SUMMARY
Nucleic acids structures are highly conserved through evolution and when self nucleic acids are aberrantly detected by Toll-Like Receptors (TLRs) they contribute to autoimmune disease. For this reason, multiple regulatory mechanisms exist to prevent response to self nucleic acids. TLR9 is a nucleic acid sensing TLR that is regulated at multiple levels including association with accessory proteins, intracellular localization and proteolytic processing. In the endolysosomal compartment TLR9 is proteolytically processed to an 80 kilodalton form (p80) and this processing is a prerequisite for activation. Here we identified a soluble form of TLR9 generated by a novel proteolytic event that cleaved TLR9 between amino acids 724–735. Similar to p80, sTLR9 was generated in endosomes. However, generation of sTLR9 was independent of the cysteine protease cathepsin B active at acidic pH, but partially dependent on cathepsin S, a protease active at neutral pH. Most importantly, sTLR9 inhibited TLR9-dependent signaling. Together, these data support a model where an intrinsic proteolytic processing mechanism negatively regulates TLR9 signaling. Proper balance between the independent proteolytic events likely contributes to regulation of TLR9 mediated innate immunity and prevention of autoimmune disease.
Keywords: Toll-like receptor, proteolysis, soluble-TLR9
INTRODUCTION
Toll-like receptors (TLRs) detect a variety of microbial structures and initiate protective innate immune responses [1, 2] from the cell surface (TLR1, TLR2, TLR4, TLR5, TLR6) [3–6], and endosomes (TLR3, TLR7, TLR8 and TLR9) [7–9]. Nucleic acids are universally conserved and detected by endosomal TLRs. These TLRs retain the ability to respond to self nucleic acids, and contribute to autoimmune disease [10].
Under normal conditions self nucleic acids are not detected by TLRs due to multiple levels of regulation including receptor localization and trafficking, association with accessory proteins, and proteolytic processing. TLR9 is primarily localized in the endoplasmic reticulum and traffics to endolysosomes [11, 12] through the Golgi [13]. [11]. This maximizes the potential to detect microbial nucleic acids and limits response to self-nucleic acids [14]. Specific motifs in the cytoplasmic tail, and association of TLR9’s transmembrane domain with UNC93B1 [14–16]regulate access to endosomes [14–16]. Inhibition of endosomal/lysosomal acidification blocks cellular response to CpG DNA [17–19] and prevents proteolytic processing of TLR9 [20].
Proteolytic cleavage between amino acids 441–470 of mouse TLR9 occurs in macrophage lysosomes and results in an 80 kDa (p80) TLR9 proteolytic fragment capable of signaling [20, 21]. Additional proteolytic events occur in dendritic cells suggesting that cell types may differ in proteolytic capacity and specificity [22, 23]. Unlike the activating proteolysis for TLR9, TLR2 and TLR4 are proteolytically processed to generate negative regulatory, soluble receptors [24, 25]. Whether similar processing occurs for endosomal TLRs has not been studied.
Here we describe a unique proteolytic cleavage event that resulted in a soluble, negative regulatory, form of TLR9 (sTLR9). The generation of sTLR9 was distinct from the active form of TLR9 since both bafilomycin A1 and cathepsin B inhibitor did not block sTLR9. sTLR9 was retained in endosome-like vesicular structures and functioned as a receptor antagonist. We propose a new model where TLR9 is differentially proteolytically processed to influence, both positively and negatively, the response to DNA.
RESULTS
The ecto-domain of TLR9 is differentially proteolytically processed
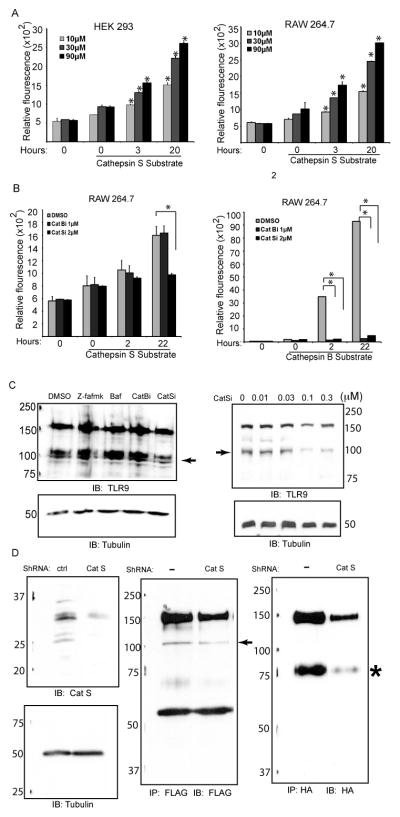
To determine whether proteolytic events occurred in addition to the one that generates the active form of TLR9 (p80), human embryonic kidney 293 (HEK 293) cells were co-transfected with N-terminally Flag tagged mouse TLR9 (Flag-TLR9) and C-terminally HA tagged mouseTLR9 (TLR9-HA). UNC93B1 was included to enhance the proteolytic processing of p80 [20]. When cell lysates were immunoprecipitated and immunoblotted for HA at the C-terminus, both full length and p80 were detected (Fig. 1A). When the same immunoprecipitates were immunoblotted against Flag, very little full length TLR9 was detected. Reactivity with full length TLR9 indicated co-immunoprecipitation and interaction between the two different tagged TLR9s. Also, the p80 form was not detected when immunoblotted for Flag, since it lacks the N-terminal 440 amino acids. To identify additional proteolytic products, TLR9 was immunoprecipitated and immunoblotted with anti-flag (N-terminus). This analysis revealed full length TLR9, a 68 KDa band that likely corresponds to the N-terminal fragment of p80, and a novel 100 kDa fragment. The 100 kDa fragment was reactive with the flag antibody, but not with the HA antibody, indicating that it contained the N-terminus, but not the C-terminus of TLR9 (Fig. 1A). To confirm the unique nature of the p80 and 100 kDa fragments, cells were immunoprecipitated simultaneously with both HA and Flag antibodies and were immunoblotted simultaneously with both HA and Flag antibodies. This permitted detection of all three forms of TLR9 i.e., full length, p80 and 100kDa fragments (Fig. 1A, middle panel). This study confirmed that TLR9 was differentially proteolytically processed to produce two forms, one form lacks the N-terminal end of ecto-domain, but contains the transmembrane and C-terminus (p80), and the other form lacks the transmembrane and cytoplasmic tail. To determine whether this proteolytic event also occurred with endogenous TLR9, and with human TLR9, lysates from HEK 293 cells stably expressing human TLR9-YFP, and human intestinal epithelial cell lines expressing endogenous TLR9 were immunoprecipitated and immunoblotted for N-terminus with anti-TLR9 antibody. This approach confirmed that the 100 kDa fragment of TLR9 was generated with human and mouse TLR9, in multiple cell types, and with endogenous protein (Fig. 1B and Fig. 1C). These data conclusively demonstrated that TLR9 was alternatively proteolytically processed at two distinct sites in the receptor ecto-domain.
