SUMMARY
Electric organs (EOs) have evolved independently in vertebrates six times from skeletal muscle (SM). The transcriptional changes accompanying this developmental transformation are not presently well understood. Mormyrids and gymnotiforms are two highly convergent groups of weakly electric fish that have independently evolved EOs: while much is known about development and gene expression in gymnotiforms, very little is known about development and gene expression in mormyrids. This lack of data limits prospects for comparative work. We report here on the characterization of 28 differentially expressed genes between SM and EO tissues in the mormyrid Brienomyrus brachyistius, which were identified using suppressive subtractive hybridization (SSH). Forward and reverse SSH was performed on tissue samples of EO and SM resulting in one cDNA library enriched with mRNAs expressed in EO, and a second library representing mRNAs unique to SM. Nineteen expressed sequence tags (ESTs) were identified in EO and nine were identified in SM using BLAST searching of Danio rerio sequences available in NCBI databases. We confirmed differential expression of all 28 ESTs using RT-PCR. In EO, these ESTs represent four classes of proteins: (1) ion pumps, including the α- and β-subunits of Na+/K+-ATPase, and a plasma membrane Ca2+-ATPase; (2) Ca2+-binding protein S100, several parvalbumin paralogs, calcyclin-binding protein and neurogranin; (3) sarcomeric proteins troponin I, myosin heavy chain and actin-related protein complex subunit 3 (Arcp3); and (4) the transcription factors enhancer of rudimentary homolog (ERH) and myocyte enhancer factor 2A (MEF2A). Immunohistochemistry and western blotting were used to demonstrate the translation of seven proteins (myosin heavy chain, Na+/K+-ATPase, plasma membrane Ca2+-ATPase, MEF2, troponin and parvalbumin) and their cellular localization in EO and SM. Our findings suggest that mormyrids express several paralogs of muscle-specific genes and the proteins they encode in EOs, unlike gymnotiforms, which may post-transcriptionally repress several sarcomeric proteins. In spite of the similarity in the physiology and function of EOs in mormyrids and gymnotiforms, this study indicates that the mechanisms of development in the two groups may be considerably different.
KEY WORDS: MEF2, electric organ, mormyrid, muscle, subtractive hybridization
INTRODUCTION
Darwin (Darwin, 1859) considered strongly discharging electric fishes as a case of special difficulty for his theory of natural selection, noting of their electric organs (EOs) that it was ‘impossible to conceive by, what steps these wondrous organs have been produced’. Nearly 100 years would pass before Lissman recorded the first ‘weakly electric’ discharges from the fish Gymnarchus niloticus and noted its importance as a solution to Darwin's difficulty (Lissman, 1951). We now know that strongly discharging electric eels have evolved from weakly electric ancestors, and that EOs evolved originally for the purposes of electrolocation (Lissmann and Machin, 1958) and electrocommunication (Mörhes, 1957; Lissmann, 1958).
In his consideration of EOs, Darwin recognized that their diversity among fishes had not arisen from a single common ancestor, but multiply through convergent evolution (Darwin, 1859). We presently know of six independent origins of EOs in fishes: torpedinoids, rajoids, mormyriforms, gymnotiforms, siluriforms and uranoscopids (Bass, 1986). In all but one family of gymnotiforms, the Apteronotidae [we note here that the exceptional Apteronotidae have a myogenic larval organ that appears early in development, but is later replaced with a neurogenic adult electric organ (see Kirschbaum, 1983)] consisting of 64 species (Albert, 2003), EOs are derived during development from skeletal muscle tissue (Bass, 1986; Bennett, 1971). There is considerable variation between lineages (reviewed by Bass, 1986; Bennett, 1971), particularly in the types of skeletal muscle (SM) that EOs originate from (e.g. eye muscles, trunk musculature, pectoral fin musculature), in the voltage of electrical discharge (∼10 mV in ‘weakly electric’ mormyrids and gymnotiforms to 600 V in the strongly electric gymnotiform Electrophorus electricus), in their innervation (either in a restricted region in the case of most mormyrids or diffusely over a single electrocyte face, as in E. electricus), and in the complexity of the electrocyte anatomy (ranging from simple sac-like electrocytes in Torpedo to those with complex stalk-like protrusions, as in mormyrids). In addition, the electrical discharge of marine species, including elasmobranchs and teleosts, is the result of acetylcholine receptor-mediated post-synaptic potentials, whereas in freshwater species, the electrical discharge results from activation of voltage-gated sodium channels restricted to the EO plasma membrane.
Despite this considerable diversity, two groups of freshwater teleosts, the gymnotiforms of South America and mormyrids of Africa, exhibit several convergently evolved traits. Gymnotiforms and mormyrids have convergently evolved two classes of tuberous electroreceptors: one type that encodes electric organ discharge (EOD) amplitude and a second that encodes timing information (Zakon, 1986; Kawasaki, 2005). Mormyrids and gymnotiforms have similar electrosensory behaviors, most famously the jamming avoidance response (Heiligenberg, 1986). The two groups also have similar electromotor discharge patterns: both groups have independently evolved short ‘pulse-type’ EODs with long intervals in between and essentially continuous, quasi-sinusoidal ‘wave-type’ discharges (Zupanc and Bullock, 2005). Unlike many other electric fish, the electrocytes that comprise the EOs of both mormyrids and gymnotiforms produce spikes on both cell faces, and have complex anatomical features such as protrusions from the innervated membrane, termed ‘stalks’ (Bennett, 1971; Bass, 1986). Convergence between mormyrid and gymnotiform EOs has even been demonstrated at the molecular level; gymnotiforms and mormyrids utilize the same sodium channel Scn4aa for producing EODs (which arose by fish-specific whole-genome duplication ca. 200–300 million years ago). Scn4aa has been modified by positive selection leading to amino acid substitutions that affect sodium channel inactivation kinetics convergently, likely contributing to electric signal variation (Arnegard et al., 2010b).
It is notable that teleost fishes have evolved a wide variety of highly specialized muscle tissues aside from EOs, including sonic muscles capable of high-frequency contraction [e.g. plainfin midshpimen and toadfishes (Rome, 2006)], and heater organs for efficient thermogenesis [e.g. billfishes and swordfishes (Block, 1994)]. In each case, a suite of anatomical and physiological adaptations is required to produce these ‘novel’ structures from muscle (Block, 1994), though the molecular factors underlying the origins of these tissues remain poorly understood in all of these cases. It may be considerably advantageous, therefore, to consider EOs as a model for such molecular and developmental processes because of the repeated evolution of EOs, particularly among gymnotiforms and mormyrids, which exhibit remarkably similar EOs.
Several studies over the past two decades have contributed to our understanding of gene expression and development in gymnotiform EOs, in particular with regard to the role of innervation (Cuellar et al., 2006; Kim et al., 2008; Kim et al., 2009; Kim et al., 2004; Unguez and Zakon, 1998a; Unguez and Zakon, 1998b; Zakon and Unguez, 1999; Patterson and Zakon, 1996; Patterson and Zakon, 1997). Comparative studies between gymnotiforms and mormyrids, however, are presently limited because no studies have described gene expression in the mormyrid EO, which is the focus of the present study.
Study design
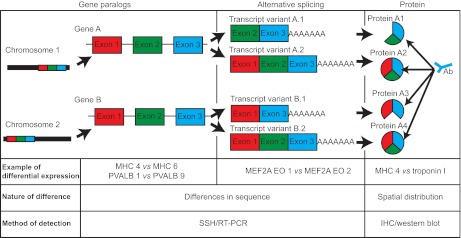
Considering the close developmental relationship between SM and EO, as well as the focus of previous studies in gymnotiforms on changes in gene expression between the two tissues, we were motivated to design a study that would simultaneously identify and determine various patterns of differential gene expression between SM and EO tissues in mormyrids (see Gallant, 2011). We expected that important differences between tissues might be due to spatial distribution (e.g. Taffarel et al., 1989), differences between the tissues expressing paralogous genes (e.g. Zakon et al., 2006) or differential expression of alternatively spliced mRNA transcripts, though no previous studies have examined such differences in EO or SM tissues.
To detect each of these patterns of differential expression, we utilized several strategies. First, we took the approach suppressive subtractive hybridization (SSH), a simple PCR-based method that selectively enriches for differentially expressed mRNAs, to identify unique patterns of gene expression in the EO and SM without a priori assumptions regarding their identities, and confirmed their differential expression using reverse transcription PCR (RT-PCR). We expected this approach to detect differences in the expression of gene paralogs, as well as differences in the expression of alternatively spliced transcripts (Fig. 1). Next, we examined the localization of protein products in SM and EO using immunohistochemistry. Because the antibodies used recognized highly conserved epitopes across a wide variety of species, the antibodies were not selective for particular paralogs or splice variants detected during SSH and RT-PCR (Fig. 1). Instead, this approach permitted our examination of SM and EO tissues for evidence of differences in spatial distribution of particular proteins in the EO. Finally, we examined the differential expression of alternatively spliced transcripts of a myogenic regulatory factor (detected above using SSH), myocyte enhancer factor 2A (MEF2A), using RT-PCR.
Fig. 1.
Study design. We examined three potential patterns of differential expression between electric organ (EO) and skeletal muscle (SM) in this study: differential expression of gene paralogs, differential expression of alternatively spliced mRNA transcripts and differential spatial expression of proteins. The first two differences were analyzed using a combination of suppressive subtractive hybridization (SSH) and reverse transcription PCR (RT-PCR) (see Materials and methods), whereas spatial distribution of proteins was analyzed using widely cross-reactive antibodies that did not recognize specific paralogs or alternative splice variants (see Materials and methods). We list examples of each pattern we detected using these methods, which are explained in more detail in the Results.
We then compare our results with previous findings in gymnotiforms, which indicates important differences in the developmental mechanisms underlying EO development. We conclude by briefly considering avenues for future comparative work between mormyrids and gymnotiforms.
MATERIALS AND METHODS
RNA isolation, subtractive hybridization and expressed sequence tag (EST) identification
All specimens used were adult Brienomyrus brachyistius [Osteoglossiformes: Mormyridae (Gill, 1862)]. Fish were housed in groups, and kept on a 12 h light:12 h dark cycle and fed live blackworms daily. All methods described are in accordance with Cornell University Animal Research and Education Committee policies. SM and EO tissues were collected from 10 adult B. brachyistius EOs. EOs were dissected by removal of the skin and muscle from the caudal peduncle, excision of the EO and spinal column, and finally removal of the spinal cord by inserting a fine pin into the vertebral column. Trunk skeletal muscle was dissected from the same 10 individuals (∼2×1×0.5 cm, caudal to operculum, dorsal to lateral line; skin removed). Tissue was immediately frozen in liquid nitrogen, then pulverized using pestle and mortar, and total RNA was extracted using Trizol solution (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. Extracted RNA was resuspended in RNase-free water (treated with diethylpyrocarbonate then autoclaved), and then stored at –80°C in aliquots until use.
