Abstract
The active regulation of spine structure and function is of fundamental importance for information storage in the brain. Many proteins involved in spine development and activity-dependent remodelling are potential or validated substrates for modification by the Small Ubiquitin-like Modifier (SUMO). The functional consequences of neuronal protein SUMOylation appear diverse and, in many cases, have not yet been determined. However, for several proteins SUMOylation has been shown to be a key regulator, which has a profound impact on spine dynamics and protein trafficking and function. Here we provide an overview of neuronal SUMOylation and discuss how greater understanding of this relatively recently discovered posttranslational modification will provide insight into the complexity of protein interactions that control synaptic activity and dysfunction.
Introduction
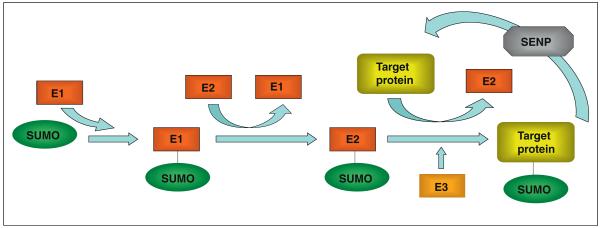
SUMOylation is the addition of a 97-amino acid peptide to the primary amine group of target lysine residues in the substrate protein by an isopeptide bond, via a pathway analogous to ubiquitination. There are four SUMO genes in the mammalian genome, termed SUMO-1–4. SUMO-2 and SUMO-3 differ by only 3 amino acids and are collectively referred to as SUMO-2/3. SUMO-1 shares 50% homology with SUMO-2/3 [1]. Although SUMO-4 mRNA expression has been reported, as yet no SUMO-4 protein has been detected raising doubts as to whether it is physiologically relevant [2]. Excellent reviews are available that detail the SUMOylation pathway (e.g. [1,3,4•]). Briefly, nascent SUMO peptides are matured by cleavage of C-terminal residues by sentrin-specific proteases (SENPs). Mature SUMO is then activated by the E1 enzyme, a heterodimer of SAE1 and SAE2 in mammals, and passed to the active site cysteine of the SUMO-specific conjugating enzyme, Ubc9. Ubc9 corresponds to an E2 ubiquitin ligase but, whereas there are many ubiquitin E2s, Ubc9 is the only SUMO E2 and is required by all three SUMO paralogues. Although in some cases it appears Ubc9 alone is sufficient for SUMOylation, E3 enzymes such as PIAS3 probably increase the substrate specificity of this process. As well as maturing nascent SUMO, SENPs actively deSUMOylate conjugated target proteins. There are six SENP proteins (SENP1–3 and SENP5–7) in mammals, which exhibit selectivity between the SUMO paralogues and have distinct subcellular localisations [11,12]. A schematic of SUMOylation is shown in Figure 1.
Figure 1.
The SUMO pathway. Schematic diagram representing the major proteins involved in SUMO conjugation/deconjugation. Mature SUMO is first activated by the E1 enzyme complex (SAE1/SAE2), before being loaded onto the E2 ligase, Ubc9. Ubc9 then directly binds to the SUMOylation motif and catalyses the formation of an isopeptide bond between the C-terminus of SUMO and the primary amine of the target lysine. This ligation reaction may also involve an E3 enzyme (e.g. PIAS3) to increase the specificity/rate of SUMOylation. See text for more details.
SUMOylation often (but not always) occurs at a consensus ψKxD/E site (where ψ is a large hydrophobic residue) that directly binds Ubc9 [5]. A region of negative charge C-terminal to the consensus site can enhance SUMOylation and this can arise from phosphorylation of nearby residues, providing a potential mechanism for the regulation of SUMOylation by phosphorylation [6]. Much of the work on SUMOylation relates to its nuclear roles, for example regulation of transcription factors and nuclear export (for reviews see [7-8]) and in mice Ubc9 knockout is embryonic lethal owing to defects in nuclear integrity and chromosomal segregation [9].
It is well established that phosphorylation and ubiquitination play essential roles in regulating spine structure and synaptic function [10-11]. SUMOylation is known to have long-term effects on neuronal function via regulation of transcription and nuclear traffic [4•,12]. More recently, however, as we discuss below, it has been shown that SUMOylation also has more rapid, extra-nuclear roles, including regulating neuronal function by influencing pathways that control synaptic structure and function.
Technical considerations—identification of SUMO substrates
Although many neuronal and synaptic proteins contain SUMOylation motifs [13] the consensus sequence is short and relatively degenerate, for example of 5884 open reading frames in Saccharomyces cerevisiae, 2799 contain consensus SUMO sites [14]. It is therefore unlikely that every protein containing this motif is a bone fide substrate. Further, the identification of physiologically SUMOylated proteins in neurones is confounded by the issues that, even for proven SUMO substrates, only a small percentage of the total substrate is modified at any one time [1] and that the SUMOylated form is usually highly transient owing to SENP activity. These problems are exacerbated by the lack of anti-SUMO antibodies for effective immunoprecipitation, which hinders the purification of endogenously SUMOylated proteins.
In part to circumvent these difficulties, alternative approaches have been applied to identify SUMO substrates. These include affinity purification of Ubc9 interactors [15,16•], affinity purification with synthetic SUMO-interaction motifs (SIM) peptides [17] and improved bio-chemical [18] and proteomic [19] techniques to identify SUMOylation sites. However, it should be noted that even with these techniques, identifying specific functionally relevant SUMOylation sites on validated target proteins still remains challenging.