Figure 1.
TLR9 is proteolytically processed to generate a unique 100 kDa polypeptide. (A) HEK 293 cells were co-transfected with Flag-mTLR9, mTLR9-HA, and UNC93B1. The lysates were immunoprecipitated for either FLAG, or HA. The immunoprecipitates were resolved by SDS-PAGE gel, transferred to nitrocellulose, and immunoblotted for either FLAG (N-Term, left panel), HA (C-Term, right panel), or both FLAG and HA simultaneously (middle panel). In the lanes where there is both Flag-mTLR9 and mTLR9-HA, individual immunoprecipitates were mixed prior to loading on the gel. ➛, full length and p100 forms of TLR9; * p80, ** N-fragment of p80. (B) Immunoblot analysis for TLR9 (N-Term) and GFP/YFP (C-Term) immunoprecipitates from HEK 293 cells without (−) or with stable expression of human TLR9-YFP (9). Membranes were immunoblotted with antibody to TLR9 (N-Term). ➛, p100. (C) Lysates from Henle 407 (Henle), T84, and Caco-2 cells were immunoprecipitated for TLR9 (N-Term) and assayed by immunoblotting for TLR9 (N-Term). ➛ p100. Data are representative of three independent experiments.
Identification of the proteolytic cleavage site to generate soluble TLR9 (sTLR9)
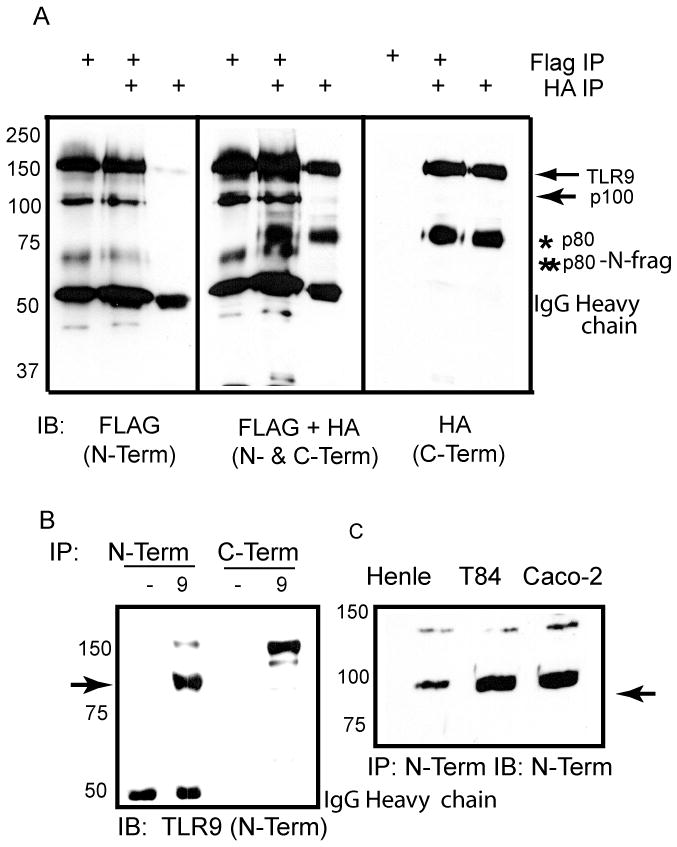
To determine where the novel proteolytic event occurred, the 100 kDa fragment was immunoprecipitated and subjected to liquid chromatography tandem mass spectrometry. Nine peptides in the ecto-domain of TLR9 were unambiguously identified (. 2A, underlined). No peptides C-terminal to amino acid 723 were observed. Since the most C-terminal peptide detected by mass spectrometry ended with amino acid 723, the cleavage site was conclusively determined to lie between amino acids 724 and 735 (Fig. 2A bold, italics). The de-glycosylated p100 form of TLR9 resolved at 75 kDa (Fig. 2B) consistent with the predicted molecular weight of TLR9 ending at amino acid 723. Alignment of TLR9 sequences from different species demonstrated that this region was highly conserved, supporting its biological significance (Fig. 2C).
Figure 2.