Total RNA was PolyA+ purified using a FastTrack MAG mRNA isolation kit (Invitrogen). PolyA+ RNA was reverse transcribed to cDNA using SuperScript III RT (Invitrogen) with oligo dT priming. SSH was then performed following the method of Rebrikov (Rebrikov, 2003), using EO as the tester and SM as the driver (EO library). A second library was also constructed using SM as the tester with EO as the driver (SM library). This SSH protocol resulted in two pools of PCR products representing mRNAs enriched in EO and SM, respectively.
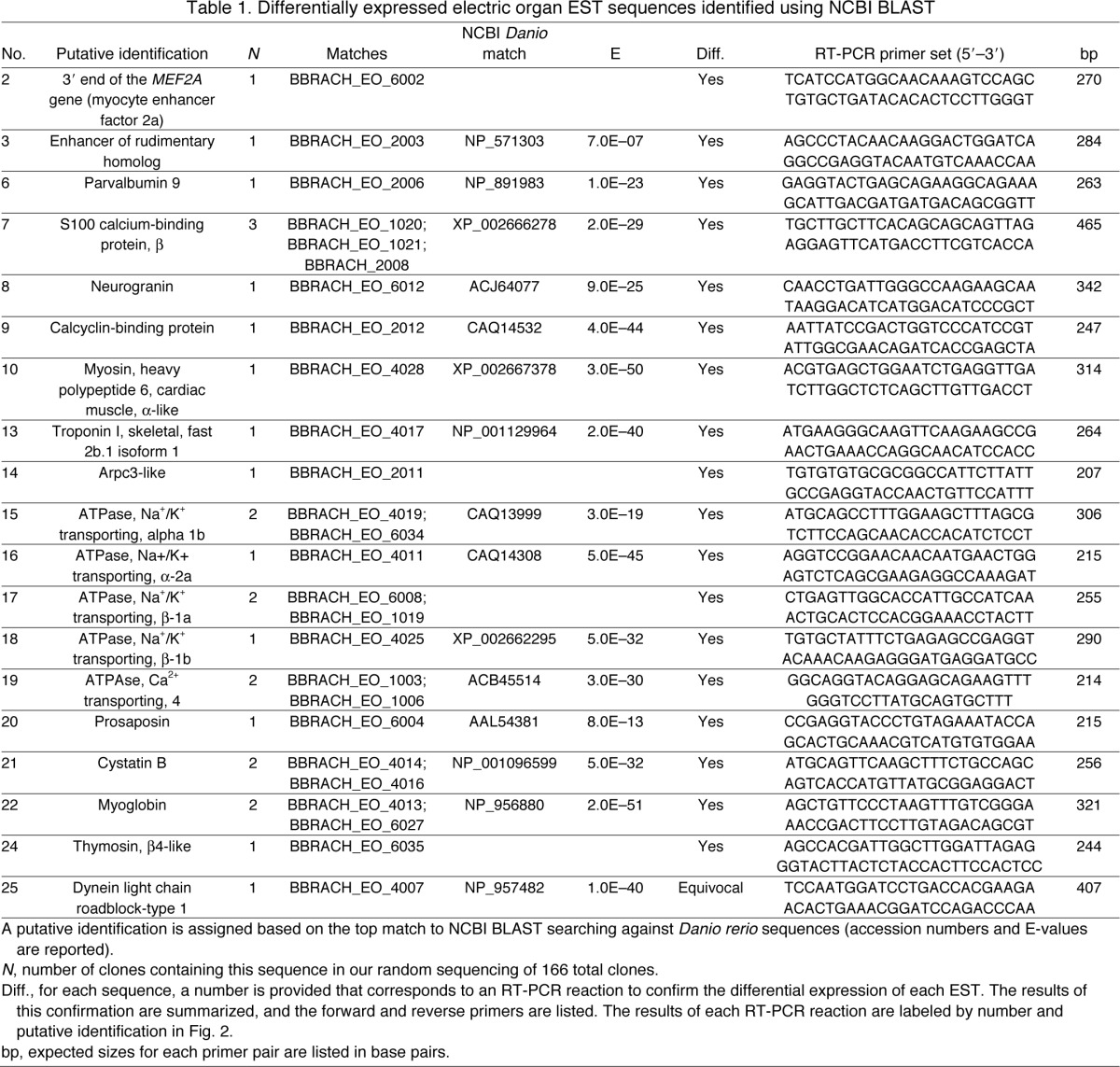
PCR products from EO and SM libraries were subsequently subcloned into chemically competent E. coli using the TOPO TA cloning system (Invitrogen), resulting in many hundreds of colonies for each library. We randomly selected 130 colonies from the EO library and 36 colonies from the SM library for sequencing. These colonies were grown overnight in LB broth containing 50 μg ml–1 ampicillin, and DNA was isolated and purified using a PureLink Miniprep kit (Invitrogen). Isolated and purified DNA was then submitted for Sanger sequencing via Cornell Core Laboratory Services, and the resulting ESTs were assigned unique numbers. EST sequences were edited automatically to remove contaminating cloning vector sequence, and then identified by performing searches against entries for Danio rerio sequences in the NCBI nr database with BLASTx and BLASTn search strategies using default parameters: Matrix BLOSUM62; gap opening cost 11; gap extension cost 1; no low-complexity filtering. For DNA sequences: match reward 1, mismatch penalty –3, non-affine gapping costs wordsize 28. The E-value threshold for matches was set to 1E–6. Sequences with significant matches to database entries are listed in Tables 1 and 2. All edited sequences were deposited in the NCBI dbEST database (Boguski et al., 1993), and are listed with their accession numbers in supplementary material Table S1.
Table 1.
Differentially expressed electric organ EST sequences identified using NCBI BLAST
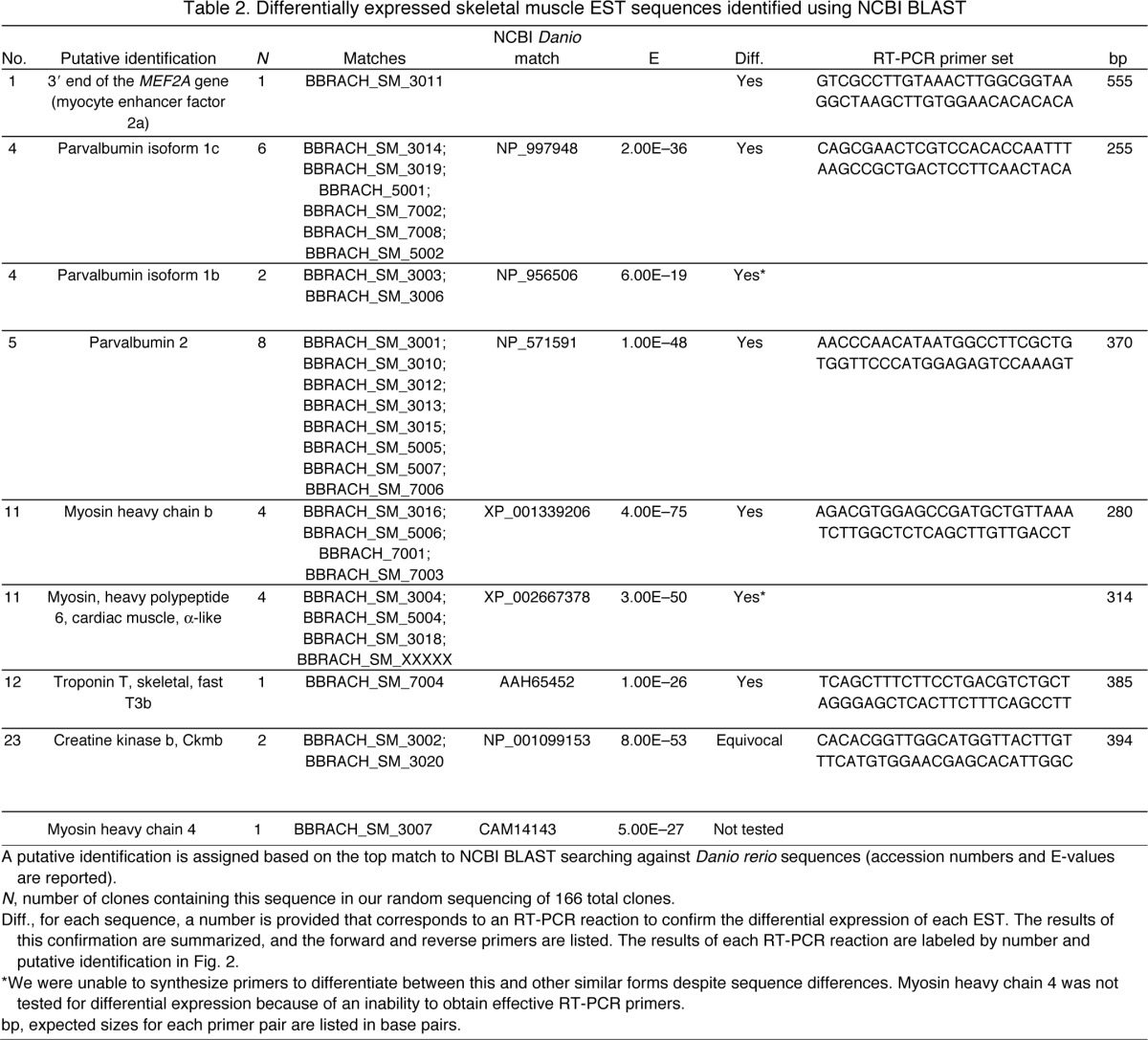
Table 2.
Differentially expressed skeletal muscle EST sequences identified using NCBI BLAST
RT-PCR
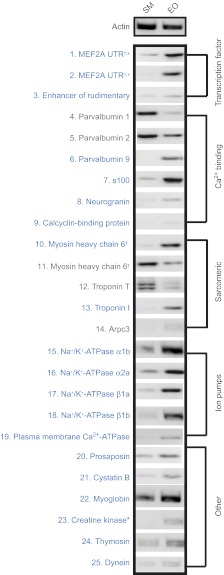
To independently confirm differential expression of identified ESTs (Tables 1, 2), primers were designed for RT-PCR. RT-PCR reactions were performed using the RT-PCR OneStep method (Invitrogen) on total RNA isolated from either EO or SM (isolated as above). Thermal cycling conditions for all primer pairs were 55°C (30 min), 94°C (1 min) followed by 25 cycles of 94°C (15 s), 60°C (30 s), 68°C (1 min) and a final extension step of 68°C (5 min). PCR products were resolved on a 1% agarose gel containing 1 μg ml–1 ethidium bromide. The results of each RT-PCR reaction are shown as negative gel images in Fig. 2.
Fig. 2.
