Abstract
Deregulation of the p16INK4a-Cdk4/6-Rb pathway is commonly detected in patients with glioblastoma multiforme (GBM) and is a rational therapeutic target. Here, we characterized the p16INK4a-Cdk4/6-Rb pathway in the Mayo panel of GBM xenografts, established from primary tissue samples from patients with GBM, and evaluated their response to PD0332991, a specific inhibitor of Cdk4/6. All GBM xenograft lines evaluated in this study had disruptions in the p16INK4a-Cdk4/6-Rb pathway. In vitro evaluation using short-term explant cultures from selected GBM xenograft lines showed that PD0332991 effectively arrested cell cycle in G1-phase and inhibited cell proliferation dose-dependently in lines deleted for CDKN2A/B-p16INK4a and either single-copy deletion of CDK4 (GBM22), high-level CDK6 amplification (GBM34), or deletion of CDKN2C/p18INK4c (GBM43). In contrast, 2 GBM lines with p16INK4a expression and either CDK4 amplification (GBM5) or RB mutation (GBM28) were completely resistant to PD0332991. Additional xenograft lines were screened, and GBM63 was identified to have p16INK4a expression and CDK4 amplification. Similar to the results with GBM5, GBM63 was resistant to PD0332991 treatment. In an orthotopic survival model, treatment of GBM6 xenografts (CDKN2A/B-deleted and CDK4 wild-type) with PD0332991 significantly suppressed tumor cell proliferation and prolonged survival. Collectively, these data support the concept that GBM tumors lacking p16INK4a expression and with nonamplified CDK4 and wild-type RB status may be more susceptible to Cdk4/6 inhibition using PD0332991.
Keywords: Cdk4, Cdk6, glioblastoma, p16INK4a, retinoblastoma protein
The p16INK4a-Cdk4-Rb pathway is disrupted in the vast majority of glioblastoma (GBM) and is a potentially interesting target for novel therapeutic strategies in this devastating disease.1,2 The p16INK4a -Cdk4-Rb axis critically regulates G1 to S phase progression. In response to mitogenic signaling, cyclin D/Cdk4, cyclinD/Cdk6, and cyclin E/Cdk2 complexes cooperate to phosphorylate the tumor suppressor retinoblastoma protein (Rb) on multiple serine and threonine residues. These phosphorylation events disrupt Rb-mediated transcriptional repression and facilitate progression into S-phase. The activity of Cdk4/6 is suppressed by INK4 family proteins, including p15INK4b, p16INK4a, p18INK4c, and p19INK4d. Overall, approximately 78% of GBM tumors have defects in the p16INK4a-Cdk4-Rb pathway; notable specific lesions include homozygous deletion of CDKN2A/2B (52%), amplification of CDK4 (18%), amplification of CDK6 (1%), and deletion or mutation of RB (12%).2 These observations highlight the critical role of the p16INK4a-Cdk4-Rb pathway in GBM and suggest that targeting this pathway might be a promising strategy to improve the therapeutic efficacy among patients with GBM.
PD0332991 is a small molecule inhibitor developed to specifically inhibit the kinase activity of Cdk4 and Cdk6. This compound has potent anti-proliferative effects in multiple tumor models, including glioma, myeloma, and lung and breast cancers,3–5 and is being tested in multiple clinical trials, including as a monotherapy for patients with recurrent GBM. To more fully understand the genetic alterations that influence response to PD0332991 treatment, efficacy was evaluated in a panel of Mayo GBM xenograft lines, which were developed and maintained through serial heterotopic transplantation in nude mice. This model system faithfully maintains the key genetic and molecular features of the original patient tumor samples and has been used extensively to investigate the efficacy of a number of novel and conventional therapies in GBM.6–9 To investigate the effects of PD0332991, the molecular status of key components of the p16INK4a-Cdk4-Rb pathway were defined in the Mayo GBM xenograft panel, and tumor lines with disparate molecular features were evaluated for response to PD0332991.
Materials and Methods
Cells and Reagents
All animal experiments were conducted with prior approval of the Mayo Institutional Animal Care and Use Committee. Each of the glioma lines used in this study were derived from primary xenograft lines established from tumors from patients with GBM and maintained by serial heterotopic passage in mice.10 As described previously, short-term explant cultures were established with flank xenografts and grown in DMEM supplemented with 2.5% fetal bovine serum, 1% penicillin, and 1% streptomycin. PD0332991 was obtained from Pfizer. Antibodies against p16 (cat# sc-468), Cdk4 (cat# sc-460), and Cdk6 (cat# sc-32501) were purchased from Santa Cruz Biotechnology, and antibodies against phospho-Rb (serine780) and total Rb protein were purchased from Cell Signaling Technologies. β-actin antibody was purchased from Sigma-Aldrich. Secondary anti-rabbit IgG and anti-mouse IgG were purchased from Cell Signaling Technologies and Pierce, respectively.
RNA Isolation and Reverse-Transcription Polymerase Chain Reaction (RT-PCR)
Total RNA was isolated from frozen xenograft tumor tissues using RNeasy kit (Qiagen). For RT-PCR, total RNA (1 μg) was reverse transcribed using random primers and reverse transcriptase (Promega) according to the manufacturer's instructions. Reverse transcription conditions were denaturing at 70°C for 5 min, cool on ice for 5 min, and extension at 37°C for 60 min. Primers used for PCR amplification were synthesized and purchased from Integrated DNA Technologies. Human-specific primers for p15INK4b, p16INK4a, p18INK4c, and p19INK4d were as described previously.11 PCR amplification was performed in a thermocycler with denaturation at 95°C for 10 min and 35 cycles of 30 s at 95°C, 30 s at 55°C, and 1 min at 72°C, followed by a final elongation at 72°C for 10 min.
Western Blotting
Cells were lysed in RIPA lysis buffer (cat# R0278, Sigma-Aldrich) supplemented with protease inhibitor cocktail (Roche). Total proteins were isolated from flash-frozen flank xenograft tissues or short-term explant cell cultures, separated by SDS-PAGE, and electro-transferred onto polyvinylidene difluoride (PVDF) membranes. Membranes were blocked in Tris-buffered saline (TBS) containing 5% milk and 0.1% Tween 20 at room temperature. All primary antibodies were incubated overnight at 4°C, followed by room temperature incubation with a secondary antibody conjugated with horseradish peroxidase for 1 h. Detection was performed with Super Signal Chemiluminescent reagent according to the manufacturer's protocol (Pierce).
Array Comparative Genomic Hybridization (aCGH)
aCGH was performed on DNA specimens using the Human Genome 244A microarray (Agilent Technologies). The labeling and hybridization steps were done according to the manufacturer's recommendations. In brief, random primers and exo-Klenow fragment (Agilent Technologies) were used to differentially label 1 μg of tumor DNA with Cy5 and 1 μg of reference DNA with Cy3. Reference DNA was from a single male control for female tumor samples and was from a single female control for male tumor samples. Labeled genomic reactions were passed over purification columns (Millipore Corporation) and hybridized at 65°C for 40 h. Microarrays were scanned in a G2565CA Microarray Scanner System (Agilent Technologies). Feature extraction was performed with Feature Extraction Software, version 10.5.1.1 (Agilent Technologies). Extracted data was imported and analyzed using Genomic Workbench, version 5.0.14 (Agilent Technologies).