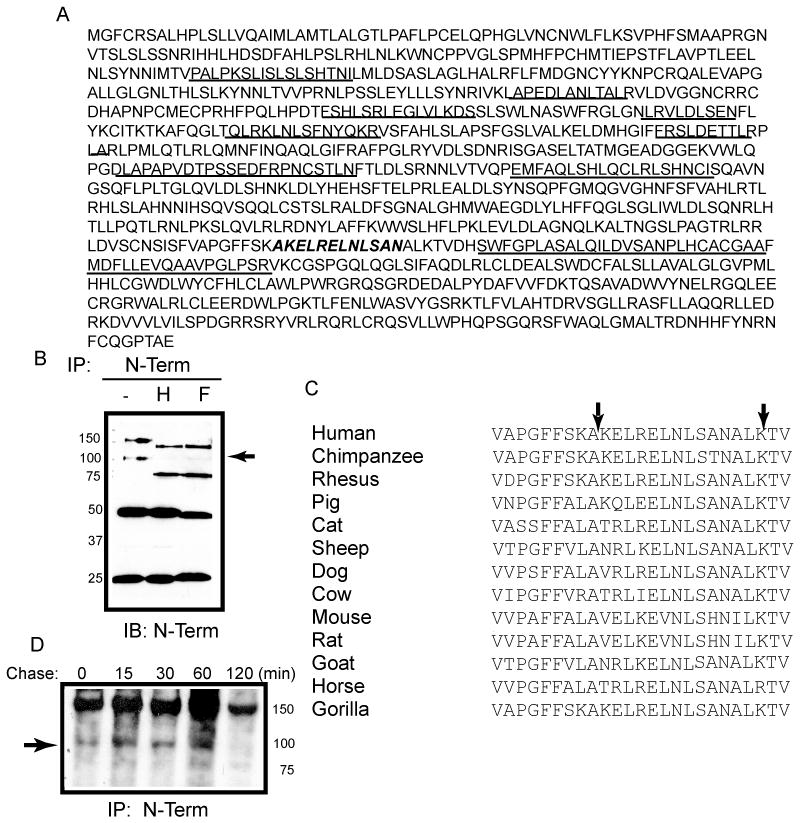
Identification of the C-terminal boundary of sTLR9 by tandem mass spectrometry. (A) Human TLR9 ecto-domain amino acid sequence with the peptides that were identified and sequenced by tandem mass spectrometry underlined and the predicted cleavage site italicized and bolded. (B) HEK 293 cells expressing TLR9-YFP were immunoprecipitated with anti-TLR9 and were either untreated (−) or treated with either endo H (H), or PNGase F (F) prior to resolving by SDS-PAGE and assaying by immunoblotting for the N-terminus (anti-TLR9). ➛ deglycosylated soluble TLR9 (C) Alignment of TLR9 LRRs 24–25 from different species. Arrows indicate the boundaries of the identified proteolytic site. (D) Pulse-chase analysis of proteolytic generation of sTLR9 in mouse embryonic fibroblasts. ➛ sTLR9. Data are representative of two independent experiments.
To determine the kinetics of sTLR9 generation, a pulse chase experiment was performed. The p100 fragment was detected as early as time 0 following a 30 minute pulse, and decreased over the 120 minute chase (Fig. 1D). Therefore, in contrast to p80 [20], the p100 fragment was rapidly generated and had a short half-life. Since the 100kDa fragment lacks the transmembrane and TIR domain of TLR9, yet retains ligand binding sites, we conclude that the 100 kDa fragment is a soluble form of TLR9 (sTLR9) [26].
Soluble TLR9 is generated and retained intra-cellularly
Soluble versions of TLR2 and TLR4 are released from cells, bind to ligand, and prevent signaling by the full length receptors [25, 27]. Therefore, we next determined the intracellular compartment in which sTLR9 was generated, and whether sTLR9 was secreted. A chimera between the ecto-domain of TLR9 and the transmembrane and cytoplasmic tail of TLR4 (TLR9–4) has previously been demonstrated to improperly traffic and fail to be proteolytically processed to the p80 active form [14]. Similarly, cells transfected with TLR9–4 contained no p80 or sTLR9 (Fig. 3A and 3B). To determine whether sTLR9 was released from cells, TLR9 was immunoprecipitated from cell supernatants. Despite detection of a known secreted protein, Flag-tagged human MD2, in cell culture supernatants (Fig. 3C) [28], we failed to detect sTLR9 in supernatants of cells that contained sTLR9 (compare cell lysate (lys) to supernatant (sup), Fig. 3D). We next asked whether neutralization of endosomes would interfere with generation of sTLR9. Treatment of cells with bafilomycin A1, an inhibitor of vacuolar-type (+) - ATPase, significantly reduced the amount of p80, as expected (Fig. 3E, right), yet had no effect on the amount of sTLR9 (Fig. 3E, left). We conclude that sTLR9 is generated and retained in the endosomal compartment but the generation of sTLR9 and p80 occur in distinct endocytic compartments.
Figure 3.
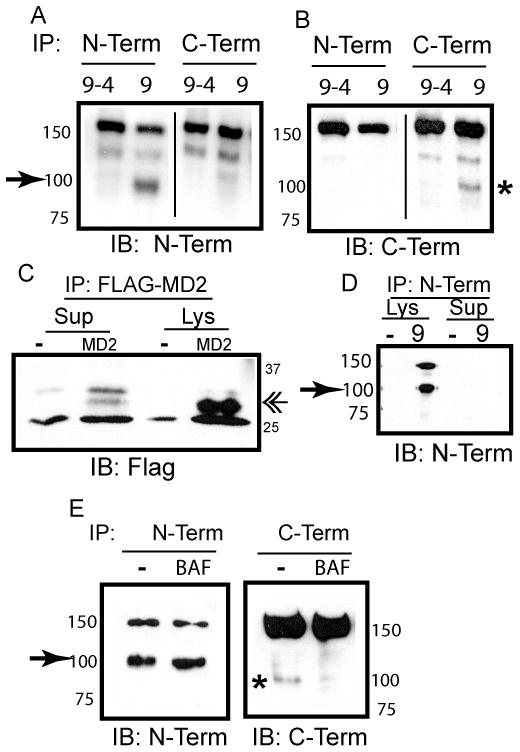
sTLR9 is generated and retained in endosomes, but independently of acidic proteases. (A) HEK 293 cells expressing TLR9–4-GFP (9–4) or TLR9-YFP (9) were immunoprecipitated for TLR9 (N-Term) or GFP/YFP (C-Term). Following resolution by SDS-PAGE and transfer to nitrocellulose, immunoprecipitates were immunoblotted for TLR9 (N-Term).➛ sTLR9 (B) As in (A), except that membranes were immunoblotted for GFP/YFP (C-Term). * p80 (C) Lysates (Lys) or cell culture supernatants (Sup) from HEK 293 cells (−) or HEK 293 cells expressing Flag-MD2 (MD2) were immunoprecipitated, and assayed by immunoblotting for Flag. «-MD2 (D) Lysates (Lys) or cell culture supernatants (Sup) from HEK 293 cells (−) or HEK 293 cells expressing TLR9-YFP (9) were immunoprecipitated for TLR9 (N-Term), and assayed by immunoblotting for TLR9 (N-Term). ➛ sTLR9 (E) HEK 293 cells stably expressing TLR9-YFP were treated with DMSO (−) or 100 nM Bafilomycin A1 (BAF) for 7 hours at 37°C. Lysates were immunoprecipitated for TLR9 (N-Term) or GFP/YFP (C-Term), and assayed by immunoblotting for TLR9 (N-Term) or GFP/YFP (C-Term). ➛ sTLR9; * p80. Data are representative of three independent experiments.