SUMOylation in neurones
SUMOylation is essential for all eukaryotic cells since knockdown or deletion of Ubc9 is lethal [9,20]. In rat brain, the mRNA levels encoding Ubc9 and SUMO-1 are spatio-temporally regulated, with the highest levels in proliferating neuronal stem cells during development and in dentate granule and hippocampal pyramidal neurones in the adult [21], implicating SUMOylation in neuronal differentiation and maturation. In many cases the identity of the SUMOylated neuronal proteins remains to be determined. However, a number of substrates have been validated and these provide insight into the multiple, and previously unsuspected, roles SUMOylation plays in neurones processes, including spine structure and function, synaptic development and plasticity.
Extranuclear protein SUMOylation in synaptogenesis
Spines are highly dynamic actin-rich dendritic protrusions that contain the postsynaptic machinery of excitatory synapses [22]. In addition to neurotransmitter receptors, spines contain scaffolding, adaptor and signalling proteins, organelles and mRNAs. The surface expression of postsynaptic neurotransmitter receptors regulates the efficiency of synaptic transmission and this plasticity is a major mechanism underlying learning, memory and cognition. Remodelling of the spine actin cytoskeleton also contributes to synaptic plasticity by altering spine shape and volume. Thus, dendritic spines are the locus of postsynaptic plasticity and processes that influence plasticity in turn regulate spine structure (for reviews on dendritic spines see [23,24]).
The formation of synapses and spines during development has been extensively studied and many signalling and cytoskeletal protein pathways are implicated [23] in including SUMOylation [21]. SUMOylation of two substrate proteins, CASK and MEF2A have been demonstrated to play key roles. CASK (calcium/calmodulin-dependent serine protein kinase) is a member of the same MAGUK protein family as PSD-95. However, unlike the postsynaptic density protein PSD-95, CASK is widely distributed throughout neurones [25] and is involved in multiple pathways (see [26]). CASK interacts with the synaptogenic factors syndecan-2 and SynCAM [27-28] and also with 4.1 protein [29] and it has been shown to be a key protein in the formation of dendritic spines [23]. CASK is SUMOylated by SUMO-1 at K679, which decreases its interaction with 4.1 protein, and results in reduced spine density and size [30]. 4.1 protein interacts with spectrin and promotes its interaction with the actin cytoskeleton. Thus, CASK SUMOylation probably affects spine formation by reducing the CASK/4.1 protein/actin complex that, in turn, disrupts the localisation and/or function of postsynaptic membrane proteins, such as syndecan-2 and SynCAM. While further work is necessary to define how SUMOylation of CASK is regulated, these observations demonstrate a central role for SUMOylation in orchestrating protein: protein interactions required for spinogenesis.
Transcription factor SUMOylation and synapse formation
Synapse formation also requires the coordinated activation of transcription factors, many of which are regulated by SUMOylation [4•]. During cerebellar development, postsynaptic granule neurones develop dendritic claws onto which mossy fibre axons form presynaptic terminals [31], a process that is regulated by SUMO-1-ylation of the transcription factor MEF2A at K403 [32]. SUMOylation inhibits MEF2A to promote dendritic claw differentiation whereas increased synaptic activity causes deSUMOylation and acetylation of K403, inhibiting synapse formation. This SUMO-acetyl switch is controlled by the dephosphorylation of S408 by calcineurin in response to activity-dependent Ca2+ influx. This regulatory system provides an elegant illustration of how, in an increasing number of pathways, SUMOylation has been shown to act as phosphorylation-regulated switch. Subsequent work showed that overexpression or knockdown of the E3 ligase, PIASx, respectively enhances or reduces dendritic claw formation by increasing or decreasing MEF2A SUMOylation [33]. Further complexity and flexibility was suggested by another study that reported MEF2A transcriptional activity is regulated by PIAS1-promoted SUMOylation of K395 [34].
Interestingly, in these examples SUMO-1-ylation of two different substrates has opposite effects on synaptogenesis. CASK SUMOylation inhibits, whereas MEF2A SUMOylation promotes synapse formation. These contrasting roles highlight the fact that posttranslational modification by SUMO, as for phosphorylation, results in outcomes which are diverse and difficult to predict, suggesting that the effects and functional consequences of SUMOylation are substrate specific rather than generalised for pathways, cells or tissues. Thus, while there are many unanswered questions about the roles of SUMOylation in neuronal development and much work still to do, it is already clear that SUMOylation is a key regulator.
SUMOylation dependent modulation of synaptic activity
Signal transduction and activity-dependent modulation of synaptic activity are the core functions of dendritic spines and changes in synaptic plasticity provide the most widely accepted cellular model of learning, memory and cognition. Synaptic plasticity can be expressed either presynaptically via, for example, changes in neurotransmitter release or postsynaptically by alternations in the number and composition of receptors in the postsynaptic density. There is accumulating evidence that protein SUMOylation plays important roles in both pre- and postsynaptic mechanisms.
Postsynaptic modulation
Long-term potentiation (LTP) and long-term depression (LTD) of synaptic responsiveness is mediated largely by the insertion or removal of AMPA receptors at the postsynaptic membrane (for reviews see [35-36]). Preliminary data have suggested that increased protein SUMOylation is required for LTP (E. Leznik et al., abstract 424.4, Society for Neuroscience Annual Meeting, Chicago, IL, October 2009) and although direct SUMOylation of AMPA receptors themselves has not been demonstrated, other proteins important in regulating neuronal and synaptic activity are SUMO targets.
