Abstract
Engineered designer nucleases can be used to efficiently modify genomic sequence in a wide variety of model organisms and cell types. Zinc finger nucleases (ZFNs), consisting of an engineered zinc finger array fused to a non-specific cleavage domain, have been extensively used to modify a broad range of endogenous genes. Here we describe protocols for engineering ZFNs targeted to specific gene sequences of interest using the Context-Dependent Assembly (CoDA) method.
Keywords: engineered zinc finger nucleases, engineering zinc finger proteins, context-dependent assembly, protein engineering, DNA-binding domains
Introduction
Engineered zinc finger nucleases (ZFNs) can be used to introduce site-specific genome alterations in a wide variety of model organisms and a range of different mammalian cell types (Rahman et al., 2011; Urnov et al., 2010). ZFNs are customizable restriction enzymes that can be used to create targeted double-stranded breaks (DSBs) in cells. ZFNs consist of an array of engineered zinc fingers fused to the non-specific cleavage domain of the FokI Type IIS restriction enzyme (Kim et al., 1996). The engineered zinc finger portion of the ZFN directs the nuclease activity to a specific location in the genome. Because the FokI domain must dimerize to cleave DNA, a pair of ZFNs must be engineered to cleave a given target site of interest. Each monomer in a ZFN dimer pair binds to a “half-site” with cleavage of the DNA induced in an intervening 5–7 bp “spacer” sequence.
The repair of ZFN-induced DSBs by normal mechanisms used by nearly all cells can be exploited to make targeted genomic alterations. For example, non-homologous end-joining mediated repair of a ZFN-induced break can lead to the efficient introduction of insertion or deletion mutants, providing a means to create frameshift and knockout mutations if introduced into coding sequences of endogenous genes (Bibikova et al., 2002; Santiago et al., 2008). In addition, ZFN-induced DSBs can also be repaired by homologous recombination with an investigator-supplied “donor template” (Bibikova et al., 2003; Urnov et al., 2005). This template consists of DNA sequences homologous to endogenous sequence surrounding the DSB but with mutations or insertions of interest also incorporated. Repair of the ZFN-induced DSB with this donor template can thus be used to introduce particular mutations or insertions into specific genomic loci.
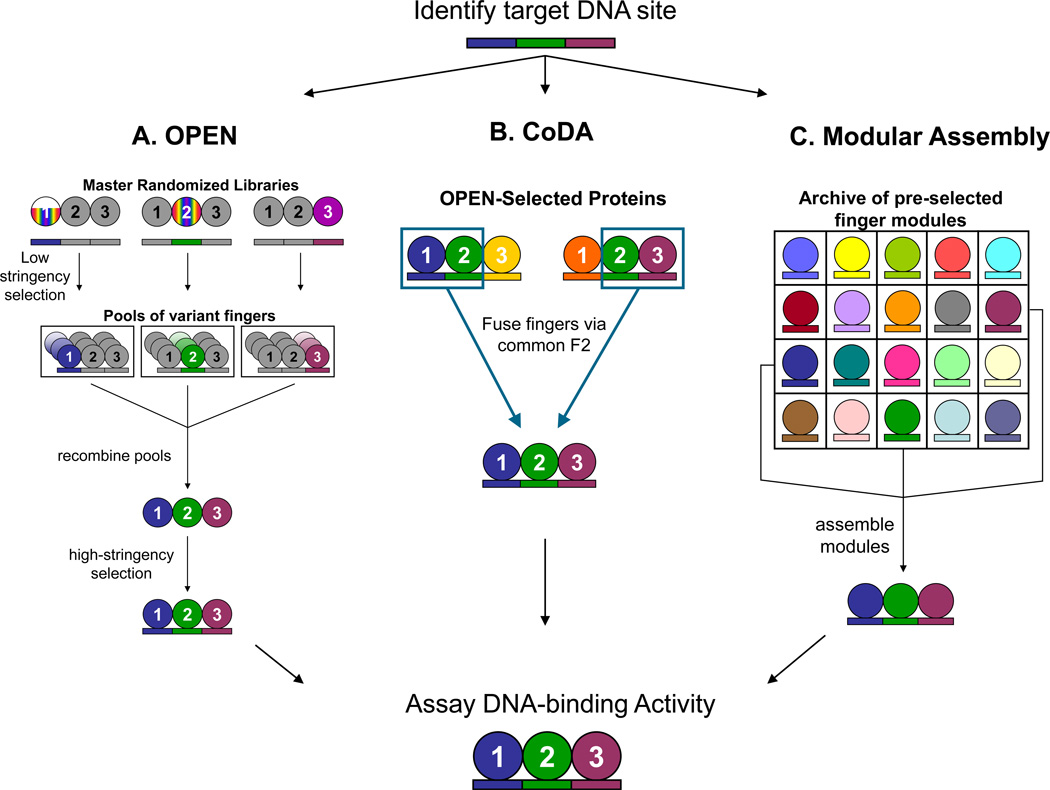
Three methods have been described (Beerli and Barbas, 2002; Maeder et al., 2008; Sander et al., 2011) for engineering zinc finger arrays required to make ZFNs (Figure 1). These methods are known as (1) modular assembly, (2) oligomerized pool engineering (OPEN), and (3) context-dependent assembly (CoDA). Each of these methods possesses different advantages and disadvantages that we discuss briefly here.