CDK4 Copy Number Analysis by Real-Time PCR
Total DNA was isolated from frozen xenograft tissues with Puregene Tissue Kit (Gentra Systems) according to the manufacture's instructions. Primers and probe sequences for CDK4 and ALB (albumin) and real-time PCR with standard curve was performed as previously described.12 The level of amplification for CDK4 in GBM was calculated as a ratio of copy number of CDK4 to that of the reference gene ALB.
CyQUANT Cell Proliferation Assay
A cell proliferation assay was performed using the CyQUANT Cell Proliferation kit (Molecular Probes) according to the manufacturer's recommendations. Cells were seeded in triplicate in 96-well plates and treated the next day with PD0332991 for 5 days. Media was then removed, and plates were stored at −80°C. The plates were thawed and lysed in CyQUANT GR dye-containing lysis buffer. After 4 min incubation at room temperature, the fluorescence intensity of the DNA binding dye was measured using a TECAN plate reader with filters appropriate for excitation at 480 nm and emission at 520 nm.
BrdU-Based Cell Proliferation Enzyme-Linked Immunosorbent Assay (ELISA)
Cellular BrdU incorporation was quantitated using a BrdU-based cell proliferation ELISA assay kit (Roche). Cells were plated in triplicate in 96-well plates overnight and then treated with graded concentrations of PD0332991 for 48 h. Subsequently, cells were incubated with 10 μM BrdU for 2 h and then fixed and processed according to manufacturer's protocol. Absorbance was read on a TECAN plate reader at 450 nm.
Flow Cytometry Analysis
Cell cycle distribution was analyzed using flow cytometry. After treatment with PD0332991, the cells were harvested, washed with phosphate-buffered saline (PBS), and fixed with ice-cold 70% ethanol/30% PBS. Cells were resuspended in PBS containing 40 μg/mL propidium iodide, 100 μg/mL RNase A, and 0.05% Triton X-100 and incubated at 37°C for 30 min. DNA content was determined using a FACScan flow cytometer system (Becton Dickinson), and results were analyzed using Modfit software (Verity Software House).
Intracranial Xenograft Model and Therapy Response Evaluation
Drug efficacy was evaluated in vivo using an orthotopic GBM xenograft model described previously.6 Mice with established intracranial xenografts from GBM6 were randomized into groups of 10 mice each and treated with drug vehicle alone versus PD0332991. PD0332991 was suspended in sodium lactate solution (Sigma-Aldrich) and administered by oral gavage daily at 150 mg/kg for 28 days. All mice used for therapy evaluation were observed daily and euthanized after they reached a moribund condition. To assess whether PD0332991 inhibits its target and cell proliferation, mice with intracranial GBM6 established as above were treated either with vehicle or with PD0332991 daily at 150 mg/kg for 6 days before euthanasia and subsequent processing for paraffin embedding.
Immunohistochemistry
Brains from mice with intracranial tumor were resected, placed in formalin, and embedded in paraffin. Ki67 immunohistochemistry was performed with a rabbit monoclonal antibody (SP6, Epitomics) as described previously.13 Similarly, phospho-Rb staining was performed with a rabbit p-Rb (Serine807/811) antibody (Cell Signaling Technologies) at a 1:75 dilution. To determine the positivity of Ki67 and p-Rb staining, digital images of 3–5 high-power fields were obtained for each tumor-bearing animal, and the fraction of area with positive staining was determined with KS400 image analysis software (Carl Zeiss). Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay was performed using an ApopTag Plus Peroxidase In Situ Apoptosis detection kit (Millipore) according to the manufacturer's protocol.
Statistical Analysis
The data were presented as either mean ± standard error of the mean or mean ± standard deviation. A 2-sample Student's t test was used to compare the measures across groups. Survival distributions were estimated using the Kaplan-Meier method. The log rank test was used to compare survival across groups. The criteria for statistical significance were taken as 2-tailed P values <.05.
Results
Molecular Abnormalities of p16INK4a -Cdk4-Rb Pathway
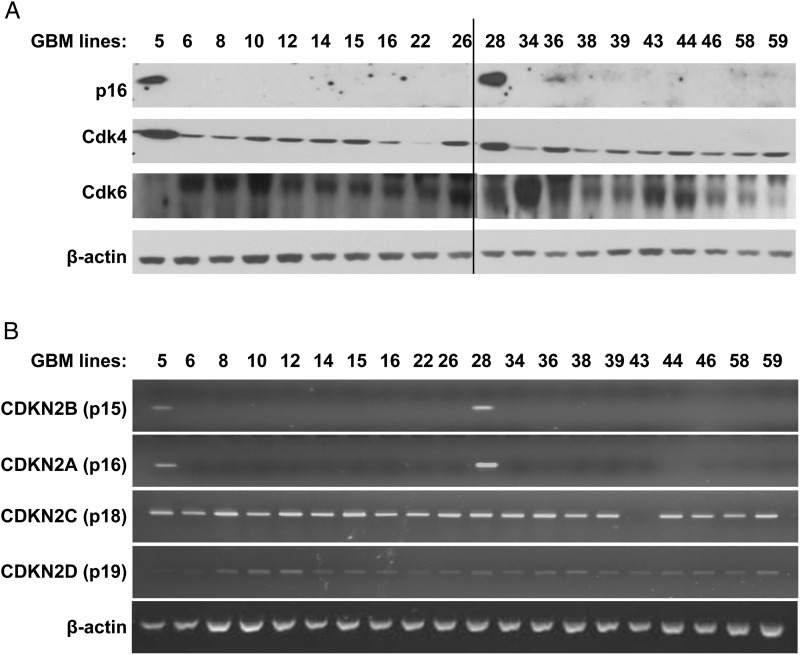
A panel of 20 GBM xenografts established directly from patient tumor samples was examined for molecular defects in components of the p16INK4a-Cdk4-Rb pathway. Expression of the Cdk inhibitors p16INK4a and p15INK4b was detected in only 2 of 20 xenograft lines (GBM5 and GBM28) (Fig. 1). In contrast, p18INK4c and p19INK4d are expressed in all the tumor lines except in GBM43, which specifically lacks p18INK4c (Fig. 1B). Western blotting of the lines for Cdk4 and Cdk6 suggests elevated expression of Cdk4 in both of the p16-expressing tumors (GBM5 and GBM28), elevated Cdk6 expression in GBM34, and reduced Cdk4 expression in GBM22 (Fig. 1A). Thus, at the RNA and protein level, all 20 lines have alterations in the p16INK4a -Cdk4-Rb pathway.
Fig. 1.
Expression analysis of INK4, Cdk4, and Cdk6 in glioblastoma multiforme (GBM) xenograft panel. Archived flank tumor specimens from 3 individual mice for each tumor line were processed for Western blotting and RT-PCR. Protein and RNA samples from each animal were pooled at equal ratio and used for analysis. (A) Western blot analysis of p16INK4a, Cdk4, and Cdk6 expression. (B) RT-PCR using human-specific primers for CDKN2A, CDKN2B, CDKN2C, and CDKN2D.