Several proteins downstream of cell surface glutamate receptors have also been identified as potential SUMO targets. One such protein is Arc/Arg3.1, (activity-related cytoskeletal-associated protein, for reviews see [37,38•]), which acts downstream of mGluR activation to cause AMPAR endocytosis in mGluR-induced LTD [39]. Arc transcription and translation are also induced by synaptic activity [40,41] it plays a role in AMPAR internalisation in homeostatic synaptic scaling [42]. Arc has two consensus SUMOylation sites and it has been proposed that SUMOylation of Arc induces its association with the cytoskeleton [38•] in response to the induction of LTP. Since Arc is required for the maintenance of LTP [43,44], this raises the possibility that SUMOylation of Arc may play a pivotal role in this process. A possible mechanism for this is the remodelling of spine actin cytoskeleton, resulting in changes in spine shape and size associated with LTP [45]. Changes in Arc function and localisation on SUMOylation could also potentially explain how this protein is integral to both AMPAR endocytosis (mGluR-dependent LTD and homeostatic plasticity) and the maintenance of surface AMPAR during late-phase LTP. However, before clear conclusions can be drawn about the possible effects of Arc SUMOylation a great deal of additional work is required, not least an unequivocal demonstration that Arc is indeed a bone fide SUMO substrate.
Kainate receptors regulate synaptic transmission and neuronal excitability [46•]. The GluK2 (formerly GluR6) kainate receptor subunit is a SUMO-1 substrate [15], which binds both Ubc9 and the SUMO E3 ligase, PIAS3, and is SUMOylated at a single C-terminal lysine (K886) in response to kainate stimulation [15]. This SUMOylation is required for agonist-induced endocytosis of GluK2-containing kainate receptors, but not for NMDA-induced KAR endocytosis. Infusion of SUMO-1 causes a specific rundown in kainate-receptor mediated excitatory postsynaptic currents (EPSCs), but has no effect on AMPA receptor-mediated EPSCs. Thus, SUMOylation is involved in the down regulation of kainate receptor surface expression but may be implicated in the up regulation of AMPARs. This potential differential regulation is consistent with the concept that SUMOylation of neuronal proteins can have varied effects, most probably depending on neuronal physiology and the temporal and spatial localisation of the proteins modified.
The regulated internalisation and degradation of neurotransmitter receptors is a crucial determinant for regulation of the receptor content at the synapse. The brain-specific AKAP450-like protein, GISP [47] has a role in the regulation of membrane protein degradation via the ESCRT (endosomal sorting complex required for transport) pathway [48] and is SUMOylated in response to LTP [16•]. GISP was originally identified as a GABAB-interactor, but there is still comparatively little known about it and therefore the SUMOylation of this protein might be an important step in the alteration of the surface levels of different synaptic proteins during synaptic plasticity.
Presynaptic modulation
Presynaptic plasticity is expressed as activity-dependent increases or decreases in levels of neurotransmitter release. Global levels of SUMOylation are increased in synaptosomes following K+ -evoked depolarisation, suggesting that presynaptic proteins undergo activity-dependent SUMOylation [49••]. At least some SUMOylated presynaptic proteins are involved in the regulation of neurotransmitter release since incorporation of SUMO-1, but not non-conjugatable SUMO-1, into synaptosomes caused a decrease in K+-evoked glutamate release whereas inclusion of SENP-1 increased release. Interestingly, these effects were reversed when kainate was used to stimulate glutamate release, implying that protein SUMOylation can either inhibit or enhance presynaptic neurotransmitter release in a stimulus-dependent manner. Although no candidate SUMOylation targets have yet been validated in the literature, numerous presynaptic proteins have SUMO consensus sequences [13]. For example, the syntaxin-1A interacting protein, tomosyn, has recently been shown to be a SUMO-2/3 substrate at K730 [50•]. Mutation of K730 enhances tomosyn inhibition of presynaptic exocytosis, suggesting that SUMOylation of this protein could regulate presynaptic exocytosis.
The G-protein coupled metabotropic glutamate receptor family (mGluRs) [51] have also been implicated in the control of synaptic plasticity by SUMO-1. Group III mGluRs (mGlu4, 6, 7 and 8) are predominantly presynaptic. They interact with the SUMO-E3 enzyme PIAS1 via their C-termini and mGluR8 is SUMOylated in cell lines [52]. Further, all group III mGluR C-termini can be SUMOylated in a bacterial assay [13]. In mGluR7 the SUMOylated lysine is K889 [53•] and in mGluR8a SUMOylation occurs at K882 and K903 [54•]. Importantly, however, despite concerted effort as yet no SUMOylated mGluRs have been detected in neurones and no functional consequences of SUMOylation have been identified, raising the question as to whether group III mGluRs are physiologically relevant SUMO substrates [53•].
mRNA transport and local protein synthesis
Protein translation in dendrites plays an important role in synaptic plasticity and in axons it is required for processes such as axonal guidance [55,56]. Local protein synthesis requires the transport of mRNA to distal processes by mRNA binding proteins. At least one of these proteins, La, is SUMOylated at K41 [57] and SUMOylation probably modulates other mRNA proteins. La SUMOylation regulates interactions with the motor proteins kinesin and dynein, which are necessary for retrograde and anterograde transport respectively [58]. These reports find that SUMOylated La binds only dynein, whereas non-SUMOylated La binds only kinesin, thus determining the direction of the axonal transport of La and its associated mRNAs [57]. This switch-like regulation is both elegant and simple, but it remains unclear how the SUMOylation of La is mediated and how SUMO interacts with other mRNA binding proteins that can influence directional regulation. Identification and characterisation of other mRNA and protein transport proteins is required for but it seems probable that SUMOylation may play a more general role in mRNA delivery.
