Abstract
Analysis of cell-free fetal DNA in maternal plasma holds great promise for the development of non-invasive prenatal genetic diagnostics. However, previous studies have been restricted to detection of fetal trisomies (1, 2) or specific, paternally inherited mutations (3), or to genotyping common polymorphisms using invasively sampled material (4). Here, we combine genome sequencing of two parents, genome-wide maternal haplotyping (5), and deep sequencing of maternal plasma to non-invasively determine the genome sequence of a human fetus at 18.5 weeks gestation. Inheritance was predicted at 2.8×106 parentally heterozygous sites with 98.1% accuracy. Furthermore, 39 of 44 de novo point mutations in the fetal genome were detected, albeit with limited specificity. Subsampling these data and analyzing a second family trio by the same approach indicate that ~300 kilobase parental haplotype blocks combined with shallow sequencing of maternal plasma are sufficient to substantially determine the inherited complement of a fetal genome. However, ultra-deep sequencing of maternal plasma is necessary for the practical detection of fetal de novo mutations genome-wide. Although technical and analytical challenges remain, we anticipate that non-invasive analysis of inherited variation and de novo mutations in fetal genomes will facilitate the comprehensive prenatal diagnosis of both recessive and dominant Mendelian disorders.
Introduction
On average, ~13% of cell-free DNA isolated from maternal plasma during pregnancy is fetal in origin (6). The concentration of cell-free fetal DNA in maternal circulation varies between individuals, increases across gestation, and is rapidly cleared postpartum (7, 8). Despite this variability, cell-free fetal DNA has been successfully targeted as a resource for non-invasive prenatal diagnosis, including the development of targeted assays for single gene disorders (3). More recently, several groups demonstrated shotgun, massively parallel sequencing of cell-free DNA from maternal plasma as a robust approach for non-invasively diagnosing fetal aneuploidies such as trisomy 21 (1, 2).
Ideally, it would be possible to non-invasively predict the whole genome sequence of a fetus to high accuracy and completeness, potentially enabling the comprehensive prenatal diagnosis of Mendelian disorders and obviating the need for invasive prenatal diagnostic procedures with their attendant risks. However, several key technical obstacles must be overcome for this goal to be achieved using cell-free DNA from maternal plasma. First, the sparse representation of fetal-derived sequences poses the challenge of detecting low frequency alleles inherited from the paternal genome as well as those arising from de novo mutations in the fetal genome. Second, maternal DNA predominates in plasma, obscuring one’s ability to assess maternally inherited variation at individual sites in the fetal genome.
Lo et al. recently showed that fetal-derived DNA is distributed evenly enough to support the inference of fetal genotypes, and furthermore demonstrated how knowledge of parental haplotypes could be leveraged to this end (4). However, their study was limited in several ways. First, the proposed method depended on the availability of parental haplotypes, but at the time of their work, no technologies existed to measure these experimentally at a genome-wide scale. Therefore, an invasive procedure, chorionic villus sampling (CVS), was used to obtain placental material for fetal genotyping. Second, parental genotypes and invasively obtained fetal genotypes were used to infer parental haplotypes, which were then used in combination with the sequencing of DNA from maternal plasma to predict the fetal genotypes. Although necessitated by the lack of genome-wide haplotyping methods, the circularity of these inferences makes it difficult to assess how well the method would perform in practice. Third, their analysis was restricted to several hundred thousand parentally heterozygous sites of common single nucleotide polymorphism (SNPs) represented on a commercial genotyping array. These common SNPs are only a small fraction of the several million heterozygous sites present in each parental genome, and include few of the rare variants that predominantly underlie Mendelian disorders (9). Fourth, no effort was made to ascertain de novo mutations in the fetal genome. As de novo mutations underlie a substantial fraction of dominant genetic disorders, their detection is critical for comprehensive prenatal genetic diagnostics. Therefore, although this study demonstrated the first successful construction of a genetic map of a fetus, it required an invasive procedure and did not attempt to determine the whole genome sequence of the fetus.
Results
We and others recently demonstrated methods for experimentally determining haplotypes for both rare and common variation at a genome-wide scale (5, 10–12). In the current study, we set out to integrate the haplotype-resolved genome sequencing of a mother, the shotgun genome sequencing of a father, and the deep sequencing of cell-free DNA in maternal plasma to non-invasively predict the whole genome sequence of a fetus (Fig. 1A). Although results associated with two such mother-father-child trios are described (“I1” – a first trio at 18.5 wks gestation, “G1” – a second trio at 8.2 wks gestation), we focus here primarily on the trio for which considerably more sequence data of each type was generated (“I1”) (Table 1).
Fig. 1.
Experimental approach. (A). Schematic of sequenced individuals in a family trio. Maternal plasma sequences were ~13% fetal-derived based on read depth at chrY and alleles specific to each parent. (B). Inheritance of maternally heterozygous alleles inferred using long haplotype blocks. Among plasma sequences, maternal-specific alleles are more abundant when transmitted (expected 50% versus 43.5%), but there is substantial overlap between the distributions of allele frequencies when taking considering sites in isolation (left histogram, yellow=shared allele transmitted, green=maternal-specific allele transmitted). Taking average allele balances across haplotype blocks (right histogram) provides much greater separation, permitting more accurate inference of maternally transmitted alleles. (C). Histogram of fractional read depth among plasma data at paternal-specific heterozygous sites. In the overwhelming majority of cases when the allele specific to the father was not detected, the opposite allele had been transmitted (96.8%, n=561,552). (D). De novo missense mutation in the gene ACMSD detected in 3 of 93 maternal plasma reads and later validated by PCR and resequencing. The mutation, which is not observed in dbSNP nor among >4,000 individuals’ coding exons sequenced as part of the NHLBI Exome Sequencing Project (http://evs.gs.washington.edu), creates a leucine-to-proline substitution at a site conserved across all aligned mammalian genomes (UCSC Genome Browser) in a gene implicated in Parkinson’s disease by genome-wide association studies (24).