RT-PCR of ESTs identified via SSH. Negative gel images of 25 RT-PCR reactions performed confirm differential expression of ESTs detected by SSH (see Tables 1, 2; supplementary material Table S1). For each RT-PCR reaction, the number listed corresponds to primers and sequence information listed in Table 1 or Table 2. RT-PCR products are grouped by putative cell functions (explained in Materials and methods). Sequences labeled in blue are upregulated in EO, sequences in gray are upregulated in SM when compared to actin control. †ESTs that matched different regions of the same Danio rerio sequence during BLAST searching (see Materials and methods). *ESTs that SSH revealed as differentially expressed in one library that were later found via RT-PCR to be differentially expressed in the opposite library. All PCR products matched expected sizes listed in Tables 1 and 2.
Immunohistochemistry
SM and EO tissues were obtained from five additional B. brachyistius as described above. For each specimen, the caudal peduncle or ∼1 cm of axial muscle posterior to the operculum and dorsal to the lateral line was removed and flash frozen in liquid nitrogen-chilled isopentane (Cuellar et al., 2006), then mounted on a dry ice-chilled chuck using Cryo-M-Bed (Bright Instrument Company, Huntingdon, UK). Tissues were cut at 20 μm using a cabinet cryostat onto freshly subbed slides and dried overnight at room temperature. Sections were ringed with PAP pens and washed 3 times using a 1× PBS wash buffer containing 0.3% Triton-X 100 (PBST). Sections were blocked in 5% goat serum at 4°C for 1 h, then washed 3 times for 30 s, and then 3 times for 10 min, replacing the solution each time. Sections were incubated with primary antibodies (Table 3) diluted in wash buffer containing 5% normal goat serum for 1 h at room temperature, in a humidified box. Sections were again washed as above, and then incubated with Alexa-488-conjugated secondary antibodies (Invitrogen) diluted 1:200 in wash buffer containing 5% normal goat serum for 1 h at room temperature in a humidified box. Sections were then washed as above and shaken dry. Slides were mounted and coverslipped using VectaShield (Vector Laboratories, Burlingame, CA, USA) and stored in the dark at –20°C until analyzed. Slides were imaged on a Nikon Eclipse E600FN microscope equipped with a Diagnostic Instruments Camera and Spot 2.2 software (Diagnostic Instruments, Sterling Heights, MI, USA). Fluorescence images were captured using the Nikon FITC-HYQ Filter Set (460–500 nm excitation, 500 nm dichromatic mirror and 510–560 nm barrier filter). A digital interference contrast (DIC) and fluorescence image were captured for each field of view. The two images were overlaid using the ‘Cover Overlay’ function in Photoshop (Adobe Systems, Macintosh version 10.0.1). Brightness and contrast were adjusted in the software to compensate for exposure differences.
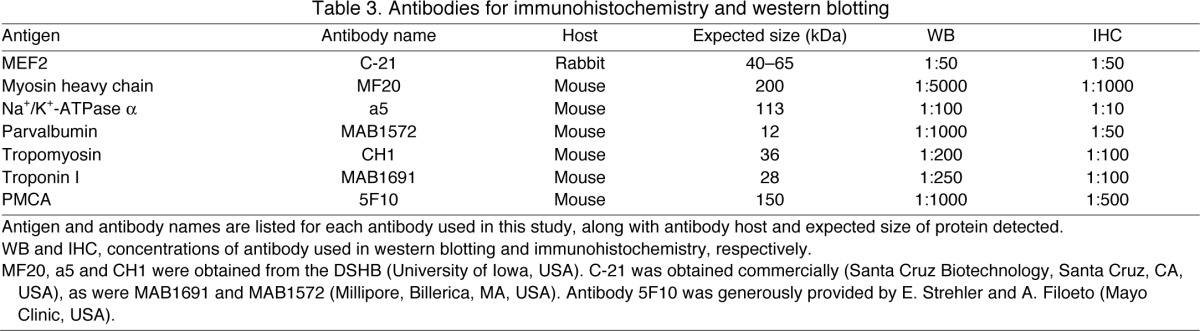
Table 3.
Antibodies for immunohistochemistry and western blotting
Western blotting
SM and EO tissues were removed from a single B. brachyistius as above. Each tissue was frozen in liquid nitrogen, and then pulverized using a dry ice-chilled pestle and mortar. Tissues were then transferred to a glass homogenizer, and protein extractions were performed using a Total Protein Extraction Kit (Millipore, Billerica, MA, USA). Protein concentrations were quantified using a Bradford assay kit (BioRad Laboratories, Hercules, CA, USA). Protein samples were aliquoted and stored at –80°C until use. Protein samples (10 μg) were combined 1:1 with Lammeli buffer (BioRad Laboratories) containing 5% β-mercaptoethanol and boiled at 95°C for 3–5 min then directly applied to 8–15% gradient gels (BioRad Laboratories) with the exception of preparations for Na+/K+-ATPase and plasma membrane Ca2+-ATPase proteins, which were applied to gels without boiling. EO protein (30 μg) was loaded for western blotting of parvalbumin. Gels were run at 180 V until Bromothymol Blue reached the end of the gel. Polyvinylidene fluoride (PVDF) membranes were wetted for 15 s in methanol, and then submerged in ddH2O for 2 min. Proteins were transferred to a PVDF membrane using a Mini PROTEAN3 System (BioRad Laboratories) for 1.5 h at 100 V in a cold room under constant circulation to prevent overheating, except parvalbumin, which because of its small size was transferred for only 45 min at 100 V. Following transfer, PVDF membranes were blocked in 5% non-fat dried milk for 1 h at room temperature on an orbital shaker, then rinsed three times in PBST briefly. Primary antibody solution (5 ml) diluted in PBST containing 5% bovine serum albumin (BSA) was then directly applied the PVDF membrane, at room temperature with constant shaking for 1 h. Membranes were then rinsed quickly three times in a large volume of PBST, followed by three 10 min washes in PBST. HRP-conjugated secondary antibodies (see Table 3 for concentrations) diluted in PBST containing 5% BSA were then applied to the membrane and incubated for 1 h at room temperature. Membranes were again rinsed quickly 3 times in a large volume of PBST, followed by three 10 min washes in PBST. Membranes were then treated with Amersham ECL reagent (GE Healthcare, Hatfield, UK), then exposed to film for 1–10 min.
MEF2 cloning
A 3′ UTR of the transcription factor MEF2A was differentially expressed in EO tissues (Table 1, Fig. 1). In fishes and other vertebrates, there are four known MEF2 genes, designated MEF2a–d. We attempted to obtain coding sequence of all MEF2 mRNAs expressed in EO and SM. Degenerate primers were developed to conserved sequences in fishes and other vertebrates for exons 1 (encoding amino acid sequence MGRKKIQI) and exon 6 (RKPDLRV) of MEF2a, c and d. The forward primer was 5′-CCGAATTCATGGGRMGGAARAAGATWCAGATCA-3′ and the reverse primer was 5′-CCAAGCTACTCTCARGTCTGGCTTGCG-3′. Additional degenerate primers were generated for conserved residues in the more divergent MEF2b gene (EMQLKVK and KTDMQSWEEQSQGA). Degenerate primers for MEF2B were 5′-GAATTCGARATGCARYTSAAAGTVARA-3′ (forward) and 5′-AAGCTCTCTGRTCCTCCCAGCTCTGCATG-3′ (reverse). mRNA from EO and SM was reverse transcribed using an oligodT primer and the Superscript III reverse transcriptase protocol. The degenerate primer sets were used to amplify MEF2 cDNAs from SM or EO cDNA using the FailSafe PCR system. Cycling conditions for PCR were 94°C (3 min) followed by five cycles of 95°C (45 s), 50°C (45 s), 72°C (3min) and 35 cycles of 95°C (45 s), 55°C (45 s), 72°C (3 min), followed by a final extension step of 72°C for 10 min.
The resulting amplified cDNAs were subcloned using the TOPO TA vector as described above. Ten colonies from SM and 10 colonies from EO were selected for sequencing using both the T7 forward and T3 reverse primers of the TOPO PCR2.1 vector. These sequences were edited by visual inspection, and then aligned using CLC Sequence Viewer (CLC Bio, Aarhus, Denmark) to generate consensus sequences. Each MEF2 consensus sequence was then identified by BLASTn and BLASTx alignment to published D. rerio MEF2 sequences. The edited consensus sequences were then deposited in the NCBI GenBank database. Expression differences for each detected transcript of MEF2A were further evaluated between SM and EO by performing RT-PCR using primers generated to sequences conserved in all MEF2A transcripts detected in SM and EO The forward primer was 5′-AGTACAACGAGCCACATGAGAGCA-3′ and the reverse primer was 5′-TTGACAAAGCCGTTTCCTGCACTG-3′. In addition, we determined the relative expression of two alternatively spliced forms that we detected in EO tissue, designated EO1 and EO2, by using RT-PCR. The EO1-specific forward primer was 5′-AGCCCTGAACCCGATGACTGTTT-3′ and the EO2-specific forward primer was 5′-CCAGACGCCTCCTATGTCCTCA-3′. Both reactions used the reverse primer described above. Thermal cycling conditions for all reactions (including actin control) were 94°C (30 s) followed by 27 cycles of 94°C (30 s), 55°C (30 s), 68°C (1 min). PCR products were resolved on a 2% agarose gel containing 1 μg ml–1 ethidium bromide.
RESULTS
Identification of differentially expressed transcripts between EO and SM
We obtained cDNA sequences from 152 of 166 selected clones after SSH. Of these sequences, 117 were from EO and 35 were from SM. The average length was 577 bp for all sequences. The cDNA sequence of each clone was given a unique identifier and submitted to dbEST maintained by the NCBI. The individual accession numbers for all sequences (HO702384–HO702394; HO702396–HO702414; HO702416–HO702420; HO702424–HO702463; HO702465–HO702542) are included in supplementary material Table 1. Of these sequences no D. rerio matches were identified for 88 clones from EO and nine clones from SM using either BLASTn or BLASTx search strategies, likely due to 3′ UTR bias associated with the reverse transcription priming strategy (Brooks et al., 1995). However, we were able to putatively identify 28 differentially expressed sequences: 19 were enriched in the EO library and nine were enriched in the SM library. The matches of these identified EST sequences to D. rerio entries in the NCBI databases are summarized for EO in Table 1 and SM in Table 2.
To confirm differential expression of these identifiable ESTs, we performed RT-PCR using primers generated from each EST sequence (Tables 1, 2). The results of each RT-PCR reaction, and the forward and reverse primers used are listed for each EST in Tables 1 and 2. Additionally, gel images of each RT-PCR reaction are presented in Fig. 2. For the purposes of later discussion, we assigned putative functional roles based on the known function of these genes in skeletal muscle: transcription factor, Ca2+ binding, sarcomeric proteins, ion pump and other (Fig. 2). ESTs that were upregulated in EO are shown in blue, ESTs upregulated in SM are shown in grey.