Cathepsin S is necessary for generation of sTLR9
Since bafilomycin A1 treatment did not reduce the abundance of sTLR9, we next asked whether generation of sTLR9 was dependent on cathepsin S, a protease active at neutral pH. Cathepsin S activity was measured by cleavage of a fluorescently labeled substrate. Once cleaved, the fluorophores were dequenched and fluorescence was measured. When lysates from HEK 293 or RAW 264.7 cells were incubated with cathepsin S substrate for 0, 3, or 20 hours, there was a time- and substrate concentration-dependent increase in fluorescence (Fig. 4A). These data confirmed that cathepsin S was active in both cell types. We confirmed the activity of cathepsin S and B inhibitors using the same assay. Cathepsin S enzyme activity was inhibited by cathepsin S inhibitor but not by cathepsin B inhibitor (Fig 4B). However, both cathepsin B inhibitor and cathepsin S inhibitor blocked cathepsin B activity. Generation of p80 and sTLR9 was assessed following treatmet with Z-FA-fmk, bafilomycin, or inhibitors to cathepsin B or cathepsin S. Cathepsin S inhibitor blocked the generation of sTLR9 while neither cathepsin B inhibitor nor Z-FA-fmk reduced sTLR9 (Fig. 4C). The reduction in sTLR9 by cathepsin S inhibitor was concentration dependent (Fig. 4C). Since there was some non-specificity of the inhibitors, we used a more specific shRNA mediated knockdown to address the role of cathepsin S in generation of soluble TLR9. Stable cell lines for cathepsin S shRNA had reduced abundance of full length TLR9; however, these cells also had reduced abundance of both p80 and sTLR9 (Fig. 4D). Therefore we concluded that generation of sTLR9 was dependent, at least in part, on the activity of cathepsin S.
Figure 4.
Generation of sTLR9 is regulated by cathepsin S. (A) The lysates from RAW 264.7 and HEK 293 cells were incubated with 10, 30, or 90 μM of quenched fluorescent cathepsin S substrate. Some lysates were incubated without substrate to determine background fluorescence (0, no substrate). At 0, 3, and 20 hours fluorescence was measured. Increased fluorescence indicated cleavage of the substrate. (B) Cathepsin S and cathepsin B activity were measured as in (A) except the RAW 264.7 cells were incubated with DMSO control, 2μM cathepsin S inhibitor or 1μM cathepsin B inhibitor and fluorescence was measured at 0, 2, and 22 hours. (C) HEK293 cells stably expressing TLR9-YFP were incubated for six hours with 10μM Z-FA-fmk, 100nM Bafilomycin A1, 10μM cathepsin B inhibitor or 30nM cathepsin S inhibitor (left panel), or with different concentrations of cathepsin S inhibitor (right panel). Whole cell lysates were resolved by SDS-PAGE, transferred to nitrocellulose, and immunoblotted for TLR9 (N-Term) and tubulin. ➛ sTLR9. (D) Whole cell lysates of HEK 293 cells stably tranduced with control or cathepsin S shRNA were resolved by SDS-PAGE and analyzed by immunoblot analysis with anti-cathepsin S, or tubulin (left panel). Cathepsin S shRNA or control HEK 293 cells were transfected with UNC93B1 and flag-mTLR9 (middle panel) or UNC93B1 and mTLR9-HA (right panel). The lysates were immunoprecipitated for either FLAG (middle panel) or HA (right panel). The immunoprecipitates were resolved by SDS-PAGE gel, transferred to nitrocellulose, and immunoblotted for either FLAG (N-Term, middle panel) or HA (C-Term, right panel). Data are representative of three independent experiments.
sTLR9 negatively regulates TLR9 signaling
To determine whether sTLR9 inhibited TLR9 signaling, a plasmid encoding sTLR9 was generated encompassing amino acids 1–723. Confocal microscopy analysis of sTLR9 tagged at the C-terminus with yellow fluorescent protein (sTLR9-YFP) demonstrated both a basket-weave and punctate pattern, consistent with endoplasmic reticulum and endosomal localization (Fig. 5A). When co-expressed with full length TLR9, sTLR9 retained dimerization activity since it was co-immunoprecipitated with full length TLR9 (Fig. 5B). Expression of sTLR9-GFP resulted in the full-length soluble form, and several proteolytic fragments suggesting this form of TLR9 was not stable. These data are consistent with the rapid generation and loss of sTLR9 observed by pulse chase analysis (Fig. 2D). To determine whether sTLR9 also bound to CpG DNA, a biotinylated CpG DNA pull down experiment was performed. N-terminal immunoprecipitation demonstrated the presence of both full length and sTLR9 in B cells (Fig. 5C, N-Term IP lanes). As expected, only in cells incubated with 3′ biotinylated CpG DNA, biotin pull-down precipitated full length TLR9 (Fig. 5C). In the same lysate, detection of sTLR9 confirmed that this proteolytic fragment also associated with CpG DNA (Fig. 5C). Since sTLR9 bound to CpG DNA and associated with full-length TLR9, we next asked if it was a dominant-negative inhibitor of TLR9 signaling. Increasing amounts of sTLR9 inhibited TLR9 signaling in a concentration dependent manner (Fig. 5D). This effect was specific since TLR5 signaling was not affected. Together these studies support a model where two distinct proteolytic processing events occur in the ecto-domain of TLR9, one generates an active form of the receptor (p80) and the other generates a negative regulator of TLR9 signaling (sTLR9).
