Abstract
Background
Sensitivity to epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs) and frequency of activation mutations in EGFR is lower in Caucasian than Asian non small-cell lung cancer (NSCLC) patients. Increased EGFR gene copy numbers evaluated by fluorescence in situ hybridization (FISH) has been reported as predictor of clinical benefit from EGFR-TKIs in Caucasian NSCLC patients. This study was carried out to verify whether EGFR FISH had similar performance in Japanese patients.
Methods
A cohort of 44 Japanese patients with recurrent NSCLC after surgery was treated with gefitinib 250 mg daily. The cohort included 48% females and 52% never-smokers; 73% had prior chemotherapy and 57% had stage III-IV at the time of surgery. Adenocarcinoma was the most common histology (86%). FISH was performed using the EGFR/Chromosome Enumeration Probe 7 and PathVysion DNA probes (Abbott Molecular). Specimens were classified as FISH positive when showing gene amplification or high polysomy (≥4 copies of the gene in ≥40% of tumor cells). Tumor response to gefitinib was assessed by RECIST for 33 patients with measurable diseases.
Results
Twenty-nine tumors (66%) were EGFR FISH+ and 23 (53%) were HER2 FISH+. Overall response rate was 52%, representing 65% of EGFR FISH+ patients and 29% of EGFR FISH+ patients (p = 0.0777). Survival was not impacted by the EGFR FISH (p = 0.9395) or the HER2 FISH (p = 0.0671) status. EGFR FISH= was significantly associated with HER2 FISH+ (p = 0.015) and presence of EGFR mutation (p = 0.0060). EGFR mutation significantly correlated with response (p < 0.0001) and survival after gefitinib (p = 0.0204). EGFR and HER2 FISH status were not associated with KRAS mutation.
Conclusion
Frequency of EGFR FISH+ status was higher and its predictive power for TKI sensitivity was lower in this Japanese cohort than in Western NSCLC cohorts. These findings support differences in the mechanisms of EGFR pathway activation in NSCLC between Asian and Caucasian populations. Confirmation of these results in larger cohorts is warranted.
Keywords: FISH, EGFR, HER2, KRAS, Biomarkers, NSCLC, Tyrosine inhibitors
Tumor dependence on specific molecular pathways may identify the best target for therapy exploration. Activation of the epidermal growth factor receptor (EGFR)-related signaling pathways drives numerous cancer-promoting processes, such as cell proliferation, apoptosis inhibition, angiogenesis, cell adhesion, and motility and invasion, and also controls development of drug resistance.1 Therefore, anti-EGFR approaches (antibodies directed against the extracellular domain and small inhibitors of the tyrosine kinase activity) have been one of the most successful examples of molecular target therapy in human solid neoplasias, mainly in non small-cell lung cancer (NSCLC), head and neck, pancreatic and colorectal carcinomas.2
Targeted therapies are expected to be effective when the targeted molecule is a major player in the tumor cellular processes, which usually does not occur in all patients with any specific solid tumor. Strategies for patient selection for targeted therapy are almost universally considered to be necessary but are not fully implemented, even for anti-EGFR therapies. In NSCLC, causally associated with EGFR activation are mutations in the adenosine triphosphate-binding site of the tyrosine kinase domain that sustain abnormal response to the ligand,3,4 activate multiple signaling transduction pathways5,6 and selectively activate AKT and signal transducers and activators of transcription signaling.6,7 Increased gene copy numbers is also a well known mechanism for activation of EGFR-related pathways in gliomas,8 breast,9 colon,10 head and neck cancers,11and NSCLC.12
In NSCLC, at least three molecular markers have been consistently associated with sensitivity or resistance to EGFR-TKIs (tyrosine kinase inhibitors): mutations and amplification/ overrepresentation of the EGFR gene3–5,12–14 and mutation in the KRAS genes.15–18 The impact on survival has been extensively investigated for activating EGFR mutations, 19 and less for the EGFR gene copy numbers12,14,20,21 or for the KRAS mutations16,22 and results among studies have not been totally concordant. Distinct technologies have been used to identify mutations and genomic gain and part of the discrepancies among results from different studies may due to technical factors. However, other factors such as smoking status, gender, and ethnicity have been demonstrated to impact sensitivity to EGFR-TKIs. Patients of Eastern Asian origin have significantly better clinical outcome to EGFR-TKIs than western populations23,24 but reasons for these differences are not completely understood. The most important factor so far accounting for this finding is that the Asian NSCLC patients including Japan, have high incidence of activating EGFR mutations.4,25
This study aimed to verify the role of EGFR genomic gain as a marker for sensitivity to gefinitib in a Japanese cohort using fluorescence in situ hybridization (FISH), a technology proved to be successful for prediction of outcome to EGFR TKIs in some Caucasian NSCLC populations. In addition, the study aimed to compare EGFR genomic gain with two other gefitinib-related markers, activating mutations in EGFR and resistant mutations in KRAS, which were previously investigated in this cohort.13
PATIENTS AND METHODS
Description of Patient Population and Definition of Effectiveness of Gefitinib Treatment
From a population of NSCLC patients who underwent surgery between 1999 and 2003 in the Aichi Cancer Center Hospital in Nagoya, Japan, 75 had recurrent disease and were treated with 250 mg/daily of gefitinib for recurrent disease. From those, response to treatment could not be evaluated in 6 cases, tumor material was not available in 24 cases, and FISH analyses failed in 4 cases. Thus, the current study reports on 44 patients, all of whom provided consent for the study.
Tumor materials obtained at initial tumor resection for these 44 NSCLC cases had been previously investigated for EGFR and KRAS mutations.13,16 Tumor response to gefitinib treatment was evaluated for 33 patients eliminating 11 patients who did not have measurable diseases. Tumor response was judged according to the RECIST, without requirement of confirmation of tumor response no less than 4 weeks apart. The length of gefitinib therapy was used as a surrogate for disease free survival and overall survival was calculated form the start of gefitinib administration to death from any cause or the most recent date on which the patient was known to be alive.