Regulation of neuronal excitability
In addition to modulation of the pre- and postsynaptic response, synaptic properties and neuronal networks are influenced by more general changes in neuronal excitability. Neuronal excitability is governed by the activity of multiple membrane proteins, such as inhibitory and excitatory receptors and voltage gated ion channels, including the Kv2.1 voltage-gated potassium channel. Recently, SUMOylation of Kv2.1 has been implicated in the regulation of hippocampal excitability [59••]. Inclusion of SUMO-1 in the recording electrode during current clamp experiments increased the rate of action potential firing, whereas SENP1 had the opposite effect. SUMO-1 increased excitability by decreasing the voltage sensitivity of the delayed inwardly rectifying current, IDR. Kv2.1, a component of the IDR, was identified as a SUMO-1 substrate by a FRET-based technique, and SUMOylation at a single consensus site, K470, was shown to affect the voltage sensitivity of the channel. This study suggests another pathway by which SUMOylation can regulate neuronal activity, although the physiological role of this modulation remains unknown. However, it is interesting to note that this SUMO-1 dependent increase in neuronal excitability correlates well with the proposed role of SUMO-1 in LTP induction, as both would lead to an increase in synaptic activity. Additionally, two other K+-channels, K2P [60] (but see [61]) and Kv1.5 in heart [62], have been reported to be SUMO substrates, suggesting that SUMOylation may play a general role in K+-channel regulation.
Conclusion
Neuronal protein SUMOylation is a relatively new and exciting area of research. Steadily increasing numbers of putative and validated nuclear and non-nuclear SUMO targets are being identified and some of these proteins are involved in spine and synapse formation, function and modulation (see Figure 2 and Table 1). Directly analogous to other post-translational modifications, such as phosphorylation and ubiquitination, SUMOylation impacts on a wide range of neuronal and synaptic processes. It is becoming increasingly clear that SUMO modification can have global (cellular function and signalling pathway) effects that are not necessarily easily reconciled with the subtle protein specific effects. Therefore, because the field is still developing it is not yet possible to draw a coherent model for the overarching roles of SUMO in neuronal function, if indeed there is one.
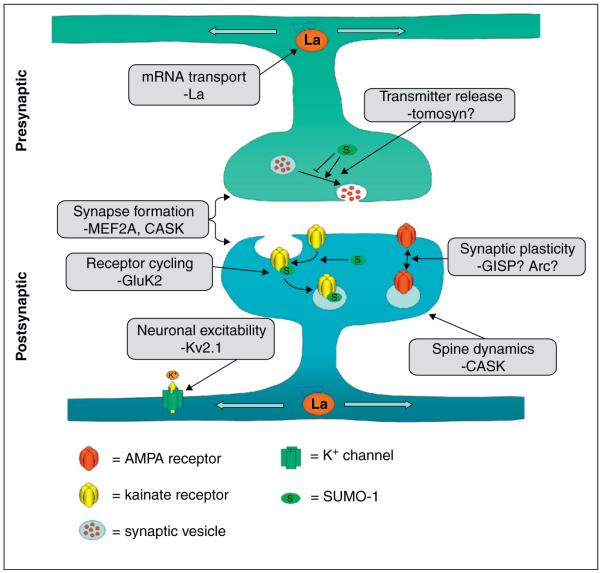
Figure 2.
The many roles of neuronal SUMOylation. SUMOylation has been implicated in a wide variety of synaptic functions, from spine development to modulation of neurotransmitter release. In some cases, it is only known that SUMOylation affects process, as is the case in neurotransmitter release, but no target proteins have been identified. One potential presynaptic target is tomosyn, a syntaxin 1A binding protein. More is known about SUMOylation in the postsynapse, where substrates have been identified which are involved in spine formation, for example MEF2A and CASK, and synaptic modulations, for example GluK2. Modulation of potassium channels by SUMOylation is also likely to be a mechanism by which SUMOylation modulates neuronal function. There are also hints that SUMO-1 modification is a regulator of LTP, but there are, as yet, no definite substrates identified. As can be seen from this figure, the picture is very incomplete and there is, as yet, no all-encompassing general role for SUMO-1 modification at synapses. References to all the processes illustrated in this figure can be found in the text.