Figure 5.
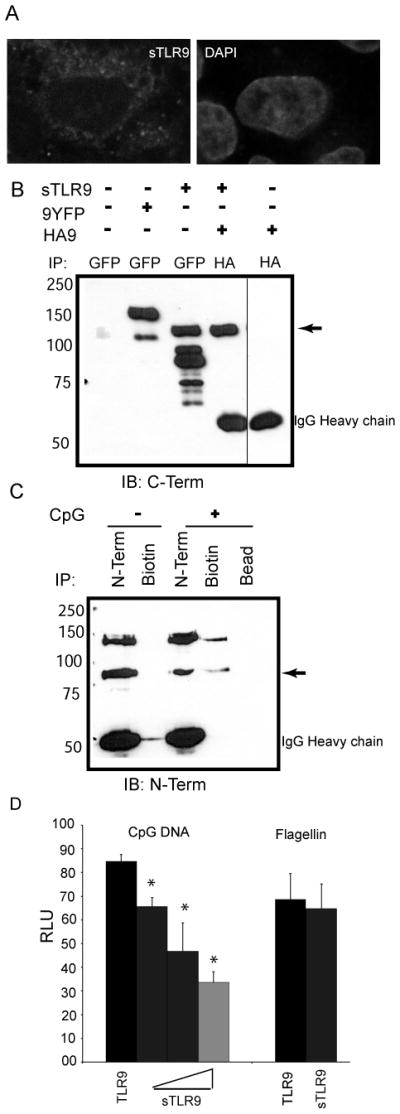
sTLR9 binds to CpG DNA and negatively regulates TLR9 signaling. (A) HeLa cells were transfected with sTLR9 tagged with YFP at the C-terminal end, fixed, mounted in anti-fade with DAPI, and imaged by confocal laser scanning microscopy. (B) HEK 293 cells were transfected with full length TLR9-YFP (9YFP), sTLR9-YFP (sTLR9), N-terminally HA tagged-TLR9 (HA9) or sTLR9-YFP and HA-TLR9. Lysates were immunoprecipitated for GFP/YFP or HA as indicated and assayed by immunoblotting for GFP/YFP. ➛ sTLR9-YFP. (C) A human B cell line was treated with media (−) or 3′ biotinylated CpG DNA (+) for 15 min, lysed, and immunoprecipitated for TLR9 (N-Term), biotin, or incubated with beads alone (Bead). Immunoprecipitates were analyzed by immunoblotting for TLR9 (N-term). ➛ sTLR9-YFP. (D) HEK 293 cells were transfected with a plasmid encoding full length TLR9 either without (black bar) or with increasing concentrations of a plasmid encoding TLR9 amino acids 1–723 (sTLR9) (grey bars), plus an NF-kB-luciferase reporter. After overnight stimulation with 2 μg/mL CpG DNA, or 100 ng/mL flagellin as a control, cells were lysed, and assayed for luciferase activity. Data are representative of three independent experiments.
DISCUSSION
We have discovered a novel mechanism of negative regulation for an endosomal TLR through proteolytic processing of the ectodomain to generate a soluble, ligand binding, form of the receptor. Generation of soluble TLR9 shares some similarities with a previously described proteolytic event, but the regulatory mechanisms are distinct. We found that, unlike the active form, neutralization of endosomal pH had no impact on the generation of sTLR9. Instead, cathepsin S inhibitor blocked the generation of sTLR9 and enhanced signaling. Cathepsin S is primarily expressed in epithelial cells and antigen presenting cells [29, 30], and is found in both early and late endosomes [30]. Targeting of TLR9 to specific endosomal compartments, with different protease repertoires, will likely determine the amount of activating or inhibiting proteolytic processing. A similar mechanism may exist for other nucleic acid-sensing TLRs, including TLR7, and suggests that analysis of total full length abundance of nucleic acid sensing TLRs is inadequate to determine the magnitude of signaling responses.
Once generated, sTLR9 is retained intra-cellularly where it binds ligand and functions as a receptor antagonist. Proteolytic processing also regulates cytokine production through generation and shedding of soluble cytokine receptors, which can abrogate or promote cytokine signaling [33, 34]. Soluble forms of TLR2 and TLR4 (sTLR2, sTLR4) are also secreted and act as decoy receptors [24, 25, 27]. In these studies we show that sTLR9 inhibited TLR9 signaling, yet, was retained intracellularly. Therefore, we propose that sTLR9 is a unique, cell-intrinsic, nucleic acid-sensing TLR antagonist.
In summary, we provide a new mechanism for negative regulation TLR9 signaling. TLR9 is proteolytically processed in the ecto-domain to generate a soluble, ligand-binding competitor. Changes in proteolytic activity, which alter the relative amounts of the two proteolytically processed forms, may contribute to autoimmune inflammatory disease. The proteases responsible for generating the active and inhibitory forms of TLR9 are exciting candidates to target for therapeutically modulating nucleic acid receptor signaling.