EGFR and HER2 Fluorescence In Situ Hybridization Assays
Formalin-fixed, paraffin-embedded tumor blocks were sectioned at 4 µm and submitted to dual-color FISH assays using the Locus Specific Indicator EGFR Spectrum- Orange/CEP 7 SpectrumGreen and the PathVysion DNA Probe Kit (HER2 SpectrumOrange/CEP 17 SpectrumGreen Vysis/Abbott Molecular) following protocol previously described. 12 Briefly, slides were deparaffinized in CitriSolv (Fisher Scientific) and washed in 100% ethanol for 5 minutes. The slides were then sequentially incubated in 2× SSC (saline sodium citrate) at 75°C for 13 to 18 minutes, digested in 0.25 mg/ml Proteinase K/2× SSC at 45°C for 14 to 18 minutes, washed in 2× SSC for 5 minutes and dehydrated in ethanol series. Probes were applied according to the manufacturer instructions to the selected hybridization areas, which were covered and sealed. DNA denaturation was performed in dry oven for 15 minutes at 80°C and hybridization was allowed to occur for 20 hours at 37°C in a humidified chamber. Posthybridization washes were performed consecutively in 2× SSC/0.3% NP-40 at 72°C and 2× SSC for 2 minutes each. Following dehydration in ethanol, chromatin was counterstained with 4’ = 6-diamidino-2- phenylindole (DAPI) (0.3 µg/ml in antifade Vectashield mounting medium, Vector Laboratories). Analysis was performed on epifluorescence microscopes using single interference filters sets for green, red (Texas red) and blue (DAPI) as well as dual (red/green) and triple (blue, red, green) band pass filters. For documentation, images were acquired using charged-coupled device camera with Z-stacking and merged using dedicated software (CytoVision, Applied Imaging).
At least 50 tumor nuclei were analyzed in tumor areas selected using the correspondent HE stained slide as a guide. Scoring system followed previous publications.12 According to the frequency of tumor cells with specific number of copies of the gene and the CEP targets, the tumors were initially classified into six FISH categories (disomy, low and high trisomy, low and polysomy, and gene amplification) and finally grouped into two strata: (a) FISH negative including disomy to low polysomy tumors, which basically have ≥4 copies of the gene in <40% of cells; and (b) FISH positive including tumors with high polysomy (≥4 copies in ≥40% of cells) and gene amplification (defined by a ratio gene/chromosome per cell ≥2, presence of small or nonenumerable clusters of the gene signal or ≥15 copies of the gene signal in ≥10% of the analyzed cells).
Statistical Analysis
For comparisons of proportions, the Pearson’s χ2 test or the Fisher’s exact test was used. Nonparametric Wilcoxon rank sum test or Kruskal-Wallis test was used to compare the difference in continuous variables. The Kaplan-Meier method was used to estimate the probability of survival as a function of time, and survival differences between groups were analyzed by the log-rank test. The two-sided significance level was set at p < 0.05. All analyses were performed using SAS version 9.1 (SAS Institute Inc, Cary, NC) software.
RESULTS
Clinical and demographical characteristics are summarized in Table 1. The patients were evenly split between males and females, never or ever smokers and with early or advanced stage disease. Adenocarcinoma histology and poorly or moderately differentiated histologic grade were prevalent. Most patients had not received prior chemotherapy. Median disease free interval after surgery was 375 days, median survival after gefitinib treatment was 562 days, and 66% of patients were alive at the time of last follow up.
TABLE 1.
Population Characteristics
| Variable | Categories | Statistics |
|---|---|---|
| Age (years) | Median | 60.9 × 10.3 |
| Range | 38–79 | |
| Gender | Male | 23 (52.3%) |
| Female | 21 (47.7%) | |
| Smoking | Never | 23 (52.3%) |
| Ever | 21 (47.7%) | |
| Histology | Adenocarcinoma | 38 (86.4%) |
| Nonadenocarcinoma (SqC, LC)a | 6 (13.6%) | |
| Differentiation | Poor | 10 (26.3%) |
| Moderate | 22 (57.9%) | |
| Well | 6 (15.8%) | |
| Not determined | 6 | |
| Stage | Early (I–II) | 19 (43.2%) |
| Advanced (III–IV) | 25 (56.8%) | |
| Prior chemotherapy | Yes | 12 (27.3%) |
| No | 32 (72.7%) | |
| Survival after surgery (days) | Median | 2081 |
| Range | 250–2655 | |
| Tumor response (RECIST) | Yes | 17 (52%) |
| No | 16 (48%) | |
| Disease free interval (days) | Median | 375 |
| Range | 99–1818 | |
| Survival after gefitinib (days) | Median | 562 |
| Range | 69–724 | |
| Death | Dead | 15 (34.1%) |
| Alive | 29 (65.9%) |
SqC, Squamous cell carcinoma; LC, Large cell carcinoma.
EGFR FISH and mutation status in relation to demographics are summarized in Table 2. While EGFR mutation was associated with female gender, never-smoking status, and adenocarcinoma histology, none of these was related with EGFR-FISH status.
TABLE 2.