Table 1.
Individuals sequenced, type of starting material, and final fold-coverage of the reference genome after discarding PCR or optical duplicate reads. GA, gestational age.
| Individual | Sample | Depth of coverage |
|---|---|---|
| Mother (I1-M) | Plasma (5 ml, GA 18.5 wk) | 78 |
| Whole blood (<1 mL) | 32 | |
| Father (I1-P) | Saliva | 39 |
| Offspring (I1-C) | Cord blood at delivery | 40 |
In brief, the haplotype-resolved genome sequence of the mother (“I1-M”) was determined by first performing shotgun sequencing of maternal genomic DNA from blood to 32-fold coverage (coverage = median-fold coverage of mapping reads to the reference genome after discarding duplicates). Next, by sequencing complex haploid subsets of maternal genomic DNA while preserving long-range contiguity (5), we directly phased 91.4% of 1.9 × 106 heterozygous SNPs into long haplotype blocks (N50 of 326 kilobases (kbp)). The shotgun genome sequence of the father (“I1-P”) was determined by sequencing of paternal genomic DNA to 39-fold coverage, yielding 1.8 × 106 heterozygous SNPs. However, paternal haplotypes could not be assessed because only relatively low molecular weight DNA obtained from saliva was available. Shotgun DNA sequencing libraries were also constructed from 5 mL of maternal plasma (obtained at 18.5 wks gestation), and this composite of maternal and fetal genomes was sequenced to 78-fold non-duplicate coverage. The fetus was male, and fetal content in these libraries was estimated at 13%. To properly assess the accuracy of our methods for determining the fetal genome solely from samples obtained non-invasively at 18.5 wks gestation, we also performed shotgun genome sequencing of the child (“I1-C”) to 40-fold coverage via cord blood DNA obtained after birth.
Our analysis comprised four parts: (1) predicting the subset of ‘maternal-only’ heterozygous variants (homozygous in the father) transmitted to the fetus; (2) predicting the subset of ‘paternal-only’ heterozygous variants (homozygous in the mother) transmitted to the fetus; (3) predicting transmission at sites heterozygous in both parents; (4) predicting sites of de novo mutation – that is, variants occurring only in the genome of the fetus. Allelic imbalance in maternal plasma, manifesting across experimentally determined maternal haplotype blocks, was used to predict their maternal transmission (Fig. 1B). The observation (or lack thereof) of paternal alleles in shotgun libraries derived from maternal plasma was used to predict paternal transmission (Fig. 1C). Finally, a strict analysis of alleles rarely observed in maternal plasma, but never in maternal or paternal genomic DNA, enabled the genome-wide identification of candidate de novo mutations (Fig. 1D). Fetal genotypes are trivially predicted at sites where the parents are both homozygous (for the same or different allele).
We first sought to predict transmission at ‘maternal-only’ heterozygous sites. Given the fetal-derived proportion of ~13% in cell-free DNA, the maternal-specific allele is expected in 50% of reads aligned to such a site if it is transmitted, versus 43.5% if the allele shared with the father is transmitted. However, even with 78-fold coverage of the maternal plasma “genome”, the variability of sampling is such that site-by-site prediction results in only 64.4% accuracy (Fig. 2). We therefore examined allelic imbalance across blocks of maternally heterozygous sites defined by haplotype-resolved genome sequencing of the mother (Fig. 1B). As anticipated given the haplotype assembly N50 of 326 Kb, the vast majority of experimentally defined maternal haplotype blocks were wholly transmitted, with partial inheritance in a small minority of blocks (0.6%, n=72) corresponding to switch errors from haplotype assembly and to sites of recombination. We developed a Hidden Markov model (HMM) to identify likely switch sites and thus more accurately infer the inherited alleles at maternally heterozygous sites (Fig. 3 and 4, and SOM Materials and Methods). Using this model, accuracy of the inferred inherited alleles at 1.1 × 106 phased, ‘maternal-only’ heterozygous sites increased from 98.6% to 99.3% (Table 2). Remaining errors were concentrated among the shortest maternal haplotype blocks (fig. S1), which provide less power to detect allelic imbalance in plasma data as compared with long blocks. Among the top 95% of sites ranked by haplotype block length, prediction accuracy rose to 99.7%, suggesting that remaining inaccuracies can be mitigated by improvements in haplotyping.
Fig. 2.
Accuracy of fetal genotype inference from maternal plasma sequencing. Accuracy is shown for paternal-only heterozygous sites, and for phased maternal-only heterozygous sites, either using maternal phase information (black) or instead predicting inheritance on a site-by-site basis (gray).
Fig. 3.
HMM-based predictions correctly predict maternally transmitted alleles across ~1 Mbp on chromosome 10, despite site-to-site variability of allelic representation among maternal plasma sequences (red).
Fig. 4.
