Abstract
Type 1 diabetes results from T cell–mediated β-cell destruction. The HLA-A*24 class I gene confers significant risk of disease and early onset. We tested the hypothesis that HLA-A24 molecules on islet cells present preproinsulin (PPI) peptide epitopes to CD8 cytotoxic T cells (CTLs). Surrogate β-cell lines secreting proinsulin and expressing HLA-A24 were generated and their peptide ligandome examined by mass spectrometry to discover naturally processed and HLA-A24–presented PPI epitopes. A novel PPI epitope was identified and used to generate HLA-A24 tetramers and examine the frequency of PPI-specific T cells in new-onset HLA-A*24+ patients and control subjects. We identified a novel naturally processed and HLA-A24–presented PPI signal peptide epitope (PPI3–11; LWMRLLPLL). HLA-A24 tetramer analysis reveals a significant expansion of PPI3–11-specific CD8 T cells in the blood of HLA-A*24+ recent-onset patients compared with HLA-matched control subjects. Moreover, a patient-derived PPI3–11-specific CD8 T-cell clone shows a proinflammatory phenotype and kills surrogate β-cells and human HLA-A*24+ islet cells in vitro. These results indicate that the type 1 diabetes susceptibility molecule HLA-A24 presents a naturally processed PPI signal peptide epitope. PPI-specific, HLA-A24–restricted CD8 T cells are expanded in patients with recent-onset disease. Human islet cells process and present PPI3–11, rendering themselves targets for CTL-mediated killing.
The hallmark of type 1 diabetes is the destruction of insulin-producing β-cells in the pancreas. The processes and immune cells involved are not fully understood as yet, but there is an emerging view that CD8 T cells have a major role in β-cell death. Evidence for this includes the fact that CD8 T cells are the predominant population of lymphocytes infiltrating the islets in newly diagnosed type 1 diabetic patients (1). Moreover, their frequency is inversely correlated with insulin positivity, and once insulin content within an islet is lost, the number of CD8 T cells declines (2). Several studies show an increased expression of HLA class I on islet cells in type 1 diabetic patients (1,3,4), which implies an enhanced susceptibility to CD8 T cell–mediated killing. Finally, in a previous study, we recapitulated the CD8 T-cell pathway of selective β-cell death in vitro by showing that T-cell clones generated from the blood of a patient, which recognize a peptide of preproinsulin (PPI) presented by HLA-A2*0201, can mediate specific β-cell killing (5).
More recent, further weight has been added to the importance of the CD8 T-cell autoantigen/HLA class I pathway by the discovery in a large-scale single nucleotide polymorphism study of allelic forms of HLA class I genes that confer significant type 1 diabetes risk (6). Within these alleles, HLA-A*24 has a strong disease-predisposing effect (odds ratio ∼1.5) and also is significantly associated with a younger age of diabetes onset (6). The HLA-A*24 supertype is present in 12–20% of Caucasian and ∼60% of Japanese populations, with HLA-A*2402 being the most common variant (7,8).
We hypothesized that the presence of HLA-A24 molecules confers risk for type 1 diabetes through presentation of epitopes from key β-cell autoantigens and that recognition of these by CD8 cytotoxic T cells (CTLs), when displayed on the β-cell surface, contributes to β-cell death. We therefore examined the HLA-A24 peptide ligandome of a surrogate human β-cell line generated by transfection of HLA-A*24 and INS genes into an inert HLA-negative carrier cell. We elected to study PPI as the index autoantigen, since there is evidence that proinsulin is the major target of the autoimmune response in children developing type 1 diabetes (9). In the current study, we identify a new epitope naturally processed and presented from the signal peptide of PPI and examine the prevalence of circulating PPI-specific CD8 T cells in newly diagnosed, HLA-A*24+ type 1 diabetic patients. Of import, we show the generation of CD8 T-cell clones specific for the HLA-A24–PPI complex and use cytotoxicity assays to examine the natural processing and presentation of PPI by human islet cells. These data highlight the importance of the PPI signal peptide in shaping the β-cell–specific autoreactive T-cell repertoire in type 1 diabetes and elucidate potential mechanisms through which HLA class I molecules might contribute to the pathogenesis of the disease.
RESEARCH DESIGN AND METHODS
Transfected cell lines.
PPI cDNA was inserted between the BamHI and EcoRI sites in pcDNA6/myc-His B vector (Invitrogen, Paisley, Scotland, U.K.) for expression in mammalian cell lines. HLA-A*2402 was cloned from Epstein Barr virus–transformed B cells and inserted between BamHI and EcoRI sites in pcDNA3.1 vector (Invitrogen). K562 cells were cultured in RPMI-1640 medium supplemented with 10% heat-inactivated FCS and 1% penicillin/streptomycin solution (all Invitrogen) at 37°C, 5% CO2, and transfected using Effectene (QIAGEN, Hilden, Germany) to yield K562-PPI, K562-A24, and K562-A24-PPI cell lines that were maintained in the presence of appropriate selection media (Blasticidin 10 μg/mL and Geneticin 0.7 mg/mL). Expression of HLA class I molecules was confirmed by flow cytometry using anti–HLA-ABC antibody (W6/32; AbD Serotec, Oxford, U.K.). Proinsulin was detected in supernatant of overnight cultures by ELISA (DRG International, Marburg, Germany). Next, 5 × 109 cells from the relevant cell line were harvested and washed with PBS, and cell pellets were stored at −80°C.
Affinity purification and mass spectrometry.
Immunoaffinity purification of HLA class I molecules and peptide extraction were carried out as previously described (5). Peptides were resuspended in 100/0.1 v/v water/formic acid and analyzed by an online nano high-performance liquid chromatography–mass spectrometry system (10). Next, 15-µL injections were added onto a precolumn (Reprosil, 15 mm × 100 µm) and eluted via an analytical column (Reprosil, 20 cm × 50 μm), and peptides were resolved using a gradient of 30 min to 50% solvent B (10/90/0.1 v/v/v water/acetonitrile/formic acid). The analytical column was drawn to a tip of ∼5 μm internal diameter, and the tip functioned as the electrospray needle of the nanoelectrospray source of an LTQ FT Ultra mass spectrometer (Thermo Scientific, Bremen, Germany). All tandem mass spectra were searched against the human International Protein Index database with the database search program MASCOT version 2.2.04. Peptides with MASCOT scores <40 were discarded.