EGFR FISH and Mutation Status According to Demographics
| EGFR FISH | EGFR Mutation | ||||||
|---|---|---|---|---|---|---|---|
| Variable | Categories | Positive | Negative | p | Positive | Negative | p |
| Age (years) | Median | 61.0 | 62.0 | 61.0 | 61.0 | ||
| Gender | Male | 15 (65%) | 8 | p = 0.9193 | 11 (48%) | 12 | p = 0.0536 |
| Female | 14 (67%) | 7 | 16 (76%) | 5 | |||
| Smoking | Never | 15 (67%) | 8 | p = 0.9193 | 18 (78%) | 5 | p = 0.016 |
| Ever | 14 (65%) | 7 | 9 (43%) | 12 | |||
| Histology | Adenocarcinoma | 25 (66%) | 13 | p = 0.9664 | 26 (68%) | 12 | p = 0.0151 |
| Nonadenocarcinoma (SqC, LC)a | 4 (67%) | 2 | 1 (17%) | 5 | |||
FISH, fluorescence in situ hybridization; EGFR, epidermal growth factor receptor.
SqC, Squamous cell carcinoma; LC, Large cell carcinoma.
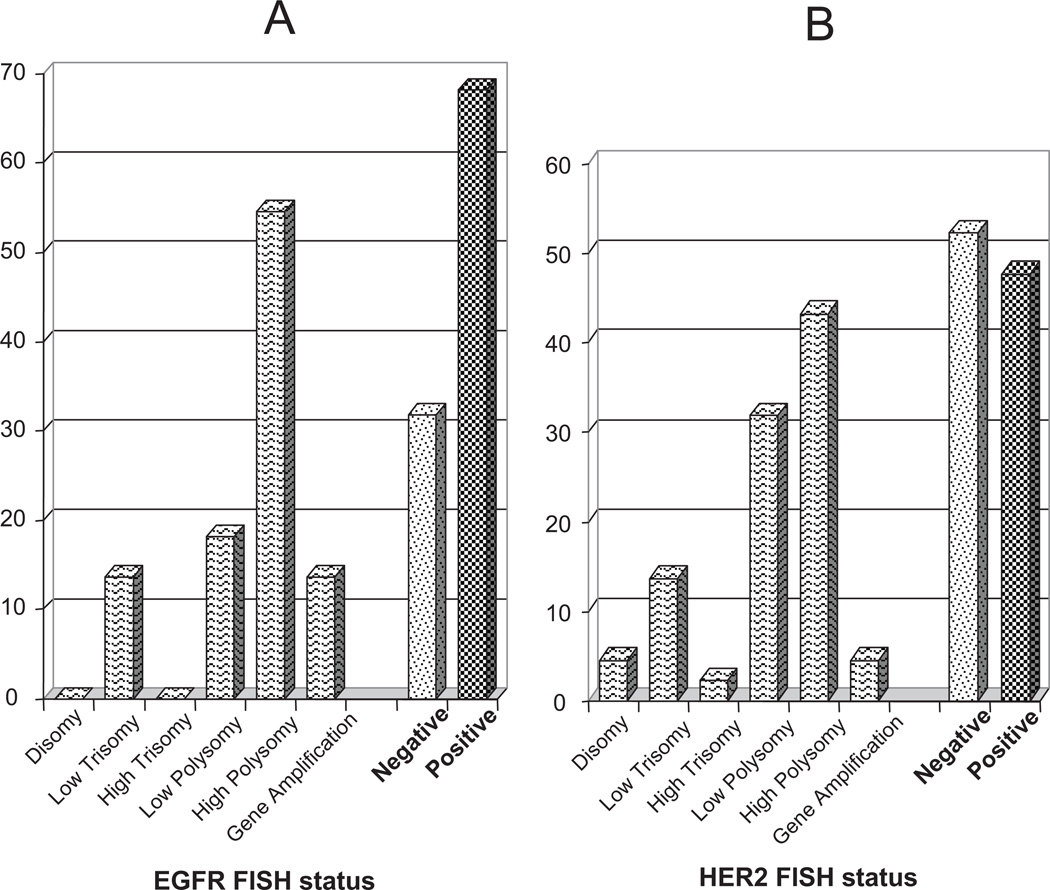
Distribution of patients through the FISH categories is illustrated in Figure 1A for the EGFR gene and Figure 1B for the HER2 gene. The majority of tumors (29 cases [66%]) were EGFR FISH positive, predominantly due to a large representation of tumors with high polysomy (23 cases, 52%, Figure 2A) rather than gene amplification (6 cases, 14%, Figure 2B). Also, a high number of tumors (23 cases, 53%) were positive for HER2 FISH, of which 21 cases (48%) were represented by high polysomy and only 2 cases (5%) by gene amplification (illustrated in Figure 2C). EGFR and HER2 patterns were significantly associated (p = 0.015): 19 cases (43%) of tumors were positive and 11cases (25%) were negative for both genes, while 14 cases (32%) had discordant patterns; EGFR FISH positives were more likely to be HER2 FISH positives (19/29 = 66%) than EGFR FISH negatives (4/15 = 27%).
FIGURE 1.
Frequencies of tumors with distinct categories for the epidermal growth factor receptor-fluorescence in situ hybridization (EGFR-FISH) (A) and the HER2 FISH (B) assays. Negative category includes disomy to low polysomy. Positive category includes high polysomy and gene amplification.