Using the combination of a SSH strategy and RT-PCR, we are able to demonstrate the differential expression of ESTs with different putative roles in the EO. We detected the differential expression of two ESTs that matched D. rerio genes encoding transcription factors (MEF2A and enhancer of rudimentary homolog, ERH), six genes encoding Ca2+-binding proteins (parvalbumins 1, 2, 9; S100; neurogranin; calcyclin), four ESTs that matched D. rerio genes encoding sarcomeric proteins (myosin heavy chain MHC, troponin T, troponin I and arpc3), and five ESTs that matched D. rerio genes encoding ion pump proteins (Na+/K+-ATPase α-1b, α-2a and Na+/K+-ATPase β-1a, β-1b, and plasma membrane Ca2+-ATPase 4). We also identified additional ESTs encoding prosaposin, cystatin B, myoglobin, creatine kinase, thymosin and dynein.
Spatial distribution of proteins is similar but not identical between SM and EO
Cuellar and colleagues described the existence of several mRNAs present in the EO of gymnotiforms (Cuellar et al., 2006), but their corresponding proteins were not found using either immunohistochemistry or western blotting, suggesting the existence of a post-transcriptional mechanism of regulating gene expression. Given the similar identities of the mRNAs detected by Cuellar and colleagues (Cuellar et al., 2006) and the differentially expressed ESTs described in this study, we wished to determine whether protein products like those differentially expressed ESTs described above were translated in mormyrid EO. We determined protein presence and distribution in the mormyrid EO and SM using widely cross-reactive antibodies for a subset of seven proteins: MHC, Na+/K+-ATPase, plasma membrane Ca2+-ATPase, MEF2, troponin I, parvalbumin and tropomyosin (Table 3).
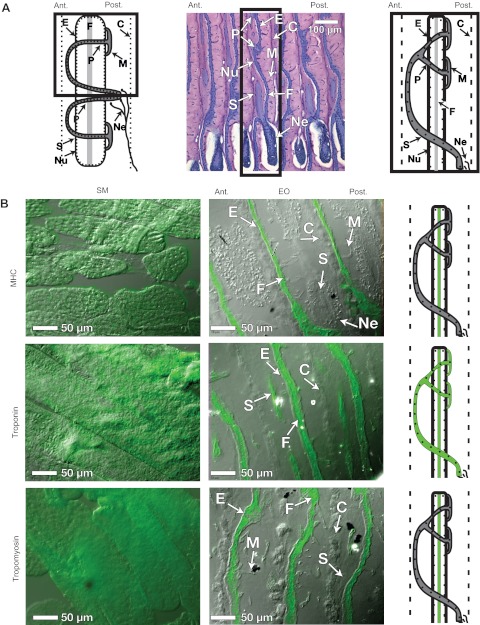
Because of its unique histology, we digress briefly to summarize the major anatomical features of the mormyrid EO. For a more detailed and thorough review of EOs in a comparative context, we refer the reader to the excellent reviews of Bass and Bennett (Bass, 1986; Bennett, 1971). Shown in Fig. 3A are illustrations to orient the reader to the major anatomical features of the EO. The electrocytes of B. brachyistius are doubly penetrating with posterior innervation (Dpp) (see Sullivan et al., 2000; Hopkins, 1999a). The 7 μm sagittal section in the center of Fig. 3A was made from a plastic-embedded specimen and stained with Toluidine Blue (see Gallant et al., 2011). After Toluidine Blue staining, each electrocyte appears as a blue stripe surrounded by loose, pink-stained connective tissue, and is separated from neighboring electrocytes by a connective tissue septum on the anterior and posterior side. Each electrocyte face contains multiple nuclei, and the center of the electrocyte contains filamentous material between the anterior and posterior membranes, which has been shown in greater detail using electron microscopy (Bass et al., 1986). Based on these micrographs, Bass and colleagues concluded that this was a disorganized layer of myofilaments, rather than the typical parallel arrangement of myofilaments found in SM (Bass et al., 1986). Another key feature of the mormyrid electrocyte is an elaborate stalk system: microstalklets emerge from the posterior face of each electrocyte, fuse and pass through penetrations to the anterior side. Further fusion into a larger diameter stalk occurs on the anterior side. The stalk passes briefly posterior once more, where it is innervated.
Fig. 3.
Anatomy of EO and sarcomeric protein localization in EO and SM. (A) A brief overview of the major anatomical features of the electric organ. Left, diagram of a single Brienomyrus brachyistius electrocyte, which have doubly penetrating stalks with posterior innervation (see Sullivan et al., 2000; Hopkins, 1999a). Center, a single 7 μm sagittal section made from a plastic-embedded specimen, stained with Toluidine Blue (see Gallant et al., 2011). Right, illustration of a single electrocyte as it appears in the Toluidine Blue-stained EO, and in B. Boxes indicate comparable areas in each of the illustrations. (B) Immunohistochemistry was performed using primary antibodies for three sarcomeric proteins: myosin heavy chain (MHC), troponin I and tropomyosin, which are listed in Table 3. The images presented in this figure are overlays of DIC and fluorescence images (see Materials and methods). Illustrations are provided to summarize the localization of each protein. For all images, anterior is left, posterior is right. Abbreviations: E, electrocytes; C, connective tissue septa; Nu, nuclei; F, myofilament material between the anterior and posterior membranes; M, microstalklets; P, penetrations; S, stalk; Ne, motor neuron.
We tested for three sarcomeric proteins found in our SSH screen using immunohistochemistry with antibodies listed in Table 3 to identify the composition of the central filamentous material found in the electrocyte. As expected, we found that MHC (Fig. 3B) was uniformly distributed across SM muscle fibers. In the EO, expression of MHC was confined to a single vertical strip between the anterior and posterior membranes of the electrocyte. We detected no MHC expression in the stalk system. Tropomyosin (Fig. 3B) staining was also widespread in SM tissues, but it was restricted to a center filament between the anterior and posterior faces of the electrocyte, like that of MHC. No staining was observed in stalk materials. Troponin I (Fig. 3B) staining was abundant in myofibrils of the SM, and was distributed similarly to MHC in both EO and SM, running in a longitudinal filament between the anterior and posterior faces. Contrasting with MHC, troponin I is also clearly visible in the stalk system.
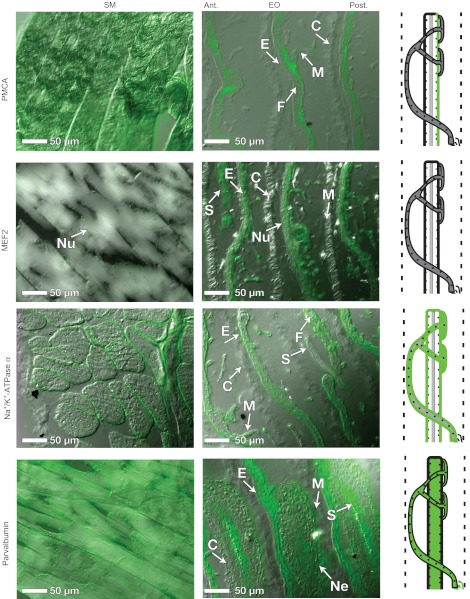
We also performed immunohistochemistry to localize additional proteins like those identified in our SSH screen. Plasma membrane Ca2+-ATPase (Fig. 4) staining was visible in the sarcolemma of SM, but in EO staining was present in both electrocyte faces and the stalk system. MEF2 (Fig. 4) staining was localized to nuclei in both SM and EO tissues. An increased number of larger, more spherical nuclei (vs SM) are visible in both the stalks and electrocytes in the EO. Na+/K+-ATPase α (Fig. 4) staining was localized to the sarcoplasm in SM and to the anterior and posterior faces of the electrocyte. Again, no staining of the stalk system was visible. Parvalbumin staining (Fig. 4) is abundant in SM cytoplasm, as well as in EO electroplasm, stalks, and in motor neurons innervating the electrocyte.
Fig. 4.
Protein localization in EO and SM. Immunohistochemistry was performed using primary antibodies for four additional proteins: plasma membrane Ca2+-ATPase (PMCA), myocyte enhancing factor 2 (MEF2), the α subunit of the Na+/K+-ATPase and parvalbumin, which are listed in Table 3. The images presented in this figure are overlays of DIC and fluorescence images (see Materials and methods). Illustrations are provided to summarize the localization of each protein. For all images, anterior is left, posterior is right. Abbreviations: E, electrocytes; C, connective tissue septa; Nu, nuclei; F, myofilament material between the anterior and posterior membranes; M, microstalklets; P, penetrations; S, stalk; Ne, motor neuron. See Fig. 3 for an overview of anatomical features in the EO.
To summarize, we found evidence that proteins like the seven differentially expressed ESTs described above (MHC, Na+/K+-ATPase, plasma membrane Ca2+-ATPase, MEF2, troponin I, tropomyosin and parvalbumin) are translated in both EO and SM using immunohistochemistry (Figs 3, 4). We emphasize that the antibodies utilized (Table 3) recognized highly conserved epitopes across a wide variety of species, and therefore were not selective for specific paralogs detected during SSH and RT-PCR above, and were not expected to exhibit a differential presence in SM and EO. Our results therefore should be viewed as demonstrating the presence of all seven proteins in both EO and SM, as well as their spatial distribution in muscle fibers vs electrocytes. The spatial distribution was generally quite similar (i.e. ion pumps were localized to plasma membrane, transcription factors in nuclei, etc.), with some notable exceptions in the distribution of proteins involved in SM contraction (i.e. MHC, troponin I and tropomyosin).
Western blotting reveals differences in abundance of some proteins between SM and EO
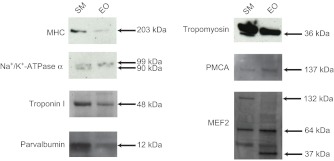
Western blots were also performed to confirm that the selected antibodies recognized proteins of the expected size. In addition, western blots allowed some insight into the relative amounts of protein between tissues, assuming the epitope was equally well recognized in both tissue types. Overall, western blotting results (Fig. 5) indicate that proteins detected using immunohistochemistry (Figs 3, 4) recognized proteins of expected sizes (Table 1).
Fig. 5.
Western blots of proteins from EO and SM. For each of seven proteins, western blotting was also performed using 10 μg SM and EO protein extracts (see Materials and methods). For parvalbumin, 3x the protein was loaded for EO than for SM. Size markers are listed to the right; expected sizes for each antibody are listed in Table 3 along with primary antibody concentrations.
For all western blots, 10 μg of protein homogenate was loaded for both SM and EO, except for parvalbumin, where 30 μg of EO homogenate was required to detect a band. Assuming that the antibodies recognized the epitopes with the same affinity, this suggests a 3- to 4-fold reduction in the concentration of parvalbumin in the EO. Comparing the intensity of bands between tissues, MHC, parvalbumin and tropomyosin were also more concentrated in SM than in EO, whereas Na+/K+-ATPase α, plasma membrane Ca2+-ATPase and MEF2 seemed to be in approximately equal concentrations in the two tissues.