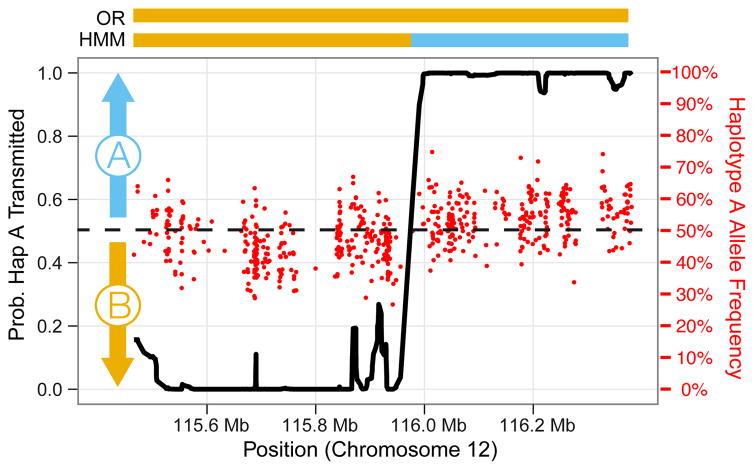
HMM-based detection of recombination events and haplotype assembly switch errors. A maternal haplotype block of 917 Kbp on chromosome 12q is shown, with red points representing the frequency of haplotype A alleles among plasma reads, and the black line indicating the posterior probability of transmission for haplotype A computed by the HMM at each site. A block-wide odds ratio test (OR) predicts transmission of the entire haplotype B, resulting in incorrect prediction at 272 of 587 sites (46.3%). The HMM predicts a switch between chromosomal coordinates 115,955,900 and 115,978,082, and predicts transmission of haplotype B alleles from the centromeric end of the block to the switch point, and haplotype A alleles thereafter, resulting in correct predictions at all 587 sites. All three overlapping informative clones support the given maternal phasing of the SNPs adjacent to the switch site, suggesting that the switch predicted by the HMM results from a maternal recombination event rather than an error of haplotype assembly.
Table 2.
Number of sites and accuracy of fetal genotype inference from maternal plasma sequencing (percentage of transmitted alleles correct out of all predicted) by parental genotype and phasing status. Sites later determined by trio sequencing (including the offspring) to have poor genotype quality scores or genotypes that violated Mendelian inheritance were discarded the purpose of evaluating accuracy (14,000 maternal-only, 32,233 paternal-only, and 480 shared heterozygous sites, or 1.5% of all sites).
| Individual | Site | Other parental genotype | Number of Sites | Accuracy |
|---|---|---|---|---|
| Mother (I1-M) | Heterozygous, phased | Homozygous | 1,064,255 | 99.3% |
| Heterozygous | 576,242 | 98.7%† | ||
|
| ||||
| Heterozygous, not phased | All | 121,425 | N.D. | |
|
| ||||
| Father (I1-P) | Heterozygous | Homozygous | 1,134,192 | 96.8% |
| Heterozygous | 631,721 | N.D. | ||
Among biparentally heterozygous sites, accuracy was assessed only where the offspring was homozygous (48.8%, n=631,721), allowing the “true” transmitted alleles to be unambiguously inferred from trio genotypes.
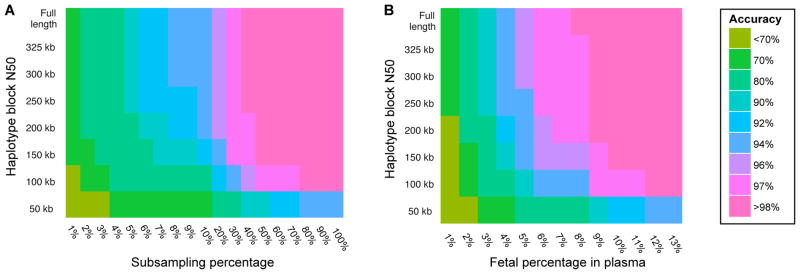
We performed simulations to characterize how the accuracy of haplotype-based fetal genotype inference depended upon haplotype block length, maternal plasma sequencing depth, and the fraction of fetal-derived DNA. To mimic the effect of less successful phasing, we split the maternal haplotype blocks into smaller fragments to create a series of assemblies with decreasing contiguity. We then subsampled a range of sequencing depths from the pool of observed alleles in maternal plasma, and predicted the maternally contributed allele at each site as above (Fig. 5A). The results suggest that inference of the inherited allele is robust to either decreasing sequencing depth of maternal plasma, or to shorter haplotype blocks, but not both. For example, using only 10% of the plasma sequence data (median depth = 8X) in conjunction with full-length haplotype blocks, we successfully predicted inheritance at 94.9% of ‘maternal-only’ heterozygous sites. We achieved nearly identical accuracy (94.8%) at these sites when highly fragmented haplotype blocks (N50 = 50 Kb) were used with the full set of plasma sequences. We next simulated decreased proportions of fetal DNA in the maternal plasma by spiking in additional depth of both maternal alleles at each site and subsampling from these pools, effectively diluting away the signal of allelic imbalance used as a signature of inheritance (Fig. 5B). Again, we found the accuracy of the model to be robust to either lower fetal DNA concentrations or shorter haplotype blocks, but not both.
Fig. 5.
Simulation of effects of reduced coverage, haplotype length, and fetal DNA concentration on fetal genotype inference accuracy, defined as the percentage of sites at which the inherited allele was correctly identified out of all sites where prediction was attempted. Heatmaps of accuracy after in silico fragmentation of haplotype blocks and (A) shallower sequencing of maternal plasma, or (B) reduced fetal concentration among plasma sequences.
We next sought to predict transmission at ‘paternal-only’ heterozygous sites. At these sites, when the father transmits the shared allele, the paternal-specific allele should be entirely absent among the fetal-derived sequences. If instead the paternal-specific allele is transmitted, it will on average constitute half the fetal-derived reads within the maternal plasma “genome” (~5 reads given 78-fold coverage, assuming 13% fetal content). To assess these, we performed a site-by-site log-odds test; this amounted to taking the observation of one or more reads matching the paternal-specific allele at a given site as evidence of its transmission, and conversely the lack of such observations as evidence of non-transmission (Fig. 1C). In contrast to maternal-only heterozygous sites, this simple site-by-site model was sufficient to correctly predict inheritance at 1.1 × 106 paternal-only heterozygous sites with 96.8% accuracy (Table 2). We anticipate that accuracy could likely be improved upon by deeper sequence coverage of the maternal plasma (fig. S2), or alternatively by taking a haplotype-based approach if high molecular weight genomic DNA from the father is available.
