Abstract
Medulloblastoma is diagnosed histologically; treatment depends on staging and age of onset. Whereas clinical factors identify a standard- and a high-risk population, these findings cannot differentiate which standard-risk patients will relapse and die. Outcome is thought to be influenced by tumor subtype and molecular alterations. Poor prognosis has been associated with isochromosome (i)17q in some but not all studies. In most instances, molecular investigations document that i17q is not a true isochromosome but rather an isodicentric chromosome, idic(17)(p11.2), with rearrangement breakpoints mapping within the REPA/REPB region on 17p11.2. This study explores the clinical utility of testing for idic(17)(p11.2) rearrangements using an assay based on fluorescent in situ hybridization (FISH). This test was applied to 58 consecutive standard- and high-risk medulloblastomas with a 5-year minimum of clinical follow-up. The presence of i17q (ie, including cases not involving the common breakpoint), idic(17)(p11.2), and histologic subtype was correlated with clinical outcome. Overall survival (OS) and disease-free survival (DFS) were consistent with literature reports. Fourteen patients (25%) had i17q, with 10 (18%) involving the common isodicentric rearrangement. The presence of i17q was associated with a poor prognosis. OS and DFS were poor in all cases with anaplasia (4), unresectable disease (7), and metastases at presentation (10); however, patients with standard-risk tumors fared better. Of these 44 cases, tumors with idic(17)(p11.2) were associated with significantly worse patient outcomes and shorter mean DFS. FISH detection of idic(17)(p11.2) may be useful for risk stratification in standard-risk patients. The presence of this abnormal chromosome is associated with early recurrence of medulloblastoma.
Keywords: FISH, idic(17)(p11.2), i17q, medulloblastoma, pediatric oncology
Medulloblastoma is the most common malignant brain tumor in children and accounts for approximately 20% of all central nervous system (CNS) tumors.1,2 Despite its relatively high incidence, the biology of this tumor is poorly understood. The World Health Organization classifies all medulloblastomas as grade IV given their potential for aggressive behavior. However, these tumors vary widely in terms of associated clinical outcomes. Risk stratification strategies that place patients into standard- and high-risk groups show 5-year survival rates of roughly 86(±9)% and 40%, respectively.3,4 Factors associated with increased risk include age <3 years, residual disease following surgical resection, and metastatic disease at presentation. However, clinical staging does not always accurately predict tumor behavior in the individual patient. Current treatment strategies, while often curative, frequently result in significant lifelong debilitation, and the limited predictive power of the 2-tier stratification system likely significantly contributes to overall patient morbidity, given that some patients with less aggressive tumors are likely overtreated.5 Histologic classification has also been shown to correlate well with patient outcome.6–8 However, the value of such schemes is limited due to the relative rarity of anaplasia in medulloblastoma,9 limited tumor sampling, and heterogeneous tumor histology. There is currently an effort in the field to subclassify medulloblastomas based on molecular profiling (as most recently summarized by Taylor et al10); however, these new schemes have yet to be translated into clinical practice.
Molecular and cytogenetic studies have uncovered several markers associated with patient outcome independent of clinical staging.9,11–14 More recently, changes at the genomic level have been implicated, with gain of 6q being associated with a poor prognosis and loss of 6q or monosomy 6 (also noted to have alterations in the Wnt signaling pathway) having the opposite effect.15,16 Isochromosome (i)17q, the most frequent cytogenetic abnormality seen in medulloblastoma (estimated in 20%–50% of cases), has also been associated with a poor prognosis;12,16–19 however, not all studies have found this association to be significant.20,21 This alteration has also been reported to be correlated with higher-grade tumors and anaplasia;22,23 as such, it remains unclear whether this represents a statistically independent prognostic marker.
Isochromosome 17q is usually not a true isochromosome with rearrangement breakpoints at the centromere but is most often a chromosome with 2 centromeres (isodicentric) in close proximity and rearrangement breakpoints mapping to the low copy repeat (LCR) REPA/REPB region of chromosome 17p11.2; the resulting idic(17)(p11.2) has been proposed to be a tumorigenic event.24,25 Several groups have verified this site as the most common rearrangement breakpoint hotspot using a comparative genomic hybridization array.18,26,27 The REPA/REPB region is comprised primarily of LCRs that are prone to non-allelic homologous recombination (NAHR) and thus deletions, duplications, and inversions.28,29 The mechanism of rearrangement leading to i17q, based on NAHR, was described by Barbouti et al,30 although subsequent models have also been proposed.31 Thus, while i17q is seen in other malignancies,32 it appears that this rearrangement may be specific to medulloblastomas when considering primary brain tumors. As such, it may be a useful diagnostic marker, as it is not present in histologically similar tumors, such as CNS primitive neuroectodermal tumors (PNETs) and atypical teratoid/rhabdoid tumors (AT/RTs).33
The broader category of tumors revealing i17q has been studied for some time; however, no investigation regarding the specific idic(17)(p11.2) chromosomal rearrangement has been performed in a clinically relevant setting to date. Furthermore, studies have shown that i17q may be heterogeneous, potentially representing the result of tumor evolution, while idic(17)(p11.2) is a reproducible, possibly tumorigenic event occurring by a specific mechanism. We hypothesized that idic(17)(p11.2) is a unique biological event the presence of which may have clinical ramifications. Current techniques commonly used for genomic and molecular analyses can be either expensive or time consuming, require fresh tissue, or cannot distinguish tumor from nontumor tissue. Clinically, the gold standard technique is to identify i17q by karyotype analysis, although other methods, including fluorescent in situ hybridization (FISH), are becoming routine. The identification of i17q and other biomarkers are becoming more common in pathologic assessment of medulloblastoma. However, clinical implementation of an i17q assay is not currently standard of care, as the threshold of convincing data has yet to be reached.
In this study, we apply a novel FISH assay to identify both idic(17)(p11.2) and other forms of i17q on formalin-fixed, paraffin-embedded samples. In a blinded fashion, we first demonstrated that this assay is both sensitive and specific for the identification of idic(17)(p11.2) and other forms of i17q in specimens archived for as long as 25 years. This test was then performed on 58 consecutive cases from the archives of Washington University in St. Louis, and the presence of idic(17)(p11.2) and i17q in medulloblastomas was compared with clinical outcome in high- and standard-risk patients. Our data demonstrate that the presence of idic(17)(p11.2) substratifies standard risk (SR) patients by identifying those that are more likely to have a tumor recurrence. Our findings are also in agreement with previous reports that i17q is a marker of poor outcome, independent of histology.