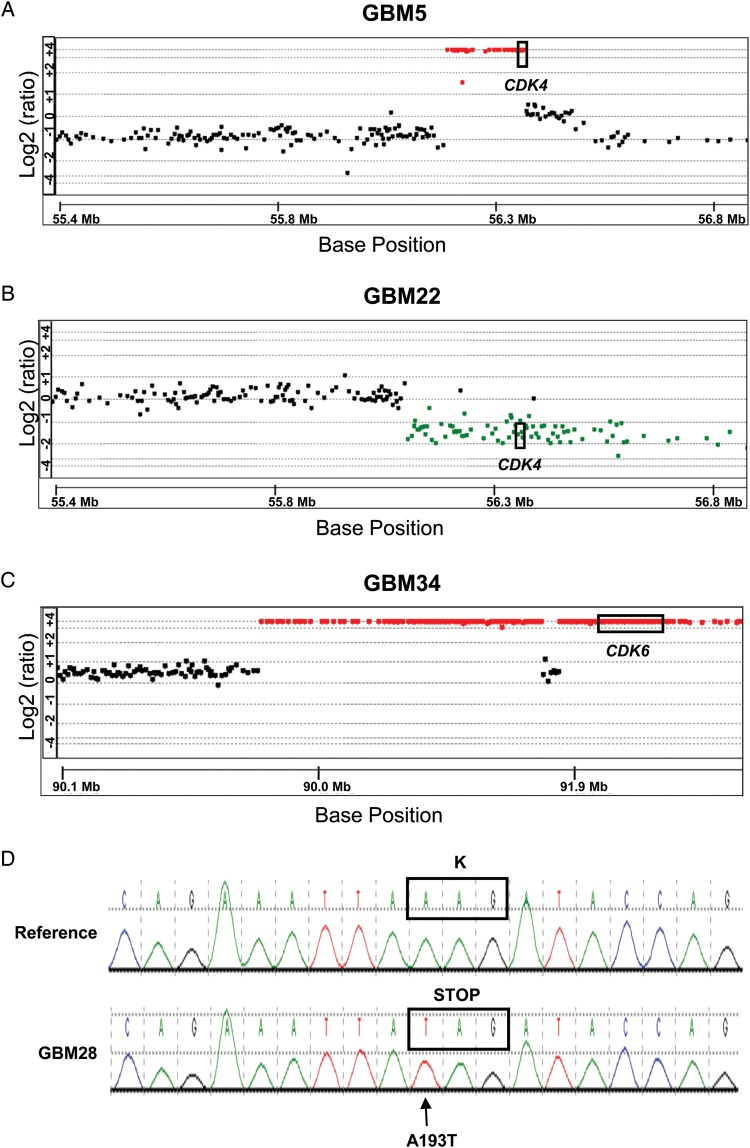
The nature of the genetic lesions associated with altered expression within the p16INK4a-Cdk4-Rb axis was evaluated by aCGH and Sanger sequencing. This analysis identified homozygous deletions of the CDKN2A/B locus in all 18 xenograft lines lacking p16INK4a expression by RT-PCR (Table 1). In contrast, for the p16INK4a-expressing lines, a hemizygous deletion of CDKN2A/B was identified in GBM5, and no deletion within the CDKN2A/B locus was identified in GBM28. Also consistent with the RT-PCR results, homozygous deletion of CDKN2C was identified only in GBM43 (Table 1). Evaluation of the CDK4 locus across the lines revealed high-level CDK4 amplification in GBM5 and a single copy loss of CDK4 in GBM22 (Fig. 2A and B). CDK6 resides on chromosome 7; trisomy of chromosome 7 was observed in all but 2 lines (GBM12 and GBM28), which had the usual 2 copies of chromosome 7 (Table 1). Of interest, high-level CDK6 amplification was observed in GBM34 in addition to homozygous CDKN2A/B deletion (Fig. 2C and Table 1). RB status was assessed in both p16-intact tumors by DNA sequencing: GBM5 was wild-type for RB, whereas GBM28 harbors a bi-allelic missense mutation in RB exon 2 (A193 → T) that encodes for a K68 to stop codon change (Fig. 2D). In summary, 18 lines have CDKN2A/B deletion, GBM5 has CDK4 amplification, GBM22 has CDK4 hemizygous deletion, GBM34 has CDK6 amplification, and GBM28 has an RB truncation mutation.
Table 1.
Genetic features of glioblastoma multiforme (GBM) lines
| GBM lines | CDKN2A | CDKN2B | CDKN2C | CDKN2D | CDK4 | CDK6 | CCND1 | CCND2 | CCND3 | RB1 |
|---|---|---|---|---|---|---|---|---|---|---|
| GBM5 | hemi del | hemi del | amp | gain | amp | gain | ||||
| GBM6 | homo del | homo del | gain | gain | gain | |||||
| GBM8 | homo del | homo del | gain | gain | gain | |||||
| GBM10 | homo del | homo del | gain | gain | gain | hemi del | loss | |||
| GBM12 | homo del | homo del | gain | gain | ||||||
| GBM14 | homo del | homo del | loss | loss | gain | loss | ||||
| GBM15 | homo del | homo del | gain | gain | loss | |||||
| GBM16 | homo del | homo del | loss | gain | gain | gain | loss | gain | loss | |
| GBM22 | homo del | homo del | gain | gain | hemi del | gain | loss | loss | loss | |
| GBM26 | homo del | homo del | gain | gain | hemi del | |||||
| GBM28 | loss | loss | ||||||||
| GBM34 | homo del | homo del | amp | |||||||
| GBM36 | gain | |||||||||
| GBM38 | homo del | homo del | gain | loss | loss | hemi del | ||||
| GBM39 | homo del | homo del | loss | gain | loss | |||||
| GBM43 | homo del | homo del | homo del | gain | gain | |||||
| GBM44 | homo del | homo del | loss | gain 2 | ||||||
| GBM46 | homo del | homo del | gain | gain | gain | gain 2 | loss | |||
| GBM58 | gain | |||||||||
| GBM59 | homo del | homo del | gain | gain | gain | loss | loss |
Abbreviations: Homo del, homozygous deletion of the gene; Hemi del, hemizygous deletion of the gene; Amp, amplification of the gene; Gain, gain (trisomy) of the chromosome or chromosomal region containing the gene; Gain 2, gain greater than trisomy of the chromosome or chromosomal region containing the gene; Loss, loss of the chromosome or chromosomal region containing the gene.
Fig. 2.
Copy number and sequence alternations detected in the CDK4, CDK6 and RB genes in selected GBM lines. The genomic locations of CDK4 and CDK6 genes are indicated by the boxes. (A) aCGH results illustrating CDK4 amplification on a background of chromosome 12 q-arm deletion in GBM5. (B) aCGH results illustrating CDK4 deletion (a single copy loss) in GBM22. (C) aCGH results illustrating CDK6 amplification on a background of chromosome 7 q-arm gain in GBM34. In addition, a large segment surrounding the gene is also amplified. (D) DNA sequencing analysis of RB1 gene in GBM28 identified an A to T transversion at nucleotide 193, resulting in K68 to stop codon change. The reference sequence is shown in the upper panel.