We next considered transmission at sites heterozygous in both parents. We predicted maternal transmission at such shared sites phased using neighboring ‘maternal-only’ heterozygous sites in the same haplotype block. This yielded predictions at 576, 242/631, 721 (91.2%) of shared heterozygous sites with an estimated accuracy of 98.7% (Table 2). Although we did not predict paternal transmission at these sites, we anticipate that analogous to the case of maternal transmission, this could be done with high accuracy given paternal haplotypes. We note that shared heterozygous sites primarily correspond to common alleles (fig. S3), which are less likely to contribute to Mendelian disorders in non-consanguineous populations.
De novo mutations in the fetal genome are expected to appear within the maternal plasma sequences as ‘rare alleles’ (Fig. 1D), similar to transmitted paternal-specific alleles. However, the detection of de novo mutations poses a much greater challenge: unlike the 1.8 × 106 paternally heterozygous sites defined by sequencing the father (of which ~50% are transmitted), the search space for de novo sites is effectively the full genome, throughout which there may be only ~60 sites given a prior mutation rate estimate of ~1 × 10−8 (13). Indeed, whole genome sequencing of the offspring (“I1-C”) revealed only 44 high-confidence point mutations (‘true de novo sites’; Table S1). Taking all positions in the genome at which at least one plasma-derived read had a high-quality mismatch to the reference sequence, and excluding variants present in the parental whole genome sequencing data, we found 2.5 × 107 candidate de novo sites, including 39 of the 44 true de novo sites. At baseline, this corresponds to sensitivity of 88.6% with a signal-to-noise ratio of 1-to-6.4×105.
We applied a series of increasingly stringent filters (fig. S4) intended to remove sites prone to sequencing or mapping artifacts. Removing alleles also found in at least one read among any other individual sequenced in this study, known polymorphisms from dbSNP (release 135), and sites adjacent to 1–3mer repeats reduced the number of candidate de novo sites to 1.8 × 107. Further requiring at least 2 independent supporting reads, removing sites with excessively many reads supporting the alternate allele (uncorrected P < 0.05, per-site one-sided binomial test using fetal-derived fraction of 13%), and requiring supporting base quality scores summing to at least 105 brought the total number of candidate to 3,884, including 17 true de novo sites. This candidate set is substantially depleted for sites of systematic error, and is instead likely dominated by errors originating during PCR, as even a single round of amplification with a proofreading DNA polymerase with an error rate of 1 × 10−7 would introduce over 300 candidate sites. This ~2,800-fold improvement in signal-to-noise ratio reduced the candidate set to a size accessible to validation by targeted methodologies [e.g. an order of magnitude fewer than the number of candidate de novo sites requiring validation in a previous study involving pure genomic DNA from parent-child trios within a nuclear family (14)], particularly if only candidate mutations predicted to be pathogenic were validated [e.g. only 33 of the 3,884 candidate sites were predicted to be protein-altering].
Discussion
In summary, we have demonstrated noninvasive prediction of the whole genome sequence of a human fetus through the combination of haplotype-resolved genome sequencing (5) of a mother, shotgun genome sequencing of a father, and deep sequencing of maternal plasma. Of note, the types and quantities of materials used were consistent with those routinely collected in a clinical setting (Table 1). To replicate these results, we repeated the full experiment for a second trio (“G1”) from which maternal plasma was collected earlier in the pregnancy, at 8.2 weeks after conception. Both the overall sequencing depth and the fetal-derived proportion were each lower relative to the first trio (by 28% and 51%, respectively), resulting in an average of fewer than four fetal-derived reads per site. Nevertheless, we achieved 95.7% accuracy for prediction of inheritance at maternal-only sites, consistent with accuracy obtained under simulation with data from the first trio (Fig. 5). These results underscore the importance of specific technical parameters in determining performance, namely the length and completeness of haplotype-resolved sequencing of parental DNA, and the depth and complexity of sequencing libraries derived from low starting masses of plasma-derived DNA (less than 5 ng for both I1 and G1 in our study).
There remain several key avenues for improvement. First, although we predicted inheritance at 2.8 × 106 heterozygous sites with high accuracy (98.2% overall), there were 7.5 × 105 sites for which we did not attempt prediction (Table 2). These include 6.3 × 105 sites heterozygous in both parents for which we could not assess paternal transmission, and 1.2 × 105 “maternal-only” heterozygous sites which were not included in our haplotype assembly. The former are in principle accessible but require haplotype-resolved (rather than solely shotgun) sequencing of paternal DNA, which was not possible here with either the I1 or G1 trio due to unavailability of high molecular weight DNA from each father. The latter are also in principle accessible but require improvements to haplotyping technology to enable phasing of SNPs residing within blocks of relatively low heterozygosity as well as within segmental duplications. More generally, despite recent innovations from our group and others (5, 10–12), there remains a critical need for genome-wide haplotyping protocols that are at once robust, scalable, and comprehensive. Significant reductions in cost, along with standardization and automation, will be necessary for compatibility with large-scale clinical application.
Second, although we were successful in detecting nearly 90% of de novo single nucleotide mutations by deep sequencing of maternal plasma, this was with very low specificity. The application of a series of filters resulted in a ~2,800-fold gain in specificity at a ~2-fold cost in terms of sensitivity. However, there is clearly room for improvement if we are to enable the sensitive and specific prenatal detection of potentially pathogenic de novo point mutations at a genome-wide scale, a goal that will likely require deeper than 78-fold coverage of the maternal plasma “genome” (15) in combination with targeted validation of potentially pathogenic candidate de novo mutations.