Table 1. Neuronal SUMO substrates.
| Substrate | SUMOylation demonstrated in neurones? | Effect/predicted function |
|---|---|---|
| MEF2A | Yes | Regulation of dendritic claw formation |
| CASK | No | Regulation of spine density of width |
| GluK2 | Yes | Agonist-evoked endocytosis |
| Kv2.1 | Yes | Increased neuronal excitability |
| GISP | Yes | Possible involvement in synaptic plasticity |
| Type III mGluRs | No | N/A |
| Tomosyn | No | Disinhibition of neurotransmission? |
| La | Yes | Direction of mRNA transport |
| Arc | No | Unknown |
As is often the case in emergent areas of study, at first sight several reports on the affects of neuronal SUMOylation appear contradictory. For example, SUMOylation has a negative effect on presynaptic neurotransmitter release, whereas SUMOylation of Kv2.1 increases neuronal excitability. However, as has happened for similar apparent discrepancies with phosphorylation, we anticipate that many of these will be resolved as a fuller and more detailed understanding of neuronal and synaptic SUMOylation emerges. Intriguingly, as far as we can currently ascertain, this diversity of function is mainly associated with SUMO-1, which seems to be the main form of activity-dependent SUMOylation in the synapse. SUMOylation by SUMO-2 seems to have a general role in neuroprotection during stress (beyond the scope of this review, but see [63-65]), suggesting a division of function within the SUMO family. It should be noted, however, that two SUMO-1 knockout mice display largely normal phenotypes [66-67], suggesting that compensation may occur between the SUMO pathways.
Future directions
It is already evident that, like other posttranslational modifications, SUMOylation fulfils distinct roles under different conditions and in different areas of the cell. Deeper understanding will require investigation of individual substrates and specifically how their individual regulation by SUMOylation contributes to the larger, more complex interplay that orchestrates neuronal function and dysfunction. In particular, there is a need to define how SUMOylation is temporally and spatially regulated, to identify and validate specific targets and determine how transient SUMOylation regulates their localisation, function and protein interactions. Clearly these are formidable undertakings and will require, among other things, increased understanding the interplay between SUMOylation and other posttranslational modifications and how the SUMO machinery, including Ubc9 or SENPs are regulated.
Given the number of potential and validated targets, SUMOylation clearly represents a major neuronal signalling pathway. Despite significant recent advances, basic questions remain to be addressed, illustrating that understanding and appreciation of neuronal SUMOylation is still at an early stage. For example, how and under what circumstances SUMOylation is involved in mechanisms such as spine development and maturation, synapse formation and synaptic plasticity are still unknown, and relatively few synaptic proteins have been confirmed as SUMO targets. As the field expands and matures we expect that, like phosphorylation, it will become increasing clear that SUMOylation plays a complex and subtle but nonetheless vital role in neuronal physiology and pathology.
Acknowledgements
We are grateful to the MRC, BBSRC, Wellcome Trust and the ERC for funding. We thank Dr Kevin Wilkinson for reading and commenting on the manuscript.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Hay RT. SUMO: a history of modification. Mol Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 2.Su HL, Li SS. Molecular features of human ubiquitin-like SUMO genes and their encoded proteins. Gene. 2002;296:65–73. doi: 10.1016/s0378-1119(02)00843-0. [DOI] [PubMed] [Google Scholar]
- 3.Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- 4•.Hannoun Z, Greenhough S, Jaffray E, Hay RT, Hay DC. Posttranslational modification by SUMO. Toxicology. 2010;278:288–293. doi: 10.1016/j.tox.2010.07.013. [Excellent and comprehensive overview of SUMOylation.] [DOI] [PubMed] [Google Scholar]
- 5.Sampson DA, Wang M, Matunis MJ. The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J Biol Chem. 2001;276:21664–21669. doi: 10.1074/jbc.M100006200. [DOI] [PubMed] [Google Scholar]
- 6.Yang SH, Galanis A, Witty J, Sharrocks AD. An extended consensus motif enhances the specificity of substrate modification by SUMO. EMBO J. 2006;25:5083–5093. doi: 10.1038/sj.emboj.7601383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seeler JS, Dejean A. Nuclear and unclear functions of SUMO. Nat Rev Mol Cell Biol. 2003;4:690–699. doi: 10.1038/nrm1200. [DOI] [PubMed] [Google Scholar]
