Abstract
Background. Oseltamivir resistance in A(H1N1)pdm09 influenza is rare, particularly in untreated community cases. Sustained community transmission has not previously been reported.
Methods. Influenza specimens from the Asia–Pacific region were collected through sentinel surveillance, hospital, and general practitioner networks. Clinical and epidemiological information was collected on patients infected with oseltamivir-resistant viruses.
Results. Twenty-nine (15%) of 191 A(H1N1)pdm09 viruses collected between May and September 2011 from Hunter New England (HNE), Australia, contained the H275Y neuraminidase substitution responsible for oseltamivir resistance. Only 1 patient had received oseltamivir before specimen collection. The resistant strains were genetically very closely related, suggesting the spread of a single variant. Ninety percent of cases lived within 50 kilometers. Three genetically similar oseltamivir-resistant variants were detected outside of HNE, including 1 strain from Perth, approximately 4000 kilometers away. Computational analysis predicted that neuraminidase substitutions V241I, N369K, and N386S in these viruses may offset the destabilizing effect of the H275Y substitution.
Conclusions This cluster represents the first widespread community transmission of H275Y oseltamivir-resistant A(H1N1)pdm09 influenza. These cases and data on potential permissive mutations suggest that currently circulating A(H1N1)pdm09 viruses retain viral fitness in the presence of the H275Y mutation and that widespread emergence of oseltamivir-resistant strains may now be more likely.
(See the editorial commentary by Fry and Gubareva, on pages 145-7.)
Oseltamivir (Tamiflu®) is the most commonly used drug for the treatment or prophylaxis of influenza, and has been widely used in Japan, North America, and Europe. Oseltamivir has also been stockpiled by many countries as part of pandemic preparedness and was widely used during the 2009 influenza A(H1N1) pandemic, when it was shown to improve clinical outcomes in infants, adults, and pregnant women [1–6]. Since the emergence of the pandemic H1N1 2009 virus (A[H1N1]pdm09), oseltamivir resistance has been detected at a frequency of approximately 1%, with the majority of resistant viruses being isolated from immunocompromised patients undergoing oseltamivir treatment [7–9]. Virtually all of these resistant viruses have contained the histidine (H) to tyrosine (Y) substitution at position 275 of the neuraminidase (NA; N1 numbering—the same substitution is referred to as H274Y based on N2 numbering), which confers highly reduced oseltamivir sensitivity in vitro [10]. This substitution was also present in the oseltamivir-resistant variant of the prepandemic seasonal A(H1N1) subtype that emerged in Europe and then spread globally in less than a year in 2008 [11, 12]. This demonstrated the capacity of an A(H1N1) virus to overcome the inherently detrimental fitness effect of the H275Y substitution [13, 14] and transmit readily between individuals in the absence of drug selection pressure. Clinical data have shown that the effectiveness of oseltamivir was significantly reduced during the treatment of the previously circulating seasonal A(H1N1) H275Y variants [15–18].
Transmission of oseltamivir-resistant H275Y A(H1N1)pdm09 strains to date has been limited or unsustained. Most commonly, transmission has occurred in closed, near-contact settings, such as hospital wards [19–21], or after prolonged close contact in community settings such as a long train journey or at a school camp [22, 23]. Oseltamivir-resistant H275Y variants were detected at a low frequency (<1%) among community cases during the 2010–2011 northern hemisphere influenza season [24, 25] and, based on our testing, this continues to be the case for most countries in the southern hemisphere during the current 2011 influenza season. In contrast, between June and August 2011, in the Hunter New England region around Newcastle, Australia, we detected a significantly increased frequency of H275Y oseltamivir-resistant A(H1N1)pdm09 viruses in community patients, of whom only 1 patient had been treated with oseltamivir [26]. Following our initial brief report [26], here we describe the virological and epidemiological aspects of the cluster of H275Y oseltamivir-resistant A(H1N1)pdm09 influenza and discuss potentially permissive NA substitutions that may have enabled the A(H1N1)pdm09 virus to retain viral fitness in the presence of the H275Y mutation.
MATERIALS AND METHODS
Sample Collection
Newcastle (population 273 805) is a regional coastal city in the state of New South Wales (NSW), and the sixth largest city in Australia, providing tertiary referral specialist services for northern NSW. It is located approximately 120 kilometers north of the NSW capital, Sydney (Figure 1). The Hunter New England (HNE) health district, of which Newcastle is the major city, stretches from Lake Macquarie in the south to the Queensland border. The HNE health district is served by Hunter Area Pathology Services (HAPS), which receive specimens for influenza testing from emergency departments, intensive care units, and general practitioners. Respiratory specimens (swabs and nasopharyngeal aspirates) that were positive for influenza A or B by nucleic acid testing at HAPS in 2011 were referred weekly to the World Health Organization (WHO) Collaborating Centre for Reference and Research on Influenza (WHO CCRRI) for further virological analysis. A(H1N1)pdm09 viruses from the HNE health district were compared with viruses obtained from WHO National Influenza Centres that form part of the WHO Global Influenza Surveillance and Response System (GISRS), and other laboratories both within and outside of Australia.
Figure 1.
Map of the Hunter New England (HNE) health district and Australia. HNE health district is colored in red.
To enable the review of oseltamivir-resistant viruses in the HNE health district, the NSW Chief Health Officer granted an ethics waiver and authorized the investigation under the NSW Public Health Act 2010 as an urgent public health investigation. For all patients with oseltamivir-resistant viruses, hospital records were reviewed and treating medical practices contacted to obtain details of clinical symptoms and outcomes, antiviral and concomitant treatment administered during the current and previous treatment episodes, and medical history. Patients infected with oseltamivir-resistant influenza viruses were interviewed using a structured questionnaire to gather data on patient demographics, medical history of immune-compromising conditions, influenza vaccination status in 2011, and antiviral treatment history.