Figure 1. Publicly available methods for engineering zinc finger arrays.
A. The Oligomerized Pool Engineering (OPEN) method utilizes master randomized libraries in which up to 95 different zinc fingers have been selected to bind to a 3 bp subsite at a specific position within a 3-finger array. Appropriate combinatorial libraries composed of randomly recombined pools for a 9 bp target site of interest are constructed and then interrogated using a bacterial two-hybrid selection system to yield three-finger proteins selected to bind the 9bp site of interest.
B. The Context Dependent Assembly (CoDA) method takes context-dependent effects between neighboring fingers into account by identifying three-finger arrays selected in the bacterial two-hybrid system that share a common middle finger. These fingers can then be combined, via the common middle finger, to yield a three-finger protein targeted to the 9 bp site of interest.
C. Modular Assembly utilizes an archive of single-finger modules that have been characterized to bind to a specific 3 bp DNA site. These modules are then combined to generate a 3-finger protein.
With modular assembly (Figure 1C), individual pre-selected fingers are joined together to create a zinc finger array (Beerli and Barbas, 2002). This method relies on archives of pre-selected individual zinc fingers and assumes that the behaviors of fingers in an array are independent of one another. The advantage of this method is the ease with which it can be practiced - arrays can be engineered in as little as a week. However, the failure rate of this method for constructing pairs of three-finger ZFNs has been reported to be very high (>94%) (Kim et al., 2009; Ramirez et al., 2008). In addition, three-finger arrays produced by modular assembly have also been shown to have low affinities, low specificities, and low activities as ZFNs in cells (Cornu et al., 2008; Hurt et al., 2003; Ramirez et al., 2008). Various detailed protocols for practicing modular assembly have been previously described (Carroll et al., 2006; Gonzalez et al., 2010; Wright et al., 2006).
The OPEN method, unlike modular assembly, explicitly accounts for the context-dependent activities of individual zinc fingers in a three-finger array (Maeder et al., 2008; Maeder et al., 2009). The method relies on the creation of a combinatorial library of multi-finger arrays derived from “pools” of pre-selected fingers for individual three bp “subsites”. Three pools are randomly recombined together to create a library and then a bacterial two-hybrid selection system is used to identify specific library members that bind to a target site of interest (Figure 1A). The OPEN method is highly efficient and has yielded ZFNs that function efficiently in zebrafish, plants, and human somatic and pluripotent stem cells (Foley et al., 2009; Maeder et al., 2008; Townsend et al., 2009; Zhang et al., 2010; Zou et al., 2009). Although OPEN represents a substantially simpler protocol to practice over previously described selection-based methods, it remains lengthy and challenging for most laboratories to practice. The method takes approximately 2 months to complete and requires a substantial investment of time (typically 6 to 12 months) to successfully master. A detailed protocol for practicing OPEN has been previously described (Maeder et al., 2009).
The CoDA method is an assembly-based method that attempts to account for the context-dependent activities of adjacent zinc fingers in an array (Sander et al., 2011). With CoDA, three-finger arrays are engineered by assembling together fingers that have been pre-selected to function well together. As shown in Figure 1B, an amino-terminal finger (F1) is joined to a particular middle finger (F2) with which it is known to work well. A third carboxy-terminal finger (F3) is then added that is known to work well with the same F2. The resulting three-finger arrays are then screened for DNA-binding activity using a bacterial two-hybrid assay. This method is as simple to practice as modular assembly but is also highly efficient with approximately 50% of ZFN pairs showing activities in zebrafish and plants (Curtin et al., 2011; Sander et al., 2011).
Strategic Planning
Researchers interested in engineering their own ZFNs face the initial challenge of choosing from among the three methods described above. In general, we recommend the use of CoDA as a first-line approach. CoDA is simple to practice and has a high success rate for yielding functional ZFNs. CoDA ZFNs have been used successfully in zebrafish and plants and unpublished data from our lab shows that these nucleases also function in transformed human cell lines. The one exception to our recommendation of CoDA as the initial method of choice is for those investigators requiring ZFNs with the highest possible activities and specificities. Although CoDA does account for context-dependent effects between adjacent fingers, in our experience the OPEN method generally yields ZFNs with higher activities. Presumably OPEN ZFNs also possess higher specificities than those made by CoDA because the selection process interrogates a larger number of combinations of fingers and identifies those arrays that can find their target site in the context of competing E. coli genomic DNA.
The CoDA method will not yield ZFNs for every target gene of interest. CoDA currently has a targeting range of approximately 1 in every 500 bps of random DNA sequence. If one is unable to identify ZFNs using CoDA, we recommend that users attempt to use OPEN, which has a targeting range of approximately 1 in every 200 bps of random DNA sequence. If no potential OPEN ZFN target sites can be identified or if OPEN selections fail to yield ZFNs, users can consider using modular assembly with the caveat that the success rate may be low.