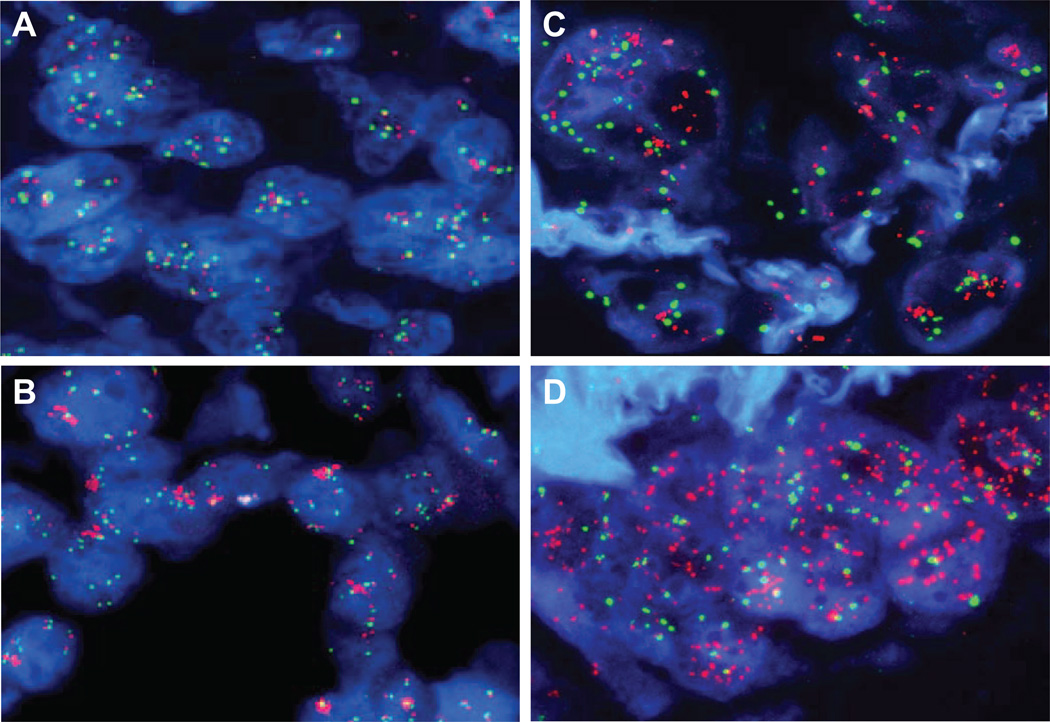
FIGURE 2.
Hybridization of the non small-cell lung cancer (NSCLC) sections with the epidermal growth factor receptor EGFR/CEP7 (A, B, D) and the PathVysion probe set (C) showing EGFR high polysomy (A), EGFR clustered gene amplification (B), HER2 gene amplification (C) and EGFR amplification as double minutes (D).
Overall, the specimens with amplification of the EGFR or HER2 genes exhibited clusters of loosely associated signals (Figures 2B, C) indicating that the amplification occurred as homogenously staining regions. However, one specimen displayed EGFR gene amplification as numerous, diffuse signals mimicking the extrachromosomal double minutes (Figure 2D). Heterogeneity for both EGFR and HER FISH patterns was common, with tumor foci showing nuclei with high copy numbers (including gene amplification) interspaced with nuclei with low copy numbers.
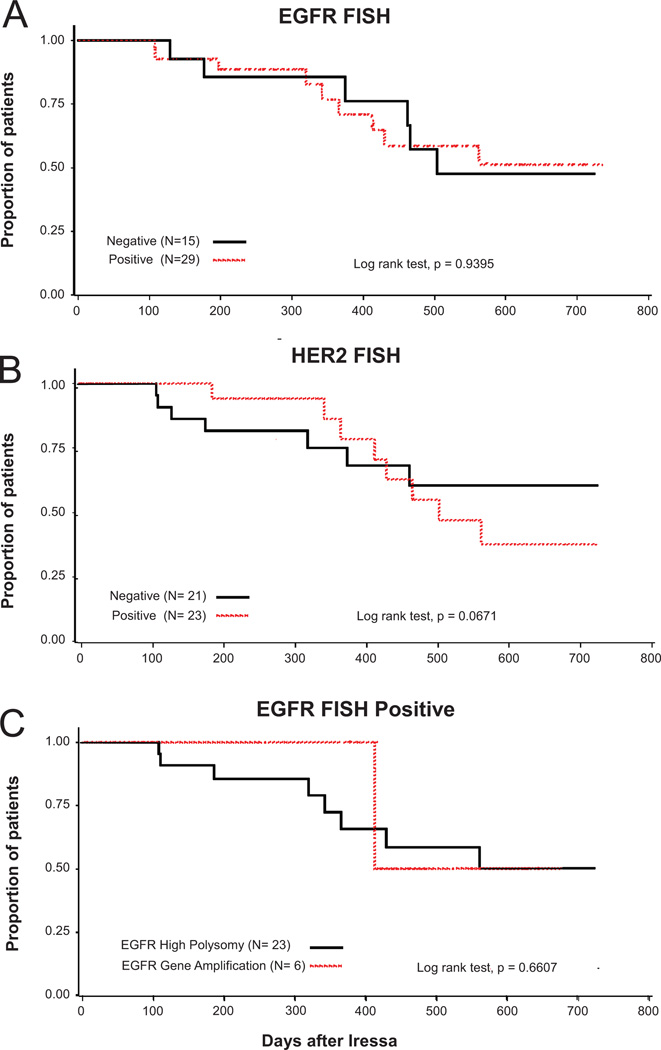
The association between FISH patterns and response to the gefitinib treatment for 33 patients with measurable diseases is shown in Table 3. Response to gefitinib was marginally higher in EGFR FISH positive (65%) than negative (29%) patients (p = 0.0777). Patients with EGFR gene amplification had a trend towards better benefit (response in 4 of 4 = 100%) than patients with high polysomy (response in 9 of 16 = 56%). HER2 FISH positive pattern trended no impact, including 47% of responders (p = 0.4426). Response rate was 62% of patients with EGFR and HER2 FISH positive tumors, in 45% of patients with EGFR or HER2 FISH positive tumors, and in 44% of patients EGFR and HER2 FISH negative tumors. Time to treatment failure (TTF) was not significantly associated with EGFR or HER2 FISH positivity (Table 4). Overall survival was not associated with patterns of EGFR FISH (p = 0.93) or HER2 FISH (p = 0.69), as shown in Figure 3A, B. EGFR FISH+ patients with high polysomy (score 5) and true gene amplification (score 6) did not differ regarding survival (p = 0.6607; Figure 3C).
TABLE 3.