Materials and Methods
Validation Study Samples
Seventeen patients were selected from the archives of Texas Children's Hospital (TCH) based on a diagnosis of medulloblastoma and the presence of a known karyotype. These patients were selected by the staff of TCH to have a proportion of i17q-positive and -negative cases based on cytogenetic analysis.
Selected tissue blocks from these cases were provided from the archives of TCH without patient identifiers. Slides were prepared for hematoxylin and eosin, immunohistochemistry (IHC), and FISH analyses. The FISH analysis was conducted first in a blinded fashion (ie., no knowledge of tumor subtype, karyotype, or clinical data). Subsequently, all slides were reviewed by 2 of the authors (A.P. and G.B.W.) to verify the diagnosis of medulloblastoma. IHC stains for INI-1 (BAF-47) were also performed to rule out the possibility of a misdiagnosed AT/RT.
Study Set Cohort
Patients were selected based on a diagnosis of medulloblastoma from the archives of Washington University in accordance with an approved institutional review board protocol (#201104083). Patient identification was made from the electronic records database for the years 1989 to 2008. All cases identified in the search were enrolled in this study. A review of all medical, radiologic, and pathologic reports was conducted. SR patients were identified by a documented lack of residual disease, a lack of drop metastases in brain and spinal MRI status postsurgical resection, and negative CSF cytology. Only a single case in this cohort had residual disease <1.5 cm2. This patient underwent a second surgical resection, resulting in complete resection, and is subsequently still in remission. High-risk (HR) patients typically had either residual disease or drop metastases (as reported in postsurgical or radiologic reports). Patient age was not used as an identifier of high risk given conflicting literature.34–36
Exclusion Criteria (Study Set)
Selected patients had to have complete surgical pathology reports and medical records (including staging spinal MRI). Furthermore, patients were required to have either clinical follow-up until recurrence or a minimum of 5 years if in remission (disease-free).
Histology
Histologic variants were recorded for each case based on the surgical pathology reports and slide review. The desmoplastic variant required a nodular growth pattern demonstrating intranodular maturation (presence of neuropil and neurocytic cells) with reticulin-rich internodular regions composed of primitive-appearing cells. Anaplasia was defined by a more than just focal presence of enlarged cells with bizarre hyperchromatic nuclei (anaplastic features) and/or oval vesicular nuclei with macronucleoli (large cell features).
Molecular Subtyping
Molecular subtyping was conducted by IHC analysis. Established clinical histologic protocols were used to identify Wnt activation and sonic hedgehog (SHH) signaling alterations (identified by nuclear β-catenin and internodular cytoplasmic GAB1 reactivity).37 An attempt was also made to classify groups C and D based on the protocols published by Northcott et al38 using NPR3 and KCNA1 antibodies to identify groups C and D, respectively. Unfortunately, we found NPR3 to lack adequate sensitivity for reproducible antigen detection and KCNA1 to lack specificity for the group D subtype. For these reasons, neither of these latter IHC stains was used in the subsequent analyses. All cases were also screened for MYC amplification using commercially available, clinically validated FISH probes (Abbott Molecular).39
FISH Assay
Human BAC and fosmid DNA-based FISH probes flanking the REPA/REPB region of chromosome 17p11.2 were fluorescently labeled as previously described using a direct-labeling method.39,40 The assay was performed with clones RP11-970O14 (GenBank accession number AC005722) and CTC-457L16 (AC003957), mapping to regions proximal (centromeric-green) and distal (telomeric-red) to the chromosome 17p11.2 breakpoint region. For cases suspected to carry an i17q with breakpoints outside the REPA/REPB region, a second probe set was used and contained the telomeric 17p11.2 probe (above) paired with a probe on 17q24.3, utilizing BAC RP11-169I9 (AC060771). These cases showed a loss of one set of flourophores per cell with the test probes (see Fig. 1D). The labeled probes were hybridized to slides containing deparaffinized tumor tissue samples, using previously published protocols.39,40
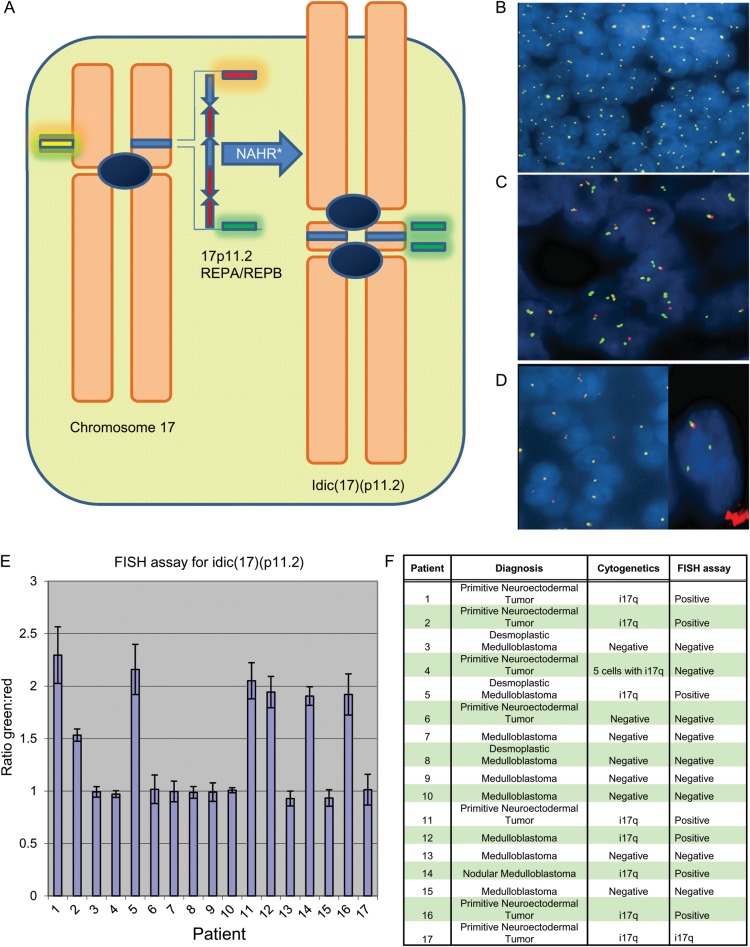
Fig. 1.