Basic Protocol 1: Identifying and Synthesizing CoDA ZFN Targets
This section describes in detail how to practice the Context Dependent Assembly (CoDA) for engineering zinc finger proteins. CoDA reagents are currently capable of targeting one site in every 500 bp of random sequence, a range sufficient for generating non-homologous end joining (NHEJ) frame shift mutations for most genes in many organisms. CoDA ZFs are designed using zinc finger domains that have been selected to work well together. The resulting DNA sequences encoding CoDA ZF arrays are less than 300 bp in length and can be synthesized and subsequently cloned into standardized expression plasmids for testing binding affinity in the bacterial two-hybrid (B2H) system or for use as site-specific nucleases.
Materials
DNA sequence of genomic region to target with ZFNs
A computer with internet access
-
1)Open a web browser and visit http://www.repeatmasker.org. Paste the sequence for the genomic region of interest into the text box. Select your host genome from the drop-down menu labeled “DNA source.” Leave the other options as their default settings and click the “submit sequence” button. If the output states “No repetitive sequences were detected” continue to step 2. If repeat elements are detected, consider varying the target region.Repeat elements will not be identified if the submitted sequence is shorter than the repeat element. To ensure that the repeat elements are identified submit the genomic sequence that includes several hundred base pairs on both sides of your region of interest.Use genomic sequence (not CDS) to ensure the ZFN targets are present in adjacent sequence.It is also recommended to use actual DNA sequence obtained from the host organism to ensure there are no polymorphic variations from the published sequence.
-
2)Redirect the web browser to the ZiFiT web server (http://zifit.partners.org). Click ZiFiT on the menu below the Zinc Finger Consortium banner then select “Design Zinc Finger Nucleases” under the section labeled “CoDA (Context Dependent Assembly)”. Paste either the FASTA or raw DNA sequence into the text box labeled “Sequence” and click the “Submit” button to scan your sequence for CoDA ZFN sequences. White space and numbers are ignored. Genomic sequence entered should be limited to 50,000 bps or less.By default, ZiFiT defines any sequences in uppercase as exons and sequences in lowercase as introns. This can be used later to aid in picking targets. This option can be deactivated by unchecking the box labeled “Exon/Intron Case Sensitivity” below the lower right corner of the sequence box.
-
3)ZiFiT displays targets in a condensed format comprising individual target sequences flanked by their start and stop positions and labeled according to their FASTA identifier (if provided), the spacer length, and an index number. Scroll through the ZiFiT output in the main window to view CoDA targets within the submitted sequence. Expand information for individual targets by clicking the “+” adjacent to the label.Alternatively, select targets by clicking the blue, green, and gold bars representing targets color coded by spacer size (5bp=Blue; 6bp=Green; 7bp=Gold) above the red bars (thin bar = introns; thick bar = exon) from the graphic summary pop-up window. Pop-ups must be enabled in your web browser for this feature to work. Use of the back button can disrupt the link between the popup and the main window. Restore functionality by closing the popup and selecting the ZiFiT link from the menu.
-
4)Select the link labeled “ZF DNA Sequence” under the table of an expanded target to display the DNA sequences encoding each of the CoDA arrays. Order each zinc finger sequence (<300bp each) through a commercial gene synthesis provider.Use the following criteria to maximize success rates 1) Choose target sites where each of the two half-sites are composed of two or three GNN subsites 2) Avoid targets with half-sites that are composed of four or more T’s 3) Click on the color-coded array in the double-strand DNA target sequence to query ZiFDB and determine whether a zinc finger array for a half-site has already been assayed for DNA-binding activity. Use arrays that have been previously shown to activate three-fold or more in the bacterial two-hybrid assay. 4) Pick targets early in coding sequence to maximize likelihood of generating a knockout mutation. 5) Pick target sites within 100bp of the location of the alteration to be introduced by homologous recombination. 6) Pick two or three ZFN target sites per gene of interest.