Third, our analyses focused exclusively on single nucleotide variants, which are by far the most common form of both non-pathogenic and pathogenic genetic variation in human genomes (16, 17). Clinical application of non-invasive fetal genome sequencing will require more robust methods for detecting other forms of variation, e.g. insertion-deletions, copy number changes, repeat expansions, and structural rearrangements. Ideally, techniques for the detection of other forms of variation could derive from short sequencing reads in a manner that is directly integrated with experimental methods and algorithms for haplotype-resolved genome sequencing.
The ability to non-invasively sequence a fetal genome to high accuracy and completeness will undoubtedly have profound implications for the future of prenatal genetic diagnostics. Although individually rare, when considered collectively, the ~3,500 Mendelian disorders with a known molecular basis (18) contribute substantially to morbidity and mortality (19). Currently, routine obstetric practice includes offering a spectrum of screening and diagnostic options to all women. Prenatal screening options have imperfect sensitivity and focus mainly on a small number of specific disorders, including trisomies, major congenital anomalies, and specific Mendelian disorders. Diagnostic tests, generally performed through invasive procedures such as chorionic villus sampling and amniocentesis, also focus on specific disorders and confer risk of pregnancy loss that may inversely correlate with access to high-quality care. Noninvasive, comprehensive diagnosis of Mendelian disorders early in pregnancy would provide vastly greater amounts of information to expectant parents, with the greater accessibility inherent to a noninvasive test and without tangible risk of pregnancy loss. The less tangible implication of incorporating this level of information into prenatal decision-making raises many ethical questions that must be considered carefully within the scientific community and on a societal level. A final point is that as in other areas of clinical genetics, our capacity to generate data is outstripping our ability to interpret it in ways that are useful to physicians and patients. In other words, although the non-invasive prediction of a fetal genome may be technically feasible, its interpretation – even for known Mendelian disorders – will remain a paramount challenge (20).
Materials and Methods
Whole-genome shotgun library preparation and sequencing
Genomic DNA was extracted from whole blood, as available, or alternatively from saliva, with the Gentra Puregene Kit (Qiagen), or OrageneDx (DNA genotek), respectively. Purified DNA was fragmented by sonication with a Covaris S2 instrument (Covaris). Indexed shotgun sequencing libraries were prepared using the Kapa Library Preparation Kit (Kapa Biosystems), following the manufacturer’s instructions. All libraries were sequenced on HiSeq 2000 instruments (Illumina) using paired-end 101 bp reads with an index read of 9 bp.
Maternal plasma library preparation and sequencing
Maternal plasma was collected by standard methods and split into 1 ml aliquots which were individually purified with the Qiaamp Circulating Nucleic Acids kit (Qiagen). DNA yield was measured with a Qubit fluorometer (Invitrogen). Sequencing libraries were prepared with the ThruPlex-FD kit (Rubicon Genomics), comprising a proprietary series of end-repair, ligation, and amplification reactions. Index read sequencing primers compatible with the WGS and fosmid libraries from this study were included during sequencing of maternal plasma libraries to permit detection of any contamination from other libraries. The percentage of fetal-derived sequences was estimated from plasma sequences by counting alleles specific to each parent as well as sequences mapping specifically to the Y chromosome (fig. S5).
Maternal haplotype resolution via clone pool dilution sequencing
Haplotype-resolved genome sequencing was performed essentially as previously described (5), with minor updates to facilitate processing in a 96-well format. Briefly, high molecular weight DNA was mechanically sheared to mean size ~38 kbp using a Hydroshear instrument (DigiLab), with the following settings: volume=120 ul, cycles=20, speed code=16. Sheared DNA was electrophoresed through 1% Low Melting Point UltraPure agarose (Invitrogen) with the buffer (0.5X TBE) chilled to 16 °C, using the following settings on a BioRad ChefDR-II pulsed field instrument: 170V, initial A=1, final A=6. After running for 17 h, lanes containing size markers (1 kbp extension ladder, Invitrogen) were excised, stained with SYBR Gold dye (Invitrogen), and placed alongside the unstained portion of the gel on a blue light transilluminator. The band between 38–40 kbp was then excised, melted for 10 min in a 70 °C water bath, spun at 15,000 rpm to pellet debris, and incubated at 47 °C for 1 h with 0.5 units beta-agarase (Promega) per 200 mg gel to digest the agarose. Sheared, size-selected DNA was precipitated onto Ampure XP beads (Beckman Coulter) as follows: 100 ul of beads in the supplied buffer were supplemented with additional binding buffer (2.5M NaCl + 20% PEG 8000) to match the volume of the digested gel and DNA. The beads and buffer were then gently mixed with the DNA/agarase reaction mixture, pelleted and rinsed following the manufacturer’s directions, and finally eluted into 60 ul H2O. DNA was next end-repaired with the End-IT kit (Epicentre), cleaned up by precipitation onto 30ul Ampure XP beads supplemented with 70ul additional binding buffer, and eluted into 12ul H2O. Ligation to the fosmid vector backbone pCC1Fos and clone packaging were conducted as previously described using the CopyControl Fosmid Construction Kit (Epicentre). A single bulk infection per maternal sample was performed using each phage library and each was then split by dilution into 1.5 ml cultures (LB + 12.5ug/ml chloramphenicol) across a deep-well 96-well plate. The resulting master culture was grown overnight at 37 °C shaking at 225 rpm. The following day, subcultures were made by in 96-well plates by adding 200ul inoculum from each master culture well into fresh outgrowth media (LB + 12.5ug/ml chloramphenicol + 1X final autoinduction solution) to a final volume of 1.5ml per well. After overnight outgrowth (37 °C, 225 rpm shaking), clone pool DNA was extracted by alkaline lysis mini-preparation in 96 well plates, following standard procedures. Indexed Illumina sequencing libraries were prepared in sets of 96 using the Nextera library preparation kit as previously described (21), followed by library pooling and size selection to 350–650 bp.