Response to PD0332991 Relative to p16-Cdk4-Rb Status
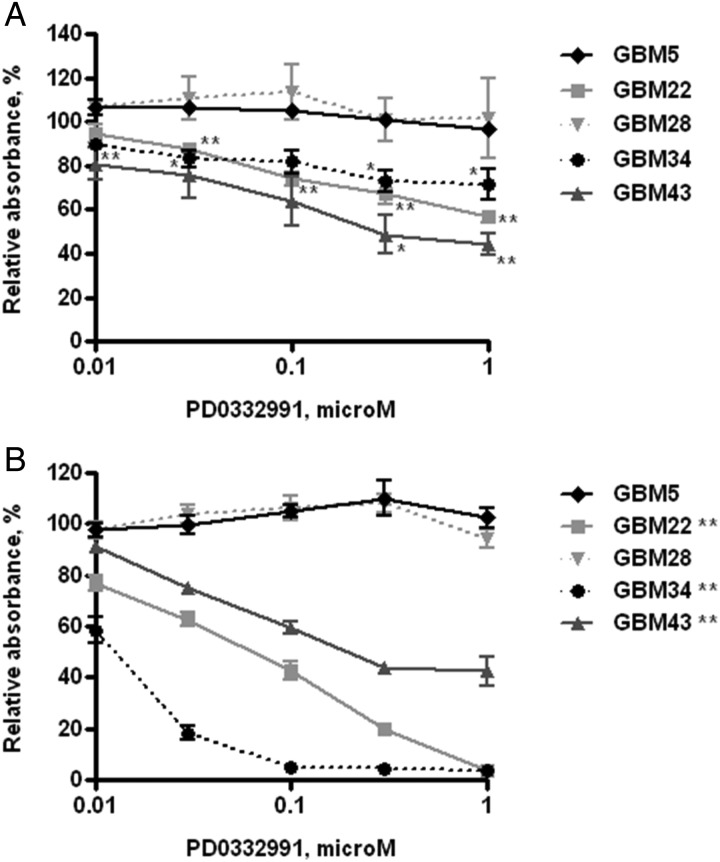
Based on the molecular characterization, specific tumor lines with distinct genetic lesions in the p16INK4a-Cdk4/6-Rb pathway were selected for evaluation of response to the Cdk4/6 inhibitor PD0332991. Specifically, 2 lines with intact p16INK4a were selected, as well as GBM5 (CDK4 amplified) and GBM28 (RB truncation mutant), and 3 lines with homozygous deletion of CDKN2A/p16INK4a GBM22 (hemizygous deletion of CDK4), GBM34 (amplification of CDK6), and GBM43 (homozygous deletion of CDKN2C/p18INK4c). Short-term explant cultures from each of the xenograft lines were treated with graded concentrations of PD0332991 for 5 days and subjected to CyQUANT cell proliferation assay, which measures total cellular nucleic acid as a marker for cell number. Of the p16-deleted lines, treatment with 1 μM of PD0332991 effectively suppressed cell proliferation with a reduction in absorbance to 56.6 ± 4.8% (P< .001), 71.3 ± 12.1% (P< .05), and 44.3 ± 8.2% (P< .01) of control in GBM22, GBM34, and GBM43, respectively (Fig. 3A). In contrast, PD0332991 had no impact on cell proliferation in either p16-intact cell line, GBM5, or GBM28 at any dose level.
Fig. 3.
In vitro sensitivity of PD0332991 in GBM xenograft lines. (A) Effects of PD0332991 on cell proliferation evaluated using short-term explant cultures by CyQUANT assay following 5-day drug incubation. Error bars represent the standard error of the mean from 3 independent experiments with each treatment analyzed in triplicate and normalized to control treatment for a given experiment. Data sets marked with asterisks are significantly different from control (DMSO treated) as assessed by Student's t-test. *P < .05 and **P < .01. (B) Effects of PD0332991 on BrdU incorporation examined by BrdU ELISA cell proliferation assay following 48-hour drug incubation. Results presented as mean ± SEM from three independent experiments with each treatment analyzed in triplicate and normalized to control treatment for a given experiment. Difference between drug treatment and control was compared using Student's t-test. **P < .01 represents significant difference at every dose of treatment vs. control.
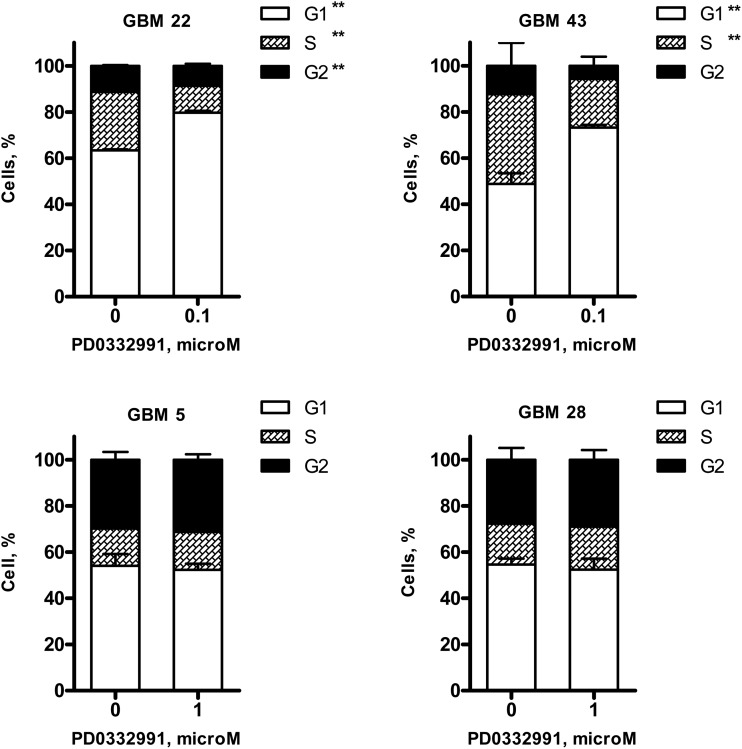
The impact of drug treatment on cell proliferation was interrogated more specifically by BrdU incorporation and by cell cycle distribution analysis. Cells were treated with PD0332991 for 48 h, pulse labeled with BrdU, and then processed in a BrdU ELISA assay to measure changes in cell proliferation. For each of the p16-deleted lines, PD0332991 significantly inhibited BrdU incorporation in a dose-dependent manner with maximal suppression of BrdU uptake at 1 μM. At this dose level, PD0332991 significantly suppressed BrdU incorporation to 3.9 ± 2.7% (P < .001), 3.6 ± 0.6% (P < .001), and 42.6 ± 7.0% (P < .001) of the control in GBM22, GBM34, and GBM43, respectively (Fig. 3B). In contrast, the p16INK4a expressing cell lines were highly resistant to PD0332991 treatment with no suppression of BrdU incorporation in either GBM5 or GBM28 at any dose level. To further evaluate the effects of PD0332991 on cell cycle, cell content was analyzed using flow cytometry (Fig. 4). In 2 sensitive tumor lines, treatment with 100 nM PD0332991 for 24 h resulted in significant accumulation of cells in G1 (GBM22: 79.7 ± 0.8% treated vs. 63.4 ± 0.5% control cells in G1 phase, P< .001; GBM43: 70.6 ± 1.1% treated vs. 48.9 ± 4.6% control cells in G1 phase, P< .001). In contrast, a 10-fold higher concentration of PD0332991 (1 μM) had no significant effect on cell cycle distribution in the 2 p16-intact lines, GBM5 (52.3 ± 2.7% treated vs. 54.0 ± 5.1% control, P = .478), or GBM28 (52.4 ± 4.7% treated vs. 54.6 ± 2.6% control, P = .348). Thus, in p16INK4a deleted tumor lines, PD0332991 promoted a G1 arrest associated with reduced BrdU incorporation. In contrast, p16INK4a expressing/CDK4-amplified GBM5 and p16INK4a expressing/RB mutant GBM28 were insensitive to PD0332991 treatment.