Finally, we note slight differences in the sizes of protein bands between Na+/K+-ATPase α and MEF2A. One band detected in SM for Na+/K+-ATPase α (90 kDa) was not present in EO, and a second band (99 kDa) was present in both EO and SM. Western blotting for MEF2 protein indicated the presence of a large (132 kDa) form in SM that was absent in EO, and a smaller form (37 kDa) that was absent in SM.
MEF2A is differentially expressed in EO vs SM
MEF2 is an important transcription factor regulating the development of muscle fibers (Black and Olson, 1998). In vertebrates, there are four MEF2 genes, designated MEF2a–d. The detection of differentially expressed, conserved 3′ UTRs of the MEF2A transcription factor (Fig. 2), and evidence of potentially different MEF2 proteins from western blotting (Fig. 5) prompted us to clone the coding sequence of transcribed MEF2 genes to determine whether this important transcription factor is differentially expressed between SM and EO.
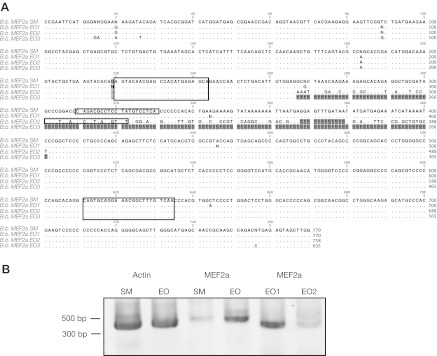
We successfully cloned transcripts of MEF2 genes in EO and SM: based upon alignment to NCBI D. rerio sequences, we determined that these sequences matched MEF2A (Fig. 6) and MEF2C (three transcripts, data not shown). We were unable to detect MEF2B or MEF2D expression in SM or EO. Sequencing revealed three different MEF2A transcripts: one unique MEF2A transcript was expressed in SM (Fig. 6A; B.b. MEF2a SM1) and three unique MEF2A transcripts were expressed in EO (Fig. 6A; B.b. MEF2a EO1, 2 and 3). One of the forms (B.b. MEF2a 3) lacked an approximately 100 bp region, while B.b. MEF2a EO1 and B.b. MEF2a EO2 contained this region, but had divergent sequences over the region between 267 and 401 bp. This region corresponds to exon 5 of the D. rerio MEF2a gene. All MEF2 sequences detected have been uploaded to NCBI GenBank with the accession numbers JN107727–JN107733. To determine the relative abundance of the three MEF2A alternative transcripts in EO, RT-PCR was performed using primers generated to conserved the sequence of all MEF2A transcripts sequenced (primer locations shown Fig. 6A by large boxes). In addition, we used RT-PCR to quantify the relative abundance of the two alternative transcripts B.b. MEF2a EO1 and B.b. MEF2a EO2 in EO tissue (primer locations shown Fig. 6A by small boxes).
Fig. 6.
Differential expression of MEF2A. The discovery of differentially expressed MEF2A 3′ UTRs in EO prompted further subcloning and sequencing of all MEF2 genes expressed in EO and SM (see Materials and methods). (A) Subcloning revealed multiple sequences of MEF2a expressed in SM (B.b. MEF2a SM1) and EO (B.b. MEF2a EO1–3). These sequences were largely similar except for a region from 264–401 bp that was variable, corresponding to exon 5 of MEF2a in D. rerio. One of the transcripts (B.b. MEF2a EO3) completely lacked this region (gray squares indicate ‘missing’ bases). Using MEF2a-specific primers (large boxed regions), we amplified all MEF2A transcripts expressed in EO to determine which of these sequence variants were expressed in EO and SM. In addition, we developed primers to amplify alternative transcript-specific forms (small boxed regions). (B) RT-PCR comparing actin and MEF2A expression reveals the upregulation of MEF2A in EO. Transcript-specific RT-PCR is consistent with B.b. MEF2a EO1 expressed in high abundance and EO2 expressed in low abundance in EO. Using the above primers, 27 cycles failed to produce an EO3-sized splice variant.
A band at ∼400 bp (Fig. 6B) is consistent with B.b. MEF2a EO1 (expected size 414 bp) and B.b. MEF2a EO2 (expected size 402 bp), rather than B.b. MEF2a EO3 (expected size 280 bp). Using the same cycling parameters, we detected a very faint band at this expected size in SM (Fig. 6B), indicating that MEF2a expression is higher in SM than in EO. Our examination of the alternative transcripts B.b. MEF2a EO1 and B.b. MEF2a EO2 in EO revealed that the B.b. MEF2a EO1 form was more abundantly expressed than the B.b. MEF2a EO2 type in EO tissues; however, B.b. MEF2a EO2 was still detectable. To verify the specificity of these reactions, the final PCR products from both transcript-specific reactions were sequenced.
DISCUSSION
SSH results
In this study, we have shown evidence for the differential expression of 28 genes between mormyrid EO and SM. We suspect that these 28 genes represent only a fraction of the total number of differentially expressed genes between SM and EO. We base this conclusion on the fact that (1) roughly the same proportion of new differentially expressed ESTs were found in each batch of randomly selected clones submitted for sequencing, and (2) most transcripts (particularly those in the EO) were represented only once.
Our strategy to identify these differentially expressed genes utilized a novel approach as applied to EO gene discovery, SSH. Previous approaches to characterizing the molecular differences between SM and EO in gymnotiforms have largely relied on a priori assumptions regarding the identities of specific genes that might be differentially expressed (Patterson and Zakon, 1996; Patterson and Zakon, 1997; Cuellar et al., 2006; Kim et al., 2008; Kim et al., 2009; Kim et al., 2004; Unguez and Zakon, 1998a; Unguez and Zakon, 1998b; Zakon and Unguez, 1999). Unlike these prior approaches, our use of the SSH technique allows for the construction of a dataset for which fewer assumptions have been made. As such, our dataset contains evidence of differential expression in closely related genes (e.g. parvalbumin and myosin paralogs expressed in SM and EO), which may not have been detected using a candidate gene approach.
What types of genes are differentially expressed between EO and SM?
Below, we consider some of the differentially expressed genes identified in B. brachyistius in the context of what is known about their function in SM and EO tissues in other vertebrates. We conclude by briefly considering these data in terms of the growing molecular and developmental data that are becoming available for weakly electric fish.
Transcription factors
Our SSH approach revealed two differentially expressed transcription factors in the adult EO: ERH, and the 3′ UTR of the MEF2a gene. ERH is a highly conserved gene among vertebrates (Gelsthorpe et al., 1997; Pogge von Strandmann et al., 2001). Studies have implicated its function in transcriptional regulation for pyridine biosynthesis (Wojcik et al., 1994) and cell cycle repression (Gelsthorpe et al., 1997). More recently, ERH was found to directly interact with a DNA replication factor (Lukasik et al., 2008; however, little is ultimately known about the role of this gene in development.
Following the early specification of muscle progenitors, the combined actions of the muscle regulatory factors (MRFs) MyoD and MEF2 are responsible for the transcriptional activation of muscle-specific genes (Black and Olson, 1998). Indeed, MEF2a has been demonstrated to be crucial in the development of posterior somites in fish, from which EOs originate in mormyrids (Denizot et al., 1982). Knockdown of MEF2a in D. rerio induces apoptosis of posterior somites during development (Wang et al., 2006), whereas MEF2c/d controls thick filament assembly (Hinits and Hughes, 2007). Following muscle development, MEF2s continue to interact with signaling pathways (e.g. MAPK and Ca2+ signaling) to regulate gene expression in response to changes in electrical activity (Black and Olson, 1998). Differential expression of these transcription factors may therefore provide an attractive hypothesis to explain physiological and morphological differences between EOs and skeletal muscle. A series of studies (Kim et al., 2004: Kim et al., 2008; Kim et al., 2009) examined the transcriptional patterns and roles of several MRFs and their co-factors including MyoD, Myogenin, MYF5, MRF4, MEF2C, ID1 and ID2 in the gymnotiform Sternopygus macurus. These studies found little difference in expression levels between SM and EO, and that several of the myogenic regulatory factors cloned from S. macurus retain their ability to induce normal muscle development in mouse cultured myoblasts (Kim et al., 2008; Kim et al., 2009).
We found no evidence of differential expression of any of these previously analyzed MRF genes in mormyrids, but did detect differential expression of the 3′ UTR of the MEF2a gene in our subtractive hybridization screen. We were able to secondarily verify this using RT-PCR, indicating that both UTRs were upregulated in EO, and one appeared to be exclusively expressed in EO (Fig. 2, Table 1).
Our immunohistochemical and western blotting utilized the C-21 antibody, which was developed to the C-terminus of the human MEF2A protein, and recognizes primarily the MEF2A and, to a lesser extent, MEF2C and D (product description, Santa Cruz Biotechnology, Santa Cruz, CA, USA). This antibody permitted the detection of proteins in western blot that matched the expected sizes (Table 3, 40–65 kDa) of D. rerio MEF2A protein forms, with an additional large fragment at 132 kDa that could not be identified. Immunohistochemistry revealed the expected localization of this protein; namely, in the nuclei of both SM and EO tissues. While this antibody was not specific enough to delineate between different MEF2 proteins expressed, taken with our cloning results (discussed below), we are confident that the antibody detected the expression of MEF2A and MEF2C proteins.
We developed degenerate primers to clone a protein-coding portion of this gene from SM and EO. Utilizing this strategy, we determined that MEF2a and MEF2c genes are expressed in EO and SM, and were unable to detect the expression of MEF2b or MEF2d genes, which is consistent with the results of our immunohistochemical survey above.
We detected evidence of alternative splicing in MEF2A in B. brachyistius as well; we detected the existence of alternative splice variants of the MEF2a gene, which varied in their inclusion of a sequence homologous to the 5th exon of the D. rerio sequence of MEF2a (Fig. 6). As they are not full-length transcripts, we have denoted these forms numerically according to the tissue they are expressed in. B.b. MEF2a EO1 and B.b. MEF2a SM1 have identical sequences across the region cloned, whereas B.b. MEF2a EO2 has an alternative sequence in the region homologous to D. rerio exon 5, and B.b. MEF2a EO3 lacks this exon. Various RT-PCR reactions performed on these different alternatively spliced forms confirmed that EO tissues more abundantly express MEF2a overall. Additional RT-PCR reactions showed that EO tissues more abundantly express the EO1 form vs the EO2 form; however, although low in abundance, the EO2 form is detectable. We were unable to detect significant expression of the EO3 form in EO, despite our ability to clone it from EO tissue originally.