Virological Analysis
Viruses were isolated from original clinical samples and repassaged in Madin–Darby canine kidney cells (ATCC CCL-34). Antigenic characterization was performed using a hemagglutination-inhibition assay [27]. All virus isolates were analyzed for oseltamivir, zanamivir, and peramivir sensitivity using a fluorescence-based NA inhibition assay [28], followed by hemagglutinin (HA) and NA sequence analysis of those strains with elevated inhibitory concentration (IC50) values (the concentration of drug required to inhibit the NA activity by 50%). Where virus isolates could not be obtained, original specimens were analyzed for the presence or absence of the H275Y NA substitution using a pyrosequencing assay [29]. The HA and NA genes were fully sequenced using standard techniques on an ABI 3500XL sequencer. Sequences from oseltamivir-resistant strains were uploaded to a publically available influenza sequence database (GISAID; www.gisaid.org), and given accession numbers EPI334765-334790 and 335634-335637. Maximum likelihood phylogenetic trees were constructed with PhyML (http://www.atgc-montpellier.fr/phyml) using HKY85 as the nucleotides substitution model with bootstrapping (Geneious Pro 5.1.4 software).
Computational Structural Analysis
Computational structural analysis of combinations of candidate permissive mutations in the NA was conducted using FoldX [31], an empirical all-atom force field that allows the calculation of protein stability changes upon mutation by approximating the free energy of unfolding through weighted terms. To validate the calculations, more than 1000 mutations representative of most structural environments were analyzed and achieved a global correlation of 0.8 with experimentally measured thermodynamic data. In this work, the A(H1N1)pdm09 NA crystal structure (PDB:3nss) was minimized with the RepairPDB function, then mutations were introduced separately or as cumulative combinations using FoldX in YASARA [32], where the stochastic side chain minimization procedure is repeated 5 times for each mutant and the averages are taken as final predicted free-energy change.
Results
Frequency of Oseltamivir Resistance
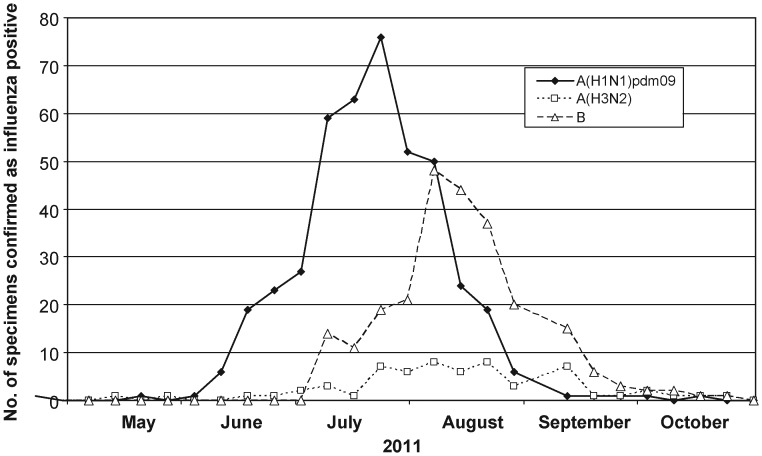
From the HNE health district, 2673 respiratory tract specimens were collected for influenza testing by nucleic acid testing (NAT) at HAPS in the period 20 May 2011 to 28 October 2011 (23 weeks) from a population of 874 644 (0.3%). Of the 749 influenza NAT-positive specimens, 439 (59%) were A(H1N1)pdm09 viruses (Figure 2) and 191 had sufficient volume to enable antiviral susceptibility analysis by the WHO CCRRI. Twenty-nine (15%) of these 191 A(H1N1)pdm09 viruses were found to contain the H275Y NA substitution by either pyrosequencing of original specimens (n = 18) or conventional NA sequencing of isolates following an NA inhibition assay (n = 11). H275Y A(H1N1)pdm09 viruses were obtained from patients between 31 May and 19 August 2011 (Figure 3). The frequency of resistance over time was 4/51 (8%) in June, 20/85 (24%) in July, 4/45 (8%) in August, and 0/4 (0%) in September by which time influenza activity had become very low in the region (Figure 2). Twenty-six of the 29 patients infected with oseltamivir-resistant viruses lived in 5 adjacent local municipalities (Cessnock, Lake Macquarie, Maitland, Newcastle, Port Stephens) within 50 kilometers of Newcastle. The other 3 patients lived in rural towns 90, 150, and 490 kilometers away from Newcastle.
Figure 2.
Number of laboratory-confirmed influenza positive specimens detected in the Hunter New England health district during 2011.
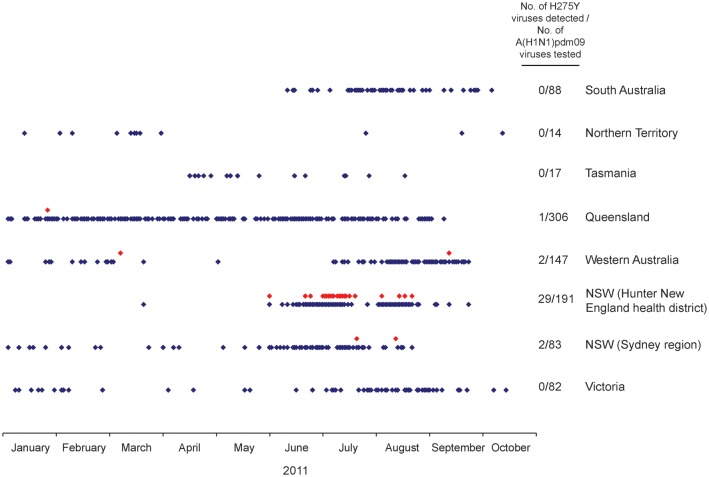
Figure 3.