Tumor Response in Relation to EGFR FISH, HER2 FISH, EGFR Mutation and KRAS Mutation Status
| Patients | Tumor response | ||||||
|---|---|---|---|---|---|---|---|
| Molecular marker | Categories | n | % | PR (%) | SD | PD | p |
| EGFR | Positive (+) | 20 | 61 | 13 (65%) | 1 | 6 | p = 0.0777 |
| Negative (−) | 13 | 39 | 4 (29%) | 0 | 9 | ||
| HER2 | Positive (+) | 17 | 52 | 8 (47%) | 0 | 9 | p = 0.4426 |
| Negative (−) | 16 | 48 | 9 (56%) | 1 | 6 | ||
| EGFR and HER2 | +/+ | 13 | 39 | 8 (62%) | 0 | 5 | p = 0.2451 |
| +/− | 7 | 21 | 5 (71%) | 1 | 1 | ||
| −/+ | 4 | 12 | 0 (0%) | 0 | 4 | pa | |
| −/− | 9 | 27 | 4 (44%) | 0 | 5 | ||
| EGFR mutation | Positive (+) | 20 | 61 | 17 (85%) | 1 | 2 | p = 0.0001 |
| Negative (−) | 13 | 39 | 0 (0%) | 0 | 13 | ||
| EGFR FISH and EGFR mutation | +/+ | 16 | 48 | 13 (81%) | 1 | 2 | p = 0.0029 |
| +/− | 4 | 12 | 0 (0%) | 0 | 4 | ||
| −/+ | 4 | 12 | 4 (100%) | 0 | 0 | pa | |
| −/− | 9 | 27 | 0 (0%) | 0 | 9 | ||
| KRAS mutation | Positive (+) | 4 | 13 | 0 (0%) | 0 | 4 | p = 0.0995 |
| Negative (−) | 26 | 87 | 14 (54%) | 1 | 11 | ||
| EGFR FISH and KRAS mutation | +/+ | 0 | 0 | 0 (0%) | 0 | 0 | pa |
| +/− | 17 | 57 | 10 (59%) | 1 | 6 | ||
| −/+ | 4 | 13 | 0 (0%) | 0 | 4 | pa | |
| −/− | 9 | 30 | 4 (44%) | 0 | 5 | ||
FISH, fluorescence in situ hybridization; EGFR, epidermal growth factor receptor; PR, partial response; PD, progressive disease.
p value could not be calculated because of blank cells.
TABLE 4.
Time to Treatment Failure According to EGFRFISH, HER2 FISH, EGFR Mutation and KRAS Mutation Status
| Molecular marker | Categories | Patients | TTF after Gefitinib (Days) Median |
p | |
|---|---|---|---|---|---|
| n | % | ||||
| EGFR | Positive (+) | 29 | 66 | 169 | 0.722 |
| Negative (−) | 15 | 34 | 97 | ||
| HER2 | Positive (+) | 23 | 53 | 121 | 0.1815 |
| Negative (−) | 21 | 47 | 144 | ||
| EGFR and HER2 | +/+ | 19 | 43 | 169 | 0.0179 |
| +/− | 10 | 23 | 118 | ||
| −/+ | 4 | 9 | 56 | ||
| −/− | 11 | 25 | 144 | ||
| EGFR mutation | Positive (+) | 27 | 61 | 311 | <0.0001 |
| Negative (−) | 17 | 39 | 83 | ||
| EGFR FISH and EGFR mutation | +/+ | 22 | 50 | 182 | <0.0001 |
| +/− | 7 | 16 | 67 | ||
| −/+ | 5 | 11 | 916 | ||
| −/− | 10 | 23 | 83 | ||
| KRAS mutation | Positive (+) | 5 | 12 | 87 | 0.0248 |
| Negative (−) | 36 | 88 | 146 | ||
| EGFR FISH and KRAS mutation | +/+ | 1 | 2 | 113 | 0.0767 |
| +/− | 25 | 61 | 169 | ||
| −/+ | 4 | 10 | 57 | ||
| −/− | 11 | 25 | 144 | ||
FISH, fluorescence in situ hybridization; EGFR, epidermal growth factor receptor; TTF, time to treatment failure.
FIGURE 3.
Effect on survival from the day of initiating gefitinib treatment in recurrent non small-cell lung cancer (NSCLC) after surgery by epidermal growth factor receptor fluorescence in situ hybridization (EGFR FISH) status (A), HER2 FISH status (B), and EGFR high polysomy and gene amplification (C).