- 8.Heun P. SUMOrganization of the nucleus. Curr Opin Cell Biol. 2007;19:350–355. doi: 10.1016/j.ceb.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 9.Nacerddine K, Lehembre F, Bhaumik M, Artus J, Cohen-Tannoudji M, Babinet C, Pandolfi PP, Dejean A. The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev Cell. 2005;9:769–779. doi: 10.1016/j.devcel.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 10.Saneyoshi T, Fortin DA, Soderling TR. Regulation of spine and synapse formation by activity-dependent intracellular signaling pathways. Curr Opin Neurobiol. 2010;20:108–115. doi: 10.1016/j.conb.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mabb AM, Ehlers MD. Ubiquitination in postsynaptic function and plasticity. Annu Rev Cell Dev Biol. 2010;26:179–210. doi: 10.1146/annurev-cellbio-100109-104129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilkinson KA, Nakamura Y, Henley JM. Targets and consequences of protein SUMOylation in neurons. Brain Res Rev. 2010;64:195–212. doi: 10.1016/j.brainresrev.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilkinson KA, Nishimune A, Henley JM. Analysis of SUMO-1 modification of neuronal proteins containing consensus SUMOylation motifs. Neurosci Lett. 2008;436:239–244. doi: 10.1016/j.neulet.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson ES. Protein modification by SUMO. Annu Rev Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 15.Martin S, Nishimune A, Mellor JR, Henley JM. SUMOylation regulates kainate-receptor-mediated synaptic transmission. Nature. 2007;447:321–325. doi: 10.1038/nature05736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16•.Kantamneni S, Wilkinson KA, Jaafari N, Ashikaga E, Rocca D, Rubin P, Jacobs SC, Nishimune A, Henley JM. Activity-dependent SUMOylation of the brain-specific scaffolding protein GISP. Biochem Biophys Res Commun. 2011;409:657–662. doi: 10.1016/j.bbrc.2011.05.060. [In this study the authors demonstrate the activity-dependent SUMOylation of GISP in neurones, suggesting it has a role in chem-LTP.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tanaka N, Saitoh H. A real-time SUMO-binding assay for the analysis of the SUMO-SIM protein interaction network. Biosci Biotechnol Biochem. 2010;74:1302–1305. doi: 10.1271/bbb.100082. [DOI] [PubMed] [Google Scholar]
- 18.Tatham MH, Rodriguez MS, Xirodimas DP, Hay RT. Detection of protein SUMOylation in vivo. Nat Protoc. 2009;4:1363–1371. doi: 10.1038/nprot.2009.128. [DOI] [PubMed] [Google Scholar]
- 19.Matic I, Schimmel J, Hendriks IA, van Santen MA, van de Rijke F, van Dam H, Gnad F, Mann M, Vertegaal AC. Site-specific identification of SUMO-2 targets in cells reveals an inverted SUMOylation motif and a hydrophobic cluster SUMOylation motif. Mol Cell. 2010;39:641–652. doi: 10.1016/j.molcel.2010.07.026. [DOI] [PubMed] [Google Scholar]
- 20.Hayashi T, Seki M, Maeda D, Wang W, Kawabe Y, Seki T, Saitoh H, Fukagawa T, Yagi H, Enomoto T. Ubc9 is essential for viability of higher eukaryotic cells. Exp Cell Res. 2002;280:212–221. doi: 10.1006/excr.2002.5634. [DOI] [PubMed] [Google Scholar]
- 21.Watanabe M, Takahashi K, Tomizawa K, Mizusawa H, Takahashi H. Developmental regulation of Ubc9 in the rat nervous system. Acta Biochim Pol. 2008;55:681–686. [PubMed] [Google Scholar]
- 22.Tada T, Sheng M. Molecular mechanisms of dendritic spine morphogenesis. Curr Opin Neurobiol. 2006;16:95–101. doi: 10.1016/j.conb.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 23.Nimchinsky EA, Sabatini BL, Svoboda K. Structure and function of dendritic spines. Annu Rev Physiol. 2002;64:313–353. doi: 10.1146/annurev.physiol.64.081501.160008. [DOI] [PubMed] [Google Scholar]
- 24.Hotulainen P, Hoogenraad CC. Actin in dendritic spines: connecting dynamics to function. J Cell Biol. 2010;189:619–629. doi: 10.1083/jcb.201003008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsueh YP, Sheng M. Regulated expression and subcellular localization of syndecan heparan sulfate proteoglycans and the syndecan-binding protein CASK/LIN-2 during rat brain development. J Neurosci. 1999;19:7415–7425. doi: 10.1523/JNEUROSCI.19-17-07415.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsueh YP. The role of the MAGUK protein CASK in neural development and synaptic function. Curr Med Chem. 2006;13:1915–1927. doi: 10.2174/092986706777585040. [DOI] [PubMed] [Google Scholar]
- 27.Hsueh YP, Yang FC, Kharazia V, Naisbitt S, Cohen AR, Weinberg RJ, Sheng M. Direct interaction of CASK/LIN-2 and syndecan heparan sulfate proteoglycan and their overlapping distribution in neuronal synapses. J Cell Biol. 1998;142:139–151. doi: 10.1083/jcb.142.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Biederer T, Sara Y, Mozhayeva M, Atasoy D, Liu X, Kavalali ET, Sudhof TC. SynCAM, a synaptic adhesion molecule that drives synapse assembly. Science. 2002;297:1525–1531. doi: 10.1126/science.1072356. [DOI] [PubMed] [Google Scholar]
- 29.Biederer T, Sudhof TC. CASK and protein 4.1 support F-actin nucleation on neurexins. J Biol Chem. 2001;276:47869–47876. doi: 10.1074/jbc.M105287200. [DOI] [PubMed] [Google Scholar]
- 30.Chao HW, Hong CJ, Huang TN, Lin YL, Hsueh YP. SUMOylation of the MAGUK protein CASK regulates dendritic spinogenesis. J Cell Biol. 2008;182:141–155. doi: 10.1083/jcb.200712094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Flavell SW, Cowan CW, Kim TK, Greer PL, Lin Y, Paradis S, Griffith EC, Hu LS, Chen C, Greenberg ME. Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science. 2006;311:1008–1012. doi: 10.1126/science.1122511. [DOI] [PubMed] [Google Scholar]