Variant calling
Reads were split by index, allowing up to edit distance of 3 to the known barcode sequences, and then mapped to the human reference genome sequence (hg19) using bwa v0.6.1 (5). After removing PCR duplicate read pairs using the Picard toolkit (http://picard.sourceforge.net/), local realignment around indels, variant discovery, quality score recalibration and filtering to 99% estimated sensitivity among known polymorphisms was performed using the Genome Analysis Toolkit (22) using “best practices” parameters provided by the software package’s authors (http://www.broadinstitute.org/gsa/wiki/index.php).
Of note, there was no evidence of uniparental disomy or a numerical chromosomal abnormality; the former would have manifested as a large volume of chromosome-specific Mendelian errors (fig. S6), whereas the latter would have been detected by chromosome-specific read-depth imbalance (fig. S5).
Haplotype assembly
Reads were split per dilution pool by barcode, and a sliding-window read depth measure was used to infer clone positions (5). Using custom scripts, clone pool reads were re-genotyped against heterozygous SNPs ascertained by shotgun sequencing, and overlapping clones from different pools were assembled into haplotype blocks with a custom implementation of the HapCUT algorithm (23).
Inference of the fetal genome sequences
A Hidden Markov model (HMM) was constructed to infer the inherited maternal allele at each maternal-specific heterozygous site. The model’s latent state defines which of the two phased maternal haplotype blocks is inherited at each site, with a third state representing a between-block region at which phase is unknown. The HMM emits allele counts at each phased site, with probabilities given by binomial distribution parameterized as follows: if the maternally inherited allele is identical to the paternal (homozygous) allele at a given “maternal-only” heterozygous site, the probability of observing k such alleles among N total reads with fetal percentage F is
where the first term in the second binomial parameter represents the expected allele balance in the maternally-derived DNA in the maternal plasma, the second term represents the expected contribution of the paternal allele via the fetus, and the third term represents the expected contribution of the inherited maternal allele via the fetus.
If the inherited maternal allele and the paternal allele differ at a given site, the probability of observing k inherited maternal alleles simplifies to
Inferred transitions within phased blocks represent either true recombination events or switch errors in maternal phasing. Transition probabilities within phased blocks were held constant at 10−5; changing this parameter did not substantially affect either the number of inferred transitions within blocks nor the final accuracy. Finally, the most probable path through the observed data was determined using the Viterbi algorithm for inference of the latent state at each site, corresponding to a prediction of the inherited maternal allele. Prediction accuracy was determined by comparing the predicted to actual inheritance determined from the offspring’s genotype.
Inheritance at “paternal-only” heterozygous sites was predicted using a binomial model. At each such site, either the paternal-specific allele or the allele shared with the mother can be transmitted. Let F represent the fetal DNA concentration in the maternal plasma and N represent the depth at a given site. If the paternal-specific allele is transmitted, we expect to observe it in N × F/2 times in the maternal plasma. Similarly, if the paternal-specific allele is not transmitted, we expect to observe it 0 times. The likelihoods of observing K such alleles from N total under each inheritance models were compared, and prediction was determined by choosing the model that yielded a higher likelihood.
At each shared heterozygous site (i.e., heterozygous in both parents), the maternally contributed allele was predicted based on the inferred inheritance of the block in which the site is situated, as determined by “maternal-only” heterozygous sites within the same block. In the rare event that a block was identified to be partially inherited, either due to a real recombination event or a switch error in phasing, the inferred inheritance of the nearest “maternal-only” heterozygous site within the block was used to assign a prediction.
True de novo mutations in each offspring were identified from the trio shotgun whole gneome sequences as follows: starting with all sites called as heterozygous in the offspring and homozygous reference in both parents, known variants were removed (dbSNP v135 and 1000 Genomes Pilot 1 sites), as were sites with low coverage in either parent (< 15 reads for the I1 trio, < 10 reads for the G1 trio). Candidate de novo alleles present at high quality positions in at least one read in any other individual (Phred-scaled base quality >= 10 and mapping quality >= 20) were removed. Finally, a minimum variant quality score threshold of 230 was applied. De novo mutations were validated by PCR and direct capillary sequencing (table S1, table S4).
Downsampling methodology
The effect of reduced fetal contribution to the maternal plasma sequences was investigated by diluting the fetal-specific sequences in silico and reanalyzing the modified data. Simulated dilution of fetal content was carried out as follows. At each maternal-specific heterozygous site, alleles A and B were observed with counts NA and NB among the full dataset, with NTOTAL = NA + NB. For a given dilution coefficient D/F where 0 < D < F, the total pool of observed counts was diluted by first increasing NTOTAL by a factor of F/D, with additional counts allocated by assigning each new allele randomly to NA or NB with equal probability, and then sampling counts from the temporarily expanded pool by discarding each allele from NA and NB with probability 1 − D/F. Updated counts and fetal content estimates were used as input into the Hidden Markov model described above. Reduced coverage within plasma data was separately simulated by subsampling a portion of the observed counts at each site. For a given proportion S, each observed base was discarded with probability 1 − S. Updated counts were then used as input into the Hidden Markov model as described.
Supplementary Material
Figure S1. Accuracy of maternal transmission inference as a function of haplotype block size.
Figure S2. Inference accuracy for paternal transmission at paternal-only heterozygous sites, as a function of plasma shotgun sequencing depth (median=78-fold).