Support Protocol 1 (optional): Bacterial two-hybrid (B2H) Assay to Quantify DNA-Binding Activities of Zinc Finger Arrays
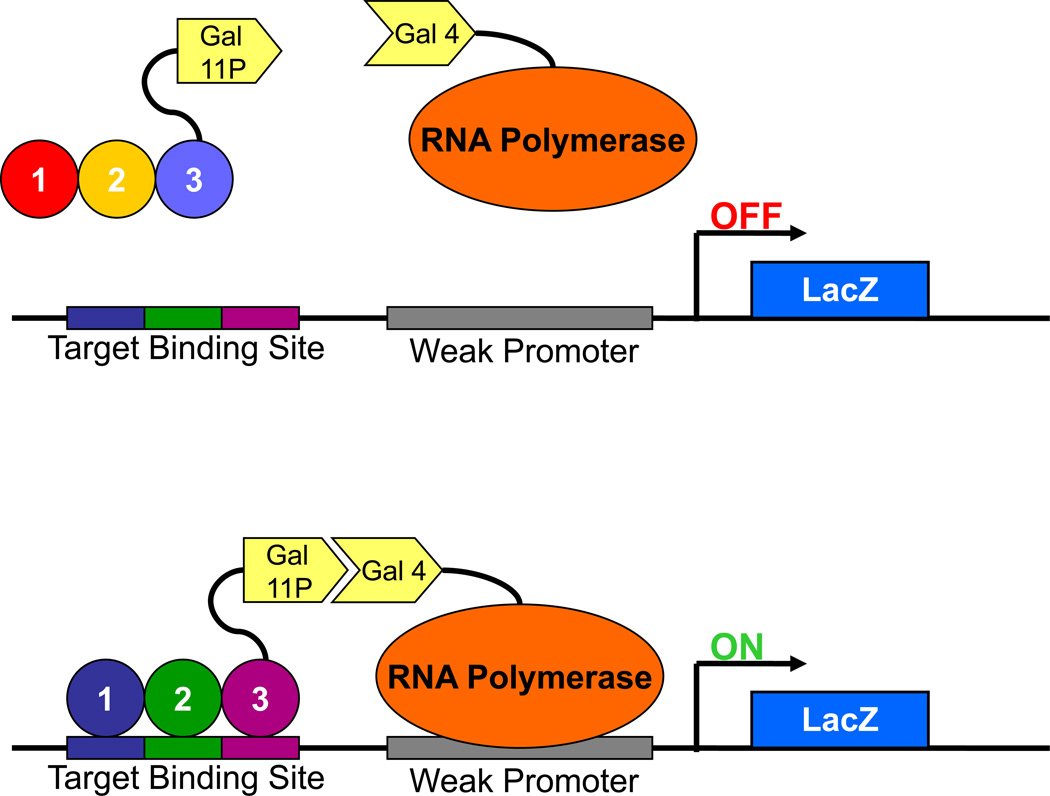
To confirm that a zinc finger array can bind to its target site, zinc finger arrays are expressed in B2H reporter strains and assayed for their abilities to activate expression of a beta-galactosidase reporter gene. The B2H reporter strain harbors a single-copy plasmid with the zinc finger array target site positioned upstream of a weak promoter which, in turn, controls expression of a lacZ reporter gene (encoding beta-galactosidase). Binding of the zinc finger array (fused to a Gal11P protein fragment) to its target site will recruit RNA polymerase to the promoter, leading to an increase in expression of beta-galactosidase (Figure 2). To perform this experiment, a B2H reporter strain is transformed with two plasmids; one expressing a zinc finger array-Gal11P fusion protein and one expressing an RNA polymerase-alpha-Gal4 hybrid protein. Because the lacZ gene encodes β-galactosidase, its expression can be measured by a simple quantitative assay. By comparing β-galactosidase expression in the presence and absence of a given zinc finger array, a “fold-activation” value can be calculated which can guide the choice of which arrays to carry forward for testing as ZFNs.
Figure 2.
Bacterial Two-Hybrid Reporter System. A DNA target site of interest is positioned upstream of a weak promoter driving lacZ expression. In the absence of a zinc finger protein that binds to this target site, lacZ is not expressed. However, when a fusion protein consisting of Gal11P and a zinc finger capable of binding the target site are introduced, they will recruit the RNA polymerase complexes containing alpha-Gal4 fusions to the weak promoter via interaction between the Gal11P and Gal4 proteins. This recruitment results in increased lacZ expression, thereby enabling a quantitative read-out of the zinc finger protein’s ability to bind the target site.
Materials
pGP-FF plasmid (plasmid, restriction map, and full sequence are freely available online from Addgene; see http://www.addgene.org/zfc)
Synthesized zinc finger plasmid (from above Basic Protocol 1)
XbaI restriction enzyme (NEB)
BamHI-HF restriction enzyme (NEB)
QIAquick Gel Extraction Kit (Qiagen)
Quick Ligation Kit (NEB)
Chemically competent XL1-Blue cells (Stratagene)
Carbenicillin
LB agar
LB broth
QIAprep Spin Miniprep Kit (Qiagen)
Primer OK61: 5'-GGGTAGTACGATGACGGAACCTGTC-3'
pBAC-LacZ plasmid (plasmid, restriction map, and full DNA sequence are freely available online from Addgene; http://www.addgene.org/zfc)
BsaI restriction enzyme (NEB)
dCTP nucleotide
Cloned Pfu Polymerase and associated 10X reaction buffer (Stratagene)
10X Annealing Buffer (see Reagents and Solutions)
Chemically Competent Bacterial Strain Transformax Epi300 (Epicentre)
Chloramphenicol
Chemically Competent Bacterial Strain KJBAC1 (available from Addgene)
Expand High Fidelity PCR Kit (Roche Applied Science)
Primer OK5: 5’- AAAATAGGCGTATCACGAGGCCCT -3’
Primer OK163: 5’- CGCCAGGGTTTTCCCAGTCACGAC -3’
1M MgCl2
Solution A + Glycerol (see Reagents and Solutions)
96-well microtiter plates
pAC-Kan-alpha-Gal4 plasmid (available from Addgene)
Kanamycin
2ml assay blocks (Corning)
10mM ZnSO4
500mM IPTG
Microtiter plate reader with temperature control option (for example, Biorad Model 680 Microplate Reader)
Popculture reagent (Novagen, cat. no. 71092)
R-lysozyme (30,000 units/ml) and associated dilution buffer (Novagen, cat. no. 71110)
Lysis Master Mix (see Reagents and Solutions)
Z-buffer with β-mercaptoethanol (see Reagents and Solutions)
4mg/ml ONPG
Construct the zinc finger-Gal11P fusion protein plasmid
-
1)Digest 1 µg of pGP-FF plasmid as follows. Combine the following reagents in a microcentrifuge tube (total reaction volume, 20 µl):
- 1 µg of pGP-FF plasmid
- 0.5 µl XbaI (10U)
- 0.5 µl BamHI-HF (10U)
- 2 µl NEB Buffer 4
- Sterile water to final volume of 20 µl
Digest for 3 hours at 37C and gel isolate the backbone (~3.5kb) using QIAquick gel extraction kit.