(A) Schematic of chromosome 17 and the formation of idic(17)(p11.2). The figure on the left represents the archetypal chromosome, as well as the location of the probes from the FISH assay. The REPA/REPB region is located in chromosome 17p11.2, and is the likely site of NAHR, resulting in the isodicentric chromosome. The FISH probes flank this region. The figure on the right represents idic(17)(p11.2), as well as the resulting fluorophore hybridization locations. (B) An example of a negative test in this assay, with a 1:1 ratio of green:red signals, which frequently appears as yellow fusion signals due to the close proximity of the two probes. (C) An example of a positive test result. There is a 3:1 ratio of green:red flourophores, with a doubling of the green signals and loss of the red signal in the isodicentric chromosome. Occasionally, the chromosomal breakpoint within chromosome 17p11.2 is close enough to the centromeric green hybridization location that the doubling effect is not seen, resulting in a 2:1 green:red signal ratio. (D) A 1:1 green:red fluorophore ratio can be seen in i17q with breakpoints centromeric to the REPA/REPB region, but the presence of only one pair of signals in most nuclei is suspicious for a rearrangement. In such cases, i17q is confirmed with a second FISH probe pairing with the green fluorescently labeled probe hybridizing to distal chromosome 17q (right image). (E) Validation set results. ImageJ was used to analyze 17 cases from TCH in a blinded fashion. Thousands of green and red signals were counted per case, resulting in the ratios shown. Those with statistically significant increased ratios were considered positive. Most positive cases had ratios in the 1.5–2.5 range, while all the negative cases had overall ratios very close to 1.0. (F) Validation set results (unblinded). Patient 4 is the false negative in our test. Patient 17 had an abnormal signal pattern (1:1 ratio of signals, but a single signal set per cell as seen in Fig. 1D), suggestive of i17q but a normal ratio by our assay. i17q was confirmed with a second set of 17p and 17q probes.
Signal Quantification
An approximate ratio of green:red flourophores was achieved by scoring 200 cells/signal clusters, and the presence of 2–3:1 signal ratios in greater than 20% of the cells was considered a positive test. Signals were also scored with the imageJ imaging software41 for an unbiased quantitative analysis (see Supplementary data and Fig. 1E).
Results
Validation Assay
Seventeen cases from TCH with a diagnosis of medulloblastoma and a known karyotype were retrieved for FISH studies. All patient information, including clinical history, surgical pathology, and karyotype, was blinded during the study. FISH probes flanking the REPA/REPB region and a telomeric 17q probe were used to identify i17q in cases with and without the common rearrangement, respectively (Fig. 1). After analysis of the FISH data was completed, surgical pathology reports and karyotypes were reviewed. The FISH-based assay was able to correctly identify the presence or absence of i17q in 16 cases, with the one exception being a false negative (Fig. 1E and F). Eight of the cases were identified as positive in the assay: 7 with the characteristic isodicentric pattern and 1 with i17q with breakpoints outside the REPA/REPB region. In this case, the ratio of green:red was 1:1 using the REPA/REPB probe set, but the signal number was aberrant with one copy of each per cell. Subsequent FISH with a control probe on 17q confirmed the presence of i17q. Therefore, using karyotyping as the gold standard, our test sensitivity for FISH was 88%, while the specificity was 100%.
Study Set
Identified from the archives of Washington University were 58 consecutive medulloblastomas with an associated minimum of 5 years clinical follow-up, complete surgical pathology records, and adequate formalin-fixed tumor material in paraffin-embedded tissue blocks (Table 1 in the Supplementary data). The mean and median cases were from the year 2000. The mean follow-up time was 120 months (10 y). FISH was performed on all cases using the REPA/REPB probes for the common rearrangement. Interpretable results were obtained in 55 cases (95%). When i17q was suspected due to aberrant signal numbers per cell but the initial test was negative because of rearrangement breakpoints outside the REPA/REPB region, the paired 17p/17q probes were used for i17q confirmation. These findings were then compared and correlated with patient outcome, histologic type, and tumor staging. Treatment information was available in a majority of cases. Seventy percent of cases in the SR group with information available received the current recommended dose of 23.4 Gy of cranial and spinal radiation.
Table 1.
Multivariate analysis of suspected prognostic variables across all patients
| Variable | Value | Standard error | P | Hazard ratio | Hazard ratio lowerbound (95%) | Hazard ratio upperbound (95%) |
|---|---|---|---|---|---|---|
| idic(17)(p11.2) | −1.592 | 0.560 | .004 | 0.204 | 0.068 | 0.610 |
| MYC amplification | −1.299 | 0.628 | .039 | 0.273 | 0.080 | 0.935 |
| Risk group | −2.878 | 0.482 | <.0001 | 0.056 | 0.022 | 0.145 |
| Desmoplastic histology | 0.629 | 0.522 | .228 | 1.875 | 0.675 | 5.214 |
A Cox proportional hazards model was used to compare suspected prognostic variables. The “value” represents the regression coefficient. As expected, the best predictor of outcome (measuring DFS) is clinical risk stratification. Other significant independent predictors are the presence of idic(17)(p11.2) (P = .004) and MYC amplification (P = .039). Note: anaplastic histology was not included because these rare tumors were almost exclusive to the HR patient group, and thus would not appear as an independent variable. Similarly, Wnt activation was rare and almost exclusive to the SR group.
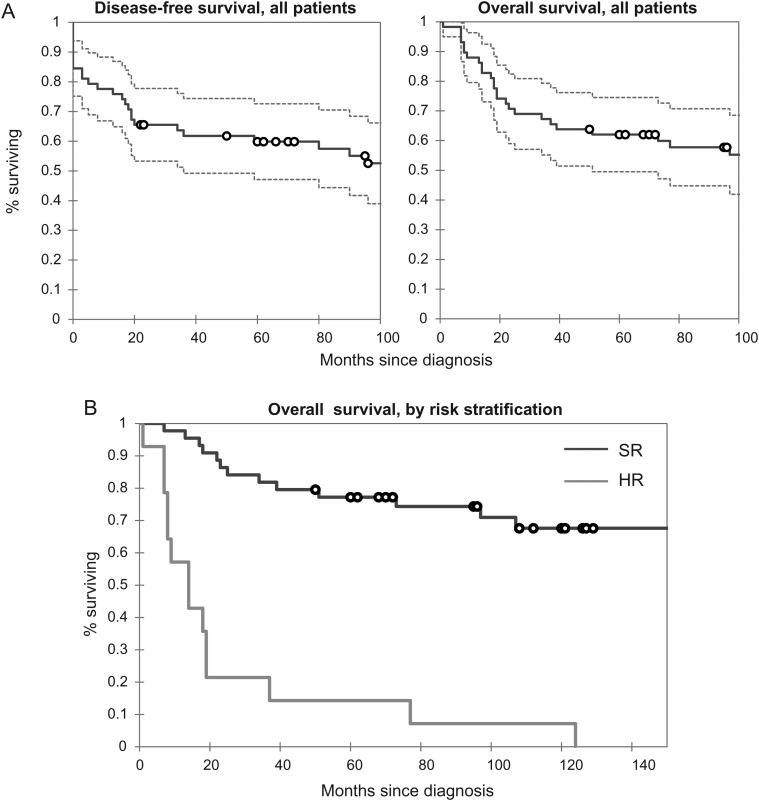
Our medulloblastoma study cohort had 5-year overall survival (OS) and disease-free survival (DFS) rates of 60% and 53%, respectively, similar to prior reports.1,3,15,42–44 Overall survival in the cohort is represented in Fig. 2A. Based on clinical staging, 44 patients (76%) were considered SR, while 14 (24%) were in the HR group. The 5-year OS in the SR group was 77%, consistent with the current literature,3 despite the changes in treatment regimen over the years encompassed in this study (1989–2008). The 5-year DFS in the SR group was 69%. The HR group had a 5-year OS of only 15%. Kaplan–Meier analyses for both HR and SR groups show a clear difference between the 2 groups (log rank, Wilcoxon P< .001) (Fig. 2B). Seventeen cases (29%) were diagnosed as the desmoplastic medulloblastoma variant, while only 4 (7%) qualified for the anaplastic and/or large cell designation, frequencies compatible with the literature.9,35,45 Within the first 2 years of diagnosis, 3 patients died of treatment-related complications, including sepsis, stroke, and secondary brain tumor (glioblastoma multiforme). The mean and median ages of our cohort at presentation were 10.4 and 7.5 years, respectively, with a range of 3 months to 35 years. Only 3 patients presented after the second decade of life. Ten (17%) of our patients were under the age of 3. Of these, 5 presented with metastases or unresectable disease, an observation compatible with other reports.46 Therefore, while comprising less than a quarter of all patients, this subset accounted for half of all presentations with metastatic or unresectable disease. A favorable outcome was seen in patients without these high-risk features.