MATERIALS AND METHODS
Reagent and plasmids
The following antibodies and reagents were used: human TLR9 and α-tubulin (eBioscience), mouse and human TLR9 (Imgenex), GFP (also detects YFP) (Invitrogen/Molecular Probes for immunoprecipitation and BD Clontech for immunoblotting), phospho- and total-p38 (Cell Signaling Technologies), biotin and Flag (Sigma), hemagglutinin for immuoprecipitation (HA) (ABM), HA for immunoblotting (Roche), cathepsin S (R&D) and secondary antibodies (Southern Biotech); Endoglycosidase H and Peptide N-glycosidase F (New England Biolabs); CpG DNAs (Eurofins MWG operon) 10104: 5′-TCGTCGTTTCGTCGTTTTGTCGTT-3′ and 2006 3′ Biotin: 5′-TCGTCGTTTTGTCGTTTTGTCGTT-3′; S. typhimurium flagellin (Invivogen); Z-fafmk Cathepsin B ( CA-074 Me) and Cathepsin S inhibitor (Calbiochem), and Bafilomycin A1 (Tocris); 5X NF-κB-luciferase (Stratagene), and pSV-β-galactosidase (Promega). Mouse TLR9 was cloned into p3xFLAG-CMV-9 (Sigma). Soluble human TLR9 was cloned into pEYFP using the following primers: Forward: 5′-ATA TAT CTC GAG ATG GGT TTC TGC CGC AGC G- 3′, Reverse: 5′-ATA TAT GGA TCC GCC TCC TTG GCC TTG G- 3′. TLR9–4 was previously described [16].
Cell culture and transfection
All cell lines except BJAB were cultured in DMEM with 2 mM L-glutamine, 50 U/ml penicillin, 50 μg/ml streptomycin, 10 mM Hepes, 1 mM sodium pyruvate and 10% low endotoxin FBS (complete DMEM). The human B cell line, BJAB, was cultured in complete RPMI 1640 medium. Cells routinely tested negative for mycoplasma by PCR. TLR9 stable cells were generated in HEK 293 cells and responded to CpG DNA as determined by 5X NF-κB-luciferase reporter assay. For transient transfection, cells were plated in 6 well plates and the following day were transfected with 4 μg DNA and 8 μl TransIT. After 36 hours the cells were lysed and immunoprecipitated with the indicated antibodies.
Retroviral transduction
Phoenix cells were transfected with MIGR2-mTLR9-HA. Viral supernatant was collected and 0.5 ml added to cells in 12 well plates with 8 μg/ml polybrene and centrifugation at 1811 × g for 90 minutes at 32°C. After 48 hours, cells were lysed and immunoprecipitated with anti-HA antibody.
Luciferase reporter assay
HEK 293 cells were transfected as previously described [13] and treated with CpG DNA or flagellin for 8 hours. Data were analyzed by determining the fold induction compared to unstimulated cells.
Immunoblotting
HEK 293 cells stably expressing TLR9-YFP were treated as indicated, washed once in PBS, and lysed for 15 minutes at 4°C in lysis buffer (137 mM NaCl, 20 mM Tris pH7.4, 1mM EDTA, 0.5% TritonX-100, protease inhibitor cocktail (Sigma) and 100 mM phenylmethylsulphonyl fluoride). Total protein was determined for clarified lysates using the BCA protein assay (Bio-Rad) and 2.0 – 2.5 mg of total protein was used for immunoprecipitation. Note that YFP is a variant of GFP and is recognized by the GFP anitbody. 25 μl of whole cell lysate was immunoblotted for anti-tubulin.
TLR9 deglycosylation
GFP immunoprecipitates from HEK 293 cells stably expressing TLR9-YFP were divided into three equal portions and were either untreated, or treated with endo H or PNGase F according to the manufacturer instructions (New England Biolabs) for two hours at 37°C and the reactions were stopped by adding 6X SDS-PAGE reduced sample buffer. The samples were analyzed by immunoblotting as described above.
Identification of the cleavage site for sTLR9
1×107 HEK 293 cells stably expressing TLR9-YFP per immunoprecipitation reaction (10 total) were lysed in one ml of lysis buffer each and immunoprecipitated with 5 μg/ml anti-TLR9 overnight at 4°C on a rotator. Protein A/G Sepharose was added for one hour at 4°C. After washing, each reaction was sequentially resuspended in 2X SDS-PAGE reduced sample buffer and boiled together for 5 minutes. The combined sample was separated by 8% SDS-PAGE and submitted to the Cornell Proteomics and Mass Spectrometry Core Laboratories Center for gel extraction, trypsin digestion and nano-liquid chromatography tandem mass spectrometry. Identified peptides had a confidence of >99%.
Pulse-chase
Mouse embryonic fibroblasts transiently transfected with Flag-tagged mouse TLR9 were cultured in cysteine and methionine free medium (Gibco) for one hour then pulsed for an additional hour with 0.1 mCi 35S-cysteine and 35S- methionine (Perkin-Elmer). The cells were washed two times in 1× HBSS and chased in complete medium for indicated times. The cells were lysed and immunoprecipitated with anti-FLAG and protein A/G sepharose beads for 4 hours. The immunoprecipitates were washed four times, eluted by boiling in 2X SDS-PAGE reduced sample buffer and separated by SDS-PAGE. The gel was soaked in enhancer (1M sodium salicylate) for 30 minutes and then visualized by autoradiography.
Protease enzyme activity
HEK 293 and RAW 264. 7 cells were lysed in RIPA lysis buffer (1× PBS, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS) and the pH was adjusted to pH 6.0 using 2N HCl. 100μl of the lysate was aliquotted into each well of a 96 well black walled plate (Costar). Cathepsin B and S substrate (Calbiochem) were added at the indicated concentration and the enzyme activity was measured by fluorometry at the indicated times(Gemini EM Microplate Reader, Molecular Devices). Substrate activity was calculated using the following excitation and emission criteria: Cathepsin B substrate (Excitation: 370, Emission: 440), Cathepsin S substrate (Excitation: 340, Emission: 405).
Cathespin s knockdown in HEK 293. Cathepsin S
shRNA and control shRNA were from Sigma-Aldrich (Catalog # SHCLNG-NM_004079). HEK 293 cells were transfected with the shRNAs and the cells were selected in 0.5mg/mL puromycin. Single cell clonal selection was performed and the knockdown of cathepsin S was confirmed by immunoblot.
Confocal microscopy
HeLa cells were transfected with fluorescently labeled sTLR9 on a cover slip in a 24 well plate. After 48 hours the cells were fixed and imaged using a Leica SP5 confocal microscope.