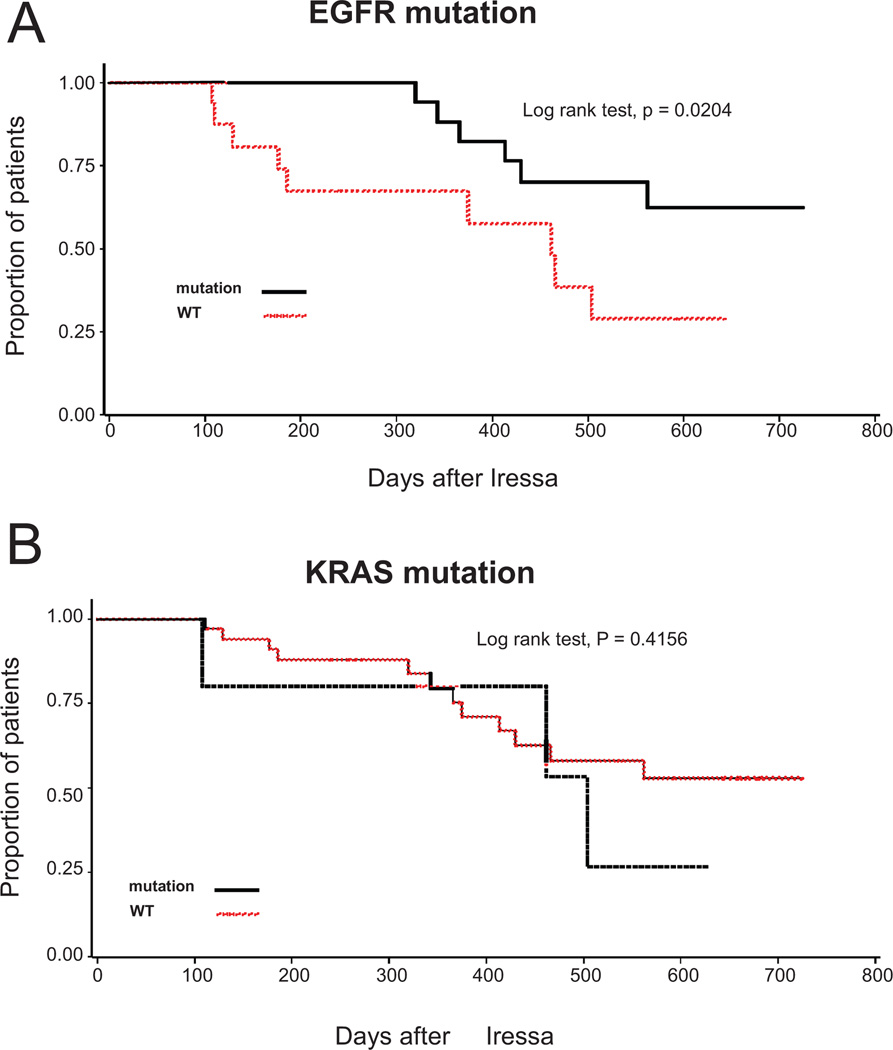
Among these 44 NSCLC patients, 27 (61%) had activating mutations in the tyrosine kinase domain of the EGFR gene and, among 41 who were tested for KRAS mutations, 5 (12%) had point mutations in codons 12 or 13. Table 3 also shows tumor response according to presence or absence of EGFR and KRAS mutations, both individually and in combination with EGFR FISH. EGFR mutation was significantly associated with tumor response (p < 0.0001) and prolonged TTF (p < 0.0001) or survival (p = 0.02; Figure 4A and Table 4). EGFR FISH positivity was significantly associated with presence of EGFR mutation (p = 0.0060). Patients with EGFR mutation were more likely to be EGFR FISH positive (22/27 = 81%) than patients with wild type EGFR (7/17 = 41%). EGFR mutations were present in all 6 tumors with EGFR gene amplification and in 16 out of 23 tumors with EGFR high polysomy (70%). Response rate was 81% of 16 cases positive for both EGFR FISH and mutation and all 4 EGFR FISH negative/EGFR mutation positive cases responded to gefitinib (Table 3).
FIGURE 4.
Effect on survival from the day of initiating gefitinib treatment in recurrent non small-cell lung cancer (NSCLC) after surgery by epidermal growth factor receptor (EGFR) activating mutation (A) and KRAS mutation (B) status.
Conversely, none of the 4 patients with KRAS mutation (none of whom were EGFR FISH positive) or of the 13 patients with EGFR wild type (4 of whom were EGFR FISH positive) benefited from gefitinib treatment. Presence of KRAS mutation was significantly associated with TTF (p = 0.0248) but not with lack of response (p = 0.0995) or overall survival (p = 0.4156, Figure 4B).
DISCUSSION
The EGFR FISH positive status had a borderline association to response of gefitinib treatment, but no impact on survival in this cohort of Japanese NSCLC patients. These results do not support a predictive role of the established EGFR FISH assay to gefitinib sensitivity in Japanese NSCLC patients. This observation contrasts with previous findings in Caucasian NSCLC populations obtained by our group12,20,21 and others,14 that had identified EGFR genomic gain by FISH as a significant predictor of outcome to EGFR-TKIs. In the current study, EGFR mutation was highly predictive of both response and survival to gefitinib. Lack of predictive value of EGFR FISH or EGFR gene copy numbers as assessed by quantitative polymerase chain reaction have also been reported by Korean17 and Japanese26 groups. Therefore, there seems to be ethnic differences as to whether EGFR genomic gain is predictive for response or survival after geftinib treatment.
The clinical and demographical characteristics of this Japanese cohort were distinctive, including high proportion of female, never smokers, early stage disease, no prior chemotherapy, and adenocarcinomas. Unselected cohorts of Asian origin usually have higher frequency of females (40%27) and never smokers (40%27) than Caucasians (34% for females, 9% for never smokers according to Kobrinsky et al.28). In addition, this cohort had one of the highest reported frequencies of EGFR FISH+ tumors (68%) and EGFR mutations (61%). Taken only studies that evaluated gene copy numbers by FISH with identical or similar scoring criteria, the frequency of EGFR FISH+ tumors ranged from 44 to 48% in Asian patients17,26,29 and from 32 to 45% in Caucasian NSCLCs.14,21 EGFR activating mutations are well known to be more prevalent in Asian (40–50% of adenocarcinomas27,30) than Caucasian NSCLCs (10% of adenocarcinomas25). Altogether, these findings substantiate the interesting hypothesis that there are ethnicity-associated molecular peculiarities in NSCLC.