- 32.Shalizi A, Gaudilliere B, Yuan Z, Stegmuller J, Shirogane T, Ge Q, Tan Y, Schulman B, Harper JW, Bonni A. A calcium-regulated MEF2 sumoylation switch controls postsynaptic differentiation. Science. 2006;311:1012–1017. doi: 10.1126/science.1122513. [DOI] [PubMed] [Google Scholar]
- 33.Shalizi A, Bilimoria PM, Stegmuller J, Gaudilliere B, Yang Y, Shuai K, Bonni A. PIASx is a MEF2 SUMO E3 ligase that promotes postsynaptic dendritic morphogenesis. J Neurosci. 2007;27:10037–10046. doi: 10.1523/JNEUROSCI.0361-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Riquelme C, Barthel KK, Liu X. SUMO-1 modification of MEF2A regulates its transcriptional activity. J Cell Mol Med. 2006;10:132–144. doi: 10.1111/j.1582-4934.2006.tb00295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shepherd JD, Huganir RL. The cell biology of synaptic plasticity: AMPA receptor trafficking. Annu Rev Cell Dev Biol. 2007;23:613–643. doi: 10.1146/annurev.cellbio.23.090506.123516. [DOI] [PubMed] [Google Scholar]
- 36.Kessels HW, Malinow R. Synaptic AMPA receptor plasticity and behavior. Neuron. 2009;61:340–350. doi: 10.1016/j.neuron.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shepherd JD, Bear MF. New views of Arc, a master regulator of synaptic plasticity. Nat Neurosci. 2011;14:279–284. doi: 10.1038/nn.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38•.Bramham CR, Alme MN, Bittins M, Kuipers SD, Nair RR, Pai B, Panja D, Schubert M, Soule J, Tiron A, et al. The Arc of synaptic memory. Exp Brain Res. 2010;200:125–140. doi: 10.1007/s00221-009-1959-2. [Good review of Arc function which also mentions the author’s (so far unpublished) observations on Arc SUMOylation, showing that SUMOylation affects Arc localisation and cytoskeletal interaction.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Waung MW, Pfeiffer BE, Nosyreva ED, Ronesi JA, Huber KM. Rapid translation of Arc/Arg3.1 selectively mediates mGluR-dependent LTD through persistent increases in AMPAR endocytosis rate. Neuron. 2008;59:84–97. doi: 10.1016/j.neuron.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Link W, Konietzko U, Kauselmann G, Krug M, Schwanke B, Frey U, Kuhl D. Somatodendritic expression of an immediate early gene is regulated by synaptic activity. Proc Natl Acad Sci USA. 1995;92:5734–5738. doi: 10.1073/pnas.92.12.5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Steward O, Wallace CS, Lyford GL, Worley PF. Synaptic activation causes the mRNA for the IEG Arc to localize selectively near activated postsynaptic sites on dendrites. Neuron. 1998;21:741–751. doi: 10.1016/s0896-6273(00)80591-7. [DOI] [PubMed] [Google Scholar]
- 42.Shepherd JD, Rumbaugh G, Wu J, Chowdhury S, Plath N, Kuhl D, Huganir RL, Worley PF. Arc/Arg3.1 mediates homeostatic synaptic scaling of AMPA receptors. Neuron. 2006;52:475–484. doi: 10.1016/j.neuron.2006.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Plath N, Ohana O, Dammermann B, Errington ML, Schmitz D, Gross C, Mao X, Engelsberg A, Mahlke C, Welzl H, et al. Arc/ Arg3.1 is essential for the consolidation of synaptic plasticity and memories. Neuron. 2006;52:437–444. doi: 10.1016/j.neuron.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 44.Messaoudi E, Kanhema T, Soule J, Tiron A, Dagyte G, da Silva B, Bramham CR. Sustained Arc/Arg3.1 synthesis controls long-term potentiation consolidation through regulation of local actin polymerization in the dentate gyrus in vivo. J Neurosci. 2007;27:10445–10455. doi: 10.1523/JNEUROSCI.2883-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bosch M, Hayashi Y. Structural plasticity of dendritic spines. Curr Opin Neurobiol. 2011 doi: 10.1016/j.conb.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46•.Contractor A, Mulle C, Swanson GT. Kainate receptors coming of age: milestones of two decades of research. Trends Neurosci. 2011;34:154–163. doi: 10.1016/j.tins.2010.12.002. [Excellent review on kainate receptors.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kantamneni S, Correa SA, Hodgkinson GK, Meyer G, Vinh NN, Henley JM, Nishimune A. GISP: a novel brain-specific protein that promotes surface expression and function of GABA(B) receptors. J Neurochem. 2007;100:1003–1017. doi: 10.1111/j.1471-4159.2006.04271.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Henne WM, Buchkovich NJ, Emr SD. The ESCRT pathway. Dev Cell. 2011;21:77–91. doi: 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 49••.Feligioni M, Nishimune A, Henley JM. Protein SUMOylation modulates calcium influx and glutamate release from presynaptic terminals. Eur J Neurosci. 2009;29:1348–1356. doi: 10.1111/j.1460-9568.2009.06692.x. [First evidence that presynaptic SUMOylation can affect neurotransmitter release. The effect reported is stimulus-dependent: SUMO-1 trapping in synaptosomes decreases KCl-induced glutamate release but increases kainate-induced glutamate release.