-
2)Digest 1 µg of each of the synthesized zinc finger plasmids in separate reactions. Combine the following reagents in a microcentrifuge tube (total reaction volume, 20 µl):
- 1 µg synthesized zinc finger plasmids
- 0.5 µl XbaI (10U)
- 0.5 µl BamHI-HF (10U)
- 2 µl NEB Buffer 4
- Sterile water to final volume of 20 µl
Digest for 3 hours at 37C and gel isolate the zinc finger fragment (~280bp) using QIAquick gel extraction kit.If your zinc finger proteins contain a NotI site instead of a BamHI site for cloning into the vectors pMLM800 or pMLM802, you will need to first amplify them by PCR (CPMB UNIT 15.1) to generate a BamHI site using the following primers:- Forward: 5’- GAAAAAAATCTAGACCCGGGGAGC -3’
- Reverse: 5’- CGCGGATCCCCTCAGGTGGGTTTTTAGGTG-3’
-
3)Ligate each purified zinc finger-encoding fragment into vector backbone for 15 minutes at room temperature as follows. Combine the following reagents in a microcentrifuge tube (total reaction volume, 13 µl):
- 5 µl zinc finger fragment (or nuclease-free water for control)
- 1 µl pGP-FF backbone
- 6 µl 2X quick ligase buffer
- 1 µl T4 DNA Ligase
-
4)
Transform ligation reactions from step 3) into E. coli by adding 150 µl XL1-blue chemically competent cells to the ligation. Incubate at 4C for 5 minutes then heat shock for 1 minute at 42C and return to 4C for 1 minute. Add 700 µl of LB and shake at 37C, 250 r.p.m. for 60 minutes. Plate 300 µl of each transformation on LB/Carb plates (LB agar supplemented with 100 µg ml−1 carbenicillin) and grow overnight at 37C.
-
5)
If zinc finger transformations have greater than 10× the number of colonies of the control plates use single colonies from the zinc finger transformation plate to inocµlate 4-ml cultures of LB/Carb100µg ml-1 and grow overnight at 37C, 250 r.p.m. Purify plasmids the next day from these cultures using a QIAprep Spin Miniprep Kit.
-
6)
Verify the DNA sequence of the cloned zinc finger array fragment using sequencing primer OK61. Note that a restriction map and the full sequence of the parental plasmid pGP-FF is available freely online through Addgene (http://www.addgene.org).
Generate B2H Reporter Plasmid
-
7)Digest pBAC-LacZ plasmid. Combine the following reagents in a microcentrifuge tube (total reaction volume, 30 µl):
- 1 µg pBAC-LacZ plasmid
- 3 µl NEB Buffer 3
- 1.5 µl BsaI (5U/µl)
- Sterile water to final volume of 30 µl
Incubate at 50°C for 2 hrs. Gel purify (CPMB UNIT 2.5A or 2.6) ~11kb vector backbone and elute in 20 µl of nuclease-free water.
-
8)Create DNA overhangs by treating the digested vector backbone from step 7) with Pfu polymerase and dCTP. Combine the following reagents in a microcentrifuge tube (total reaction volume, 40 µl):
- 20 µl BsaI digested, gel purified pBAC-LacZ plasmid (from step 7, above)
- 4 µl 10X Pfu Buffer
- 4 µl 10 mM dCTP
- 2.4 µl Pfu polymerase
- 9.6 µl Sterile water
Incubate at 72°C for 15 minutes, followed by 4°C for 2 minutes. Store indefinitely at −20°C.
-
9)
Prepare target site oligonucleotides. On the ZiFiT output page, click on the link titled “ZF DNA Sequence” and order the two displayed target site oligos for each zinc finger protein to be tested.
-
10)Anneal target site oligos. Combine the following reagents in a microcentrifuge or PCR tube:
- 1 µl 10 µM top oligo
- 1 µl 10 µM bottom oligo
- 20 µl 10X annealing buffer
- 178 µl sterile water
Heat reaction to 95°C for 2 minutes. Then slowly cool to 35°C either by shutting off the heat-block and letting it cool or by programming a thermocycler to reduce temperature by 1°C per minute. Store indefinitely at −20°C.
-
11)Insert target site oligos using ligation-independent cloning into pBAC-LacZ vector backbone. Combine the following in a microcentrifuge tube (total reaction volume, 21 µl):
- 1 µl pBAC-LacZ vector backbone from step 8 above
- 9 µl annealed oligos from step 10 above
- 10 µl 2X Quick Ligase Buffer
Incubate reaction at room temperature for 15 minutes, then place on ice.