Subjects.
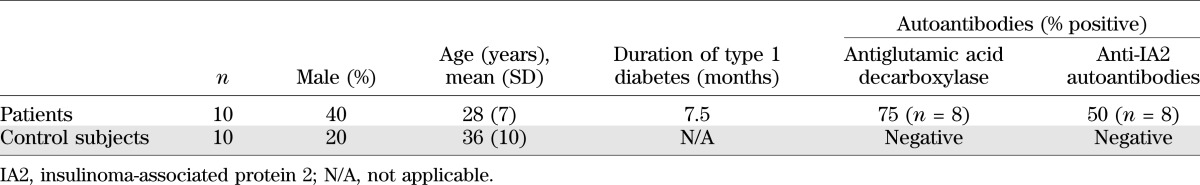
Fresh heparinized blood samples were obtained from 10 HLA-A*24+ patients with recent-onset type 1 diabetes and 10 HLA-A*24+ nondiabetic control subjects (Table 1). Studies were carried out with the approval of the local research ethics committee, and informed consent was obtained from all participants. HLA-A*24 genotype was identified by PCR sequence-specific primer sequencing. Peripheral blood mononuclear cells (PBMCs) were isolated fresh on density gradients (Lymphoprep; Nycomed, Oslo, Norway) and washed in RPMI-1640 with 1% penicillin/streptomycin and 2% human AB serum (all Invitrogen) twice before use.
TABLE 1.
Characteristics of type 1 diabetic patients and control subjects
Ex vivo tetramer analysis of CD8 T-cell responses.
HLA-A24 tetramers labeled with phycoerythrin (PE) and allophycocyanin (APC) were generated as described (5) and loaded with either PPI3–11 (LWMRLLPLL) or the immunodominant, HLA-A24–presented cytomegalovirus (CMV) peptide (IE1248–257, AYAQKIFKIL; referred to as CMV-AYA) (11,12). Patient and control subject samples were examined for the presence of tetramer-positive CD8 T cells using 3 × 106 PBMCs washed twice with PBS and stained with a live/dead violet amine reactive dye (Invitrogen) for 15 min at room temperature, followed by washing and incubation with 100 nmol/L protein kinase inhibitor (dasatinib) (13) for 30 min at 37°C, 5% CO2. Subsequently, PBMCs were stained for 15 min at 37°C, 5% CO2, with either dual-labeled (PE and APC) PPI3–11 or single-labeled (PE) CMV-AYA tetramers. After washing in PBS, cells were stained with the following surface monoclonal antibodies: anti-CD3 V500, anti-CD8 APC-H7 (both BD Biosciences, San Jose, CA), anti-CD14 Pacific Blue, and anti-CD19 Pacific Blue (both Invitrogen). The gating strategy can be found in Supplementary Fig. 1. Stained PBMC were maintained on ice and in the dark until acquisition on the same day on a BD FACSCanto II (BD Biosciences) and analyzed using FlowJo software (Tree Star, Ashland, OR).
Generation of CD8 T-cell clones.
CD8 T-cell clones specific for PPI3–11 were generated as described (5). In brief, blood monocytes were cultured in X-Vivo 15 medium (Lonza, Walkersville, MD) supplemented with 5% AB, 100 IU/mL penicillin and 100 μg/mL streptomycin (Invitrogen), 100 ng/mL interleukin (IL)-4, and 50 ng/mL granulocyte-microphage colony-stimulating factor (Biosource/Invitrogen, Paisley, Scotland, U.K.) to obtain monocyte-derived immature dendritic cells (iDCs), which were harvested on day 6 and cryopreserved. iDCs were thawed 24 h before arrival of a fresh patient sample, washed in X-Vivo/5% AB serum, and split at a ratio of 4:1 in which 4 parts were stimulated in media supplemented with tumor necrosis factor (TNF)-α (25 ng/mL; Miltenyi Biotec, Bergisch Gladbach, Germany), IL-1β (10 ng/mL; Strathmann Biotech, Hannover, Germany), IL-6 (100 ng/mL; Miltenyi Biotec), and prostaglandin E-2 (1 μg/mL; Sigma-Aldrich, Poole, U.K.) and 1 part with poly I:C (12.5 μg/mL; InvivoGen, San Diego, CA). After 24 h, matured dendritic cells (DCs) were harvested, washed, and pulsed with 10 µg PPI3–11 peptide in X-Vivo/5% AB serum for 1 h at 37°C. After extensive washing, matured DCs were cultured at a 1:30 ratio with freshly obtained immunomagnetically isolated CD8 T cells (CD8 Isolation Kit II; Miltenyi Biotec) in 48-well culture plates in X-vivo/5% AB serum supplemented with 10 ng/mL IL-7 (PeproTech, Rocky Hill, NJ), 0.1 ng/mL IL-15, 5% Cellkine (Helvetica Health Care, Geneva, Switzerland), and 25 IU/mL IL-2 (Prometheus/Novartis, San Diego, CA). On day 10, cells were harvested and stained with anti-CD3 (V500), anti-CD8 (APC H7), violet amine reactive dye (live/dead exclusion; pacific blue), and PPI3–11 tetramer (PE and APC) as appropriate. CD3+CD8 double tetramer-positive T cells were flow sorted (for gating strategy, see Supplementary Fig. 1) and seeded at one cell per well into 96-well plates containing 0.2 × 106 irradiated PBMCs in culture medium (CM; X-Vivo/5% AB serum, with IL-7 [10 ng/mL], IL-15 [0.1 ng/mL], IL-2 [100 IU/mL], and 2.5% Cellkine) and phytohemagglutinin-M (5 μg/mL; Sigma-Aldrich). Growing CD8 T-cell clones were restimulated every 15–20 days. CMV-AYA clones were generated directly ex vivo after staining of PBMCs as above using CMV-AYA tetramers.