Fig. 4.
Effects of Cdk4/6 inhibitor on cell cycle distribution in GBM xenograft lines. Short-term explant culture of GBM5, 22, 28, and 43 were treated with DMSO or Cdk4/6 inhibitor PD0332991 as indicated doses for 24 h. Cell cycle analysis was performed by PI staining followed by flow cytometry. Values are graphed as percentage of cells at different phases. PD0332991 induced G1 arrest in GBM22 and 43, but not in GBM5 and 28. Results presented as mean ± SEM from 2 independent experiments with each treatment analyzed in triplicate for a given experiment. **P < .01 vs. control treatment.
Rb Phosphorylation Is Inhibited by PD0332991 in Sensitive Lines
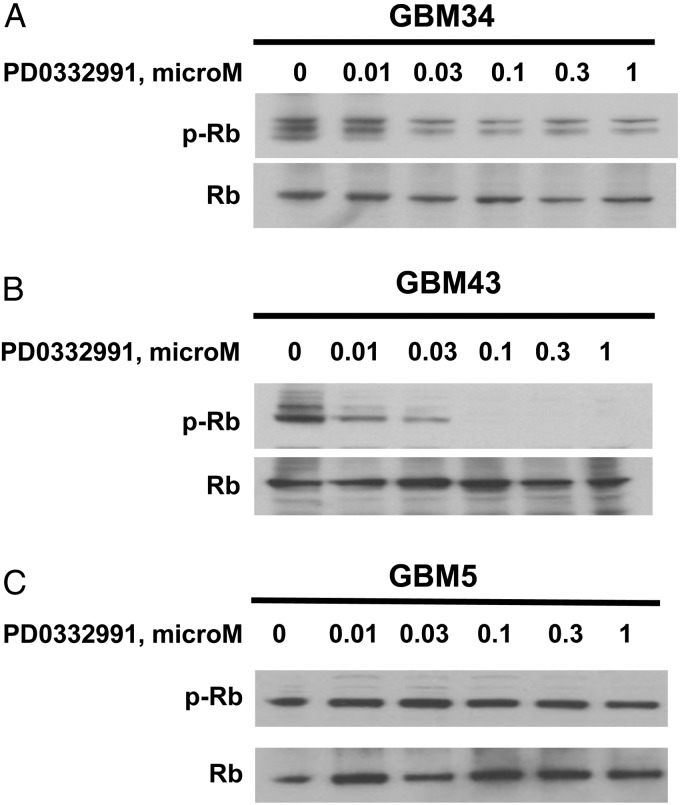
Cdk4/6-cyclin D-mediated phosphorylation of Rb drives cell cycle progression by promoting release of the E2F transcription factor. Therefore, the effects of PD0332991 treatment on Rb phosphorylation were examined by Western blotting. In the sensitive GBM43 line, PD0332991 led to marked suppression of phospho-Rb levels with increasing dose of treatment (Fig. 5B). Phospho-Rb signal was similarly suppressed after PD0332991 treatment in sensitive lines GBM22 and GBM34 (data not shown and Fig. 5A). However, in the resistant cell line GBM5 harboring CDK4 amplification, treatment with 1 μM PD0332991 did not significantly impact Rb phosphorylation (Fig. 5C). Rb protein was not detectable in resistant line GBM28, which was identified with a truncation mutation in RB. Thus, phospho-Rb is efficiently inhibited by PD0332991 in p16INK4a deficient cell lines, whereas PD0332991 treatment did not change phospho-Rb level in the p16INK4a expressing/CDK4 amplified GBM5 cells.
Fig. 5.
Effects of Cdk4/6 inhibitor on Rb pathway in GBM tumor lines. Short-term explant culture cells were treated with graded doses of PD0332991 for 24 h before harvesting. Cell lysates were obtained and subjected to Western blot analysis for phospho-Rb and total Rb. PD0332991 inhibited Rb phosphorylation with increasing concentrations in GBM34 and GBM43, which is not seen in GBM5.
Testing in Additional GBM Xenograft Lines
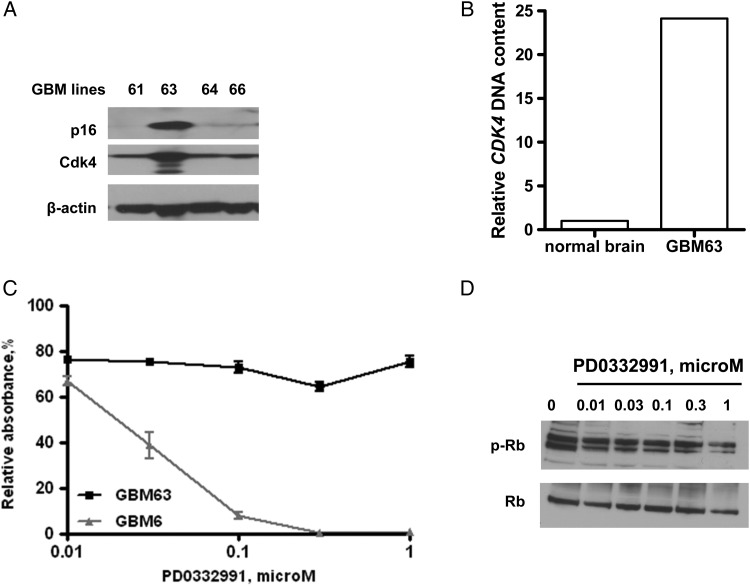
Based on the above results, additional GBM xenograft lines were screened for alterations in the p16INK4a-Cdk4-Rb axis. Among the 20 additional GBM lines screened, GBM63 was identified with intact p16INK4a expression and Cdk4 overexpression by Western blotting (Fig. 6A). Subsequent analysis of genomic DNA from GBM63 and normal human brain tissue demonstrated high-level copy number gain of CDK4 (Fig. 6B). Consistent with the results for the other CDK4 amplified line (GBM5), GBM63 line was relatively resistant to PD0332991, with a 20% reduction in BrdU incorporation at the lowest dose tested (0.01 μM) but no additional decrease in BrdU incorporation up to 1 μM PD0332991 (Fig. 6C). Similar to the BrdU results, 0.01 μM PD0332991 resulted in a minor suppression of phospho-Rb levels, but there was no further suppression at higher dose levels (Fig. 6D). In contrast, BrdU incorporation was significantly inhibited in an additional p16-deleted tumor line, GBM6 (Fig. 6C). Collectively, the data from GBM5 and GBM63 suggest that CDK4 amplification may confer resistance to PD0332991 treatment in p16 -expressing GBM lines, whereas p16-deleted tumor lines are relatively sensitive to PD0332991.