-
12)
Transform ligation into Transformax Epi300 cells as follows: Add 200 µl chemically competent Transformax Epi300 cells to ligation mix from step 11. Chill on ice for 10 minutes then heat shock at 42°C for 2 minutes, followed by 2 minutes on ice. Add 700 µl of LB and incubate at 37°C, with agitation, for 45 minutes. Plate 300 µl on LB plate supplemented with 12.5 µg/ml chloramphenicol. Incubate at 37°C overnight.
-
13)
Pick single colonies from transformation plates (from step 12) and inoculate each colony into 3 ml of LB supplemented with 12.5 µg/ml chloramphenicol. Incubate overnight, with agitation, at 37°C.
-
14)Subculture 1ml of overnight culture into 9ml of LB containing 14 µg/ml chloramphenicol and 1.1 mM arabinose. Grow cultures at 37°C, with agitation, for 5–6 hours. Harvest cells and isolate plasmid DNA using a QIAprep spin miniprep kit.Subculturing the cells containing pBAC-LacZ plasmids in arabinose induces expression of the trfA gene product, which increases the copy number of the plasmid.
Generate B2H Reporter Strain
-
15)
Transform reporter plasmids into bacterial strain KJBAC1. Combine 1 µl of miniprep reporter plasmid DNA (from step 14) with 100 µl chemically competent KJBAC1. Chill on ice for 10 minutes then heat shock at 42°C for 2 minutes, followed by 2 minutes on ice. Add 350 µl of LB and incubate at 37°C, with agitation, for 45 minutes. Plate 300 µl on LB plate supplemented with 12.5 µg/ml chloramphenicol. Incubate at 37°C overnight.
-
16)
Inoculate single colonies from transformation plates into 3 ml of LB supplemented with 12.5 µg/ml chloramphenicol. Incubate overnight, with agitation, at 37°C.
-
17)PCR amplify region of reporter plasmid bearing the target site for sequence verification. Combine the following in a PCR tube (total reaction volume, 25µl):
- 2 µl bacterial overnight culture
- 2 µl primer OK5 (10 µM)
- 2 µl primer OK163 (10 µM)
- 2.5 µl Expand Buffer 2
- 2 µl 10 mM dNTPs (2.5 mM each)
- 0.5 µl Expand Enzyme
- 14 µl Sterile water
Amplify using the following PCR protocol:1 cycle: 5 minutes 95°C (initial denaturation) 35 cycles: 30 seconds 95°C (denaturation) 30 seconds 55°C (annealing) 30 seconds 72°C (extension) 1 cycle: 5 minutes 72°C (final extension) 1 cycle: indefinitely 4°C (hold) -
18)
Sequence PCR product with primer OK163. Note that a restriction map and the full sequence of the parental reporter plasmid pBAC-LacZ is available freely online through Addgene (http://www.addgene.org).
-
19)
Subculture 1 ml of overnight culture into 50 ml of LB supplemented with 200 µl of 1M MgCl2 (final concentration of 15 mM) and 12.5 µg/ml chloramphenicol. Shake at 37°C, 250 rpm for 1 hr.
-
20)
Centrifuge cells in a sterile 50 ml conical tube at 3000–6000×g for 20 minutes.
-
21)
Resuspend cell pellet in 3ml of SolutionA + glycerol, aliquot into microcentrifuge tubes, and freeze at −80°C.
Perform Quantitative B2H Assay
-
22)
Transform pAC-Kan-alpha-Gal4 plasmid and ZF-Gal11P plasmid (from step 6 above) into B2H reporter strain competent cells (from step 22 above). In one well of a 96-well plate, combine 1 µl of ZF-Gal11P plasmid (or pGP-FF plasmid for the basal level control), 100 ng of plasmid pAC-Kan-alpha-Gal4 and 50 µl competent cells (from step 21 above). Chill on ice for 10 minutes then heat shock at 42°C for 2 minutes, followed by 2 minutes on ice. Add 200 µl of LB to each well and incubate at 37°C for 1 hr.
-
23)
Make two serial ten-fold dilutions of each transformation and spot 5µl of each dilution (100, 10−1 and 10−2) three times on LB plates supplemented with 100 µg/ml carbenicillin, 12.5 µg/ml chloramphenicol and 30 µg/ml kanamycin. Incubate at 37°C overnight.
-
24)For each zinc finger protein to be assayed, inoculate three overnight cultures (from 3 independent colonies) as well as three basal controls (pGP-FF and pAC-Kan-alpha-Gal4 plasmids transformed into the B2H reporter strain). Inoculate each colony into 1 ml of LB containing 50 µg/ml carbenicillin, 30 µg/ml kanamycin, 12.5 µg/ml chloramphenicol, 10 µM ZnSO4, and 500 µM IPTG in a 2ml 96-well block. Grow overnight at 37°C with agitation.Shaking on a platform with a small throw radius of no more than a few millimeters (for example, Microtitertron orbital shaker, Appropriate Technical Resources) will ensure uniform growth of all wells. Overnight culture time should not exceed 18 hours.
-
26)
Subculture saturated overnight cultures by diluting 25µl into 1ml of pre-warmed culture medium (same media used for overnight cultures prepared in step 22). Grow subcultures with agitation at 37°C.