Cytotoxicity assays.
HLA-A*24+ human islets were obtained with appropriate ethical approval from the King’s College Hospital Islet Transplantation Unit. As described previously (5), islet-enriched cell fractions were cultured in CMRL1066 containing 5.6 mmol/L glucose, supplemented with 10% FCS, 100 units/mL penicillin, 100 μg/mL streptomycin, 2 mmol/L l-Gln (Invitrogen), 500 IU/mL interferon (IFN)-γ, 50 IU/mL IL-1β, 2,500 IU/mL TNF-α (all Miltenyi), and 1,000 IU/mL IFN-α (Roche, Mannheim, Germany) to upregulate HLA class I expression (5). Cytotoxicity was analyzed by a nonradioactive europium hydrophilic ligand cytotoxicity assay using DELFIA Technology (PerkinElmer) as described (5). When using K562 cell lines, 1 × 106 K562-A24 cells were pulsed with 10 µg PPI3–11 for 1 h at 37°C or left unpulsed (K562-A24-PPI and K562-A24). Target cells (2.5 × 103/well) were seeded into each of the 96-well plates containing CD8 T-cell clone in CM, and after 4 h at 37°C, 20 μL supernatant was removed for measurement of the fluorescent signal (FLUOstar Omega Time-Resolved Fluorescence Reader, BMG Labtech, Offenburg, Germany). To determine maximum lysis, K562 or islets cells were treated with 20 μL lysis buffer. Spontaneous release was measured from wells containing labeled K562 or islet cells as appropriate. Specific lysis was calculated as follows: % specific lysis = (experimental-spontaneous release) × 100/(maximum-spontaneous release).
Cytokine analysis.
Clone cells (2 × 105) were seeded in duplicate into 96-well plates and incubated with 2 × 104 islet cells for 18 h at 37°C in CM. Positive control stimulation was achieved with 50 ng/mL phorbol myristic acid (PMA) and 10 µg/mL ionomycin. Supernatant (100 μL) was removed and analyzed in duplicate with a cytokine multiplex assay (Millipore, Billerica, MA), and plates were analyzed on a Luminex Flexmap 3D system (Invitrogen).
T-cell receptor sequencing.
T-cell receptor (TCR) clonotyping was performed from tetramer-sorted clone cells pelleted and resuspended in RNAlater (Ambion, Austin, TX). Total RNA was extracted (RNeasy Mini Kit, QIAGEN) and first strand synthesis of 5′ cDNA fragments prepared using the SMARTer 5′ RACE cDNA Amplification Kit (Clontech, Palo Alto, CA). PCR of the tailed cDNA was performed using a TCR β-constant region–specific primer (5′-GCT-GACCCCACTGTGCACCTCCTTCCC-3′) or α-constant region–specific primer (5′-CCAGGCCACAGCACTGTTGCTCTTGAAGTCC-3′) with Phusion High-Fidelity enzyme (Finnzymes, Oy, Finland). PCR products were purified by gel extraction, A-tailed, ligated into the pGEM-T Easy Vector System I (Promega, Madison, WI), transformed into TOP10 cells (Invitrogen), and sequences determined using the IMGT/V-QUEST tool (14).
Statistical analysis.
A two-tailed Mann-Whitney test was used for comparing the frequency of tetramer-positive cells between disease groups. A two-tailed Spearman test was used to assess the correlation between the percentage of CMV-AYA and PPI3–11-specific CD8 T cells, with P < 0.05 considered significant.
RESULTS
Identification of a novel naturally processed PPI epitope presented by HLA-A24.
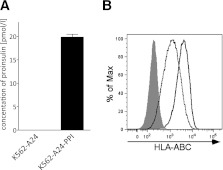
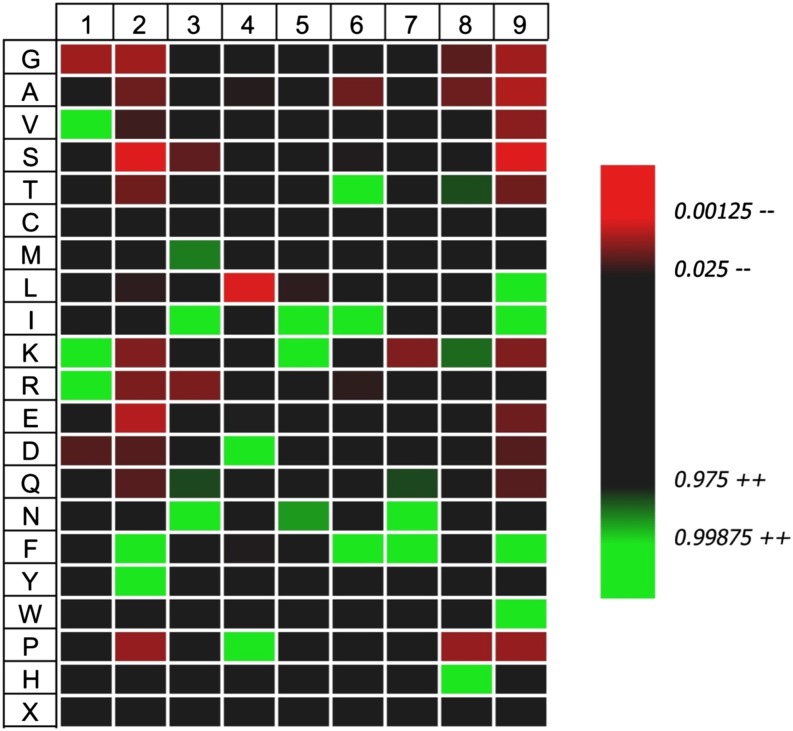
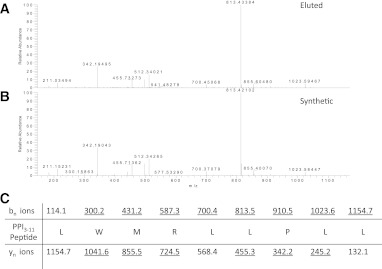
Using a previously described approach (5), we transfected the HLA-negative myelogenous leukemia cell line K562 with INS encoding human PPI and HLA-A*2402 to generate surrogate β-cells (K562-A24-PPI) secreting proinsulin and expressing HLA-A24 molecules on their surface (Fig. 1). We then affinity purified HLA-A24 from bulk cultures of these cells and acid eluted the peptide ligandome. Using stringent mass spectra matching (MASCOT scores ≥40), we identified 179 peptides of 8–12 amino acid length that represent variants of 165 unique peptides, with the vast majority (60%) being 9-mers. In accordance with earlier reports (15,16), we found a strong preference for an aromatic residue in position P2 (90% Tyr or Phe) and a nonhydrophobic amino acid at the c-terminus (89% Leu, Phe, or Ile) (Fig. 2). Within the peptide pool, we identified a sequence of PPI signal peptide (PPI3–11) (Fig. 3A) that was confirmed by the tandem mass spectrometry profile of the synthetic compound (Fig. 3B). It is intriguing that PPI3–11 has a Trp at P2; only 1.9% of 9-mer peptides we identified had a Trp at this position, and Trp at P2 is highly unusual in published algorithms of HLA-A24 binding peptides (15,16). Within our peptide pool of 9-mers, there were no peptides with nucleophilic (Ser, Thr, and Cys), acidic (Asp and Glu), or basic (His, Lys, and Arg) amino acids in P2. There was a strong preference for hydrophobic amino acids at P9, although amino acids with neutral side chains (Trp and Tyr) were also tolerated (Fig. 2). Although at other positions certain amino acids were significantly more frequently found (e.g., Ile, Lys, and Asn in P5), there were no or very few amino acids that were not tolerated at those positions. Overall, only P2 and P9 showed a restrictive amino acid pattern.