Fig. 6.
Effects of PD0332991 on additional GBM xenograft lines. (A) Western blot analysis of flank tumor specimens of additional GBM lines for p16INK4a and Cdk4 expression. (B) CDK4 copy number analysis for a GBM63 tumor and normal brain specimen. (C) Effects of PD0332991 on BrdU incorporation in GBM63 and GBM6 following 48-hour drug incubation. Results presented as mean ± SEM from three independent experiments with each treatment analyzed in triplicate for a given experiment. (D) Effects of PD0332991 treatment on Rb phosphorylation status in GBM63.
Cdk4 knockdown studies were performed to evaluate the relationship between absolute levels of Cdk4 expression and resistance to PD0332991. Transduction with 2 different lentiviral shRNAs achieved variable levels of Cdk4 knockdown and suppressed Rb phosphorylation (Supplementary material, Fig. S1A), compared with an empty vector. Of interest, despite robust Cdk4 suppression, there was no impact on the proliferative rate of GBM5 (data not shown) or the responsiveness to Cdk4 inhibition (Supplementary material, Fig. S1B). Collectively, these data suggest that absolute levels of Cdk4 kinase expression may not be the critical factor driving the resistance to PD0332991 in the CDK4-amplified GBM5 xenograft line.
PD0332991 Inhibits Cell Proliferation and Prolongs Survival of GBM Intracranial Tumor
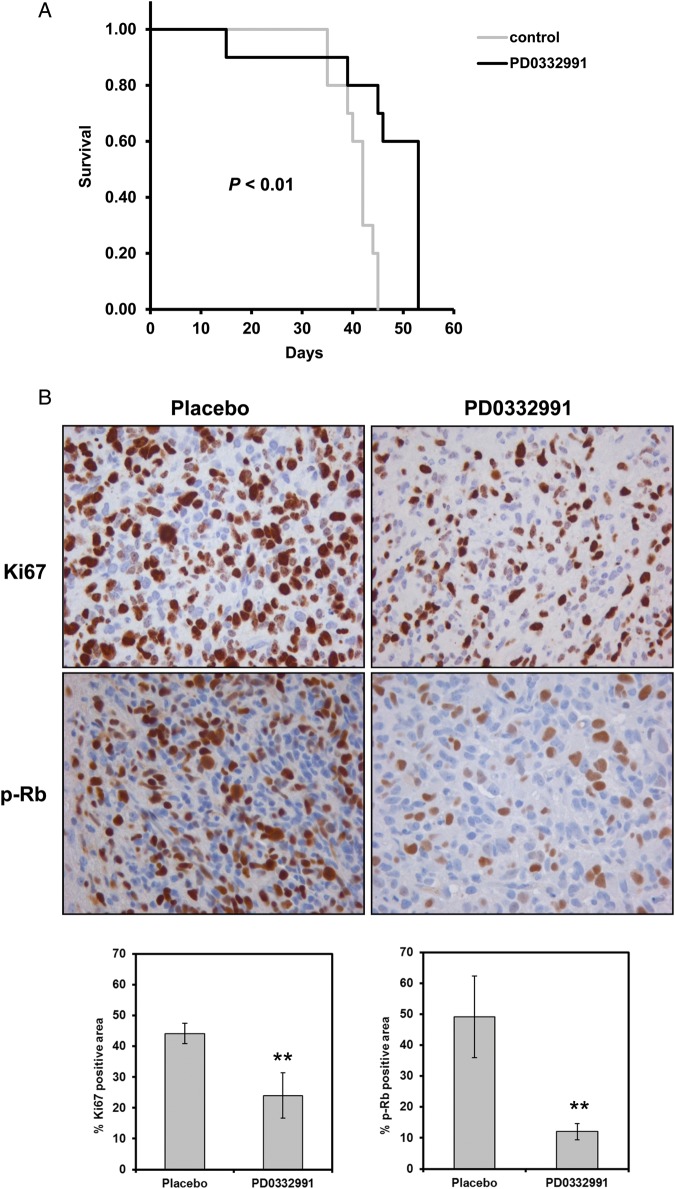
To evaluate the impact of PD0332991 treatment on survival of mice with orthotopic xenografts, a p16INK4a -deleted line GBM6 (sensitive to PD0332991 in in vitro testing) was used. In mice with established orthotopic xenografts, PD0332991 treatment resulted in a significant prolongation of survival in treated mice (median survival = 53 days), compared with placebo-treated mice (median survival = 42 days, P< .01) (Fig. 7A). Consistent with an anti-proliferative effect of PD0332991 in vivo, Ki67 staining of intracranial tumor section from placebo-treated mice (44.1 ± 3.3% Ki67 positive area) was significantly higher than Ki67 staining in the PD0332991-treated mice (24.0 ± 7.3% Ki67 positive area, P< .001) (Fig. 7B). Similarly, in the same orthotopic xenografts, PD0332991 effectively inhibited phosphorylation of Rb, the downstream target of Cdk4 (Placebo 49.2 ± 13.2% vs. PD0332991 12.1 ± 2.6% p-Rb positive area, P < .001). In contrast, no significant induction of apoptosis was observed in drug-treated mice (7.5 ± 2.3 TUNEL-positive cells per high-power field), compared with placebo-treated mice (5.6 ± 1.8 TUNEL-positive cells, P = .16) (Supplementary material, Fig. S2). Thus, similar to the in vitro studies, PD0332991-mediated suppression of Rb phosphorylation and cell proliferation was associated with a modest but significant survival benefit in GBM orthotopic xenografts.
Fig. 7.
Effects of PD0332991 on intracranial tumors. (A) Kaplan–Meier survival curves of mice with established intracranial tumor GBM6 that were randomized to therapy with placebo (n = 10) or PD0332991 (n = 10). Difference between survival curves were compared using a log-rank test and shown as P value. (B) Immunohistochemistry of Ki67 and phospho-Rb in tumor sections from GBM6 implanted mice treated with placebo or PD0332991. Mice with established intracranial tumors were randomized to therapy with placebo or PD0332991 for 6 days before euthanasia and processing of brain samples for immunostaining. Bar graphs show the effects of treatment on Ki67 labeling and phospho-Rb inhibition. Mean ± SD from 9 mice for each indicated group. **P< .001 vs. placebo group.