The discovery of multiple 3′ UTRs of the MEF2a gene, along with alternatively spliced MEF2a transcripts is of considerable interest to understanding differences between EO and SM tissues. First, as mentioned above, MEF2 genes are integral in the early stages of muscle differentiation, and the upregulation of multiple alternative transcripts of the MEF2 gene in EO tissue vs muscle suggests that this transcription factor may regulate important differences between tissues. Second, work by Black and colleagues (Black et al., 1997) demonstrated that the MEF2A 3′ UTR (which is highly conserved among vertebrates) acts as a cis-acting translational repressor, and may confer tissue-specific translational activity. Despite our efforts utilizing 3′ RACE and full-length cDNA cloning, we are presently unable to determine the association between these alternatively spliced forms and the UTR sequences detected using SSH, and plan to pursue this in a future study.
Na+/K+-ATPases
Given that EOs contain electrically excitable tissues, there has been a great deal of attention devoted to differentially expressed proteins involved in ion transport and permeability in EO vs SM (e.g. Zakon et al., 2006). In the present study, we did not detect any differentially expressed ion channels; however, we did detect differential expression of transcripts for Na+/K+-ATPase subunits α1 and α2, two Na+/K+-ATPase β-subunits, and a plasma membrane Ca2+-ATPase in EO when compared with SM (Fig. 2, Table 1; see discussion below). The differential expression of each of these was confirmed secondarily using RT-PCR, following the pattern of increased expression of all ESTs in EO vs SM tissue.
Na+/K+-ATPase proteins were detected with the antibody a5 (Table 3), the epitope of which is for the Na+/K+-ATPase α-subunit from chicken. Western blotting revealed that the relative amounts of Na+/K+ ATPase α protein in SM and EO were approximately equal (Fig. 5); however, two forms were detected in SM (99 and 90 kDa) whereas only one form (90 kDa) was recognized in EO. As expected, our immunohistochemical assay of Na+/K+-ATPase distribution indicates the localization of Na+/K+-ATPase to the plasma membrane of muscle fibers, as well as to electrocytes and their stalks. In both immunohistochemistry and western blotting, the abundance of Na+/K+-ATPase protein appears similar between SM and EO, whereas RT-PCR and SSH shows clear upregulation of specific Na+/K+-ATPase transcripts, suggesting that specific Na+/K+-ATPases may be differentially expressed between the two tissue types.
The functional consequences of differential expression of Na+/K+-ATPase subunits are presently unclear. Lowe and colleagues demonstrated that Na+/K+-ATPase subunits α1 and α2 are heterogeneously distributed between the innervated and non-innervated faces of the EO of the strongly electric gymnotiform E. electricus, using oubain sensitivity, western blotting and immunohistochemistry (Lowe et al., 2004). The two faces of E. electricus electrocytes are known to differ in terms of their specific resistance (Bennett, 1971), and therefore different α-subunits may be associated with changes in Na+ or K+ permeability between these two faces. Bell and colleagues noted similar differences in the membrane resistivity of the anterior and posterior faces of mormyrid electrocytes (Bell et al., 1976). Along these lines, because specializations for current production have led to a much lower specific resistance of electrocytes vs skeletal muscle (Bennett, 1971), we suspect that the differential expression of Na+/K+-ATPase is attributable to these specializations for current production; namely, increased ion permeability.
Intriguingly, the amount of Na+/K+-ATPase α-subunit protein did not appear to vary between SM and EO tissues (Figs 4, 5), which suggest that there may be differences between the efficiency/kinetic properties of the Na+/K+-ATPase pumps between SM and EO; specifically, EO pumps may be ‘faster’ than their SM counterparts. This is of interest given that ion pumps impose significant demands on the metabolic resources of excitable tissues (Clausen et al., 1991). Several have considered the metabolic costs of electrical signaling in both gymnotiforms (Markham et al., 2009; Salazar and Stoddard, 2008; Stoddard and Salazar, 2011) and mormyrids (Bell et al., 1976; Hopkins, 1999b), of which cation pump activity is likely to be a large component. A variety of physiological estimates have estimated that metabolic expenditure of Na+/K+-ATPases is ∼10% of the basal metabolic rate (BMR) at rest to 30–40% of the BMR during activity (Clausen et al., 1991). In gymnotiforms, diurnal fluctuation in EOD amplitude is mediated by hormonally regulated Na+ channel trafficking (Markham et al., 2009). Such fluctuation in Na+ currents in the EO would impose an estimated 30% increase in ATP consumed by electrocytes during a period of maximum EOD amplitude, as a result of Na+/K+-ATPase demand (Markham et al., 2009).
Ca2+-ATPase and Ca2+-binding proteins
We detected the upregulation of a plasma membrane Ca2+-ATPase (Table 1, Fig. 2), as well as several other proteins involved in Ca2+ binding, such as neurogranin, calcyclin-binding protein and S100. In contrast, parvalbumins were generally upregulated in SM vs EO tissue, with the exception of a transcript homologous to D. rerio parvalbumin 9. Each of these patterns of expression detected in SSH was secondarily verified using RT-PCR (Fig. 2).
Plasma membrane Ca2+-ATPase was detected using the 5F10 antibody (Table 3), which was developed to a highly conserved epitope of human PMCA4 protein between residues 719 and 738, which is conserved across all four known vertebrate plasma membrane Ca2+-ATPases (Caride, 1996), and has been demonstrated to be widely reactive across species (Caride, 1996). Western blotting suggests that plasma membrane Ca2+-ATPase concentration may be slightly increased in B. brachyistius EO compared with SM (Fig. 5). As expected, plasma membrane Ca2+-ATPase is localized to the sarcolemma in SM, and in the EO is localized only to the posterior (innervated) surface of the electrocyte and its stalk system.
Parvalbumin was detected using the MAB1572 antibody, which has an epitope against the first Ca2+-binding site in frog parvalbumins (product description, Millipore), though like the other antibodies was not selective for a particular parvalbumin protein. Western blotting showed parvalbumin to be considerably more abundant in SM than EO, requiring the loading of 3× the amount of protein in EO than SM to detect any parvalbumin protein. Parvalbumin was detected in the sarcoplasm of SM tissue and, despite its low abundance according to western blot, also in the cytoplasm of electrocytes and in the surrounding motor neuron cell bodies.
Because electrocytes do not contract, and are postsynaptic membranes, the consequence of Ca2+-binding and -transporting proteins in the EO is unclear. Our localization of Ca2+-ATPase to the posterior, innervated membrane of the electrocyte is consistent with findings by Taffarel, who also reported Ca2+-ATPase activity was localized only to the innervated face of the E. electricus electrocyte (Taffarel, 1989). Bartels reported that E. electricus electrocytes, following a depolarizing current, remain depolarized in Ca2+-free Ringer solution and are repolarized upon the addition of Ca2+ (Bartels, 1971). Bartels also demonstrated that electrocytes in a Ca2+-free Ringer solution had diminished inward K+ currents, suggesting a possible Ca2+-mediated K+ current in the repolarization of the electrocyte (Bartels, 1971). This does not, however, seem to be a common feature in all gymnotiforms; studies in Sternopygus macurus provide no evidence of Ca2+-based currents contributing to EODs (Ferrari and Zakon, 1993), despite the (relatively) long duration of the EOD pulse compared with action potentials (Ferrari and Zakon, 1993).
Related to the upregulation of plasma membrane Ca2+-ATPase is evidence for rather remarkable changes in expression of the Ca2+-binding protein parvalbumin in EO as compared with SM. Two parvalbumins, matches to D. rerio pvalb1 and pvalb2, respectively, were absent or downregulated in EO (Fig. 2, Table 2), and one transcript that matched D. rerio pvalb9 was upregulated in EO (Fig. 2, Table 1). Intriguingly, this transcript lacked the normal transcriptional start site found in all other parvalbumins (see dbEST accession number HO702432). Parvalbumin (the major allergenic compound in fish), is typically present in very high concentrations in fish muscle [as great as 1.5 mmol l–1 in some species (Gillis, 1985)], but appears to be at least 3- to 4-fold lower in mormyrid electric organ vs skeletal muscle (Fig. 5). This finding is consistent with studies by Childers and Siegel, which examined parvalbumin concentrations in SM and EO of E. electricus (Childers and Siegel, 1976). Parvalbumins are considered crucial in excitation–contraction coupling in muscle (Arif, 2009; Wilwert et al., 2006). Typically, muscles with fast relaxation rates express higher levels of parvalbumins than more powerful, slow-contracting muscles (Wilwert et al., 2006). Thus, the decreased expression of parvalbumins in EO is consistent with the fact that EOs is non-contractile. Taken together, our findings strongly indicate an important, but presently unknown role for Ca2+ in EO physiology in mormyrids.
Sarcomeric proteins
SSH identified several transcripts encoding sarcomeric proteins that were differentially expressed between SM and EO tissues. Two transcripts homologous to D. rerio MHC6 were detected; one was abundantly and exclusively expressed in EO whereas the other was predominantly but not exclusively expressed in SM. In addition, we detected downregulation of a troponin T transcript in EO vs SM, and the upregulation of a troponin I transcript in EO vs SM. These patterns of expression were all secondarily verified using RT-PCR. The detection of two MHC6 transcripts, each homologous to the same D. rerio gene and one uniquely expressed in EO, may suggest another detected instance of EO-specific alternative splicing.
MHC protein was detected using the MF20 antibody, which is widely species cross-reactive and recognizes all sarcomeric MHC proteins (DSHB Data Sheet, dshb.biology.uiowa.edu). Western blotting showed a protein of the expected size was recognized in both EO and SM tissues. The intensity of the MHC band was greater in SM than in EO, suggesting either that the abundance of MHC was greater in SM than in EO or that the epitope was less well recognized in EO. Immunohistochemistry revealed MHC was distributed, as expected, throughout the cytoplasm of SM, but was localized to a central filament that ran longitudinally through the center of each electrocyte in EO, but was not found in electrocyte stalks.
Troponin I was detected using the MAB1691 antibody, which recognizes cardiac and skeletal bovine troponin I at amino acids 87–93 (Millipore product information). Western blotting indicates that troponin I may be slightly more abundant in SM vs EO. Immunohistochemically, troponin I was distributed as expected throughout the cytoplasm of SM tissue, and was found in the central filament of each electrocyte in the EO, as well as in electrocyte stalks.
Finally, we also examined tropomyosin protein distribution, though we did not detect transcripts representing this protein in our SSH screens. Tropomyosin was identified using the CH1 antibody, which broadly recognizes tropomyosin from cardiac and skeletal muscle in a variety of species. Western blotting revealed bands of the expected size in both tissues, and greater expression in SM than in EO. Immunohistochemistry revealed that tropomyosin, as expected for other sarcomeric proteins, was localized to the sarcoplasm of SM tissue. In EO, tropomyosin was distributed in the central filament of each electrocyte, and was not present in electrocyte stalks.