FIG. 1.
Generation and characterization of surrogate β-cell line. The myelogenous leukemia cell line K562 is transfected with HLA-A*2402 and INS genes to express PPI (A), such that immunoreactive insulin is detectable in culture supernatants by ELISA, and HLA-A24 (B), shown here detected by flow cytometry using a pan–HLA-ABC monoclonal antibody W6/32. Expression of major histocompatibility complex class I molecules on K562-A24-PPI cells (solid line) was slightly higher than in the K562-A24 cells (dashed line). Isotype control staining was similar in both cell lines; gray filled curve shows K562-A24-PPI isotype staining.
FIG. 2.
Distribution of amino acids at all positions including both major anchors (P2 and P9) for all 107 9-mer peptides eluted from HLA-A*24+ K562 cells expressing PPI compared with the amino acid frequency of the human proteome. Data shown as a heatmap in which increased or decreased amino acid frequencies are drawn as a gradient of green or red shades, respectively. Only amino acids that show a significant difference in distribution from the theoretical human proteome are shown (P < 0.05). Heatmap was created using iceLogo software (21). Trp at P2 is highly unusual for HLA-A24 but does not show as significantly upregulated in iceLogo since it is also relatively rare in the human proteome (1.4%). (A high-quality color representation of this figure is available in the online issue.)
FIG. 3.
A: Tandem mass spectrometry analysis of collision-induced dissociation revealing the tandem mass spectrum of a peptide of mass (577.86 m/z) in fraction 33. B: The correct identity of the peptide was proven by tandem mass spectrometry of the synthetic compound. C: Amino acid sequence of the peptide with the expected b- and y-fragment ions. Observed fragment ions are underlined. This confirms PPI3–11 (LMWRLLPLL) as naturally processed and presented by HLA-A24 on K562-A24-PPI cells. (A high-quality color representation of this figure is available in the online issue.)
Detection of PPI3–11-specific CD8 T cells by HLA-A24 tetramer analysis.
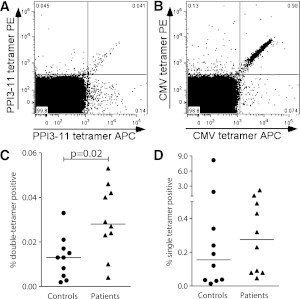
We used PPI3–11- and CMV-AYA–loaded HLA-A24 tetramers to assess the frequencies of circulating autoreactive and antiviral CD8 T cells, respectively, in recent-onset HLA-A*24+ patients with type 1 diabetes and in nondiabetic HLA-A*24+ control subjects (Table 1). To address the issue of signal to noise being unfavorable in the detection of rare autoreactive T cells, we used a dual-labeled tetramer approach for identifying PPI3–11-specific CD8 T cells similar to that used in the quantum dot studies of Velthuis et al. (17) for quantification of HLA-A2–restricted autoreactive T cells. Dual-stained PPI3–11 and CMV-AYA tetramer-positive CD8 T cells were clearly distinguishable (Fig. 4A and B and Supplementary Fig. 1). Tetramer-positive CD8 T cells reactive to PPI3–11 were significantly more frequent in recent-onset patients with type 1 diabetes compared with control subjects (P = 0.02) (Fig. 4C). In contrast, CMV-AYA–specific CD8 T cells were detected at similar levels in patients and control subjects (P = NS) (Fig. 4D). There was no significant correlation between the frequencies of PPI3–11- and CMV-AYA–specific CD8 T cells in patients and control subjects.
FIG. 4.
Representative dual tetramer staining (PE- and APC-labeled) of PBMCs from a patient with type 1 diabetes. Gated, live CD3+CD8 cells are shown stained with PPI3–11-loaded (A) and CMV-AYA–loaded (B) tetramers. C: Percentage of CD8 T cells that are dual tetramer positive when stained with PPI3–11-loaded reagents in 10 patients with recent onset type 1 diabetes and 10 control subjects, all bearing HLA-A*24. Horizontal lines represent medians, and levels are significantly higher in patients than in control subjects. D: Staining with dual tetramers loaded with CMV-AYA. Percentage dual tetramer-positive cells is similar in patients and control subjects.