Fig. 2.
Study set results. (A) Disease-free and overall survival in 58 consecutive cases from BJH, illustrated in a Kaplan–Meier plot. The mean DFS and OS in all patients are 112.2 and 114.5 months (9 y), respectively, reflecting the good response most patients have to treatment. (B) Comparison of overall survival in the high-risk (HR) and standard-risk (SR) groups (log-rank, Wilcoxon P< .001). The HR group had a mean survival time of 25.8 months (95% CI, 8.3–43.7 months), compared with 152.9 months (95% CI, 127.6–178.4 months) for the SR group.
Molecular Subtypes and Known Prognostic Variables
Six patients (14%) showed nuclear β-catenin activity, consistent with Wnt activation. One of these patients recurred shortly after resection, but a postsurgical spinal survey was not completed, barring this patient from the SR group. The remaining 5 cases are currently in remission. Despite this, Wnt activation was not significantly associated with better outcome in the SR group, although there is a noticeable trend (see Supplementary material, Fig. S1A). The inability to reach statistical significance is likely due to a paucity of Wnt-activated patients in a cohort that has good outcome overall.
SHH alterations were identified in 20 cases, comprising 21% of HR patients and 39% of the SR group. Despite overrepresentation in the SR group, there is no significant difference in outcome between the SHH and other molecular variants (see Supplementary material Fig. S1B), consistent with the current literature.
MYC amplification was identified in 5 patients within our cohort. Two were in the HR group (14%) and 3 were in the SR group (6.8%). Despite a paucity of MYC-amplified cases, those with this feature had significantly worse outcomes than those without it (see Supplementary material, Fig. S1C).
Isochromosome 17q FISH
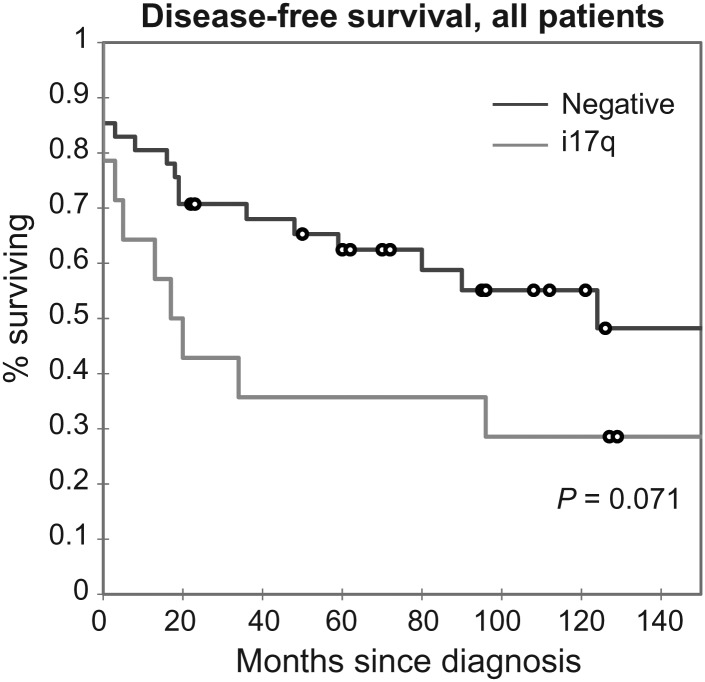
Chromosome 17 abnormalities were identified in 14 (24%) cases. Of these, 10 (71%) had the common idic(17)(p11.2) pattern resulting from breakpoints in the REPA/REPB LCR region, consistent with prior studies.16 Three cases had i17q with chromosomal breakpoints centromeric to the common rearrangement, and one showed 17p loss. These cases had 5-year OS and DFS rates of 50% and 36%, respectively. In comparison, cases that were negative for chromosome 17 alterations had 5-year OS and DFS rates of 63% and 52%, respectively. These data (for the cases without chromosome 17 abnormalities) included cases with mortality due to complications rather than disease recurrence. Figure 3 illustrates a Kaplan–Meier plot comparing outcome in i17q-positive and -negative cases. While a trend in outcome is observed in this analysis, with i17q-positive cases having a poor prognosis, statistical significance is not attained (log-rank, Wilcoxon P= .085, .071). Nonetheless, this trend appears consistent with recently published results.15,16 Mean DFS was 50.3 months (95% CI, 19.9–80.6 months) for patients with i17q compared with 111.4 months (95% CI, 84.4–138.4 months) for patients lacking i17q. Limited cases with i17q presence precluded comparisons of overall survival (see Supplementary material, Fig. S2).
Fig. 3.
Association of i17q in medulloblastoma with DFS. Similar to other published studies, i17q was associated with more frequent recurrences and shorter disease-free intervals. Although a trend is observed showing a worse outcome for positive cases, it approaches but does not meet statistical significance (log-rank, Wilcoxon P = .085, .071, respectively). Mean DFS was shorter in the positive group at 50.3 months (CI, 19.9–80.6 months) compared with 111.4 months (CI, 84.4–138.4 months) in the i17q-negative group. Assessing the effect of i17q on OS was difficult given the number of (see Supplementary material, Fig. S2).
