length polymorphism analysis for determination of fecal bifidobacterium spp. in healthy subjects. Microbiol Immunol. 2004;48:1–6. doi: 10.1111/j.1348-0421.2004.tb03481.x. [DOI] [PubMed] [Google Scholar]
- Hayashi H, Takahashi R, Nishi T, Sakamoto M, Benno Y. Molecular analysis of jejunal, ileal, caecal and recto-sigmoidal human colonic microbiota using 16S rRNA gene libraries and terminal restriction fragment length polymorphism. J Med Microbiol. 2005;54:1093–1101. doi: 10.1099/jmm.0.45935-0. [DOI] [PubMed] [Google Scholar]
- Holt JG.Genus StreptococcusIn: Bergy DH, Holt JG, Krieg NR, Sneath PH (eds).Bergey's Manual of Determinative Bacteriology Lippincott Williams & Wilkins: Baltimore, MD; 1994. p523 [Google Scholar]
- Huson DH, Auch AF, Qi J, Schuster SC. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen LJ, Julien P, Kuhn M, von Mering C, Muller J, Doerks T, et al. eggNOG: automated construction and annotation of orthologous groups of genes. Nucleic Acids Res. 2008;36:D250–D254. doi: 10.1093/nar/gkm796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LR.(ed). (2006Physiology of the Gastrointestinal Tract4 edn.Academic Press: San Diego, CA [Google Scholar]
- Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaassens ES, Boesten RJ, Haarman M, Knol J, Schuren FH, Vaughan EE, et al. Mixed-species genomic microarray analysis of fecal samples reveals differential transcriptional responses of bifidobacteria in breast- and formula-fed infants. Appl Environ Microbiol. 2009;75:2668–2676. doi: 10.1128/AEM.02492-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunin V, Raes J, Harris JK, Spear JR, Walker JJ, Ivanova N, et al. Millimeter-scale genetic gradients and community-level molecular convergence in a hypersaline microbial mat. Mol Syst Biol. 2008;4:198. doi: 10.1038/msb.2008.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa K, Itoh T, Kuwahara T, Oshima K, Toh H, Toyoda A, et al. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 2007;14:169–181. doi: 10.1093/dnares/dsm018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leplae R, Hebrant A, Wodak SJ, Toussaint A. ACLAME: a CLAssification of Mobile genetic Elements. Nucleic Acids Res. 2004;32:D45–D49. doi: 10.1093/nar/gkh084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leps J, Smilauer P.2003Multivariate Analysis of Ecological Data using CANOCO Cambridge University Press; Cambridge, UK. [Google Scholar]
- Ng SKC, Hamilton IR. Lactate metabolism by Veillonella parvula. J Bacteriol. 1971;105:999–1005. doi: 10.1128/jb.105.3.999-1005.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse B, Jellema RH, van der Werf MJ. Quenching of microbial samples for increased reliability of microarray data. J Microbiol Methods. 2006;64:207–216. doi: 10.1016/j.mimet.2005.04.035. [DOI] [PubMed] [Google Scholar]
- Pryde SE, Duncan SH, Hold GL, Stewart CS, Flint HJ. The microbiology of butyrate formation in the human colon. FEMS Microbiol Lett. 2002;217:133. doi: 10.1111/j.1574-6968.2002.tb11467.x. [DOI] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raes J, Bork P. Molecular eco-systems biology: towards an understanding of community function. Nat Rev Microbiol. 2008;6:693–699. doi: 10.1038/nrmicro1935. [DOI] [PubMed] [Google Scholar]
- Raes J, Foerstner KU, Bork P. Get the most out of your metagenome: computational analysis of environmental sequence data. Curr Opin Microbiol. 2007;10:490–498. doi: 10.1016/j.mib.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Rajilić-Stojanović M, Heilig HGHJ, Molenaar D, Kajander K, Surakka A, Smidt H, et al. Development and application of the human intestinal tract Chip (HITChip), a phylogenetic microarray: absence of universally conserved phylotypes in the abundant microbiota of young and elderly adults. Environ Microbiol. 2009;11:1736–1751. doi: 10.1111/j.1462-2920.2009.01900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajilić-Stojanović M, Smidt H, de Vos WM. Diversity of the human gastrointestinal tract microbiota revisited. Environ Microbiol. 2007;9:2125–2136. doi: 10.1111/j.1462-2920.2007.01369.x. [DOI] [PubMed] [Google Scholar]