Frequency of detection of oseltamivir-resistant H275Y variants in Australia in 2011. A red dot represents the sample date of an individual oseltamivir-resistant A(H1N1)pdm09 H275Y case, and a blue dot indicates the sample date of an individual oseltamivir-sensitive A(H1N1)pdm09 case detected in the states and territories of Australia in 2011.
In comparison, only 5 oseltamivir-resistant H275Y viruses were detected out of 737 A(H1N1)pdm09 viruses tested (frequency, 0.7%) from the rest of Australia during 2011 (Figure 3). Two of these strains from January and March 2011 were from other Australian states (Queensland and Western Australia) and were isolated from hospitalized immunocompromised patients undergoing oseltamivir treatment. However, 2 oseltamivir-resistant H275Y viruses from elsewhere in NSW and 1 from Western Australia were collected during or after the period of oseltamivir-resistant virus detection in the HNE health district. One of the NSW oseltamivir-resistant viruses was detected in July 2011 from a child in Sydney, NSW, 120 kilometers south of Newcastle, while the other was detected in August 2011 from an infant in Orange, NSW, a town approximately 380 kilometers west of Newcastle. The Western Australian oseltamivir-resistant virus was detected in September 2011, 3 weeks after the last detected case in the HNE health district, and in a location near Perth, approximately 4000 kilometers west of Newcastle. All 3 of these resistant viruses were taken from otherwise healthy children who had not been treated with oseltamivir. Neither they nor their family members had traveled recently to Newcastle, and in the Western Australian case, there had been no recent contact with anyone from NSW.
Patient Details for HNE Health District Oseltamivir-Resistant Cases
Seventeen (59%) of the 29 patients infected with oseltamivir-resistant viruses were female and 3 (10%) were pregnant. Three (10%) identified themselves as Aboriginal. The age range was 4 months to 62 years (median, 31 years) compared with a median age of 29 years for persons infected with oseltamivir-sensitive influenza A(H1N1)pdm09. No cases resided in institutional care or aged care facilities. Cough was reported by 25 (86%) cases and fever by 22 (76%) cases. One patient was asymptomatic, but was swabbed and tested for influenza due to contact with a laboratory-proven A(H1N1)pdm09 case. Six (21%) cases reported a history of asthma; none had a history of chronic obstructive pulmonary disease; 6 (21%) were current smokers with an average pack-year history of 20 (range, 1–62) and none were immunosuppressed. Three (10%) cases had received trivalent-inactivated influenza vaccine in 2011. Importantly, only 1 patient infected with an oseltamivir-resistant virus had received oseltamivir prior to their specimen being collected (in this case, received 4 days prior to specimen being taken). Eleven (38%) were prescribed oseltamivir after specimen collection. Seven (24%) patients required hospital admission with a mean length of stay of 2.3 days (range, 1–7 days). There were no intensive care admissions and no fatal outcomes. The median number of days absent from usual duties was 5. Of the 29 patients infected with an oseltamivir-resistant virus, 5 pairs of cases were epidemiologically linked. Four households had 2 affected cases each, while a fifth pair of cases shared a short car journey. No other links could be identified during interviews with cases. Detailed outcomes were not collected for persons infected with oseltamivir-sensitive influenza.
Virological Analysis of Oseltamivir-Resistant Viruses
For the viruses that were isolated in cell culture, the oseltamivir-resistant H275Y strains (n = 11) had a mean (±1 SD) oseltamivir IC50 of 205.3 ± 23.5 nM that was 513-fold higher than the mean IC50 of oseltamivir-sensitive isolates (n = 3579) (Table 1). The H275Y isolates also demonstrated elevated peramivir IC50 values (16.6 ± 1.5 nM) that were approximately 80-fold higher than sensitive strains, but were fully sensitive to zanamivir (Table 1). The oseltamivir-resistant viruses were also resistant to the adamantane class of influenza antiviral drugs based on M2 sequencing, having the characteristic S31N substitution. These H275Y variants, including those from the 3 patients that received the influenza vaccine in 2011, were antigenically indistinguishable from the oseltamivir-sensitive viruses and were well inhibited by ferret serum samples raised against the current A(H1N1)pdm09 vaccine strain A/California/7/2009 in a hemagglutination-inhibition assay (Supplementary Table 1).
Table 1.
NA Inhibitor Sensitivity of Hunter New England Health District Oseltamivir-Resistant H275Y Viruses
|
Zanamivir |
Oseltamivir Carboxylate |
Peramivir |
||||
|---|---|---|---|---|---|---|
| Virus | Mean IC50 ± SD (nM) | Fold Diffa | Mean IC50 ± SD (nM) | Fold Diffa | Mean IC50 ± SD (nM) | Fold Diffa |
| Mean of HNE health district H275Y A(H1N1) pdm09 viruses (n = 11) | 0.3 ± 0.02 | 1 | 205.3 ± 23.5 | 513 | 16.6 ± 1.5 | 83 |
| Mean of HNE health district sensitive A(H1N1) pdm09 viruses (n = 65) | 0.4 ± 0.2 | 1 | 0.5 ± 0.3 | 1 | 0.2 ± 0.1 | 1 |
| Mean of all sensitive A(H1N1)pdm09 viruses (n = 3579) | 0.3 ± 0.2 | … | 0.4 ± 0.3 | … | 0.2 ± 0.1b | … |
| A/Newcastle/2/2011 | 0.3 ± 0.02 | 1 | 191.1 ± 19.4 | 478 | 18.6 ± 6.8 | 93 |
| A/Newcastle/17/2011 | 0.3 ± 0.04 | 1 | 223.7 ± 14.2 | 559 | 18.0 ± 2.3 | 90 |
| A/Newcastle/37/2011 | 0.3 ± 0.1 | 1 | 247.9 ± 27.2 | 620 | 16.9 ± 0.8 | 85 |
| A/Newcastle/53/2011 | 0.2 ± 0.04 | 1 | 187.8 ± 30.0 | 470 | 13.8 ± 1.2 | 69 |
| A/Newcastle/62/2011 | 0.2 ± 0.04 | 1 | 199.9 ± 19.7 | 500 | 15.9 ± 1.7 | 79 |
| A/Newcastle/82/2011 | 0.2 ± 0.1 | 1 | 198.5 ± 36.2 | 496 | 15.2 ± 1.5 | 76 |
| A/Newcastle/85/2011 | 0.2 ± 1.1 | 1 | 226.6 ± 11.4 | 567 | 16.7 ± 1.3 | 83 |
| A/Newcastle/89/2011 | 0.3 ± 0.04 | 1 | 227.8 ± 13.1 | 569 | 16.9 ± 1.8 | 85 |
| A/Newcastle/132/2011 | 0.3 ± 0.04 | 1 | 198.1 ± 26.8 | 495 | 15.8 ± 1.5 | 79 |
| A/Newcastle/151/2011 | 0.2 ± 0.02 | 1 | 191.0 ± 46.6 | 478 | 15.9 ± 2.2 | 79 |
| A/Newcastle/129/2011 | 0.3 ± 0.03 | 1 | 165.9 ± 26.1 | 415 | 18.8 ± 2.7 | 94 |
Abbreviations: Diff, difference; HNE, Hunter New England; IC50, inhibitory concentration.