-
27)Monitor the growth of subcultures by measuring the OD600 (relative to a media-only blank) on a spectrophotometer. Harvest cells for lysis when they reach log phase (OD600 = 0.3–0.5). Record the OD600 at which each well was lysed.For best results lyse as close to and OD600 of 0.3 as possible.
-
28)Lyse log-phase cultures by mixing 100 µl of culture with 11 µl of Lysis Master Mix in a 96-well plate. Allow lysis reactions to proceed at room temperature for a minimum of 15 minutes.The activity of β-galactosidase is stable in the cell lysates for at least 18 hours when stored sealed at room temperature.
-
29)
Set up β-galactosidase assay by adding 15 µl of lysate to a 96-well microtiter plate well containing 135 µl of Z buffer with β-mercaptoethanol and 30 µl of 4 mg/ml ONPG and mixing well.
-
30)Place microtiter plate containing β-galactosidase assay reactions in a microtiter plate reader with temperature control. Incubate reactions at 28° C and take timed serial measurements of absorbance at 415 nm. Calculate the velocity of ONPG cleavage (v) by plotting A415 vs. time and calculating the slope of the line.Many microtiter plate readers can be programmed to take absorbance measurements at fixed intervals and to calculate the velocity of ONPG cleavage. We typically take measurements every 10–30 seconds. Reactions should not be allowed to proceed for more than 30 minutes as substrate can become limiting.
-
31)Calculate the units of β-galactosidase for each assay using the following formula:
- V × 1000/(OD600)
-
32)
Calculate the fold-activation mediated by each zinc finger protein tested by comparing β-galactosidase units obtained in the presence of the zinc finger-Gal11P hybrid protein with the units obtained in the presence of the Gal11P control.
Representative Data:
Target: GCAGAAGGT
CoDA Helices: F1: TKQRLEV - F2: QQTNLTR - F3: QGNTLTRColony
SampleOD600 Velocity
AVelocity
BAverage
VelocityV × (1000) /
OD600Fold
ActivationZinc Finger
Zinc Finger
Zinc Finger0.2827
0.3075
0.307551.80
59.30
59.9059.80
66.00
66.2055.80
62.65
63.055635.27
5816.18
5853.319.39 Basal
Basal
Basal0.2999
0.3075
0.29615.90
6.60
6.006.40
7.30
6.706.15
6.95
6.35585.49
645.21
612.33
Basic Protocol 2: Construction of ZFN expression vectors
In this step, each ZF array is cloned into a nuclease expression vector. Expression vectors differ based on the type of promoter, nuclease domain, and linker between the ZF and nuclease domain (see Table below). The linker determines the number of nucleotides (5, 6, or 7) that are permitted in the “spacer” sequence between the ZF target half-sites (Handel et al., 2009). The wild-type FokI nuclease monomer permits ZF monomers from the same half-site to form homodimers. This increases the likelihood that the ZFNs will cleave at unintended positions. Alternatively, the heterodimer variants require two different ZFNs to assemble on a full target site to mediate cleavage (Miller et al., 2007). However, the heterodimer variants used in the expression plasmids described below are less active than the wild-type FokI domains.
Materials
Synthesized zinc finger constructs (Basic Protocol 1)
XbaI (NEB)
BamHI (NEB)
NotI (NEB)
Quick Ligation Kit (NEB)
Nuclease free water
LB medium (Difco. Cat. No. 244620)
LB agar medium (Difco. Cat. No 244520)
QIAquick gel extraction kit (Qiagen)
QIAprep spin miniprep kit (Qiagen)
Carbenicillin (Sigma, cat. no T4625; 50 mg/ml−1 stock solution in ddH20)
LB/Carb plates (LB agar supplemented with 100 µg/ml carbenicillin.
Chemically competent Bacterial strain XL-1 Blue (recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F' proAB lacIq lacZDM15 Tn10 (TetR)]; Stratagene cat. no. 200249)
Primer OK567: 5’- CGCAAATGGGCGGTAGGCGTG -3’
- Nuclease Expression Plasmids: (see Table below for description; plasmids, restriction maps, and full DNA sequences of all of these plasmids are freely available online through Addgene: http://www.addgene.org/zfc)
-
-pST1374 (or)
-
-MLM290 and MLM292 (or)
-
-MLM800 and MLM802
Plasmid name For expression in FokI cleavage
domain typeZFN spacer
lengthpST1374 Mammalian cells,
zebrafish (RNA)Wild-type 5 or 6 pMLM290 Mammalian cells,
zebrafish (RNA)‘+’ obligate
heterodimer5 or 6 pMLM292 Mammalian cells,
zebrafish (RNA)‘−‘ obligate
heterodimer5 or 6 pMLM800 Mammalian cells,
zebrafish (RNA)‘+’ obligate
heterodimer7 pMLM802 Mammalian cells,
zebrafish (RNA)‘−‘ obligate
heterodimer7 pDW1775 Plants Wild-type 5 or 6 -
-
-
1)Digest 1 µg of each nuclease expression plasmid(s).