SSH, immunohistochemistry and western blots are all consistent with the retention of sarcomeric proteins (i.e. MHC, troponin and tropomyosin) in the mormyrid EO, which has been suggested by previous histological and ultrastructural studies in mormyrids (Bass et al., 1986). Because EOs are not contractile in mormyrids, the role of these proteins in EO physiology is not known. Sarcomeric proteins are clearly not necessary for EO function: in gymnotiforms, the transition between skeletal muscle and EO is associated with a profound down-regulation of sarcomeric proteins such as MHC, troponin and tropomyosin (Patterson and Zakon, 1996).
In mormyrids, electrocytes comprising the EO are large cells, with numerous structural components. Our immunohistochemistry data suggest that the distribution of these proteins is heterogeneous in the EO (Fig. 3), with MHC, troponin and tropomyosin contributing to a thick center layer between the two electrocyte faces. Intriguingly, stalks seem to be devoid of MHC and tropomyosin, but continue to express troponin. One hypothesis for the retention of sarcomeric proteins may be a need for additional cytoskeletal support associated with the large size and physical complexity of electrocyte cells to maintain structural integrity. This hypothesis is of considerable interest given that the structural features of the EO are crucial in shaping the EOD waveforms. For example, structural alterations in the stalk system such as the absence or presence of penetrating stalks can create additional complexity in EOD signals (Gallant et al., 2011). In addition, electrocyte membrane structure can contribute to EOD waveform shape, particularly in duration (Bass et al., 1986). Both of these features may be a substrate for sexual selection on EOD signals (Arnegard et al., 2010a).
Preliminary insights into EO development in mormyrids
EOs have evolved at least six times independently in the history of vertebrates from SM tissue. In addition, other specialized muscles, namely heater organs (Block, 1994) and sonic muscles (Rome, 2006), have also evolved in teleost fishes from skeletal muscle. In each of these cases, while some of the molecular mechanisms underlying the unique physiology have been identified (Block, 1994; Rome 2006; Zakon et al., 2006; Arnegard et al., 2010b), the developmental mechanisms that coordinate these transformations are poorly understood. In this sense, the highly convergent gymnotiforms and mormyrids provide a unique opportunity to take a comparative approach to better understand how such novelty evolves.
In both mormyrids and gymnotiforms, adult EOs originate during development from a distinct group of fully differentiated SM myofibrils (Patterson and Zakon, 1997). Thus SM-like progenitor electrocytes (cells that comprise the EO) undergo an additional process leading to a mature electrocyte phenotype. In mormyrids, this process is accompanied by transition from motoric organization of muscle fibers to a continuous tube of electrocytes parallel to the spinal cord (Denizot et al., 1982). The transition between the muscle-like progenitors and electrocytes is additionally characterized by a substantial change in cell size (Unguez and Zakon, 1998a; Unguez and Zakon, 1998b), morphology (Denizot et al., 1982) and physiology (Westby and Kirschbaum, 1977), ultimately leading to the retention of electrical excitability without generating contractile force. In gymnotiforms, sarcomeric proteins are downregulated, and EO-specific proteins, such as keratin, are upregulated (Patterson and Zakon, 1997; Zakon and Unguez, 1999). In mormyrids, histological evidence suggests that sarcomeric proteins are retained in EOs (Bass, 1986; Bass et al., 1986), and the present study has verified that at least MHC, troponin and tropomyosin expression is maintained in mormyrid EOs.
Some attempts have been made to determine potential mechanisms that may be involved in this developmental transition. Several have hypothesized that motor neurons innervating the EO facilitate the transition between skeletal muscle and EO during development (Patterson and Zakon, 1997; Unguez and Zakon, 1998b; Szabo and Kirschbaum, 1983). Experimental manipulation of innervation in mormyrids and gymnotiforms have led to opposing results: innervation appears to be essential in the development and maintenance of the EOs in gymnotiforms (Patterson and Zakon, 1997; Unguez and Zakon, 1998a; Unguez and Zakon, 1998b; Zakon and Unguez, 1999; Cuellar et al., 2006), but does not appear necessary for the development of EOs in mormyrids (Szabo and Kirschbaum, 1983). A recent study has suggested that motor neuron activity in gymnotiform EO may suppress SM gene expression via post-transcriptional regulation (Cuellar et al., 2006). Our data do not provide evidence of post-transcriptional regulation in mormyrids; we were able to detect both transcripts and proteins, several of which were found to be the ‘suppressed’ sarcomere mRNA transcripts detected previously (Cuellar et al., 2006). Considered in the light of previous denervation studies (Szabo and Kirschbaum, 1983), these findings raise the possibility that EO development in mormyrids may not be initiated through the same developmental processes. This is especially intriguing given that the gymnotiform EO can be regenerated upon injury (Cuellar et al., 2006; Kim et al., 2008; Kim et al., 2009; Kim et al., 2004; Unguez and Zakon, 1998a; Unguez and Zakon, 1998b; Zakon and Unguez, 1999; Patterson and Zakon, 1996; Patterson and Zakon, 1997), whereas the mormyrid EO cannot.
In addition to the role of innervation in this transition, a variety of experiments have examined the role of several muscle regulatory transcription factors (Kim et al., 2008); intriguingly, many are upregulated (e.g. MyoD, Myogenin, MYF5 and MRF4) whereas others (e.g. MEF2c, ID1, and ID2) are not (Kim et al., 2008). As found by Kim et al. (Kim et al., 2008), we saw no differential expression of MEF2C; however, we did detect up regulation of the muscle regulatory transcription factor MEF2A in mormyrid EO, which was not examined by Kim et al. (Kim et al., 2008). This is of considerable interest given the involvement of MEF2A in SM development in posterior somites, the developmental source of EOs (Wang et al., 2006).
To briefly conclude, we have summarized some of these major developmental and gene expression similarities and differences in Table 4. Our results indicate that the biochemical differences between SM and EO in mormyrids appears to be present at the transcriptional level. This contrasts strongly with the results of a comparable recent study (Cuellar et al., 2006), which demonstrates in gymnotiforms that many of the biochemical differences between tissues appears post-transcriptionally. In mormyrids, biochemical differences seem to manifest mainly as expression of different paralogs of a gene family (e.g. parvalbumins, MHC). Previous results (Zakon et al., 2006; Arnegard et al., 2010b) have demonstrated that paralogous sodium channel genes, resulting from ancient teleost gene duplication events, are capable of serving novel functions in the EO. Rather than building the novel EO by ‘switching off’ genes as appears to occur in gymnotiforms (Cuellar et al., 2006), one could easily imagine an alternative process by which neofunctionalized gene duplicates (normally muscle specific) could lead to a functional EO as effectively. We find these possibilities a compelling motivation for future comparative work using high-throughput transcriptomics and genomics techniques, which will doubtlessly allow for a more satisfying and comprehensive view of these processes.
Table 4.
Summary of gene expression and developmental differences between surveyed mormyrids and gymnotiforms
Supplementary Material
ACKNOWLEDGEMENTS
We thank the Cornell Core Laboratories Genomics Facility for performing all DNA sequencing presented here. We also thank A. Filoteo and E. Strehler (Mayo Clinic, Rochester, MN, USA) for their generous donation of the anti-PMCA antibody 5F10 used in this study.
FOOTNOTES
Supplementary material available online at http://jeb.biologists.org/cgi/content/full/215/14/2479/DC1
FUNDING
The work presented here was funded by the National Institutes of Health [NIMH TG T32-MH015793 and NIH TG 2T32GM007469 to J.R.G.; NIH R01-DC6206 to C,D,H,], the National Science Foundation [grant no. 0818305 to C.D.H.], and the Cornell Center for Vertebrate Genomics. Deposited in PMC for release after 12 months.
REFERENCES
- Albert J. S. (2003). Family Apterontoidae. In Checklist of the Freshwater Fishes of South and Central America (ed. Reis R. E., Kullander S. O., Ferraris C. J. J.), pp. 503-508 Porto Alegre: Edipucrs; [Google Scholar]
- Arif S. H. (2009). A Ca2+-binding protein with numerous roles and uses: parvalbumin in molecular biology and physiology. BioEssays 31, 410-421 [DOI] [PubMed] [Google Scholar]
- Arnegard M., McIntyre P. B., Harmon L. J., Zelditch M. L., Crampton W. G. R., Davis J. K., Sullivan J. P., Lavoué S., Hopkins C. (2010a). Sexual signal evolution outpaces ecomorphological and trophic divergence during electric fish species radiation. Am. Nat. 176, 335-356 [DOI] [PubMed] [Google Scholar]
- Arnegard M., Zwickl D., Lu Y. (2010b). Old gene duplication facilitates origin and diversification of an innovative communication system – twice. Proc. Natl. Acad. Sci. USA 107, 22172-22177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels E. (1971). Depolarization of electroplax membrane in calcium-free Ringer’s solution. J. Membr. Biol. 5, 121-132 [DOI] [PubMed] [Google Scholar]
- Bass A. H. (1986). Electric organs revisited: evolution of a vertebrate communication and orientation organ. In Electroreception (ed. Bullock T. H., Heiligenberg W.), pp. 13-70 New York: Wiley; [Google Scholar]
- Bass A. H., Denizot J. P., Marchaterre M. (1986). Ultrastructural features and hormone-dependent sex-differences of mormyrid electric organs. J. Comp. Neurol. 254, 511-528 [DOI] [PubMed] [Google Scholar]
- Bell C. C., Bradbury J., Russell C. J. (1976). The electric organ of a mormyrid as a current and voltage source. J. Comp. Physiol. A 110, 65-88 [Google Scholar]
- Bennett M. (1971). Electric organs. In Fish Physiology, vol. 5 (ed. Hoar W. S., Randall D. J.), pp. 347-491 New York, NY: Academic Press; [Google Scholar]
- Black B., Olson E. (1998). Transcriptional control of muscle development by myocyte enhancer factor-2 (mef2) proteins. Annu. Rev. Cell Dev. Biol. 14, 167-196 [DOI] [PubMed] [Google Scholar]