Isolation of CD8 T-cell clones.
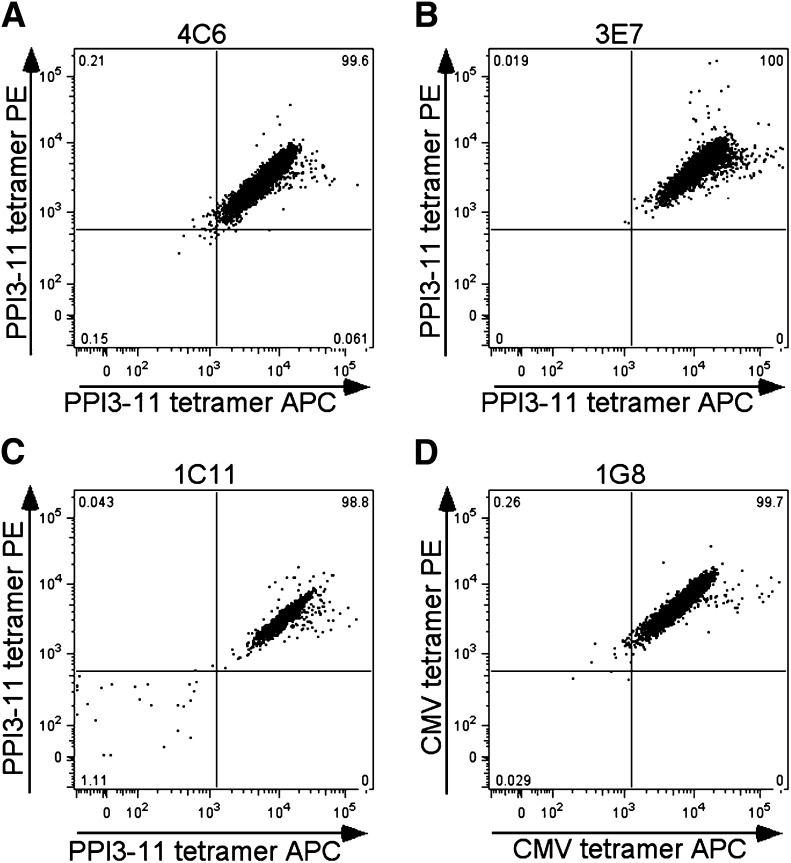
At two different time points, we attempted to isolate PPI3–11-specific CD8 T-cell clones from the same index patient with type 1 diabetes. On the first occasion, short-term peptide-stimulated PBMC lines were generated. Single cell sorting of dual PPI3–11 tetramer-positive cells on day 9, followed by expansion in vitro, yielded three clones of sufficient number to identify as dual PPI3–11 tetramer positive (1D5, 1D10, and 2B3) (Supplementary Fig. 2). Cell pellets were stored for TCR clonotyping, but these clones failed to expand further. On the second occasion, 6 months later, sufficient PBMCs from two coordinated blood draws were available to enable use of our conventional cloning strategy (5) in which autologous matured DCs pulsed with PPI3–11 are cultured with purified peripheral blood CD8 T cells. Using this strategy, a further three dual PPI3–11 tetramer-positive clones were established (4C6, 3E7, and 1C11) (Fig. 5A–C) and expanded to sufficient quantity for functional analysis. PPI3–11-specific clones did not bind irrelevant tetramer (CMV-AYA) (Supplementary Fig. 3). In addition, a CMV-AYA–specific CD8 T-cell clone was obtained by direct ex vivo single cell sorting of dual CMV-AYA HLA-24 tetramer-positive cells (Fig. 5D). TCR clonotyping established that 1D5, 1D10, and 2B3 from the first cloning and 4C6 and 3E7 (1C11 not tested) from the second cloning 6 months later all expressed the same clonotype: the TCR α-chain gene TRAV5*01 with CDR3-α sequence CAEPSGNTGKLIF and β-chain gene TRBV7–9*03 with CDR3-β sequence CASSLHHEQYF.
FIG. 5.
A–C: Representative staining of three CD8 T-cell clones generated from a single patient that are dual PPI3–11-tetramer positive and a single clone staining with CMV-AYA–loaded reagents (D).
Cytotoxic potential of PPI3–11-reactive CD8 T-cell clones.
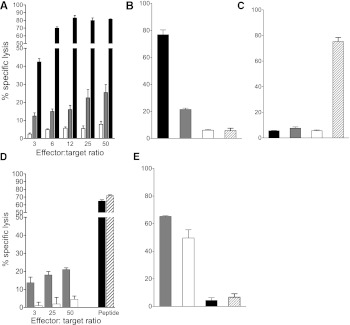
We examined the antigen-specific cytotoxic potential of PPI3–11-reactive CD8 T-cell clone 4C6 initially against K562-A24-PPI cells, which according to the elution studies, naturally process and present PPI3–11, and also against K562-A24 cells pulsed with relevant peptide. Using a range of effector-to-target ratios, killing was specific and approached a maximal ratio of 12:1 when K562-A24 cells were pulsed with PPI3–11 peptide (Fig. 6A). K562-A24-PPI cells, naturally processing and presenting PPI3–11, were also killed, albeit to a lesser extent (Fig. 6A). The specificity of 4C6 for PPI3–11 is indicated by the very low level of killing seen upon coculture of clone with K562-A24 cells pulsed with peptide only (open bars, Fig. 6A) or pulsed with an irrelevant peptide (CMV-AYA) (Fig. 6B). HLA-A24–restricted CD8 T-cell clone cells (1G8) specific for CMV-AYA did not kill K562-A24-PPI cells to any significant degree (Fig. 6C), and this low-level killing lacked specificity since it was similar to that observed when K562-A24 (unpulsed) or K562-A24-PPI cells were used as targets (Fig. 6B and C).