Interestingly, the majority of patients with the common idic(17)(p11.2) chromosomal rearrangement presented as SR patients, while those that had noncommon chromosomal breakpoints tended to fall in the HR group (3 of 4 cases). Of the 4 cases with chromosome 17 alterations not involving the REPA/REPB region, 3 presented with unresectable and progressive disease; 1 was in the SR group and has been in remission for 160 months. One HR i17q case additionally harbored MYC amplification. Unfortunately, there were insufficient case numbers to directly compare outcomes in idic(17)(p11.2) cases with those presenting with other forms of i17q.
Idic(17)(p11.2) in Standard-Risk Patients
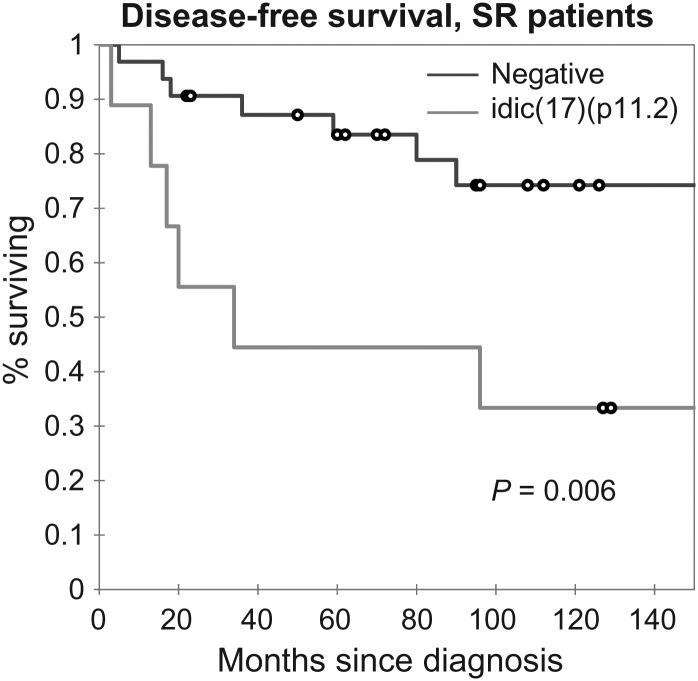
Within the subset of SR patients, those with idic(17)(p11.2) had worse outcomes than those who lacked the rearrangement, with 5-year OS and DFS of 66% and 44%, respectively, compared with 78% and 75%. Kaplan–Meier analyses between idic(17)(p11.2)-positive and -negative SR patients demonstrate a significant difference in outcome between these groups (log-rank, Wilcoxon P= .014 and .006) (Fig. 4). The mean DFS of SR patients with idic(17)(p11.2) was 63.3 months (95% CI, 25.6–101.1 months) compared with 148.8 months for the negative group (95% CI, 123.8–173.8 months).
Fig. 4.
idic(17)(p11.2) in the SR population. The presence of idic(17)(p11.2) correlated with more frequent and earlier recurrences within the SR group (log-rank, Wilcoxon P= .014, .006). The mean DFS in cases with the idic(17)(p11.2) chromosomal rearrangement was significantly shorter at 63.3 months (95% CI, 25.5–101.1 months) compared with 148.8 months (95% CI, 123.8–173.8 months) for those without.
Recent studies have suggested that patients with aberrant Wnt signaling have a favorable prognosis compared with other molecular subtypes of medulloblastoma.38 Because patients with i17q (and subsequently idic(17)(p11.2)) are molecularly distinct from this subtype, it could be argued that our findings are a result of selection bias. However, the prognostic value of idic(17)(p11.2) detection remains even if all cases with Wnt signaling abnormalities are omitted from analysis (P < .05, .02 log-rank, Wilcoxon, see Supplementary material, Fig. S3). Similarly, there was little overlap between idic(17)(p11.2)-positive cases and tumors with other molecular features—there was a single case in the SR group of the SHH subtype, and no overlap with MYC amplification. Thus, MYC amplification cannot explain the decreased DFS in idic(17)(p11.2)-positive patients.
To further demonstrate the independent prognostic significance of idic(17)(p11.2), a multivariate analysis was conducted using the Cox proportional hazards model, controlling for suspected confounding variables such as clinical risk group, MYC amplification, and desmoplastic subtype. The resulting analysis supports idic(17)(p11.2) as an independent prognostic factor (P= .004) (see Table 1).
Histologic Subtypes and i17q Status
Histologic variants of medulloblastoma are known to correlate with prognosis. Our study suggests that the desmoplastic subtype may be associated with a favorable prognosis when comparing all risk groups but that it may not be an independent prognostic variable (see Supplementary material, Fig. S4). Not surprisingly, all 4 anaplastic/large cell medulloblastomas had poor outcomes with metastatic disease at presentation, subsequent disease progression, or died of complications secondary to treatment. While this is in agreement with previous studies, we did not have enough patients in this cohort to draw any firm association between anaplasia and high-risk disease. Nevertheless, none of these patients survived beyond 2 years from diagnosis.
Of interest, none of the idic(17)(p11.2)-positive cases were identified as desmoplastic or anaplastic variants, a finding consistent with previous reports.38,47 Of the non-idic(17)(p11.2) i17q patients, one was classified as anaplastic and another as desmoplastic.
Discussion
Although numerous molecular markers have now been identified as potential aids to diagnosis and treatment of medulloblastoma, the current standard of care risk-stratification schemes rely entirely on clinical and radiologic findings. Isochromosome 17q, along with other molecular markers such as MYCC, chromosome 6q deletion/monosomy 6, and 6q duplication status have been shown to be potentially useful. However, they are not currently used routinely to improve patient stratification schemes, likely reflecting a number of limitations; many studies promoting these markers have failed to take clinical information into account, present contradictory data, and/or result in findings that are irrelevant for patient management. As an example, MYC amplification, while associated with a poor prognosis, is associated mostly with anaplastic histology, a feature already associated with poor prognosis; also, it is rare overall (5%–10% of medulloblastomas), such that it would identify only a very small percentage of cases that were not already classified as high risk.45 Even with the current clinical scheme, however, controversial issues remain. For instance, children aged ≤3 years traditionally fall into the HR category, although many clinicians do not treat them as such because they hope to avoid the unacceptable neurotoxicity of craniospinal irradiation at this age. Furthermore, there is increasing skepticism as to whether all patients <3 years should be placed into the HR category.34–36 Our data support this notion, as patients who lacked residual disease or metastases at presentation shared a favorable prognosis similar to that for SR patients >3 years of age.