a Based on comparison with mean IC50 of all sensitive A(H1N1)pdm09 viruses.
b Mean and standard deviation of peramivir IC50 values based on analysis of n = 273 isolates.
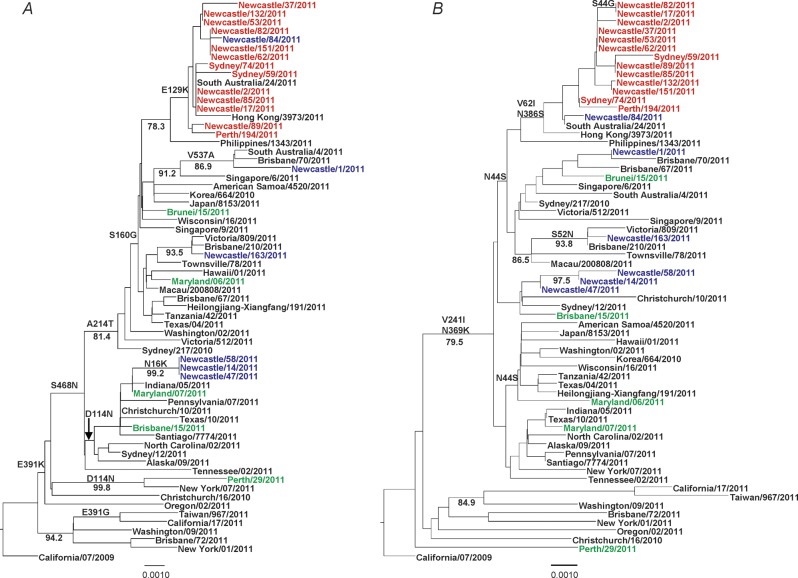
Phylogenetic analysis of the hemagglutinin and NA sequences revealed that the oseltamivir-resistant viruses from the HNE health district, the 2 other NSW viruses detected in Sydney and Orange, and the Western Australian strain from September 2011 were all highly similar (99.9% nucleotide similarity across 22 HA and 18 NA sequences) and formed a distinct subgroup in both the HA and NA phylogenetic trees (Figure 4). Apart from the H275Y NA substitution, the oseltamivir-resistant strains all contained 2 other NA amino acid substitutions, V62I and N386S, which were absent from all but 1 of the local oseltamivir-sensitive viruses (A/Newcastle/84/2011) (Figure 4). In addition to the NA substitutions, whole-genome sequence comparison of 8 oseltamivir-resistant strains and 5 oseltamivir-sensitive strains from the HNE health district revealed 3 further substitutions across 3 other segments that were specific to the resistant strains—E129K in HA, L672F in PA, and S482N in NP. Genetic comparison of the 2 viruses from each of the linked cases showed 100% nucleotide similarity across the HA and NA genes, although sequence data were not available for viruses from 2 of the paired cases. In addition, other oseltamivir-sensitive A(H1N1)pdm09 strains, including A/South Australia/24/2011 and A/Hong Kong/2973/2011, clustered phylogenetically with the resistant strains (Figure 4), sharing a high degree of genetic similarity, except for the H275Y NA substitution. Full phylogenetic trees comparing the HNE health district sequences with publically available 2011 A(H1N1)pdm09 HA and NA sequences are shown in the supplementary data (Supplementary Figure 1A and 1B).
Figure 4.
Phylogenetic trees of hemagglutinin (A) and neuraminidase (B) sequences of oseltamivir-resistant H275Y variants and oseltamivir-sensitive wild type A(H1N1)pdm09 viruses. Hunter New England health district and other NSW and Western Australian H275Y variants are shown in red, sporadic H275Y mutants from other regions in green, and Hunter New England health district sensitive viruses in blue. Amino acid mutations common to each clade and bootstrap confidence values >75 are indicated on the trees.