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50•.Williams AL, Bielopolski N, Meroz D, Lam AD, Passmore DR, Ben-Tal N, Ernst SA, Ashery U, Stuenkel EL. Structural and functional analysis of tomosyn identifies domains important in exocytotic regulation. J Biol Chem. 2011;286:14542–14553. doi: 10.1074/jbc.M110.215624. [An interesting study on a relatively unknown syntaxin-1A interacting protein, which finds that it can be SUMOylated and that mutation of the SUMOylation consensus site (K730) enhances tomosyn’s inhibition of exocytosis.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Niswender CM, Conn PJ. Metabotropic glutamate receptors: physiology, pharmacology, and disease. Annu Rev Pharmacol Toxicol. 2010;50:295–322. doi: 10.1146/annurev.pharmtox.011008.145533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tang Z, El Far O, Betz H, Scheschonka A. Pias1 interaction and sumoylation of metabotropic glutamate receptor 8. J Biol Chem. 2005;280:38153–38159. doi: 10.1074/jbc.M508168200. [DOI] [PubMed] [Google Scholar]
- 53•.Wilkinson KA, Henley JM. Analysis of metabotropic glutamate receptor 7 as a potential substrate for SUMOylation. Neurosci Lett. 2011;491:181–186. doi: 10.1016/j.neulet.2011.01.032. [This study finds that the group III mGluR, mGluR7 can be SUMOylated in vitro by both SUMO-1 and SUMO-2, at a single C-terminal lysine (K889). However this could not be deomnstrated in vivo, questioning whether group III mGluRs are relevant SUMO substrates.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54•.Dutting E, Schroder-Kress N, Sticht H, Enz R. SUMO E3 ligases are expressed in the retina and regulate SUMOylation of the metabotropic glutamate receptor 8b. Biochem J. 2011;435:365–371. doi: 10.1042/BJ20101854. [Evidence that mGluR8a can be SUMOylated and that it co-localises with SUMO1, Ubc9 and PIAS3 in retinal ganlgion cells.] [DOI] [PubMed] [Google Scholar]
- 55.Giuditta A, Kaplan BB, van Minnen J, Alvarez J, Koenig E. Axonal and presynaptic protein synthesis: new insights into the biology of the neuron. Trends Neurosci. 2002;25:400–404. doi: 10.1016/s0166-2236(02)02188-4. [DOI] [PubMed] [Google Scholar]
- 56.Martin KC. Local protein synthesis during axon guidance and synaptic plasticity. Curr Opin Neurobiol. 2004;14:305–310. doi: 10.1016/j.conb.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 57.van Niekerk EA, Willis DE, Chang JH, Reumann K, Heise T, Twiss JL. Sumoylation in axons triggers retrograde transport of the RNA-binding protein La. Proc Natl Acad Sci USA. 2007;104:12913–12918. doi: 10.1073/pnas.0611562104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hirokawa N, Takemura R. Molecular motors and mechanisms of directional transport in neurons. Nat Rev Neurosci. 2005;6:201–214. doi: 10.1038/nrn1624. [DOI] [PubMed] [Google Scholar]
- 59••.Plant LD, Dowdell EJ, Dementieva IS, Marks JD, Goldstein SA. SUMO modification of cell surface Kv2.1 potassium channels regulates the activity of rat hippocampal neurons. J Gen Physiol. 2011;137:441–454. doi: 10.1085/jgp.201110604. [This study demonstrates that SUMO-1 and SENP-1 can increase and decrease the excitability of neurones respectively. This effect is found to be due to SUMOylation of Kv2.1 at a single lysine (K470), which affects the channel’s voltage-dependence.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rajan S, Plant LD, Rabin ML, Butler MH, Goldstein SA. Sumoylation silences the plasma membrane leak K+ channel K2P1. Cell. 2005;121:37–47. doi: 10.1016/j.cell.2005.01.019. [DOI] [PubMed] [Google Scholar]
- 61.Feliciangeli S, Bendahhou S, Sandoz G, Gounon P, Reichold M, Warth R, Lazdunski M, Barhanin J, Lesage F. Does sumoylation control K2P1/TWIK1 background K+ channels? Cell. 2007;130:563–569. doi: 10.1016/j.cell.2007.06.012. [DOI] [PubMed] [Google Scholar]
- 62.Benson MD, Li QJ, Kieckhafer K, Dudek D, Whorton MR, Sunahara RK, Iniguez-Lluhi JA, Martens JR. SUMO modification regulates inactivation of the voltage-gated potassium channel Kv1.5. Proc Natl Acad Sci USA. 2007;104:1805–1810. doi: 10.1073/pnas.0606702104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Agbor TA, Taylor CT. SUMO, hypoxia and the regulation of metabolism. Biochem Soc Trans. 2008;36:445–448. doi: 10.1042/BST0360445. [DOI] [PubMed] [Google Scholar]
- 64.Bossis G, Melchior F. SUMO: regulating the regulator. Cell Div. 2006;1:13. doi: 10.1186/1747-1028-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tempe D, Piechaczyk M, Bossis G. SUMO under stress. Biochem Soc Trans. 2008;36:874–878. doi: 10.1042/BST0360874. [DOI] [PubMed] [Google Scholar]
- 66.Evdokimov E, Sharma P, Lockett SJ, Lualdi M, Kuehn MR. Loss of SUMO1 in mice affects RanGAP1 localization and formation of PML nuclear bodies, but is not lethal as it can be compensated by SUMO2 or SUMO3. J Cell Sci. 2008;121:4106–4113. doi: 10.1242/jcs.038570. [DOI] [PubMed] [Google Scholar]
- 67.Zhang FP, Mikkonen L, Toppari J, Palvimo JJ, Thesleff I, Janne OA. Sumo-1 function is dispensable in normal mouse development. Mol Cell Biol. 2008;28:5381–5390. doi: 10.1128/MCB.00651-08. [DOI] [PMC free article] [PubMed] [Google Scholar]