A new finding from our study is that the presence of idic(17)(p11.2) may identify a significant subset of SR patients who are more likely to recur or fail to respond to treatment. If the presence of idic(17)(p11.2) can predict early recurrence in SR patients, it could be used to identify patients who could potentially benefit from additional treatment, but more importantly identify patients (those negative for the rearrangement) who may potentially be treated less aggressively in an effort to reduce the long-term sequelae of cranial and spinal radiation therapy. Our FISH-based assay is both sensitive and specific for the rearrangement, relatively quick and inexpensive to perform, requires little tissue, can be performed on tissue already prepared for standard pathologic examination, and can be used in archived cases (including some cases >25 y). By isolating a specific and most common form of i17q, we hope to have clarified the prognostic value of idic(17)(p11.2), particularly in the SR patient group. It is feasible to suggest that risk stratification systems change to reflect current molecular data, as has been proposed by several groups, most recently by Ellison et al.15
What the presence of idic(17)(p11.2) means for the mechanisms and pathologic processes that result in tumor formation is vague and warrants further study. However, some associations can be made with what is known about the histologic variants of medulloblastoma. These variants are thought to reflect different underlying biological processes, as the current literature suggests that alterations of canonical cell signaling pathways are inherent to these tumors, with the desmoplastic variant typically showing alterations in SHH signaling and a subset of the “classic” type showing alterations in Wnt signaling.48–50 Our data suggest that idic(17)(p11.2) tumors are a unique medulloblastoma variant with classic histology and a potential causative mutation (the LCR-mediated NAHR), while other forms of i17q may be a result of tumor evolution. As expression microarrays have shown that many of these tumors fall into the group C and group D families of medulloblastomas, it would be interesting to see how these patients fare compared with others in the same group and whether this rearrangement is responsible for the patients who fail to respond to therapy in these groups.38
In conclusion, the presence of idic(17)(p11.2) in medulloblastoma may indicate a unique variant of the disease with classic histology. Based on a series of 58 consecutive cases, patients with this rearrangement tend to present in the SR group and are significantly more likely to recur after resection than other SR patients. The implementation of a FISH-based assay, shown here to be both sensitive and specific for the rearrangement, may be a clinically useful tool in substratification of the standard-risk population.
Supplementary Material
Supplementary material is available at Neuro-Oncology Journal online (http://neuro-oncology.oxfordjournals.org/).
Funding
This work was supported by the College of American Pathologists Foundation (Resident Research Grant) and by the Washington University Department of Pathology and Immunology.
Supplementary Material
Acknowledgments
The authors would like to thank Courtney Balantine, Todd Druley, and Rob Mitra for their helpful suggestions.
Conflict of interest statement. None declared.
References
- 1.Gurney JG, Smith MA, Bunin GR. CNS and miscellaneous intracranial and intraspinal neoplasms. In: Reis LAG, Smith MA, Gurney JG, et al., editors. Cancer Incidence and Survival among Children and Adolescents: United States SEER Program 1975–1995. Bethesda, MD: National Cancer Research Institute Cancer Surveillance Research Program, NIH Pub. No. 99-4649; 1999. pp. 51–63. Vol NIH Pub. No. 99-4649. [Google Scholar]
- 2.Gurney JG, Kadan-Lottick N. Brain and other central nervous system tumors: rates, trends, and epidemiology. Curr Opin Oncol. 2001;13(3):160–166. doi: 10.1097/00001622-200105000-00005. doi:10.1097/00001622-200105000-00005. [DOI] [PubMed] [Google Scholar]
- 3.Packer RJ, Gajjar A, Vezina G, et al. Phase III study of craniospinal radiation therapy followed by adjuvant chemotherapy for newly diagnosed average-risk medulloblastoma. J Clin Oncol. 2006;24(25):4202–4208. doi: 10.1200/JCO.2006.06.4980. doi:10.1200/JCO.2006.06.4980. [DOI] [PubMed] [Google Scholar]
- 4.Packer RJ, Sutton LN, Elterman R, et al. Outcome for children with medulloblastoma treated with radiation and cisplatin, CCNU, and vincristine chemotherapy. J Neurosurg. 1994;81(5):690–698. doi: 10.3171/jns.1994.81.5.0690. doi:10.3171/jns.1994.81.5.0690. [DOI] [PubMed] [Google Scholar]
- 5.Polkinghorn WR, Tarbell NJ. Medulloblastoma: tumorigenesis, current clinical paradigm, and efforts to improve risk stratification. Nat Clin Pract Oncol. 2007;4(5):295–304. doi: 10.1038/ncponc0794. doi:10.1038/ncponc0794. [DOI] [PubMed] [Google Scholar]
- 6.Garre ML, Cama A, Bagnasco F, et al. Medulloblastoma variants: age-dependent occurrence and relation to Gorlin syndrome–a new clinical perspective. Clin Cancer Res. 2009;15(7):2463–2471. doi: 10.1158/1078-0432.CCR-08-2023. doi:10.1158/1078-0432.CCR-08-2023. [DOI] [PubMed] [Google Scholar]
- 7.Eberhart CG, Kepner JL, Goldthwaite PT, et al. Histopathologic grading of medulloblastomas: a Pediatric Oncology Group study. Cancer. 2002;94(2):552–560. doi: 10.1002/cncr.10189. doi:10.1002/cncr.10189. [DOI] [PubMed] [Google Scholar]
- 8.Perry A. Medulloblastomas with favorable versus unfavorable histology: how many small blue cell tumor types are there in the brain? Adv Anat Pathol. 2002;9(6):345–350. doi: 10.1097/00125480-200211000-00003. doi:10.1097/00125480-200211000-00003. [DOI] [PubMed] [Google Scholar]