Computational Analysis of Potentially Permissive NA Substitutions
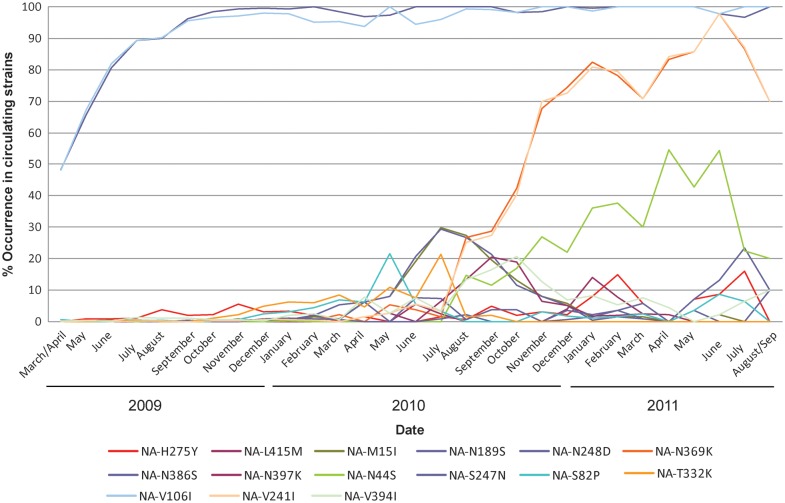
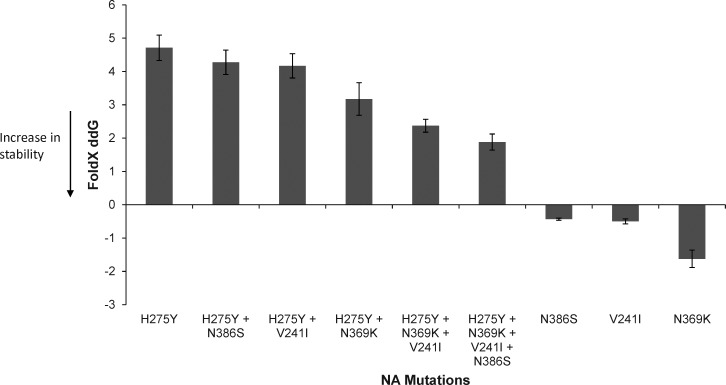
To investigate the role of potentially permissive NA mutations that may be responsible for offsetting the negative effects of the H275Y substitution, a computational analysis of NA protein stability was conducted. Since the emergence of the A(H1N1)pdm09 strain in 2009, a number of NA mutations have been described and, in some cases, have become fixed within globally circulating strains (Figure 5). Based on computational analysis, 2 of these mutations, V241I and N369K, both of which emerged in A(H1N1)pdm09 viruses in 2010 and are now present in over 80% of currently circulating strains (Figure 5), could restore approximately 50% of the protein stability that was lost as a result of the H275Y mutation (Figure 6). In addition, the HNE health district and other NSW oseltamivir-resistant viruses contained an N386S NA substitution. This substitution is less common in other A(H1N1)pdm09 viruses, but computational analysis suggested that it would also further stabilize the NA (Figure 6). Another NA substitution that has been on the rise in recent strains and was present in the oseltamivir-resistant viruses described here is N44S. It creates a new potential N-glycosylation site at position 42, changing the motif at positions 42–44 from NQN to NQS. However, this region of the NA is in the stalk and not part of the globular domain and, therefore, is not known how it would affect NA structure and stability.
Figure 5.
All human A(H1N1)pdm09 NA sequences containing date information (at least year/month) were downloaded from the National Center for Biotechnology Information influenza virus resource [41] (http://www.ncbi.nlm.nih.gov/genomes/FLU/) and GISAID (http://www.gisaid.org) and merged with the ones reported in this study (keeping only 1 per unique strain identifier). Sequences shorter than 90% of the median length are considered as fragmentary and were removed from the analysis. The resulting 8085 sequences were then aligned with the multiple sequence alignment program MAFFT [42] and mutation frequencies relative to the vaccine reference strain A/California/07/2009 counted with a custom perl script. All mutations with at least 50 occurrences since March 2009 and global frequencies >20% in any month are shown, plus L415M, which is characteristic for an outbreak with H275Y in Japan in January. Data collected in the month of March 2009 were merged with April 2009, while data collected from September 2011 were merged with August 2011 as there were less than 10 sequences in those months.
Figure 6.
Computational structural analysis of combinations of candidate permissive mutations in the neuraminidase (NA). Mutations were modeled with FoldX in the A(H1N1)pdm09 NA crystal structure (PDB:3nss) after minimization and using 5 repetitions.
Discussion
The H275Y variants detected in the HNE health district, other parts of NSW, and in Western Australia represent the largest and most widespread cluster of oseltamivir-resistant A(H1N1)pdm09 influenza identified since the virus first emerged in humans in 2009. The oseltamivir-resistant strains detected from these locations were virtually identical genetically, suggesting emergence from a single source.
The rapid global spread of oseltamivir-resistant seasonal A(H1N1) H275Y viruses during 2007–2008 [11, 12] showed that A(H1N1) influenza viruses with this substitution could retain transmissibility, even though previous in vitro and in vivo studies found the substitution reduced fitness in other A(H1N1) strains [13, 14]. A recent study demonstrated that seasonal A(H1N1) oseltamivir-resistant H275Y viruses exhibited reduced susceptibility to postvaccination antibody inhibition compared to the cocirculating oseltamivir-sensitive viruses [30], which may have contributed to the virtual replacement of the oseltamivir-sensitive seasonal A(H1N1) strain within 1 year. Based on antigenic analyses with postinfection ferret serum samples, the oseltamivir-resistant A(H1N1)pdm09 H275Y viruses reported here are antigenically similar to oseltamivir-sensitive A/California/7/2009-like A(H1N1)pdm09 strains circulating in the HNE health district and elsewhere in Australia, although future studies using postvaccination human serum should be performed [30]. The high frequency of resistance in untreated community patients suggests that they are not markedly less fit than sensitive viruses. This situation is similar to the oseltamivir-resistant seasonal A(H1N1) viruses that emerged in Norway in 2007 where, like Australia, the use of oseltamivir was low compared to countries such as Japan and the United States [31].