Discussion
The p16INK4a-Cdk4-Rb pathway controlling cell-cycle progression is commonly hyper-activated in patients with GBM and is a potentially important therapeutic target.14 In 2 recent whole genome analyses of GBM tumors, disruption of the p16-Cdk4-Rb pathway was observed in >70% of patients,1,2 and similar to these studies, all GBM xenografts evaluated in this study had disruptions in this pathway. Although deletion of CDKN2A/B occurs in 50% of primary GBM tumors, this deletion was seen in 18 of 21 xenograft lines analyzed in this study. This relative over-representation of p16 deletion has also been observed in studies of established glioma cell lines15,16 and may reflect a potential selection bias favoring p16 deletion for growth in cell culture or as xenografts. Of lines with detectable p16INK4a expression, one tumor had a truncating RB mutation and 2 tumors had high-level CDK4 amplification. Of interest, 2 xenografts were found to have co-alterations in the pathway; of the xenograft lines with CDKN2A deletion, one line also had a single-copy loss of CDK4 (GBM22) and one line had high-level CDK6 amplification (GBM34). In the context of these known genetic lesions, results from this study demonstrate a spectrum of sensitivity to the specific Cdk4/Cdk6 inhibitor PD0332991 across the panel of GBM lines, and the results from these studies provide insight into potential biomarkers that could be used to select patients for therapy.
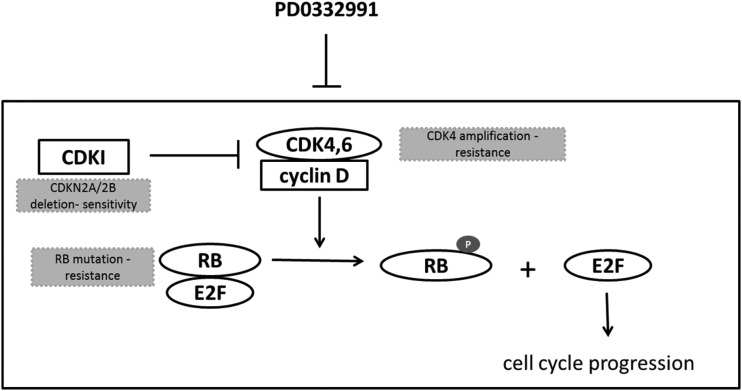
The molecular status of CDKN2A/B and RB is an important determinant of PD0332991 sensitivity (Fig. 8). P16INK4a functions as a tumor suppressor that specifically suppresses Cdk4 and Cdk6 kinase activity, and consistent with previous studies using established GBM cell lines,5,17 GBM xenograft lines deleted for CDKN2A/B were consistently sensitive to PD0332991. Also similar to studies in brain and breast cancer models,5,18 mutation of RB, the primary downstream target for Cdk4/6 activity, was associated with marked resistance to the anti-proliferative effects of PD0332991 in GBM28. Extending these previous findings, our data using primary GBM xenografts demonstrate that high-level CDK4 amplification in 2 independent xenograft lines was consistently associated with persistent Rb-phosphorylation and resistance to the anti-proliferative effects of PD0332991. In contrast, GBM34 cells with high-level CDK6 amplification/CDKN2A deletion were highly sensitive to PD0332991. This latter result is similar to the published marked sensitivity of the CCF-STTG1, a GBM cell line with CDK6 amplification and intact CDKN2A.5 Collectively, these data support a model in which response to PD0332991 is critically determined by presence of wild-type RB and possibly associated with the gene dosage of CDK4, with CDK4 amplified tumors being highly resistant to PD0332991.
Fig. 8.
p16-Cdk4-Rb signaling pathway diagram. The components that were evaluated in this study and associated with response to PD0332991 therapy are highlighted in grey color-filled boxes.
The molecular basis for selective resistance to PD0332991 in CDK4-amplified but not CDK6-amplified lines is not known. Inhibition of Rb-Ser780 phosphorylation is an accepted marker for Cdk4/6 inhibition, and the persistent Rb-phosphorylation in the CDK4-amplified lines after PD0332991 treatment is consistent with a failure to adequately inhibit Cdk4 activity in the 2 amplified lines. Our demonstration that shRNA knockdown of Cdk4 levels had essentially no impact on either the proliferation rate of untreated cells or the responsiveness to PD0332991 suggests that drug resistance in these lines is not simply attributable to unfavorable stoichiometry of the drug target relative to PD0332991 levels. Moreover, these data suggest that resistance to PD0332991 in GBM5 may not be mechanistically linked to CDK4 amplification. One explanation for these results could be that ≥1 genes co-amplified with CDK4 on chromosome 12 in GBM5 may be involved in the resistance to the therapy (Supplementary material, Table).
In contrast to the CDK4-amplified lines, 2 different CDK6-amplified lines (GBM34 from this study and CCF-STTG1 from Michaud, et al5) are highly sensitive to the anti-proliferative effects of PD0332991 in association with robust suppression of Rb phosphorylation on Ser780. Because PD0332991 is equally potent against the kinase activities of Cdk4 and Cdk6 in vitro,3 these data suggest that high-level Cdk4 versus Cdk6 expression has different biological effects within GBM cells. Consistent with this idea, CDK6 amplification is much more uncommon in GBM tumors (1%), compared with CDK4 amplification (18%). In GBM34 specifically, CDK6 amplification is observed in the context of codeletion of CDKN2A, which suggests that both hyper-activation of Cdk6 and disinhibition of Cdk4 activity through loss of p16 activity may have contributed to gliomagenesis in GBM34. In other model systems, Cdk4 and Cdk6 knockout mice have subtle differences in their phenotypes, and overexpression of Cdk4 versus Cdk6 can lead to distinct phenotypes in specific cell lines.19 Moreover, recent studies have suggested distinct substrate specificities for Cdk4 and Cdk6 activity against various phosphorylation sites in Rb protein and other protein targets.20 Although beyond the scope of the current studies, future experiments will focus on defining why CDK4 amplification, but not CDK6 amplification, is associated with resistance to PD0332991 in the GBM xenograft models.
The Mayo GBM xenograft panel has been used extensively to characterize responses to novel therapeutics. The xenograft panel preserves key molecular features of tumors, including abnormalities of the p16-Cdk4 pathway, and multiple studies have demonstrated a good correlation between treatment efficacy in the xenograft model and results in corresponding clinical trials.7,9,13 The demonstration of modest in vivo efficacy in primary GBM orthotopic models by our group and by others5 provides a reasonable rationale for clinical testing of PD0332991 in patients with GBM. Moreover, the results from this study and others support the concept of selecting only those patients for therapy with tumors harboring wild-type RB and nonamplified CDK4. As illustrated in this study, array comparative genomic hybridization (aCGH) in combination with sequencing analysis may be a useful screening tool for identifying multiple genetic abnormalities that could be used in a personalized therapeutic approach to select patients with GBM for treatment with the Cdk4/6 inhibitor PD0332991. As a first step toward a personalized approach with this drug, a clinical trial at the University of California, San Francisco, is currently accruing patients with recurrent GBM only if their tumor has detectable Rb expression by immunohistochemistry (ClinicalTrials.gov ID: NCT01227434), and the results from this study are eagerly awaited.
Supplementary Material
Supplementary material is available at Neuro-Oncology Journal online (http://neuro-oncology.oxfordjournals.org/).