CpG DNA pull-down
BJAB cells (6×107) were treated with 5μM of 3′ biotinylated CpG DNA, lysed and immunoprecipitated with anti-TLR9, anti-biotin antibodies, or beads alone for 4 hours. Protein A/G sepharose beads were added for an additional 12 hours at 4°C. After washing, 2X SDS-PAGE reduced sample buffer was added and proteins were separated on an 8% SDS-PAGE gel, transferred to nitrocellulose, and immunoblotted for TLR9 (N-terminus).
Statistical analysis
Data were analyzed using Instat, Graphpad prism software. Data were compared for significance using an unpaired t-test and were considered significant with p value of < 0.0001. Error bars represent mean ± standard deviation of triplicate samples. Data for cathepsin activity were analyzed by paired t-test.
Acknowledgments
We thank J. Baines (Cornell University) and his lab members for help with the pulse chase experiments, D. Russell for assistance with the cathepsin activity assay, G. Barton (University of CA, Berkley) for mTLR9-HA and B. Beutler (The Scripps research institute, CA) for GFP-UNC93B1. We also thank A. Marshak-Rothstein, C. Norbury and W. Rose II for suggestions on the manuscript. Supported by the National Institutes of Health (AI076588 and AI076588-S1) to CAL.
List of abbreviations
- sTLR9
soluble TLR9
- p80
80 kDa TLR9 proteolytic fragment
Footnotes
AUTHOR CONTRIBUTIONS
A.C, J.B, J.C and C.L performed experiments. A.C and C.L designed the research, analyzed the data, and wrote the paper. The authors declare no conflict of interest.
References
- 1.Janeway CA, Jr, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 2.Takeda K, Takeuchi O, Akira S. Recognition of lipopeptides by Toll-like receptors. J Endotoxin Res. 2002;8:459–463. doi: 10.1179/096805102125001073. [DOI] [PubMed] [Google Scholar]
- 3.Poltorak A, Smirnova I, He X, Liu MY, Van Huffel C, McNally O, Birdwell D, Alejos E, Silva M, Du X, Thompson P, Chan EK, Ledesma J, Roe B, Clifton S, Vogel SN, Beutler B. Genetic and physical mapping of the Lps locus: identification of the toll-4 receptor as a candidate gene in the critical region. Blood Cells Mol Dis. 1998;24:340–355. doi: 10.1006/bcmd.1998.0201. [DOI] [PubMed] [Google Scholar]
- 4.Takeuchi O, Kawai T, Sanjo H, Copeland NG, Gilbert DJ, Jenkins NA, Takeda K, Akira S. TLR6: A novel member of an expanding toll-like receptor family. Gene. 1999;231:59–65. doi: 10.1016/s0378-1119(99)00098-0. [DOI] [PubMed] [Google Scholar]
- 5.Ozinsky A, Underhill DM, Fontenot JD, Hajjar AM, Smith KD, Wilson CB, Schroeder L, Aderem A. The repertoire for pattern recognition of pathogens by the innate immune system is defined by cooperation between toll-like receptors. Proc Natl Acad Sci U S A. 2000;97:13766–13771. doi: 10.1073/pnas.250476497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hayashi F, Smith KD, Ozinsky A, Hawn TR, Yi EC, Goodlett DR, Eng JK, Akira S, Underhill DM, Aderem A. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature. 2001;410:1099–1103. doi: 10.1038/35074106. [DOI] [PubMed] [Google Scholar]
- 7.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 8.Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, Lipford G, Wagner H, Bauer S. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303:1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 9.Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K, Akira S. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 10.Leadbetter EA, Rifkin IR, Hohlbaum AM, Beaudette BC, Shlomchik MJ, Marshak-Rothstein A. Chromatin-IgG complexes activate B cells by dual engagement of IgM and Toll-like receptors. Nature. 2002;416:603–607. doi: 10.1038/416603a. [DOI] [PubMed] [Google Scholar]
- 11.Latz E, Schoenemeyer A, Visintin A, Fitzgerald KA, Monks BG, Knetter CF, Lien E, Nilsen NJ, Espevik T, Golenbock DT. TLR9 signals after translocating from the ER to CpG DNA in the lysosome. Nat Immunol. 2004;5:190–198. doi: 10.1038/ni1028. [DOI] [PubMed] [Google Scholar]
- 12.Leifer CA, Kennedy MN, Mazzoni A, Lee C, Kruhlak MJ, Segal DM. TLR9 is localized in the endoplasmic reticulum prior to stimulation. J Immunol. 2004;173:1179–183. doi: 10.4049/jimmunol.173.2.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chockalingam A, Brooks JC, Cameron JL, Blum LK, Leifer CA. TLR9 traffics through the Golgi complex to localize to endolysosomes and respond to CpG DNA. Immunol Cell Biol. 2009;87:209–217. doi: 10.1038/icb.2008.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barton GM, Kagan JC, Medzhitov R. Intracellular localization of Toll-like receptor 9 prevents recognition of self DNA but facilitates access to viral DNA. Nat Immunol. 2006;7:49–56. doi: 10.1038/ni1280. [DOI] [PubMed] [Google Scholar]