Recently, it has been shown that certain substitutions in the NA, V234M and R222Q, enabled the seasonal A(H1N1) virus to remain fully functional in the presence of the H275Y substitution [32, 33]. These permissive substitutions are not present in the A(H1N1)pdm09 oseltamivir-resistant H275Y variants reported here. Most A(H1N1)pdm09 sequences (including the strains reported here) have the nonpermissive V at position 234, and the most typical amino acid at position 222 among the A(H1N1)pdm09 NAs is N. However, 3 other NA substitutions, V241I, N369K, and the N386S, which were present in the oseltamivir-resistant strains from the cluster reported here, may offset the destabilizing effect of the H275Y substitution. The V241I and N369K substitutions are present not only in the H275Y variants reported here, but are also in viral sequences of North American and a large number of Japanese oseltamivir-resistant H275Y strains recently deposited on the GISAID sequence database (Supplementary Figure 1A and 1B). Importantly, the N369K substitution, which computationally was predicted to cause the largest change in protein stability, has previously been shown experimentally to increase NA surface expression and activity in combination with H275Y [32].
Infection with oseltamivir-resistant seasonal A(H1N1) viruses significantly reduced the effectiveness of oseltamivir during the 2008–2009 season, particularly in younger children [15–18]. For patients infected with oseltamivir-resistant viruses, fever levels were comparable between oseltamivir-treated and untreated patients, but significantly reduced in zanamivir-treated patients [15]. Mean duration of fever after the start of oseltamivir treatment was also significantly longer in persons infected with oseltamivir-resistant viruses than in those infected with oseltamivir-sensitive viruses [15, 17, 18], while treatment of the H275Y viruses with zanamivir reduced fever duration to normal levels [18]. Although the relationship between in vitro IC50 of a virus and clinical effectiveness is not well understood, it is noteworthy that the mean oseltamivir IC50 of the H275Y A(H1N1)pdm09 viruses is generally lower than that of the H275Y seasonal A(H1N1) variants [12, 34, 35], and is below the steady-state trough plasma concentrations found in children, adolescents, and adults following recommended dosing [36]. As a result, oseltamivir treatment may be more effective for the A(H1N1)pdm09 H275Y variant than has been reported for the H275Y seasonal A(H1N1) variants. More data regarding the effectiveness of oseltamivir for the treatment of A(H1N1)pdm09 H275Y viruses, and other variants with reduced oseltamivir sensitivity, are needed. Eleven of the oseltamivir-resistant HNE health district cases received oseltamivir after specimen collection but, because the resistant virus was detected after the patient had recovered and treatment had ceased, it was not possible to investigate the effectiveness of oseltamivir. The effectiveness of peramivir is also likely to be reduced for the treatment of H275Y A(H1N1)pdm09 viruses given the 80-fold increase in IC50 [37], as such current WHO guidelines state that intravenous peramivir should only be considered for the treatment of such viruses when intravenous zanamivir is not available [38]. However, recent animal data suggest that 5 days of therapy with intravenous peramivir may overcome the shift in sensitivity to peramivir caused by the H275Y substitution [39]. Given that the H275Y variant is fully susceptible to zanamivir in vitro, this drug remains the recommended alternative antiviral treatment option when oseltamivir resistance is detected [38].
Of the 29 H275Y A(H1N1)pdm09 viruses detected in the HNE health district, none caused severe illness or resulted in patients being admitted to intensive care wards. However, because detailed clinical or epidemiological information was not collected at the same time from a comparison group of patients with oseltamivir-sensitive virus infections, it was not possible to determine whether there were any differences between the types of patients infected or the types of illness caused by oseltamivir-sensitive and -resistant viruses. Another limitation is that a relatively small number of A(H1N1)pdm09 viruses were available for testing (191 from the HNE health district and 737 from the rest of Australia). Therefore, not only may the true prevalence of resistance be different to that reported here, but the spread of the genetically related oseltamivir-resistant virus may also be wider than detected.
Influenza activity in the HNE health district and the rest of Australia has now returned to baseline levels, although the detection of the variant outside the HNE health district is concerning and demonstrates its capacity to spread widely. Oseltamivir-resistant strains that were indistinguishable to those detected in the HNE health district were identified in Sydney, the largest city in Australia and a large international travel hub, and near Perth, Western Australia, on the other side of the country to Newcastle. To date, there is no indication that this oseltamivir-resistant A(H1N1)pdm09 strain has spread into the northern hemisphere, although it will be important for countries to monitor strains throughout the upcoming northern hemisphere influenza season. The most recent A(H1N1)pdm09 sequences strains deposited on GISAID at the time of writing are from the United States (Hawaii, California, Pennsylvania), and although none of them have the H275Y substitution, they all have the NA substitutions V241I and N369K, which may buffer the detrimental effect of the H275Y oseltamivir-resistance mutation. The other non-NA amino acid substitutions in the oseltamivir-resistant viruses from the cluster (HA, PA, and NP) may also play a role in viral fitness. In vitro and in vivo studies are currently in progress to analyze the fitness of the strains from this cluster and investigate the role of these potentially “permissive” mutations. However, this cluster of cases, along with the recent reports of increased detection of H275Y A(H1N1)pdm09 viruses in untreated community patients in the United Kingdom and United States [24, 40] and the large number of recent sequences with the H275Y substitution deposited on GISAID from Japan, suggests that the currently circulating A(H1N1)pdm09 may be becoming more tolerant of the H275Y mutation than it was when the strain first emerged in 2009, and that widespread emergence of oseltamivir-resistant A(H1N1)pdm09 viruses may now be more likely.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. The authors thank the laboratories and clinicians that submit specimens and isolates to the Melbourne WHO CCRRI. Oseltamivir carboxylate, the active form of the ethyl ester prodrug oseltamivir phosphate, was kindly provided by Hoffmann-La Roche Ltd, Switzerland; zanamivir was kindly provided by GlaxoSmithKline, Australia; peramivir was kindly provided by BioCryst Pharmaceuticals, Birmingham, AL. The Melbourne WHO Collaborating Centre for Reference and Research on Influenza is supported by the Australian Government Department of Health and Ageing.
Financial support. This work was supported by institutional funds.