Figure S3. Shared heterozygous sites are primarily common polymorphisms.
Figure S4. Detecting sites of de novo mutation among maternal fetal plasma sequences.
Figure S5. Coverage of autosomes and sex chromsomes and estimation of percent fetal contribution to maternal plasma sequences.
Figure S6. Lack of uniparental disomy in either trio.
Table S1. De novo point mutations identified by whole-genome shotgun sequencing in two trios.
Table S2. Individuals, sample materials, and median sequencing depth (after duplicate read removal) for second trio “G1”.
Table S3. Number of sites and accuracy of fetal genotype inference from maternal plasma sequencing for trio “G1”.
Table S4. PCR primer sequences
Acknowledgments
We thank C. Lee, B. Munson, D. Nickerson and the Northwest Genomics Center (U.W.) for assistance with sequencing; J. Langmore and E. Kamberov (Rubicon Genomics) for early access to reagents; M. McMillin (U.W.) for sample coordination; and members of the Shendure Lab for helpful discussions.
Funding: Our work was supported in part by grants from the National Institutes of Health/National Human Genome Research Institute (J.S.), a gift from the Washington Research Foundation (J.S.) and a National Science Foundation Graduate Research Fellowship (J.O.K.). E.E.E. is an investigator of the Howard Hughes Medical Institute. A portion of this research was conducted using specimens collected and stored by the Maternal Fetal Tissue Bank supported by the Department of Obstetrics & Gynecology at the University of Iowa, or by the Global Alliance to Prevent Prematurity and Stillbirth (GAPPS) Repository.
Footnotes
Author contributions: J.O.K. and J.S. conceived and designed the study. L.E.S., H.S.G., C.E.R., D.A.S, J.C.M, and M.J.B. recruited patients and provided samples. J.O.K., M.V., A.P.L., and R.Q. performed experiments. E.E.E. supervised experiments. J.O.K. and M.W.S. performed analyses. J.O.K., MW.S. and J.S. wrote the manuscript with substantial input from all of the authors. All aspects of the study were supervised by J.S.
Competing interests: J.S. is a member of the scientific advisory board or serves as a consultant for Ariosa Diagnostics, Stratos Genomics, Good Start Genetics, and Adaptive TCR. E.E.E. is on the scientific advisory boards for Pacific Biosciences, Inc., DNANexus, and SynapticDx Corp. A provisional patent application has been deposited for aspects of the methods disclosed here (J.O.K., MW.S. and J.S.).
Data and materials: The data for this study have been deposited in the database dbGaP under accession “phs000500.v1.p1”.
References
- 1.Fan HC, Blumenfeld YJ, Chitkara U, Hudgins L, Quake SR. Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proc Natl Acad Sci USA. 2008;105:16266–16271. doi: 10.1073/pnas.0808319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chiu RWK, Chan KCA, Gao Y, Lau VYM, Zheng W, Leung TY, Foo CHF, Xie B, Tsui NBY, Lun FMF, Zee BCY, Lau TK, Cantor CR, Lo YMD. Noninvasive prenatal diagnosis of fetal chromosomal aneuploidy by massively parallel genomic sequencing of DNA in maternal plasma. Proc Natl Acad Sci USA. 2008;105:20458–20463. doi: 10.1073/pnas.0810641105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dennis Lo YM, Chiu RWK. Prenatal diagnosis: progress through plasma nucleic acids. Nat Rev Genet. 2007;8:71–77. doi: 10.1038/nrg1982. [DOI] [PubMed] [Google Scholar]
- 4.Lo YMD, Chan KCA, Sun H, Chen EZ, Jiang P, Lun FMF, Zheng YW, Leung TY, Lau TK, Cantor CR, Chiu RWK. Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Sci Transl Med. 2010;2:61ra91. doi: 10.1126/scitranslmed.3001720. [DOI] [PubMed] [Google Scholar]
- 5.Kitzman JO, MacKenzie AP, Adey A, Hiatt JB, Patwardhan RP, Sudmant PH, Ng SB, Alkan C, Qiu R, Eichler EE, Shendure J. Haplotype-resolved genome sequencing of a Gujarati Indian individual. Nat Biotechnol. 2011;29:59–63. doi: 10.1038/nbt.1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nygren AOH, Dean J, Jensen TJ, Kruse S, Kwong W, van den Boom D, Ehrich M. Quantification of fetal DNA by use of methylation-based DNA discrimination. Clin Chem. 2010;56:1627–1635. doi: 10.1373/clinchem.2010.146290. [DOI] [PubMed] [Google Scholar]
- 7.Lo YM, Tein MS, Lau TK, Haines CJ, Leung TN, Poon PM, Wainscoat JS, Johnson PJ, Chang AM, Hjelm NM. Quantitative analysis of fetal DNA in maternal plasma and serum: implications for noninvasive prenatal diagnosis. Am J Hum Genet. 1998;62:768–775. doi: 10.1086/301800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lo YM, Zhang J, Leung TN, Lau TK, Chang AM, Hjelm NM. Rapid clearance of fetal DNA from maternal plasma. Am J Hum Genet. 1999;64:218–224. doi: 10.1086/302205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.MacArthur DG, Balasubramanian S, Frankish A, Huang N, Morris J, Walter K, Jostins L, Habegger L, Pickrell JK, Montgomery SB, Albers CA, Zhang ZD, Conrad DF, Lunter G, Zheng H, Ayub Q, DePristo MA, Banks E, Hu M, Handsaker RE, Rosenfeld JA, Fromer M, Jin M, Mu XJ, Khurana E, Ye K, Kay M, Saunders GI, Suner M-M, Hunt T, Barnes IHA, Amid C, Carvalho-Silva DR, Bignell AH, Snow C, Yngvadottir B, Bumpstead S, Cooper DN, Xue Y, Romero IG, Wang J, Li Y, Gibbs RA, McCarroll SA, Dermitzakis ET, Pritchard JK, Barrett JC, Harrow J, Hurles ME, Gerstein MB, Tyler-Smith C 1000 Genomes Project Consortium. A systematic survey of loss-of-function variants in human protein-coding genes. Science. 