- Reyes A, Haynes M, Hanson N, Angly FE, Heath AC, Rohwer F, et al. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466:334–338. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Valera F, Martin-Cuadrado AB, Rodriguez-Brito B, Pasiæ L, Thingstad TF, Rohwer F, et al. Explaining microbial population genomics through phage predation. Nat Rev Microbiol. 2009;7:828–836. doi: 10.1038/nrmicro2235. [DOI] [PubMed] [Google Scholar]
- Said HM. Cell and molecular aspects of human intestinal biotin absorption. J Nutr. 2009;139:158–162. doi: 10.3945/jn.108.092023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T.1989Calculating melting temperatures for perfectly matched hybrids between oligonucleotides and their target sequences Molecular Cloning – A Laboratory Manual Cold Spring Habour Laboratory Press: New York; p11–46. [Google Scholar]
- Schwiertz A, Taras D, Schäfer K, Beijer S, Bos NA, Donus C, et al. Microbiota and SCFA in lean and overweight healthy subjects. Obesity. 2010;18:190–195. doi: 10.1038/oby.2009.167. [DOI] [PubMed] [Google Scholar]
- Starrenburg MJC, Hugenholtz J. Citrate fermentation by Lactococcus and Leuconostoc spp. Appl Environ Microbiol. 1991;57:3535–3540. doi: 10.1128/aem.57.12.3535-3540.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troost FJ, van Baarlen P, Lindsey P, Kodde A, de Vos WM, Kleerebezem M, et al. Identification of the transcriptional response of human intestinal mucosa to Lactobacillus plantarum WCFS1 in vivo. BMC Genomics. 2008;9:374. doi: 10.1186/1471-2164-9-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Quince C, Faith JJ, McHardy AC, Yatsunenko T, Niazi F, et al. Organismal, genetic, and transcriptional variation in the deeply sequenced gut microbiomes of identical twins. Proc Natl Acad Sci USA. 2010;107:7503–7508. doi: 10.1073/pnas.1002355107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verberkmoes NC, Russell AL, Shah M, Godzik A, Rosenquist M, Halfvarson J, et al. Shotgun metaproteomics of the human distal gut microbiota. ISME J. 2009;3:179–189. doi: 10.1038/ismej.2008.108. [DOI] [PubMed] [Google Scholar]
- Wang M, Ahrne S, Jeppsson B, Molin G. Comparison of bacterial diversity along the human intestinal tract by direct cloning and sequencing of 16S rRNA genes. FEMS Microbiol Ecol. 2005;54:219. doi: 10.1016/j.femsec.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Wang X, Heazlewood SP, Krause DO, Florin THJ. Molecular characterization of the microbial species that colonize human ileal and colonic mucosa by using 16S rDNA sequence analysis. J Appl Microbiol. 2003;95:508–520. doi: 10.1046/j.1365-2672.2003.02005.x. [DOI] [PubMed] [Google Scholar]
- Willing BP, Dicksved J, Halfvarson J, Andersson AF, Lucio M, Zheng Z, et al. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology. 2010;139:1844–1854. doi: 10.1053/j.gastro.2010.08.049. [DOI] [PubMed] [Google Scholar]
- Wu G. Intestinal mucosal amino acid catabolism. J Nutr. 1998;128:1249–1252. doi: 10.1093/jn/128.8.1249. [DOI] [PubMed] [Google Scholar]
- Yang X, Xie L, Li Y, Wei C. More than 9,000,000 unique genes in human gut bacterial community: estimating gene numbers inside a human body. PLoS One. 2009;29:e6074. doi: 10.1371/journal.pone.0006074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoetendal EG, Booijink CC, Klaassens ES, Heilig HG, Kleerebezem M, Smidt H, et al. Isolation of RNA from bacterial samples of the human gastrointestinal tract. Nat Protoc. 2006a;1:954–959. doi: 10.1038/nprot.2006.143. [DOI] [PubMed] [Google Scholar]
- Zoetendal EG, Heilig HG, Klaassens ES, Booijink CC, Kleerebezem M, Smidt H, et al. Isolation of DNA from bacterial samples of the human gastrointestinal tract. Nat Protoc. 2006b;1:870–873. doi: 10.1038/nprot.2006.142. [DOI] [PubMed] [Google Scholar]
- Zoetendal EG, Rajilić-Stojanović M, de Vos WM. High throughput diversity and functionality analysis of the gastrointestinal tract microbiota. Gut. 2008;57:1605–1615. doi: 10.1136/gut.2007.133603. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.