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Farias JA, Fernandez A, Monteverde E, et al. Critically ill infants and children with influenza A (H1N1) in pediatric intensive care units in Argentina. Intensive Care Med. 2010;36:1015–22. doi: 10.1007/s00134-010-1853-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Creanga AA, Johnson TF, Graitcer SB, et al. Severity of 2009 pandemic influenza A (H1N1) virus infection in pregnant women. Obstet Gynecol. 2010;115:717–26. doi: 10.1097/AOG.0b013e3181d57947. [DOI] [PubMed] [Google Scholar]
- 3.Ling LM, Chow AL, Lye DC, et al. Effects of early oseltamivir therapy on viral shedding in 2009 pandemic influenza A (H1N1) virus infection. Clin Infect Dis. 2010;50:963–9. doi: 10.1086/651083. [DOI] [PubMed] [Google Scholar]
- 4.Campbell CN, Mytton OT, McLean EM, et al. Hospitalization in two waves of pandemic influenza A(H1N1) in England. Epidemiol Infect. 2010;139:1560–9. doi: 10.1017/S0950268810002657. [DOI] [PubMed] [Google Scholar]
- 5.Newsome K, Williams J, Way S, et al. Maternal and Infant Outcomes Among Severely Ill Pregnant and Postpartum Women with 2009 Pandemic Influenza A (H1N1)—United States, April 2009–August 2010. MMWR Morb Mortal Wkly Rep. 2011;60:1193–6. [PubMed] [Google Scholar]
- 6.Khandaker G, Zurynski Y, Lester-Smith D, et al. Clinical features, oseltamivir treatment and outcome in infants aged <12 months with laboratory-confirmed influenza A in 2009. Antivir Ther. 2011;16:1005–10. doi: 10.3851/IMP1848. [DOI] [PubMed] [Google Scholar]
- 7.Graitcer SB, Gubareva L, Kamimoto L, et al. Characteristics of patients with oseltamivir-resistant pandemic (H1N1) 2009, United States. Emerg Infect Dis. 2011;17:255–7. doi: 10.3201/eid1702.101724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hurt AC, Deng Y, Ernest J, et al. Oseltamivir-resistant influenza viruses circulating during the first year of the influenza A(H1N1)2009 pandemic in the Asia-Pacific region, March 2009 to March 2010. Euro Surveill. 2011:16–19770. [PubMed] [Google Scholar]
- 9.Meijer A, Jonges M, Abbink F, et al. Oseltamivir-resistant pandemic A(H1N1) 2009 influenza viruses detected through enhanced surveillance in the Netherlands, 2009–2010. Antiviral Res. 2011;92:81–9. doi: 10.1016/j.antiviral.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 10.Gubareva LV, Kaiser L, Hayden FG. Influenza virus neuraminidase inhibitors. Lancet. 2000;355:827–35. doi: 10.1016/S0140-6736(99)11433-8. [DOI] [PubMed] [Google Scholar]
- 11.Meijer A, Lackenby A, Hungnes O, et al. Oseltamivir-resistant influenza virus A (H1N1), Europe, 2007–08 season. Emerg Infect Dis. 2009;15:552–60. doi: 10.3201/eid1504.081280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hurt AC, Ernest J, Deng Y, et al. Emergence and spread of oseltamivir-resistant A(H1N1) influenza viruses in Oceania, South East Asia and South Africa. Antiviral Res. 2009;83:90–3. doi: 10.1016/j.antiviral.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 13.Herlocher ML, Truscon R, Elias S, et al. Influenza viruses resistant to the antiviral drug oseltamivir: transmission studies in ferrets. J Infect Dis. 2004;190:1627–30. doi: 10.1086/424572. [DOI] [PubMed] [Google Scholar]
- 14.Ives JAL, Carr JA, Mendel DB, et al. The H274Y mutation in the influenza A/H1N1 neuraminidase active site following oseltamivir phosphate treatment leave virus severely compromised both in vitro and in vivo. Antiviral Res. 2002;55:307–17. doi: 10.1016/s0166-3542(02)00053-0. [DOI] [PubMed] [Google Scholar]
- 15.Saito R, Sato I, Suzuki Y, et al. Reduced effectiveness of oseltamivir in children infected with oseltamivir-resistant influenza A (H1N1) viruses with His275Tyr mutation. Pediatr Infect Dis J. 2010;29:898–904. doi: 10.1097/INF.0b013e3181de9d24. [DOI] [PubMed] [Google Scholar]
- 16.Kawai N, Ikematsu H, Iwaki N, et al. Clinical effectiveness of oseltamivir for influenza A(H1N1) virus with H274Y neuraminidase mutation. J Infect. 2009;59:207–12. doi: 10.1016/j.jinf.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 17.Matsuzaki Y, Mizuta K, Aoki Y, et al. A two-year survey of the oseltamivir-resistant influenza A(H1N1) virus in Yamagata, Japan and the clinical effectiveness of oseltamivir and zanamivir. Virol J. 2010;7:53. doi: 10.1186/1743-422X-7-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawai N, Ikematsu H, Hirotsu N, et al. Clinical effectiveness of oseltamivir and zanamivir for treatment of influenza A virus subtype H1N1 with the H274Y mutation: a Japanese, multicenter study of the 2007–2008 and 2008–2009 influenza seasons. Clin Infect Dis. 2009;49:1828–35. doi: 10.1086/648424. [DOI] [PubMed] [Google Scholar]
- 19.Wolfe C, Greenwald I, Chen L. Pandemic (H1N1) 2009 and oseltamivir resistance in hematology/oncology patients. Emerg Infect Dis. 2010;16:1809–11. doi: 10.3201/eid1611.101053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moore C, Galiano M, Lackenby A, et al. Evidence of person-to-person transmission of oseltamivir-resistant pandemic influenza A(H1N1) 2009 virus in a hematology unit. J Infect Dis. 2011;203:18–24. doi: 10.1093/infdis/jiq007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen LF, Dailey NJ, Rao AK, et al. Cluster of oseltamivir-resistant 2009 pandemic influenza A (H1N1) virus infections on a hospital ward among immunocompromised patients—North Carolina, 2009. J Infect Dis. 2011;203:838–46. doi: 10.1093/infdis/jiq124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oseltamivir-resistant 2009 pandemic influenza A (H1N1) virus infection in two summer campers receiving prophylaxis—North Carolina, 2009. MMWR Morb Mortal Wkly Rep. 2009;58:969–72. [PubMed] [Google Scholar]
- 23.Le QM, Wertheim HF, Tran ND, et al. A community cluster of oseltamivir-resistant cases of 2009 H1N1 influenza. N Engl J Med. 2010;362:86–7. doi: 10.1056/NEJMc0910448. [DOI] [PubMed] [Google Scholar]
- 24.Lackenby A, Moran GJ, Pebody R, et al. Continued emergence and changing epidemiology of oseltamivir-resistant influenza A(H1N1)2009 virus, United Kingdom, winter 2010/11. Euro Surveill. 2011;16:19784. [PubMed] [Google Scholar]
- 25.WHO Global fluenza Surveillance and Response System. Summary of influenza antiviral susceptibility surveillance findings, September 2010–March 2011. http://www.who.int/influenza/gisrs_laboratory/updates/antiviral_susceptibility_20110606/en/index.html. Accessed 25 August 2011.