Funding
This work was supported by the National Institutes of Health Mayo Brain Tumor SPORE (CA108961 to J.N.S. and RO1CA159467 to T.W.) and the Mayo Clinic.
Supplementary Material
Acknowledgments
We thank the Mayo Flow Cytometry/Optical Morphology Resource and Mayo Tissue and Cell Molecular Analysis Core for expert assistance and Dr. Evanthia Galanis for independent review of the data as part of a management plan developed by the Mayo Conflict of Interest Review Board.
Conflict of interest statement. J.N.S. is the recipient of research funding from Novartis, Basilea, and Merck for work unrelated to the studies reported here. Also unrelated to this research, some of the xenograft lines used in this study have been licensed to Wyeth Pharmaceuticals. All other authors: No conflicts declared.
References
- 1.Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. doi:10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. doi:10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fry DW, Harvey PJ, Keller PR, et al. Specific inhibition of cyclin-dependent kinase 4/6 by PD 0332991 and associated antitumor activity in human tumor xenografts. Mol Cancer Ther. 2004;3:1427–1438. [PubMed] [Google Scholar]
- 4.Baughn LB, Di Liberto M, Wu K, et al. A novel orally active small molecule potently induces G1 arrest in primary myeloma cells and prevents tumor growth by specific inhibition of cyclin-dependent kinase 4/6. Cancer Res. 2006;66:7661–7667. doi: 10.1158/0008-5472.CAN-06-1098. doi:10.1158/0008-5472.CAN-06-1098. [DOI] [PubMed] [Google Scholar]
- 5.Michaud K, Solomon DA, Oermann E, et al. Pharmacologic inhibition of cyclin-dependent kinases 4 and 6 arrests the growth of glioblastoma multiforme intracranial xenografts. Cancer Res. 2010;70:3228–3238. doi: 10.1158/0008-5472.CAN-09-4559. doi:10.1158/0008-5472.CAN-09-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giannini C, Sarkaria JN, Saito A, et al. Patient tumor EGFR and PDGFRA gene amplifications retained in an invasive intracranial xenograft model of glioblastoma multiforme. Neuro Oncol. 2005;7:164–176. doi: 10.1215/S1152851704000821. doi:10.1215/S1152851704000821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang L, Clarke MJ, Carlson BL, et al. PTEN loss does not predict for response to RAD001 (Everolimus) in a glioblastoma orthotopic xenograft test panel. Clinical Cancer Research. 2008;14:3993–4001. doi: 10.1158/1078-0432.CCR-07-4152. doi:10.1158/1078-0432.CCR-07-4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clarke MJ, Mulligan EA, Grogan PT, et al. Effective sensitization of temozolomide by ABT-888 is lost with development of temozolomide resistance in glioblastoma xenograft lines. Mol Cancer Ther. 2009;8:407–414. doi: 10.1158/1535-7163.MCT-08-0854. doi:10.1158/1535-7163.MCT-08-0854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carlson BL, Grogan PT, Mladek AC, et al. Radiosensitizing effects of temozolomide observed in vivo only in a subset of O6-Methylguanine-DNA methyltransferase methylated glioblastoma multiforme xenografts. International Journal of Radiation Oncology Biology Physics. 2009;75:212–219. doi: 10.1016/j.ijrobp.2009.04.026. doi:10.1016/j.ijrobp.2009.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carlson BL, Pokorny JL, Schroeder MA, Sarkaria JN. Establishment, Maintenance, and In Vitro and In Vivo Applications of Primary Human Glioblastoma Multiforme (GBM) Xenograft Models for Translational Biology Studies and Drug Discovery. John Wiley & Sons, Inc.; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arora T, Jelinek D. Differential myeloma cell responsiveness to interferon-α correlates with differential induction of p19INK4d and cyclin D2 expression. Journal of Biological Chemistry. 1998;273:11799–11805. doi: 10.1074/jbc.273.19.11799. doi:10.1074/jbc.273.19.11799. [DOI] [PubMed] [Google Scholar]
- 12.Hostein I, Pelmus M, Aurias A, Pedeutour F, Mathoulin-Pélissier S, Coindre JM. Evaluation of MDM2 and CDK4 amplification by real-time PCR on paraffin wax-embedded material: a potential tool for the diagnosis of atypical lipomatous tumours/well-differentiated liposarcomas. The Journal of Pathology. 2004;202:95–102. doi: 10.1002/path.1495. doi:10.1002/path.1495. [DOI] [PubMed] [Google Scholar]
- 13.Sarkaria JN, Yang L, Grogan PT, et al. Identification of molecular characteristics correlated with glioblastoma sensitivity to EGFR kinase inhibition through use of an intracranial xenograft test panel. Mol Cancer Ther. 2007;6:1167–1174. doi: 10.1158/1535-7163.MCT-06-0691. doi:10.1158/1535-7163.MCT-06-0691. [DOI] [PubMed] [Google Scholar]
- 14.Ueki K, Ono Y, Henson JW, Efird JT, von Deimling A, Louis DN. CDKN2/p16 or RB alterations occur in the majority of glioblastomas and are inversely correlated. Cancer Res. 1996;56:150–153. [PubMed] [Google Scholar]
- 15.Ishii N, Maier D, Merlo A, et al. Frequent co-alterations of TP53, p16/CDKN2A, p14ARF, PTEN tumor suppressor genes in human glioma cell lines. Brain Pathology. 1999;9:469–479. doi: 10.1111/j.1750-3639.1999.tb00536.x. doi:10.1111/j.1750-3639.1999.tb00536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He J, Allen JR, Collins VP, et al. CDK4 amplification is an alternative mechanism to p16 gene homozygous deletion in glioma cell lines. Cancer Res. 1994;54:5804–5807. [PubMed] [Google Scholar]
- 17.Wiedemeyer WR, Dunn IF, Quayle SN, et al. Pattern of retinoblastoma pathway inactivation dictates response to CDK4/6 inhibition in GBM. Proc Natl Acad Sci USA. 2010;107:11501–11506. doi: 10.1073/pnas.1001613107. doi:10.1073/pnas.1001613107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dean J, Thangavel C, McClendon A, Reed C, Knudsen E. Therapeutic CDK4/6 inhibition in breast cancer: key mechanisms of response and failure. Oncogene. 2010;29:4018–4032. doi: 10.1038/onc.2010.154. doi:10.1038/onc.2010.154. [DOI] [PubMed] [Google Scholar]
- 19.Malumbres M, Sotillo R, Santamaría D, et al. Mammalian cells cycle without the D-type cyclin-dependent kinases Cdk4 and Cdk6. Cell. 2004;118:493–504. doi: 10.1016/j.cell.2004.08.002. doi:10.1016/j.cell.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 20.Takaki T, Fukasawa K, Suzuki-Takahashi I, et al. Preferences for phosphorylation sites in the retinoblastoma protein of D-type cyclin-dependent kinases, Cdk4 and Cdk6, in vitro. Journal of Biochemistry. 2005;137:381–386. doi: 10.1093/jb/mvi050. doi:10.1093/jb/mvi050. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.