- Black B., Lu J., Olson E. (1997). The mef2a 3′ untranslated region functions as a cis-acting translational repressor. Mol. Cell. Biol. 17, 2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block B. A. (1994). Thermogenesis in muscle. Annu. Rev. Physiol. 56, 535-577 [DOI] [PubMed] [Google Scholar]
- Boguski M. S., Lowe T. M., Tolstoshev C. M. (1993). dbEST – database for ‘expressed sequence tags’. Nat. Genet. 4, 332-333 [DOI] [PubMed] [Google Scholar]
- Brooks E. M., Sheflin L. G., Spaulding S. W. (1995). Secondary structure in the 3′ UTR of EGF and the choice of reverse transcripatses affect the detection of message diversity by RT-PCR. BioTechniques 19, 806-815 [PubMed] [Google Scholar]
- Caride A. J., Filoteo A. G., Enyedi A., Verma A. K., Penniston J. T. (1996). Detection of isoform 4 of the plasma membrane calcium pump in human tissues by using isoform-specific monoclonal antibodies. Biochem. J. 316, 353-359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childers S. R., Siegel F. L. (1976). Calcium-binding proteins in electroplax and skeletal muscle: comparison of the parvalbumin and phosphodiesterase activator protein of Electrophorus electricus. Biochem. Biophys. Acta 439, 316-326 [DOI] [PubMed] [Google Scholar]
- Clausen T., Van Hardeveld C., Everts M. E. (1991). Significance of cation transport in control of energy metabolisim and thermogenesis. Physiol. Rev. 71, 733-773 [DOI] [PubMed] [Google Scholar]
- Cuellar H., Kim J. A., Unguez G. A. (2006). Evidence of post-transcriptional regulation in the maintenance of a partial muscle phenotype by electrogenic cells of S. macrurus. FASEB J. 20, 2540 [DOI] [PubMed] [Google Scholar]
- Darwin C. (1859). The Origin of Species, Vol. 11, The Harvard Classics. New York: P. F. Collier & Sons; [Google Scholar]
- Denizot J. P., Kirschbaum F., Westby G. W., Tsuji S. (1982). On the development of the adult electric organ in the mormyrid fish Pollimyrus isidori (with special focus on the innervation). J. Neurocytol. 11, 913-934 [DOI] [PubMed] [Google Scholar]
- Ferrari M., Zakon H. H. (1993). Conductances contributing to the action-potential of Sternopygus electrocytes. J. Comp. Physiol. A 173, 281-292 [DOI] [PubMed] [Google Scholar]
- Gallant J. R. (2011). Mechanisms of signal diversity in mormyrid electric fish. PhD thesis, Cornell University; [DOI] [PubMed] [Google Scholar]
- Gallant J. R., Arnegard M. E., Sullivan J. P., Carlson B. A., Hopkins C. D. (2011). Signal variation and its morphological correlates in Paramormyrops kingsleyae provide insight into evolution of electrogenic signal diversity in mormyrid fish. J. Comp. Physiol. A 197, 799-817 [DOI] [PubMed] [Google Scholar]
- Gelsthorpe M., Pulumati M., McCallum C., Dang-Vu K., Tsubota S. I. (1997). The putative cell cycle gene, enhancer of rudimentary, encodes a highly conserved protein found in plants and animals. Gene 186, 189-195 [DOI] [PubMed] [Google Scholar]
- Gill T. N. (1862). On the west African genus Hemichromis and descriptions of new species in the museums of the academy and smithsonian institution. Proc. Acad. Nat. Sci. Philadelphia 14, 134-139 [Google Scholar]
- Gillis J. (1985). Relaxation of vertebrate skeletal muscle. A synthesis of the biochemical and physiological approaches. Biochim. Biophys. Acta 811, 98-145 [DOI] [PubMed] [Google Scholar]
- Heiligenberg W. (1986). Jamming avoidance responses: model systems for neuroethology. In Electroreception (ed. Bullock T. H., Heiligenberg W.). New York: John Wiley & Sons, Inc; [Google Scholar]
- Hinits Y., Hughes S. M. (2007). Mef2s are required for thick filament formation in nascent muscle fibres. Development 134, 2511-2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins C. D. (1999a). Design features for electric communication. J. Exp. Biol. 202, 1217-1228 [DOI] [PubMed] [Google Scholar]
- Hopkins C. D. (1999b). Signal evolution in electric communication. In The Design of Animal Communication (ed. Hauser M. D., Konishi M.). Cambridge, MA: MIT Press; [Google Scholar]
- Kawasaki M. (2005). Physiology of the tuberous electrosensory system. In Electroreception (ed. Bullock T. H., Hopkins C. D., Popper A. N., Fay R. R.), pp. 154-194 New York: Springer; [Google Scholar]
- Kim H. J., Archer E., Escobedo N., Tapscott S. J., Unguez G. A. (2008). Inhibition of mammalian muscle differentiation by regeneration blastema extract of sternopygus macrurus. Dev. Dyn. 237, 2830-2843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. J., Güth R., Jonsson C. B., Unguez G. A. (2009). S. macrurus myogenic regulatory factors (MRFS) induce mammalian skeletal muscle differentiation; evidence for functional conservation of MRFS. Dev. Biol. 53, 993-1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. A., Jonsson C. B., Calderone T., Unguez G. A. (2004). Transcription of myod and myogenin in the non-contractile electrogenic cells of the weakly electric fish, sternopygus macrurus. Dev. Genes Evol. 214, 380-392 [DOI] [PubMed] [Google Scholar]
- Kirschbaum F. (1983). Myogenic electric organ precedes the neurogenic organ in apteronotid fish. Naturwissenschaften 70, 205-207 [DOI] [PubMed] [Google Scholar]
- Lissmann H. W. (1951). Continuous electrical signals from the tail of a fish, Gymnarchus niloticus cuv. Nature 167, 201-202 [DOI] [PubMed] [Google Scholar]
- Lissmann H. W. (1958). On the function and evolution of electric organs in fish. J. Exp. Biol. 35, 156-191 [Google Scholar]
- Lissmann H. W., Machin K. E. (1958). The mechanisim of object location of Gymnarchus niloticus and similar fish. J. Exp. Biol. 35, 451-486 [Google Scholar]
- Lowe J., Araujo G. M., Pedrenho A. R., Nunes-Tavares N., Ribeiro M. G., Hassón-Voloch A. (2004). Polarized distribution of Na+, K+-ATPase alpha-subunit isoforms in electrocyte membranes. Biochim. Biophys. Acta 1661, 40-46 [DOI] [PubMed] [Google Scholar]
- Lukasik A., Uniewicz K. A., Kulis M., Kozlowski P. (2008). Ciz1, a p21cip1/waf1-interacting zinc finger protein and DNA replication factor, is a novel molecular partner for human enhancer of rudimentary homolog. FEBS J. 275, 332-340 [DOI] [PubMed] [Google Scholar]
- Markham M. R., McAnelly M. L., Stoddard P. K., Zakon H. H. (2009). Circadian and social cues regulate ion channel trafficking. PLoS Biol. 7, e1000203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mörhes F. P. (1957). Elektrische entladungen im dienste der revierabgrenzung bel fischen. Naturwissenschaften 44, 431-432 [Google Scholar]
- Patterson J. M., Zakon H. H. (1996). Differential expression of proteins in muscle and electric organ, a muscle derivative. J. Comp. Neurol. 370, 367-376 [DOI] [PubMed] [Google Scholar]
- Patterson J. M., Zakon H. H. (1997). Transdifferentiation of muscle to electric organ: regulation of muscle-specific proteins is independent of patterned nerve activity. Dev. Biol. 186, 115-126 [DOI] [PubMed] [Google Scholar]
- Pogge von Strandmann E., Senke S., Ryffel G. U. (2001). ERH (enhancer of rudimentary homologue), a conserved factor identical between frog and human, is a transcriptional repressor. Biol. Chem. 382, 1379-1385 [DOI] [PubMed] [Google Scholar]
- Rebrikov D. V. (2003). Identification of differential genes by suppression subtractive hybridization. In PCR Primer (ed. Dieffenback, Dveksler). Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press; [Google Scholar]
- Rome L. C. (2006). Design and function of superfast muscles: new insights into the physiology of skeletal muscle. Annu. Rev. Physiol. 68, 193-221 [DOI] [PubMed] [Google Scholar]
- Salazar V. L., Stoddard P. K. (2008). Sex differences in energetic costs explain sexual dimorphisim in the circadian rhythm modulation of the electrocommunication signal of the gymnotiform fish Brachyhypopomus pinnicaudatus. J. Exp. Biol. 211, 1012-1020 [DOI] [PubMed] [Google Scholar]
- Stoddard P. K., Salazar V. L. (2011). Energetic cost of communication. J. Exp. Biol. 214, 200-205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan J. P., Lavoue S., Hopkins C. D. (2000). Molecular systematics of the African electric fishes (mormyroidea: Teleostei) and a model for the evolution of their electric organs. J. Exp. Biol. 203, 665-683 [DOI] [PubMed] [Google Scholar]
- Szabo T., Kirschbaum F. (1983). On the differentiation of electric organs in the absence of central connections or peripheral innervation. In The Physiology of Excitable Cells (ed. Grinnell A. D., Moody W. J., Jr), pp. 451-460 New York: Alan R. Liss; [Google Scholar]
- Taffarel M., TatianaCoelho S., Teixeira-Ferreira A., Somló C., Souza W. D., Machado R. D., Vieyra A. (1989). Localization and cation dependence of a Ca or Mg-ATPase from electrocytes of Electrophorus electricus, L. J. Histochem. Cytochem. 37, 953-959 [DOI] [PubMed] [Google Scholar]
- Unguez G. A., Zakon H. H. (1998a). Phenotypic conversion of distinct muscle fiber populations to electrocytes in a weakly electric fish. J. Comp. Neurol. 399, 20-34 [PubMed] [Google Scholar]
- Unguez G. A., Zakon H. H. (1998b). Reexpression of myogenic proteins in mature electric organ after removal of neural input. J. Neurosci. 18, 9924-9935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Qian L., Dong Y., Jiang Q., Gui Y., Zhong T. P., Song H. (2006). Myocyte-specific enhancer factor 2a is essential for zebrafish posterior somite development. Mech. Dev. 123, 783-791 [DOI] [PubMed] [Google Scholar]
- Westby G. W., Kirschbaum F. (1977). Emergence and development of the electric organ discharge in the mormyrid fish, Pollimyrus isidori. J. Comp. Physiol. A 122, 251-271 [Google Scholar]
- Wilwert J. L., Madhoun N. M., Coughlin D. J. (2006). Parvalbumin correlates with relaxation rate in the swimming muscle of sheepshead and kingfish. J. Exp. Biol. 209, 227-237 [DOI] [PubMed] [Google Scholar]
- Wojcik E., Murphy A. M., Fares H., Dang-Vu K., Tsubota S. I. (1994). Enhancer of rudimentary(p1), e(r)(p1), a highly conserved enhancer of the rudimentary gene. Genetics 138, 1163-1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakon H. H. (1986). The electroreceptive periphery. In Electroreception (ed. Bullock T. H., Heiligenberg W.), pp. 103-156 New York: Wiley; [Google Scholar]
- Zakon H. H., Unguez G. A. (1999). Development and regeneration of the electric organ. J. Exp. Biol. 202, 1427-1434 [DOI] [PubMed] [Google Scholar]
- Zakon H. H., Lu Y., Zwickl D., Hillis D. M. (2006). Sodium channel genes and the evolution of diversity in communication signals of electric fishes: convergent molecular evolution. Proc. Natl. Acad. Sci. USA 103, 3675-3680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zupanc G. K. H., Bullock T. H. (2005). From electrogenesis to electroreception: an overview. In Electroreception (ed. Bullock T. H., Hopkins C. D., Popper A. N., Fay R. R.), pp. 5-46 New York: Springer; [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.