The two EGFR gene markers, activating mutation and genomic gain, were significantly correlated in this cohort. Association between EGFR gene amplification and activating mutations has been reported in NSCLC cell lines31 and clinical specimens of Caucasian12 and Asian origins.17,32 Furthermore, the selective amplification of the mutant allele was verified in the cell lines H3255, H827, PC-9, KT-2, KT-4 and Ma-1,31 as well as in Asian patients.32 These findings support the hypothesis that there is a selection of cells carrying the amplification of the mutant allele in lung tumorigenesis. Interestingly, high EGFR copy numbers due to chromosomal aneusomy or structural rearrangements (high polysomy) were also associated with mutations in this cohort and in Caucasian NSCLC.33
Status of the HER2 gene in NSCLC has been poorly explored and discrepant results have been reported in association with outcome to EGFR-TKIs.34 In this cohort, HER2 genomic gain showed up as a negative impact factor for survival after gefinitib treatment, in contrast to our previous results in an Italian cohort.34 Conversely, none of the five KRAS mutant tumors showed treatment efficacy in this study, in agreement with previously findings that KRAS mutations are primary resistance factors to EGFR-TKIs.18,35
In summary, the study showed that the EGFR FISH scoring criteria proposed for stratification of NSCLC for therapy with EGFR-TKIs was not effective in Japanese patients as in Caucasian patients. Confirmation of these results in larger cohorts is warranted and investigation of factors that may underlie distinct molecular mechanisms of activation of the EGFR pathway in these populations should be investigated.
Acknowledgments
Disclosure: Dr. Hirsch has served on advisory boards for AstraZeneca, Pfizer, Merck Serono, BMS/Imclone, Syndax, Boehringer Ingelheim, Roche, and Lilly. He has received research grants from Astra Zeneca, OSI, Genentech, Syndax, and Merck. He is also the co-inventor of a University of Colorado owned patent: EGFR FISH As a Predictive Marker for EGFR Inhibitors (patent licensed to Abbot Diagnostics). Dr. Varella-Garcia received honorarium from Abbott Molecular to speak at the Association for Molecular Pathology annual meeting in 2008 about the EGFR FISH assay. She is also a co-inventor of a patent for use of the EGFR FISH assay to select NSCLC patients for therapy. Dr. Mitsudomi has received honorarium for speaking to a professional group from AstraZeneca and Chugai Pharmaceutical. He also provided testimony in court for gefitinib. Dr. Bunn was paid an honorarium and travel expenses by GlaxoSmithKline to attend an advisory board on the MAGG A3 vaccine. A clinical trial using the vaccine is being conducted at the University of Colorado Cancer Center. Dr. Bunn is not the PI and received no funding for this trial.
REFERENCES
- 1.Hynes NE, Lane HA. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer. 2005;5:341–354. doi: 10.1038/nrc1609. [DOI] [PubMed] [Google Scholar]
- 2.Ciardiello F, Tortora G. EGFR antagonists in cancer treatment. N Engl J Med. 2008;358:1160–1174. doi: 10.1056/NEJMra0707704. [DOI] [PubMed] [Google Scholar]
- 3.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 4.Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 5.Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sordella R, Bell DW, Haber DA, et al. Gefitinib-sensitizing EGFR mutations in lung cancer activate anti-apoptotic pathways. Science. 2004;305:1163–1167. doi: 10.1126/science.1101637. [DOI] [PubMed] [Google Scholar]
- 7.Tracy S, Mukohara T, Hansen M, et al. Gefitinib induces apoptosis in the EGFRL858R non-small-cell lung cancer cell line H3255. Cancer Res. 2004;64:7241–7244. doi: 10.1158/0008-5472.CAN-04-1905. [DOI] [PubMed] [Google Scholar]
- 8.Ohgaki H, Dessen P, Jourde B, et al. Genetic pathways to glioblastoma: a population-based study. Cancer Res. 2004;64:6892–6899. doi: 10.1158/0008-5472.CAN-04-1337. [DOI] [PubMed] [Google Scholar]
- 9.Slamon DJ, Clark GM, Wong SG, et al. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 10.Moroni M, Veronese S, Benvenuti S, et al. Gene copy number for epidermal growth factor receptor (EGFR) and clinical response to antiEGFR treatment in colorectal cancer: a cohort study. Lancet Oncol. 2005;6:279–286. doi: 10.1016/S1470-2045(05)70102-9. [DOI] [PubMed] [Google Scholar]
- 11.Chung CH, Ely K, McGavran L, et al. Increased epidermal growth factor receptor gene copy number is associated with poor prognosis in head and neck squamous cell carcinomas. J Clin Oncol. 2006;24:4170–4176. doi: 10.1200/JCO.2006.07.2587. [DOI] [PubMed] [Google Scholar]
- 12.Cappuzzo F, Hirsch FR, Rossi E, et al. Epidermal growth factor receptor gene and protein and gefitinib sensitivity in non-small cell lung cancer. J Natl Cancer Inst. 2005;97:643–655. doi: 10.1093/jnci/dji112. [DOI] [PubMed] [Google Scholar]
- 13.Mitsudomi T, Kosaka T, Endoh H, et al. Mutations of the epidermal growth factor receptor gene predict prolonged survival after gefitinib treatment in patients with non-small-cell lung cancer with postoperative recurrence. J Clin Oncol. 2005;23:2513–2520. doi: 10.1200/JCO.2005.00.992. [DOI] [PubMed] [Google Scholar]
- 14.Tsao MS, Sakurada A, Cutz JC, et al. Erlotinib in lung cancer - molecular and clinical predictors of outcome. N Engl J Med. 2005;353:133–144. doi: 10.1056/NEJMoa050736. [DOI] [PubMed] [Google Scholar]