2012;335:823–828. doi: 10.1126/science.1215040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma L, Xiao Y, Huang H, Wang Q, Rao W, Feng Y, Zhang K, Song Q. Direct determination of molecular haplotypes by chromosome microdissection. Nat Meth. 2010;7:299–301. doi: 10.1038/nmeth.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan HC, Wang J, Potanina A, Quake SR. Whole-genome molecular haplotyping of single cells. Nat Biotechnol. 2011;29:51–57. doi: 10.1038/nbt.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang H, Chen X, Wong WH. Completely phased genome sequencing through chromosome sorting. Proc Natl Acad Sci USA. 2011;108:12–17. doi: 10.1073/pnas.1016725108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conrad DF, Keebler JEM, DePristo MA, Lindsay SJ, Zhang Y, Casals F, Idaghdour Y, Hartl CL, Torroja C, Garimella KV, Zilversmit M, Cartwright R, Rouleau GA, Daly M, Stone EA, Hurles ME, Awadalla P. 1000 Genomes Project, Variation in genome-wide mutation rates within and between human families. Nature Genetics. 2011;43:712–714. doi: 10.1038/ng.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roach JC, Glusman G, Smit AFA, Huff CD, Hubley R, Shannon PT, Rowen L, Pant KP, Goodman N, Bamshad M, Shendure J, Drmanac R, Jorde LB, Hood L, Galas DJ. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science. 2010;328:636–639. doi: 10.1126/science.1186802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ajay SS, Parker SCJ, Abaan HO, Fajardo KVF, Margulies EH. Accurate and comprehensive sequencing of personal genomes. Genome Research. 2011;21:1498–1505. doi: 10.1101/gr.123638.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Durbin RM, Gibbs RA, Hurles ME, McVean GA 1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stenson PD, Ball EV, Howells K, Phillips AD, Mort M, Cooper DN. The Human Gene Mutation Database: providing a comprehensive central mutation database for molecular diagnostics and personalized genomics. Hum Genomics. 2009;4:69–72. doi: 10.1186/1479-7364-4-2-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amberger J, Bocchini CA, Scott AF, Hamosh A. McKusick’s Online Mendelian Inheritance in Man (OMIM) Nucleic Acids Res. 2009;37:D793–6. doi: 10.1093/nar/gkn665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bell CJ, Dinwiddie DL, Miller NA, Hateley SL, Ganusova EE, Mudge J, Langley RJ, Zhang L, Lee CC, Schilkey FD, Sheth V, Woodward JE, Peckham HE, Schroth GP, Kim RW, Kingsmore SF. Carrier testing for severe childhood recessive diseases by next-generation sequencing. Sci Transl Med. 2011;3:65ra4. doi: 10.1126/scitranslmed.3001756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cooper GM, Shendure J. Needles in stacks of needles: finding disease-causal variants in a wealth of genomic data. Nat Rev Genet. 2011;12:628–640. doi: 10.1038/nrg3046. [DOI] [PubMed] [Google Scholar]
- 21.Adey A, Morrison HG, Asan X, Xun, Kitzman JO, Turner EH, Stackhouse B, MacKenzie AP, Caruccio NC, Zhang X, Shendure J. Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome Biol. 2010;11:R119. doi: 10.1186/gb-2010-11-12-r119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature Genetics. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bansal V, Bafna V. HapCUT: an efficient and accurate algorithm for the haplotype assembly problem. Bioinformatics. 2008;24:i153–9. doi: 10.1093/bioinformatics/btn298. [DOI] [PubMed] [Google Scholar]
- 24.Nalls MA, Plagnol V, Hernandez DG, Sharma M, Sheerin U-M, Saad M, Simón-Sánchez J, Schulte C, Lesage S, Sveinbjörnsdóttir S, Stefánsson K, Martinez M, Hardy J, Heutink P, Brice A, Gasser T, Singleton AB, Wood NW International Parkinson Disease Genomics Consortium. Imputation of sequence variants for identification of genetic risks for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet. 2011;377:641–649. doi: 10.1016/S0140-6736(10)62345-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sudmant PH, Kitzman JO, Antonacci F, Alkan C, Malig M, Tsalenko A, Sampas N, Bruhn L, Shendure J, Eichler EE 1000 Genomes Project. Diversity of human copy number variation and multicopy genes. Science. 2010;330:641–646. doi: 10.1126/science.1197005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Accuracy of maternal transmission inference as a function of haplotype block size.
Figure S2. Inference accuracy for paternal transmission at paternal-only heterozygous sites, as a function of plasma shotgun sequencing depth (median=78-fold).
Figure S3. Shared heterozygous sites are primarily common polymorphisms.
Figure S4. Detecting sites of de novo mutation among maternal fetal plasma sequences.
Figure S5. Coverage of autosomes and sex chromsomes and estimation of percent fetal contribution to maternal plasma sequences.
Figure S6. Lack of uniparental disomy in either trio.
Table S1. De novo point mutations identified by whole-genome shotgun sequencing in two trios.
Table S2. Individuals, sample materials, and median sequencing depth (after duplicate read removal) for second trio “G1”.
Table S3. Number of sites and accuracy of fetal genotype inference from maternal plasma sequencing for trio “G1”.
Table S4. PCR primer sequences