- 15.Brinkmann MM, Spooner E, Hoebe K, Beutler B, Ploegh HL, Kim YM. The interaction between the ER membrane protein UNC93B and TLR3, 7, and 9 is crucial for TLR signaling. J Cell Biol. 2007;177:265–275. doi: 10.1083/jcb.200612056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leifer CA, Brooks JC, Hoelzer K, Lopez JL, Kennedy MN, Mazzoni A, Segal DM. Cytoplasmic targeting motifs control localization of toll-like receptor 9. J Biol Chem. 2006;281:35585–35592. doi: 10.1074/jbc.M607511200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahmad-Nejad P, Hacker H, Rutz M, Bauer S, Vabulas RM, Wagner H. Bacterial CpG-DNA and lipopolysaccharides activate Toll-like receptors at distinct cellular compartments. Eur J Immunol. 2002;32:1958–1968. doi: 10.1002/1521-4141(200207)32:7<1958::AID-IMMU1958>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 18.Hacker H, Mischak H, Miethke T, Liptay S, Schmid R, Sparwasser T, Heeg K, Lipford GB, Wagner H. CpG-DNA-specific activation of antigen-presenting cells requires stress kinase activity and is preceded by non-specific endocytosis and endosomal maturation. Embo J. 1998;17:6230–6240. doi: 10.1093/emboj/17.21.6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Honda K, Ohba Y, Yanai H, Negishi H, Mizutani T, Takaoka A, Taya C, Taniguchi T. Spatiotemporal regulation of MyD88-IRF-7 signalling for robust type-I interferon induction. Nature. 2005;434:1035–1040. doi: 10.1038/nature03547. [DOI] [PubMed] [Google Scholar]
- 20.Ewald SE, Lee BL, Lau L, Wickliffe KE, Shi GP, Chapman HA, Barton GM. The ectodomain of Toll-like receptor 9 is cleaved to generate a functional receptor. Nature. 2008;456:658–662. doi: 10.1038/nature07405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park B, Brinkmann MM, Spooner E, Lee CC, Kim YM, Ploegh HL. Proteolytic cleavage in an endolysosomal compartment is required for activation of Toll-like receptor 9. Nat Immunol. 2008;9:1407–1414. doi: 10.1038/ni.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sepulveda FE, Maschalidi S, Colisson R, Heslop L, Ghirelli C, Sakka E, Lennon-Dumenil AM, Amigorena S, Cabanie L, Manoury B. Critical role for asparagine endopeptidase in endocytic Toll-like receptor signaling in dendritic cells. Immunity. 2009;31:737–748. doi: 10.1016/j.immuni.2009.09.013. [DOI] [PubMed] [Google Scholar]
- 23.Ewald SE, Engel A, Lee J, Wang M, Bogyo M, Barton GM. Nucleic acid recognition by Toll-like receptors is coupled to stepwise processing by cathepsins and asparagine endopeptidase. J Exp Med. 2011;208:643–651. doi: 10.1084/jem.20100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dulay AT, Buhimschi CS, Zhao G, Oliver EA, Mbele A, Jing S, Buhimschi IA. Soluble TLR2 is present in human amniotic fluid and modulates the intraamniotic inflammatory response to infection. J Immunol. 2009;182:7244–7253. doi: 10.4049/jimmunol.0803517. [DOI] [PubMed] [Google Scholar]
- 25.Zunt SL, Burton LV, Goldblatt LI, Dobbins EE, Srinivasan M. Soluble forms of Toll-like receptor 4 are present in human saliva and modulate tumour necrosis factor-alpha secretion by macrophage-like cells. Clin Exp Immunol. 2009;156:285–293. doi: 10.1111/j.1365-2249.2009.03854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bell JK, Mullen GE, Leifer CA, Mazzoni A, Davies DR, Segal DM. Leucine-rich repeats and pathogen recognition in Toll-like receptors. Trends Immunol. 2003;24:528–533. doi: 10.1016/s1471-4906(03)00242-4. [DOI] [PubMed] [Google Scholar]
- 27.Raby AC, Le Bouder E, Colmont C, Davies J, Richards P, Coles B, George CH, Jones SA, Brennan P, Topley N, Labeta MO. Soluble TLR2 reduces inflammation without compromising bacterial clearance by disrupting TLR2 triggering. J Immunol. 2009;183:506–517. doi: 10.4049/jimmunol.0802909. [DOI] [PubMed] [Google Scholar]
- 28.Visintin A, Mazzoni A, Spitzer JA, Segal DM. Secreted MD-2 is a large polymeric protein that efficiently confers lipopolysaccharide sensitivity to Toll-like receptor 4. Proc Natl Acad Sci U S A. 2001;98:12156–12161. doi: 10.1073/pnas.211445098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beers C, Burich A, Kleijmeer MJ, Griffith JM, Wong P, Rudensky AY. Cathepsin S controls MHC class II-mediated antigen presentation by epithelial cells in vivo. J Immunol. 2005;174:1205–1212. doi: 10.4049/jimmunol.174.3.1205. [DOI] [PubMed] [Google Scholar]
- 30.Driessen C, Bryant RA, Lennon-Dumenil AM, Villadangos JA, Bryant PW, Shi GP, Chapman HA, Ploegh HL. Cathepsin S controls the trafficking and maturation of MHC class II molecules in dendritic cells. J Cell Biol. 1999;147:775–790. doi: 10.1083/jcb.147.4.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eaton-Bassiri A, Dillon SB, Cunningham M, Rycyzyn MA, Mills J, Sarisky RT, Mbow ML. Toll-like receptor 9 can be expressed at the cell surface of distinct populations of tonsils and human peripheral blood mononuclear cells. Infect Immun. 2004;72:7202–7211. doi: 10.1128/IAI.72.12.7202-7211.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ewaschuk JB, Backer JL, Churchill TA, Obermeier F, Krause DO, Madsen KL. Surface expression of Toll-like receptor 9 is upregulated on intestinal epithelial cells in response to pathogenic bacterial DNA. Infect Immun. 2007;75:2572–2579. doi: 10.1128/IAI.01662-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Narazaki M, Yasukawa K, Saito T, Ohsugi Y, Fukui H, Koishihara Y, Yancopoulos GD, Taga T, Kishimoto T. Soluble forms of the interleukin-6 signal-transducing receptor component gp130 in human serum possessing a potential to inhibit signals through membrane-anchored gp130. Blood. 1993;82:1120–1126. [PubMed] [Google Scholar]
- 34.Mackiewicz A, Wiznerowicz M, Roeb E, Karczewska A, Nowak J, Heinrich PC, Rose-John S. Soluble interleukin 6 receptor is biologically active in vivo. Cytokine. 1995;7:142–149. doi: 10.1006/cyto.1995.1019. [DOI] [PubMed] [Google Scholar]