FIG. 6.
Cytotoxicity assays using 4C6 PPI3–11- and 1G8 CMV-AYA–specific CD8 T-cell clones. A: 4C6 PPI3–11 CTLs kill K562-A24-PPI cells naturally processing and presenting PPI3–11 (gray bar) at a range of effector-to-target ratios, with increasing killing as the ratio increases. Higher killing is observed, and peaks at a ratio of 12:1, when K562-A24 cells are pulsed with PPI3–11 (black bar). Low-level killing is seen with K562-A24 pulsed with peptide diluent only (open bar). Results are representative of three experiments, and error bars are SEMs from triplicate wells. B: 4C6 PPI3–11 CTLs kill K562-A24 cells pulsed with PPI3–11 peptide (black bar) and K562-A24-PPI naturally presenting PPI3–11 (gray bar), but only background killing of K562-A24 cells pulsed with irrelevant CMV-AYA peptide (cross-hatched bar) or peptide diluent only (open bar) is seen. Effector-to-target ratio 25:1. C: 1G8 CMV-AYA CTLs kill K562-A24 target cells pulsed with CMV-AYA peptide (cross-hatched bar), but show background killing only in the presence of K562-A24 pulsed with PPI3–11 (black bar), K562-A24-PPI naturally presenting PPI (gray bar), or K562-A24 pulsed with peptide diluent only (open bar). Effector-to-target ratio 25:1. D: 4C6 PPI3–11 CTLs kill HLA-A*24+ human islet cells naturally processing and presenting PPI3–11 (gray bar). Only low-level killing is seen with CMV-AYA CTLs (open bar). Both PPI3–11 and CMV-AYA CTLs kill human islet cells to comparable, high levels when targets are pulsed with cognate peptide (black and cross-hatched bar, respectively). Results are a single experiment, and error bars are SEMs from triplicate wells. E: There is robust, high-level killing of human HLA-A*0201+ islets by the previously characterized CTL clone 3F2, specific for PPI15–24 when presented by HLA-A2 (5) either when the islet cells are pulsed with PPI15–24 (gray bar) or when islets are naturally presenting PPI15–24 (open bar). In contrast, killing of the same HLA-A*2+ islets (unpulsed) is only at background levels in the presence of HLA-A24–restricted 4C6 PPI3–11 CTLs (black bar) and HLA-A24 1G8 CMV-AYA CTLs (cross-hatched bar). Effector-to-target ratio 50:1. Experiments represented in B, C, and E were performed in parallel to one another with the same passage of CTL.
We next examined the cytotoxic potential of the PPI3–11 CD8 T-cell clone on HLA-A*24+ human islet cells. Islet cell numbers were limiting and, therefore, a restricted range of effector-to-target ratios were used selected based on the K562 cell line killing experiments. Islet cells were killed by PPI3–11-specific CD8 T-cell clone 4C6; in contrast, CD8 T-cell clone 1G8 specific for CMV-AYA failed to kill (Fig. 6D). Both PPI3–11- and CMV-AYA–specific clones killed HLA-A*24+ islet cells to a comparable and much higher degree when the targets were pulsed with cognate peptide. The lower level of PPI3–11-mediated killing with unpulsed human islet cells is likely to reflect the heterogeneous nature of the islet cell preparations and the fact that levels of surface peptide-HLA complexes achieved via endogenous processing and presentation do not reach that attained via exogenous pulsing. Only background levels of killing were observed when HLA-A24 PPI3–11 CTL 4C6 or HLA-A24 CMV-AYA CTL 1G8 were cocultured with human HLA-A*0201+ (HLA-A*24−) islet cells (Fig. 6E).
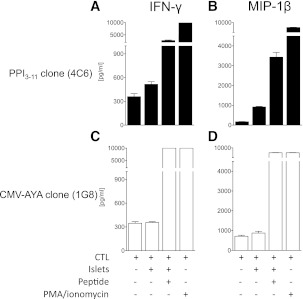
When cocultured with HLA-A*24+ human islets, the PPI3–11-specific CD8 T-cell clone 4C6 secreted effector cytokines, most notable, macrophage inflammatory protein (MIP)-1β and IFN-γ when compared with resting levels (Fig. 7), as well as TNF-α (data not shown). In contrast, the 1G8 CMV-AYA–specific CD8 clone failed to produce cytokine in response to coculture with human islets. Both clones show maximal MIP-1β and IFN-γ secretion when cocultured with cognate peptide–pulsed HLA-A*24+ islets or when stimulated with PMA/ionomycin (Fig. 7).
FIG. 7.
Cytokine production by 4C6 PPI3–11- and 1G8 CMV-AYA–specific CD8 T-cell clones after coculture with HLA-A*24+ human islet cells. 4C6 PPI3–11 CTLs make IFN-γ (A) and MIP-1β (B) in response to culture with HLA-A*24+ human islet cells. Levels are maximal when human islet cells are pulsed with PPI3–11 peptide or CTLs are stimulated with PMA/ionomycin. C and D: 1G8 CMV-AYA CTLs fail to make these cytokines in response to culture with unmanipulated human islet cells but show full cytokine production capability when cells are pulsed with cognate peptide or stimulated with PMA/ionomycin. Results are a single experiment and error bars are SEMs from duplicate wells.
DISCUSSION
The underlying basis for HLA class I gene–associated risk in type 1 diabetes is not known. In an attempt to elucidate this, in the current study we manufactured an HLA class I negative human cell line to express the type 1 diabetes–risk allele HLA-A24 and also PPI, a major β-cell–specific autoantigen targeted in the disease. We used sensitive mass spectrometry to examine the HLA-associated peptides that these cells display. A 9-mer sequence from the signal peptide region of PPI, LWMRLLPLL, was identified as naturally processed and presented by HLA-A24 under these conditions. This PPI epitope was confirmed as being recognized by CD8 T cells in the peripheral blood using HLA-A24 tetramers loaded with LWMRLLPLL. Moreover, circulating CD8 T cells recognizing this peptide were significantly more frequent in patients with type 1 diabetes than in control subjects matched for HLA-A*24. It is important that disease relevance was substantiated by the demonstration that a CD8 T-cell clone specific for LWMRLLPLL presented by HLA-A24 kills human islet cells in vitro. These results identify a new biomarker of PPI-specific autoreactivity for the HLA-A*24+ subset of type 1 diabetic patients and add weight to the notion that cytotoxic CD8 T cells have the potential to mediate β-cell death.