- 9.Brown HG, Kepner JL, Perlman EJ, et al. “Large cell/anaplastic” medulloblastomas: a Pediatric Oncology Group Study. J Neuropathol Exp Neurol. 2000;59(10):857–865. doi: 10.1093/jnen/59.10.857. [DOI] [PubMed] [Google Scholar]
- 10.Taylor MD, Northcott PA, Korshunov A, et al. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol. 2012;123(4):465–472. doi: 10.1007/s00401-011-0922-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grotzer MA, Hogarty MD, Janss AJ, et al. MYC messenger RNA expression predicts survival outcome in childhood primitive neuroectodermal tumor/medulloblastoma. Clin Cancer Res. 2001;7(8):2425–2433. [PubMed] [Google Scholar]
- 12.Gilbertson R, Wickramasinghe C, Hernan R, et al. Clinical and molecular stratification of disease risk in medulloblastoma. Br J Cancer. 2001;85(5):705–712. doi: 10.1054/bjoc.2001.1987. doi:10.1054/bjoc.2001.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bayani J, Zielenska M, Marrano P, et al. Molecular cytogenetic analysis of medulloblastomas and supratentorial primitive neuroectodermal tumors by using conventional banding, comparative genomic hybridization, and spectral karyotyping. J Neurosurg. 2000;93(3):437–448. doi: 10.3171/jns.2000.93.3.0437. doi:10.3171/jns.2000.93.3.0437. [DOI] [PubMed] [Google Scholar]
- 14.Eberhart CG, Burger PC. Anaplasia and grading in medulloblastomas. Brain Pathology. 2003;13(3):376–385. doi: 10.1111/j.1750-3639.2003.tb00037.x. doi:10.1111/j.1750-3639.2003.tb00037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ellison DW, Kocak M, Dalton J, et al. Definition of disease-risk stratification groups in childhood medulloblastoma using combined clinical, pathologic, and molecular variables. J Clin Oncol. 2010;29(11):1400–1407. doi: 10.1200/JCO.2010.30.2810. doi:10.1200/JCO.2010.30.2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pfister S, Remke M, Benner A, et al. Outcome prediction in pediatric medulloblastoma based on DNA copy-number aberrations of chromosomes 6q and 17q and the MYC and MYCN loci. J Clin Oncol. 2009;27(10):1627–1636. doi: 10.1200/JCO.2008.17.9432. doi:10.1200/JCO.2008.17.9432. [DOI] [PubMed] [Google Scholar]
- 17.Scheurlen WG, Schwabe GC, Joos S, Mollenhauer J, Sorensen N, Kuhl J. Molecular analysis of childhood primitive neuroectodermal tumors defines markers associated with poor outcome. J Clin Oncol. 1998;16(7):2478–2485. doi: 10.1200/JCO.1998.16.7.2478. [DOI] [PubMed] [Google Scholar]
- 18.Nicholson J, Wickramasinghe C, Ross F, Crolla J, Ellison D. Imbalances of chromosome 17 in medulloblastomas determined by comparative genomic hybridisation and fluorescence in situ hybridisation. Mol Pathol. 2000;53(6):313–319. doi: 10.1136/mp.53.6.313. doi:10.1136/mp.53.6.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cogen PH, McDonald JD. Tumor suppressor genes and medulloblastoma. J Neurooncol. 1996;29(1):103–112. doi: 10.1007/BF00165523. doi:10.1007/BF00165523. [DOI] [PubMed] [Google Scholar]
- 20.Biegel JA, Rorke LB, Packer RJ, et al. Isochromosome 17q in primitive neuroectodermal tumors of the central nervous system. Genes, Chromosomes & Cancer. 1989;1(2):139–147. doi: 10.1002/gcc.2870010206. doi:10.1002/gcc.2870010206. [DOI] [PubMed] [Google Scholar]
- 21.Emadian SM, McDonald JD, Gerken SC, Fults D. Correlation of chromosome 17p loss with clinical outcome in medulloblastoma. Clin Cancer Res. 1996;2(9):1559–1564. [PubMed] [Google Scholar]
- 22.Leonard JR, Cai DX, Rivet DJ, et al. Large cell/anaplastic medulloblastomas and medullomyoblastomas: clinicopathological and genetic features. J Neurosurg. 2001;95(1):82–88. doi: 10.3171/jns.2001.95.1.0082. doi:10.3171/jns.2001.95.1.0082. [DOI] [PubMed] [Google Scholar]
- 23.Pan E, Pellarin M, Holmes E, et al. Isochromosome 17q is a negative prognostic factor in poor-risk childhood medulloblastoma patients. Clin Cancer Res. 2005;11(13):4733–4740. doi: 10.1158/1078-0432.CCR-04-0465. doi:10.1158/1078-0432.CCR-04-0465. [DOI] [PubMed] [Google Scholar]
- 24.Giordana MT, Migheli A, Pavanelli E. Isochromosome 17q is a constant finding in medulloblastoma. An interphase cytogenetic study on tissue sections. Neuropathol Appl Neurobiol. 1998;24(3):233–238. doi: 10.1046/j.1365-2990.1998.00102.x. [DOI] [PubMed] [Google Scholar]
- 25.Scheurlen WG, Schwabe GC, Seranski P, et al. Mapping of the breakpoints on the short arm of chromosome 17 in neoplasms with an i(17q) Genes, Chromosomes & Cancer. 1999;25(3):230–240. doi: 10.1002/(sici)1098-2264(199907)25:3<230::aid-gcc5>3.0.co;2-e. doi:10.1002/(SICI)1098-2264(199907)25:3<230::AID-GCC5>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 26.McCabe MG, Ichimura K, Liu L, et al. High-resolution array-based comparative genomic hybridization of medulloblastomas and supratentorial primitive neuroectodermal tumors. J Neuropathol Exp Neurol. 2006;65(6):549–561. doi: 10.1097/00005072-200606000-00003. doi:10.1097/00005072-200606000-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mendrzyk F, Korshunov A, Toedt G, et al. Isochromosome breakpoints on 17p in medulloblastoma are flanked by different classes of DNA sequence repeats. Genes, Chromosomes & Cancer. 2006;45(4):401–410. doi: 10.1002/gcc.20304. doi:10.1002/gcc.20304. [DOI] [PubMed] [Google Scholar]
- 28.Carvalho CM, Lupski JR. Copy number variation at the breakpoint region of isochromosome 17q. Genome Res. 2008;18(11):1724–1732. doi: 10.1101/gr.080697.108. doi:10.1101/gr.080697.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shaw CJ, Lupski JR. Implications of human genome architecture for rearrangement-based disorders: the genomic basis of disease. Hum Mol Genet. 2004:R57–R64. doi: 10.1093/hmg/ddh073. 13 Spec No 1 doi:10.1093/hmg/ddh073. [DOI] [PubMed] [Google Scholar]