- 15.Eberhard DA, Johnson BE, Amler LC, et al. Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non-small-cell lung cancer treated with chemotherapy alone and in combination with erlotinib. J Clin Oncol. 2005;23:5900–5909. doi: 10.1200/JCO.2005.02.857. [DOI] [PubMed] [Google Scholar]
- 16.Endoh H, Yatabe Y, Kosaka T, et al. PTEN and PIK3A expression is associated with prolonged survival after gefitinib treatment in EGFR-mutated lung cancer patients. J Thorac Oncol. 2006;1:629–634. [PubMed] [Google Scholar]
- 17.Han SW, Kim TY, Jeon YK, et al. Optimization of patient selection for gefitinib in non-small cell lung cancer by combined analysis of epidermal growth factor receptor mutation, K-ras mutation, and Akt phosphorylation. Clin Cancer Res. 2006;12:2538–2544. doi: 10.1158/1078-0432.CCR-05-2845. [DOI] [PubMed] [Google Scholar]
- 18.Pao W, Wang TY, Riely GJ, et al. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med. 2005;2:e17. doi: 10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uramoto H, Mitsudomi T. Which biomarker predicts benefit from EGFR-TKI treatment for patients with lung cancer? Br J Cancer. 2007;96:857–863. doi: 10.1038/sj.bjc.6603665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirsch FR, Varella-Garcia M, Bunn PA, Jr, et al. Molecular predictors of outcome with gefitinib in a phase III placebo-controlled study in advanced non-small-cell lung cancer. J Clin Oncol. 2006;24:5034–5042. doi: 10.1200/JCO.2006.06.3958. [DOI] [PubMed] [Google Scholar]
- 21.Hirsch FR, Varella-Garcia M, McCoy J, et al. Increased epidermal growth factor receptor gene copy number detected by fluorescence in situ hybridization associates with increased sensitivity to gefitinib in patients with bronchioloalveolar carcinoma subtypes: a Southwest Oncology Group Study. J Clin Oncol. 2005;23:6838–6845. doi: 10.1200/JCO.2005.01.2823. [DOI] [PubMed] [Google Scholar]
- 22.Marchetti A, Martella C, Felicioni L, et al. EGFR mutations in non-small-cell lung cancer: analysis of a large series of cases and development of a rapid and sensitive method for diagnostic screening with potential implications on pharmacologic treatment. J Clin Oncol. 2005;23:857–865. doi: 10.1200/JCO.2005.08.043. [DOI] [PubMed] [Google Scholar]
- 23.Calvo E, Baselga J. Ethnic differences in response to epidermal growth factor receptor tyrosine kinase inhibitors. J Clin Oncol. 2006;24:2158–2163. doi: 10.1200/JCO.2006.06.5961. [DOI] [PubMed] [Google Scholar]
- 24.Shigematsu H, Gazdar AF. Somatic mutations of epidermal growth factor receptor signaling pathway in lung cancers. Int J Cancer. 2006;118:257–262. doi: 10.1002/ijc.21496. [DOI] [PubMed] [Google Scholar]
- 25.Shigematsu H, Lin L, Takahashi T, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst. 2005;97:339–346. doi: 10.1093/jnci/dji055. [DOI] [PubMed] [Google Scholar]
- 26.Sone T, Kasahara K, Kimura H, et al. Comparative analysis of epidermal growth factor receptor mutations and gene amplification as predictors of gefitinib efficacy in Japanese patients with nonsmall cell lung cancer. Cancer. 2007;109:1836–1844. doi: 10.1002/cncr.22593. [DOI] [PubMed] [Google Scholar]
- 27.Kosaka T, Yatabe Y, Endoh H, et al. Mutations of the epidermal growth factor receptor gene in lung cancer: biological and clinical implications. Cancer Res. 2004;64:8919–8923. doi: 10.1158/0008-5472.CAN-04-2818. [DOI] [PubMed] [Google Scholar]
- 28.Kobrinsky NL, Klug MG, Hokanson PJ, et al. Impact of smoking on cancer stage at diagnosis. J Clin Oncol. 2003;21:907–913. doi: 10.1200/JCO.2003.05.110. [DOI] [PubMed] [Google Scholar]
- 29.Sasaki H, Endo K, Okuda K, et al. Epidermal growth factor receptor gene amplification and gefitinib sensitivity in patients with recurrent lung cancer. J Cancer Res Clin Oncol. 2008;134:569–577. doi: 10.1007/s00432-007-0320-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tokumo M, Toyooka S, Kiura K, et al. The relationship between epidermal growth factor receptor mutations and clinicopathologic features in non-small cell lung cancers. Clin Cancer Res. 2005;11:1167–1173. [PubMed] [Google Scholar]
- 31.Okabe T, Okamoto I, Tamura K, et al. Differential constitutive activation of the epidermal growth factor receptor in non-small cell lung cancer cells bearing EGFR gene mutation and amplification. Cancer Res. 2007;67:2046–2053. doi: 10.1158/0008-5472.CAN-06-3339. [DOI] [PubMed] [Google Scholar]
- 32.Takano T, Ohe Y, Sakamoto H, et al. Epidermal growth factor receptor gene mutations and increased copy numbers predict gefitinib sensitivity in patients with recurrent non-small-cell lung cancer. J Clin Oncol. 2005;23:6829–6837. doi: 10.1200/JCO.2005.01.0793. [DOI] [PubMed] [Google Scholar]
- 33.Daniele L, Macri L, Schena M, et al. Predicting gefitinib responsiveness in lung cancer by fluorescence in situ hybridization/chromogenic in situ hybridization analysis of EGFR and HER2 in biopsy and cytology specimens. Mol Cancer Ther. 2007;6:1223–1229. doi: 10.1158/1535-7163.MCT-06-0719. [DOI] [PubMed] [Google Scholar]
- 34.Cappuzzo F, Varella-Garcia M, Shigematsu H, et al. Increased HER2 gene copy number is associated with response to gefitinib therapy in epidermal growth factor receptor-positive non-small-cell lung cancer patients. J Clin Oncol. 2005;23:5007–5018. doi: 10.1200/JCO.2005.09.111. [DOI] [PubMed] [Google Scholar]
- 35.Massarelli E, Varella-Garcia M, Tang X, et al. KRAS mutation is an important predictor of resistance to therapy with epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancer. Clin Cancer Res. 2007;13:2890–2896. doi: 10.1158/1078-0432.CCR-06-3043. [DOI] [PubMed] [Google Scholar]