The peptides we eluted and sequenced from the K562-A24-PPI cell line are entirely consistent with the previously published motif (15,16), indicating that our approach yields peptides naturally processed and presented by HLA-A24. It is intriguing that LWMRLLPLL derives from the signal peptide region that acts as a presequence targeting proinsulin to the endoplasmic reticulum. Signal peptide epitopes are unusual in relation to human disease, but this is the second time that this phenomenon has been observed in the context of type 1 diabetes. Other signal peptide epitopes have also been suggested to play a role in type 1 diabetes, including the prediction that PPI8–16 would bind DQB1*06020DQA1*0102 with high affinity (18). In a previous study that uses a similar approach, we showed that the signal peptide epitope PPI15–24 is naturally presented by HLA-A2 (5). In that study, we also provided evidence that epitope presentation from the signal peptide may not follow a conventional processing route (i.e., it does not require proteasome cleavage and import into the endoplasmic reticulum via transporter associated with processing). Future studies will be required to establish whether the same holds for LWMRLLPLL. For both PPI15–24 and PPI3–11, we confirmed presentation by human islet cells, using CTL lines as probes and cytotoxicity as the read out. This finding rules out the possibility that the discovery of signal peptide epitopes is solely an artifact of using a surrogate β-cell line in the discovery process.
This is the first time that PPI3–11 has been described as being naturally processed and presented by HLA-A24 molecules. An earlier report by Chang et al. (19) identified PPI3–11 as being capable of binding to HLA-B15, HLA-B8, and HLA-A2 using HLA class I assembly assays, although of interest, binding to HLA-A24 could not be demonstrated. Toma et al. (20) tested the binding affinity and CD8 T-cell response toward the 10-mer PPI2–11, which was identified as a potential epitope of HLA-A24 using a binding algorithm (20). Despite extensive searching for oxidized and nonoxidized forms, we did not identify this 10-mer as being naturally processed and presented by K562-A24-PPI cells. Moreover, the same group only rarely detected responses to PPI2–11 in newly diagnosed patients with type 1 diabetes using a sensitive IFN-γ enzyme-linked immunospot assay. It remains unclear, therefore, whether a nested set of peptides, similar to the combination of epitopes (PPI15–24 and PPI17–24) that we described previously for HLA-A2, also exists in this region for HLA-A24. Although our approach requires a complex series of techniques that includes the challenge of cloning epitope-specific CD8 T cells, the advantage of this strategy is that the novel HLA-A24–presented PPI3–11 epitope is confirmed as naturally processed and presented by both surrogate β-cells and human HLA-A*24+ islet cells.
The disease relevance of the PPI3–11 epitope is further bolstered by the increased number of circulating PPI3–11-specific CD8 T cells that we detected in recent-onset type 1 diabetic patients when compared with control subjects. Since both groups bear HLA-A*24+, there is a strong implication that this expansion is directly associated with either disease development or disease predisposition. The cytotoxicity studies that we conducted in vitro, although using a single CD8 T-cell clonotype from a single patient, offer the proof of concept that a CTL response to PPI3–11, in type 1 diabetic patients with HLA-A*24+, is a direct contributor to β-cell death and, hence, to disease pathogenesis. Future studies on the HLA-A*24+ subset of patients and at-risk subjects will be able to use the PPI3–11-A24 tetramer reagents that we describe here to track, enumerate, and phenotype PPI-reactive CD8 CTLs and establish whether there are temporal links to decline in β-cell mass.
On two separate occasions, 6 months apart, we generated PPI3–11-specific T-cell clones with the same antigen-specific TCR clonotype. This evidence of prolonged existence of circulating, identical, disease-related clonotypes is of considerable interest. Again, the finding resonates with our previous work, in which we showed that PPI15–24-specific CD8 T cells with identical TCR clonotypes could be isolated from fresh blood samples obtained from the same patient 1.5 years apart (5). The data imply that there may be ancestral clones that are long lived and likely to be dominant in the immune response of an individual patient. It is tempting to speculate that this quality may be a component of the mechanism(s) through which HLA-A24, and comparable disease-risk HLA molecules, render an individual susceptible to disease or an early age of onset. It is plausible that molecules such as HLA-A24 bias the selection of T-cell repertoires, either during positive selection in the thymus or during priming in the periphery, toward a configuration that favors presentation of autoantigenic epitopes that are in turn dominantly and abundantly presented by the β-cell. PPI signal peptide has properties that fit this description, being generated in tune with metabolic demands on the β-cell and being cotranslated with HLA class I molecules at the intracellular location that is critical for their peptide loading.
ACKNOWLEDGMENTS
D.K. is in receipt of a PhD Studentship from the U.K. Department of Health via the National Institute for Health Research Comprehensive Biomedical Research Centre Award to Guy’s and St. Thomas’ National Health Service Foundation Trust in partnership with King’s College London. R.R.K. is in receipt of a Diabetes U.K. PhD Studentship. The study was supported by grants from the Juvenile Diabetes Research Foundation (7-2005-877 and 1-2007-1803 to M.P.). M.P. receives funding via the EU FP7 EU Framework 7 Large-Scale Focused Collaborative Research Project on Natural Immunomodulators as Novel Immunotherapies for Type 1 Diabetes.
No potential conflicts of interest relevant to this article were reported.