- 30.Barbouti A, Stankiewicz P, Nusbaum C, et al. The breakpoint region of the most common isochromosome, i(17q), in human neoplasia is characterized by a complex genomic architecture with large, palindromic, low-copy repeats. Am.J. Human Genetics. 2004;74(1):1–10. doi: 10.1086/380648. doi:10.1086/380648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCabe MG, Ichimura K, Pearson DM, et al. Novel mechanisms of gene disruption at the medulloblastoma isodicentric 17p11 breakpoint. Genes, Chromosomes & Cancer. 2009;48(2):121–131. doi: 10.1002/gcc.20625. doi:10.1002/gcc.20625. [DOI] [PubMed] [Google Scholar]
- 32.Fioretos T, Strombeck B, Sandberg T, et al. Isochromosome 17q in blast crisis of chronic myeloid leukemia and in other hematologic malignancies is the result of clustered breakpoints in 17p11 and is not associated with coding TP53 mutations. Blood. 1999;94(1):225–232. [PubMed] [Google Scholar]
- 33.Gilbertson RJ, Ellison DW. The origins of medulloblastoma subtypes. Annu Rev Pathol. 2008;3:341–365. doi: 10.1146/annurev.pathmechdis.3.121806.151518. doi:10.1146/annurev.pathmechdis.3.121806.151518. [DOI] [PubMed] [Google Scholar]
- 34.Squire SE, Chan MD, Marcus KJ. Atypical teratoid/rhabdoid tumor: the controversy behind radiation therapy. J Neurooncol. 2007;81(1):97–111. doi: 10.1007/s11060-006-9196-z. doi:10.1007/s11060-006-9196-z. [DOI] [PubMed] [Google Scholar]
- 35.Grill J, Sainte-Rose C, Jouvet A, et al. Treatment of medulloblastoma with postoperative chemotherapy alone: an SFOP prospective trial in young children. Lancet Oncol. 2005;6(8):573–580. doi: 10.1016/S1470-2045(05)70252-7. doi:10.1016/S1470-2045(05)70252-7. [DOI] [PubMed] [Google Scholar]
- 36.Jakacki RI, Feldman H, Jamison C, Boaz JC, Luerssen TG, Timmerman R. A pilot study of preirradiation chemotherapy and 1800 cGy craniospinal irradiation in young children with medulloblastoma. Int J Radiat Oncol Biol Phys. 2004;60(2):531–536. doi: 10.1016/j.ijrobp.2004.03.027. doi:10.1016/j.ijrobp.2004.03.027. [DOI] [PubMed] [Google Scholar]
- 37.Ellison DW, Dalton J, Kocak M, et al. Medulloblastoma: clinicopathological correlates of SHH, Wnt, and non-SHH/Wnt molecular subgroups. Acta Neuropathol. 2011;121(3):381–396. doi: 10.1007/s00401-011-0800-8. doi:10.1007/s00401-011-0800-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Northcott PA, Korshunov A, Witt H, et al. Medulloblastoma comprises four distinct molecular variants. Journal of Clinical Oncology : Official Journal of the American Society of Clinical Oncology. 2011;29(11):1408–1414. doi: 10.1200/JCO.2009.27.4324. doi:10.1200/JCO.2009.27.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fuller CE, Perry A. Fluorescence in situ hybridization (FISH) in diagnostic and investigative neuropathology. Brain Pathol. 2002;12(1):67–86. doi: 10.1111/j.1750-3639.2002.tb00424.x. doi:10.1111/j.1750-3639.2002.tb00424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bien-Willner GA, Stankiewicz P, Lupski JR, Northup JK, Velagaleti GV. Interphase FISH screening for the LCR-mediated common rearrangement of isochromosome 17q in primary myelofibrosis. Am. J. Hematology. 2005;79(4):309–313. doi: 10.1002/ajh.20366. doi:10.1002/ajh.20366. [DOI] [PubMed] [Google Scholar]
- 41.Abramoff MD, Magelhaes PJ, Ram SJ. Image Processing with ImageJ. Biophotonics International. 2004;11(7):36–42. [Google Scholar]
- 42.Taylor RE, Bailey CC, Robinson K, et al. Results of a randomized study of preradiation chemotherapy versus radiotherapy alone for nonmetastatic medulloblastoma: The International Society of Paediatric Oncology/United Kingdom Children's Cancer Study Group PNET-3 Study. J Clin Oncol. 2003;21(8):1581–1591. doi: 10.1200/JCO.2003.05.116. doi:10.1200/JCO.2003.05.116. [DOI] [PubMed] [Google Scholar]
- 43.Gajjar A, Chintagumpala M, Ashley D, et al. Risk-adapted craniospinal radiotherapy followed by high-dose chemotherapy and stem-cell rescue in children with newly diagnosed medulloblastoma (St Jude Medulloblastoma-96): long-term results from a prospective, multicentre trial. Lancet Oncol. 2006;7(10):813–820. doi: 10.1016/S1470-2045(06)70867-1. doi:10.1016/S1470-2045(06)70867-1. [DOI] [PubMed] [Google Scholar]
- 44.Zeltzer PM, Boyett JM, Finlay JL, et al. Metastasis stage, adjuvant treatment, and residual tumor are prognostic factors for medulloblastoma in children: conclusions from the Children's Cancer Group 921 randomized phase III study. J Clin Oncol. 1999;17(3):832–845. doi: 10.1200/JCO.1999.17.3.832. [DOI] [PubMed] [Google Scholar]
- 45.Ellison D. Classifying the medulloblastoma: insights from morphology and molecular genetics. Neuropathology and Applied Neurobiology. 2002;28(4):257–282. doi: 10.1046/j.1365-2990.2002.00419.x. doi:10.1046/j.1365-2990.2002.00419.x. [DOI] [PubMed] [Google Scholar]
- 46.Lafay-Cousin L, Bouffet E, Hawkins C, Amid A, Huang A, Mabbott DJ. Impact of radiation avoidance on survival and neurocognitive outcome in infant medulloblastoma. Curr Oncol. 2009;16(6):21–28. doi: 10.3747/co.v16i6.435. doi:10.3747/co.v16i6.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lamont JM, McManamy CS, Pearson AD, Clifford SC, Ellison DW. Combined histopathological and molecular cytogenetic stratification of medulloblastoma patients. Clin Cancer Res. 2004;10(16):5482–5493. doi: 10.1158/1078-0432.CCR-03-0721. doi:10.1158/1078-0432.CCR-03-0721. [DOI] [PubMed] [Google Scholar]
- 48.Eberhart CG, Tihan T, Burger PC. Nuclear localization and mutation of beta-catenin in medulloblastomas. J Neuropathol Exp Neurol. 2000;59(4):333–337. doi: 10.1093/jnen/59.4.333. [DOI] [PubMed] [Google Scholar]
- 49.Pietsch T, Waha A, Koch A, et al. Medulloblastomas of the desmoplastic variant carry mutations of the human homologue of Drosophila patched. Cancer Res. 1997;57(11):2085–2088. [PubMed] [Google Scholar]
- 50.Koch A, Waha A, Tonn JC, et al. Somatic mutations of Wnt/wingless signaling pathway components in primitive neuroectodermal tumors. Int J Cancer. 2001;93(3):445–449. doi: 10.1002/ijc.1342. doi:10.1002/ijc.1342. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.