- 26.Hurt AC, Hardie K, Wilson NJ, et al. Community transmission of oseltamivir-resistant A(H1N1)pdm09 influenza. N Engl J Med. 2011;365:2541–2. doi: 10.1056/NEJMc1111078. [DOI] [PubMed] [Google Scholar]
- 27.WHO Global Influenza Surveillance Network. Manual for the laboratory diagnosis and virological surveillance of influenza. http://whqlibdoc.who.int/publications/2011/9789241548090_eng.pdf. Accessed 7 December 2011. [Google Scholar]
- 28.Hurt AC, Barr IG, Hartel G, et al. Susceptibility of human influenza viruses from Australasia and South East Asia to the neuraminidase inhibitors zanamivir and oseltamivir. Antiviral Res. 2004;62:37–45. doi: 10.1016/j.antiviral.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 29.Deng YM, Caldwell N, Hurt A, et al. A comparison of pyrosequencing and neuraminidase inhibition assays for the detection of oseltamivir-resistant pandemic influenza A(H1N1) 2009 viruses. Antiviral Res. 2011;90:87–91. doi: 10.1016/j.antiviral.2011.02.014. [DOI] [PubMed] [Google Scholar]
- 30.Wu WL, Lau SY, Chen Y, et al. The 2008–2009 H1N1 influenza virus exhibits reduced susceptibility to antibody inhibition: Implications for the prevalence of oseltamivir resistant variant viruses. Antiviral Res. 2012;93:144–53. doi: 10.1016/j.antiviral.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hurt AC, Holien JK, Parker M, et al. Oseltamivir resistance and the H274Y neuraminidase mutation in seasonal, pandemic and highly pathogenic influenza viruses. Drugs. 2009;69:2523–31. doi: 10.2165/11531450-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 32.Bloom JD, Nayak JS, Baltimore D. A computational-experimental approach identifies mutations that enhance surface expression of an oseltamivir-resistant influenza neuraminidase. PLoS ONE. 2011;6:e22201. doi: 10.1371/journal.pone.0022201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bloom JD, Gong LI, Baltimore D. Permissive secondary mutations enable the evolution of influenza oseltamivir resistance. Science. 2010;328:1272–5. doi: 10.1126/science.1187816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gubareva L, Nguyen HT, Sleeman K, et al. Comprehensive assessment of the drug susceptibility of 2009 H1N1 influenza viruses [abstract O-821] Program and abstracts of the International Society for Influenza and other Respiratory Virus Diseases: Options for the Control of Influenza VII. 2010;18 [Google Scholar]
- 35.Sheu TG, Deyde VM, Okomo-Adhiambo M, et al. Surveillance for neuraminidase inhibitor resistance among human influenza A and B viruses circulating worldwide from 2004 to 2008. Antimicrob Agents Chemother. 2008;52:3284–92. doi: 10.1128/AAC.00555-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oo C, Barrett J, Hill G, et al. Pharmacokinetics and dosage recommendations for an oseltamivir oral suspension for the treatment of influenza in children. Paediatr Drugs. 2001;3:229–36. doi: 10.2165/00128072-200103030-00005. [DOI] [PubMed] [Google Scholar]
- 37.Memoli MJ, Hrabal RJ, Hassantoufighi A, et al. Rapid selection of oseltamivir- and peramivir-resistant pandemic H1N1 virus during therapy in 2 immunocompromised hosts. Clin Infect Dis. 2010;50:1252–5. doi: 10.1086/651605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.WHO. WHO guidelines for pharmacological management of pandemic influenza A(H1N1) 2009 and other influenza viruses. Revised February 2010. http://www.who.int/csr/resources/publications/swineflu/h1n1_guidelines_pharmaceutical_mngt.pdf. Accessed 7 December 2011. [PubMed] [Google Scholar]
- 39.Abed Y, Boivin G, Yoshida R, et al. Parenteral peramivir treatment for oseltamivir-resistant 2009 pandemic influenza A H1N1 viruses. J Infect Dis. 2011;204:1641–2. doi: 10.1093/infdis/jir610. [DOI] [PubMed] [Google Scholar]
- 40.Storms AD, Gubareva LV, Su ST, et al. Oseltamivir-resistant pandemic (H1N1) 2009 virus infections, United States, 2010–11. Emerg Infect Dis. 2012;18:308–11. doi: 10.3201/eid1802.111466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bao Y, Bolotov P, Dernovoy D, et al. The influenza virus resource at the National Center for Biotechnology Information. J Virol. 2008;82:596–601. doi: 10.1128/JVI.02005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008;9:286–98. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.