D.K. designed and performed experiments, analyzed data, conceived ideas, oversaw research, and wrote the manuscript. R.R.K., M.E., R.J.E., M.G.K., A.d.R., M.E., and G.C.H. performed experiments. J.P. and C.M.D. recruited and characterized patients. A.S. performed experiments, conceived ideas, and oversaw research. P.A.v.V. designed and performed experiments, analyzed data, conceived ideas, and oversaw research. M.P. designed and performed experiments, analyzed data, conceived ideas, oversaw research, and wrote the manuscript. M.P. is the guarantor of this work and, as such, had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
The authors are grateful to Amanda Bishop and Nicola McLintock (University of Bristol, London, U.K.) and Samantha Gillman (Guy’s Hospital, London, U.K.) for their help with the sample collection, and to all patients and control subjects for their blood donation. The authors also are grateful to Professor John Todd (Cambridge University) for discussions on HLA class I associations in type 1 diabetes and Professor David Price (Cardiff University) for advice on TCR clonotyping.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db11-1520/-/DC1.
REFERENCES
- 1.Bottazzo GF, Dean BM, McNally JM, MacKay EH, Swift PG, Gamble DR. In situ characterization of autoimmune phenomena and expression of HLA molecules in the pancreas in diabetic insulitis. N Engl J Med 1985;313:353–360 [DOI] [PubMed] [Google Scholar]
- 2.Willcox A, Richardson SJ, Bone AJ, Foulis AK, Morgan NG. Analysis of islet inflammation in human type 1 diabetes. Clin Exp Immunol 2009;155:173–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Foulis AK, Farquharson MA, Hardman R. Aberrant expression of class II major histocompatibility complex molecules by B cells and hyperexpression of class I major histocompatibility complex molecules by insulin containing islets in type 1 (insulin-dependent) diabetes mellitus. Diabetologia 1987;30:333–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Itoh N, Hanafusa T, Miyazaki A, et al. Mononuclear cell infiltration and its relation to the expression of major histocompatibility complex antigens and adhesion molecules in pancreas biopsy specimens from newly diagnosed insulin-dependent diabetes mellitus patients. J Clin Invest 1993;92:2313–2322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Skowera A, Ellis RJ, Varela-Calviño R, et al. CTLs are targeted to kill beta cells in patients with type 1 diabetes through recognition of a glucose-regulated preproinsulin epitope [corrected in: J Clin Invest 2009;119:2844]. J Clin Invest 2008;118:3390–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nejentsev S, Howson JM, Walker NM, et al. Wellcome Trust Case Control Consortium Localization of type 1 diabetes susceptibility to the MHC class I genes HLA-B and HLA-A. Nature 2007;450:887–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Middleton D, Williams F, Meenagh A, et al. Analysis of the distribution of HLA-A alleles in populations from five continents. Hum Immunol 2000;61:1048–1052 [DOI] [PubMed] [Google Scholar]
- 8.Williams F, Meenagh A, Maxwell AP, Middleton D. Allele resolution of HLA-A using oligonucleotide probes in a two-stage typing strategy. Tissue Antigens 1999;54:59–68 [DOI] [PubMed] [Google Scholar]
- 9.Achenbach P, Koczwara K, Knopff A, Naserke H, Ziegler AG, Bonifacio E. Mature high-affinity immune responses to (pro)insulin anticipate the autoimmune cascade that leads to type 1 diabetes. J Clin Invest 2004;114:589–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meiring HD, van der Heeft E, ten Hove GJ, de Jong APJM. Nanoscale LC-MS(n): technical design and applications to peptide and protein analysis. J Sep Sci 2002;25:557–568 [Google Scholar]
- 11.Kuzushima K, Hayashi N, Kimura H, Tsurumi T. Efficient identification of HLA-A*2402-restricted cytomegalovirus-specific CD8(+) T-cell epitopes by a computer algorithm and an enzyme-linked immunospot assay. Blood 2001;98:1872–1881 [DOI] [PubMed] [Google Scholar]
- 12.Burrows SR, Elkington RA, Miles JJ, et al. Promiscuous CTL recognition of viral epitopes on multiple human leukocyte antigens: biological validation of the proposed HLA A24 supertype. J Immunol 2003;171:1407–1412 [DOI] [PubMed] [Google Scholar]
- 13.Lissina A, Ladell K, Skowera A, et al. Protein kinase inhibitors substantially improve the physical detection of T-cells with peptide-MHC tetramers. J Immunol Methods 2009;340:11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brochet X, Lefranc MP, Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res 2008;36(Web Server issue):W503–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kubo RT, Sette A, Grey HM, et al. Definition of specific peptide motifs for four major HLA-A alleles. J Immunol 1994;152:3913–3924 [PubMed] [Google Scholar]
- 16.Maier R, Falk K, Rötzschke O, et al. Peptide motifs of HLA-A3, -A24, and -B7 molecules as determined by pool sequencing. Immunogenetics 1994;40:306–308 [DOI] [PubMed] [Google Scholar]
- 17.Velthuis JH, Unger WW, Abreu JR, et al. Simultaneous detection of circulating autoreactive CD8+ T-cells specific for different islet cell-associated epitopes using combinatorial MHC multimers. Diabetes 2010;59:1721–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cucca F, Lampis R, Congia M, et al. A correlation between the relative predisposition of MHC class II alleles to type 1 diabetes and the structure of their proteins. Hum Mol Genet 2001;10:2025–2037 [DOI] [PubMed] [Google Scholar]
- 19.Chang L, Kjer-Nielsen L, Flynn S, et al. Novel strategy for identification of candidate cytotoxic T-cell epitopes from human preproinsulin. Tissue Antigens 2003;62:408–417 [DOI] [PubMed] [Google Scholar]
- 20.Toma A, Laïka T, Haddouk S, et al. Recognition of human proinsulin leader sequence by class I-restricted T-cells in HLA-A*0201 transgenic mice and in human type 1 diabetes. Diabetes 2009;58:394–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Colaert N, Helsens K, Martens L, Vandekerckhove J, Gevaert K. Improved visualization of protein consensus sequences by iceLogo. Nat Methods 2009;6:786–787 [DOI] [PubMed] [Google Scholar]