- ZFN targets with spacer lengths of 5 or 6 nucleotides
- 0.5 µl XbaI (10U)
- 0.5 µl BamHI-HF (10U)
- 2 µl NEB Buffer 4
- 17 µl nuclease free water
- ZFN targets with spacer lengths of 7 nucleotides
- 0.5 µl XbaI (10U)
- 1 µl NotI (10U)
- 2 µl NEB Buffer 3
- 17 µl nuclease free water
- Digest for 3 hours at 37C and gel isolate (CPMB UNIT 2.6) the nuclease backbone (~5.5kb) using QIAquick gel extraction kit.
-
2)Digest 1 µg of each of the synthesized zinc finger plasmids.
- ZFN targets with 5 or 6 nucleotides
- 0.5 µl XbaI (10U)
- 0.5 µl BamHI-HF (10U)
- 2 µl NEB Buffer 4
- 17 µl nuclease free water
- ZFN targets with 7 nucleotides
- 0.5 µl XbaI (10U)
- 1 µl NotI (10U)
- 2 µl NEB Buffer 3
- 17 µl nuclease free water
- Digest for 3 hours at 37C and gel isolate (CPMB UNIT 2.6) the fragment encoding the zinc finger fragment (~280bp) using a QIAquick gel extraction kit.
-
3)Ligate each purified zinc finger fragment with the corresponding vector backbone for 15 minutes at room temperature.
- 3 µl zinc finger fragment (use nuclease free water for ligation control)
- 2 µl nuclease expression vector backbone
- 6 µl 2X quick ligase buffer
- 1 µl T4 DNA Ligase
-
4)
Transform ligation reaction by adding 6 µl of the ligation reaction to 60 µl XL1-blue chemically competent cells. Incubate at 4C for 5 minutes then heat shock for 1 minute at 42C and return to 4C for 1 minute. Add 300 µl of LB and shake 37C, 250 r.p.m. for 60 minutes. Plate 300 µl of each transformation on LB/Carb plates (LB agar supplemented with 100 µg ml−1 carbenicillin) and grow overnight at 37C.
-
5)
If zinc finger transformations have greater than 10× the number of colonies of the control plates use single colonies from the zinc finger transformation plate to inoculate 4-ml cultures of LB/Carb100 µg ml-1 and grow overnight at 37C, 250r.p.m. Purify plasmids using a QIAprep Spin Miniprep Kit.
-
6)
Sequence-verify the portion of the plasmid encoding the zinc finger array using sequencing primer OK567.
REAGENTS AND SOLUTIONS
10X Annealing Buffer
- To prepare 1ml of 10X annealing buffer, combine the following sterile solutions:
- 400 µl 1M Tris, pH8
- 200 µl 1M MgCl2
- 100 µl 5M NaCl
- 20 µl 0.5M EDTA, pH8
- 280 µl water
Store indefinitely at −20°C.
Solution A+Glycerol
- To prepare 300ml of Solution A + Glycerol, combine the following sterile solutions:
- 3 ml 1 M MnCl2
- 15 ml 1 M CaCl2
- 60 ml 50 mM MES, pH6.3 (with KOH)
- 45 ml 100% glycerol
- 177 ml water
Store at 4°C wrapped in aluminum foil to protect from light. Solution is usable until it acquires a brown discoloration.
Lysis Master Mix
R-Lysozyme is supplied at 30,000 U/µl and can be diluted to 400U/µl and stored indefinitely at −20°C. To prepare lysis master mix, dilute R-Lysozyme to 4 U/µl and make a 10:1 mixture of Popculture reagent to diluted lysozyme. Prepare lysis master mix fresh before each use.
Z-buffer with β-mercaptoethanol
- To prepare 1 L of Z-buffer, dissolve the following in water to a final volume of 1L:
- 16.1 g Na2HPO4-7H2O
- 5.5 g NaH2PO4-H2O
- 0.75 g KCl
- 0.246 g MgSO4-7H2O
Filter sterilize and store indefinitely at room temperature. Prepare Z-buffer with β-mercaptoethanol fresh before use by adding 2.7µl of β-mercaptoethanol per 1 ml Z-buffer.
Commentary
Background Information
-
A)
Application: Zinc finger nucleases can be used to cleave specific targets within complex genomes. Subsequent repair of these double-strand breaks via the imperfect non-homologous end joining (NHEJ) repair pathway generates frame-shift mutations that can be used to abolish gene function. CoDA ZFNs have been shown to generate NHEJ -induced mutations in less than 1% to as high as 19% in zebrafish or plant somatic cells and these mutations can be passed on through the germ line to progeny. Alternatively, the homologous recombination pathway can be exploited by providing an exogenous donor template to introduce specific base changes, insertions and deletions. It has also been shown that short fragment of dsDNA can be inserted at the site of a break via NHEJ.
-
B)Critical Parameters and Troubleshooting
-
1)It is critical to submit genomic DNA sequence to ZiFiT. Submitting non-adjacent sequence, such as cDNA, may result in the identification of a site that is formed only after intron splicing and does not actually exist as contiguous sequence in genomic sequence.
-
2)The heterodimeric FokI variants have been specifically engineered to function only with their obligate partners. It is critical that one half-site is cloned into "+" vector and the other into the "−" vector.
-
3)The amount of ZFN DNA/RNA required to efficiently generate double strand breaks in cells can vary by target and ZFN pair. Using too little DNA/RNA can lead to suboptimal rates of ZFN-induced gene modification. Using too much DNA/RNA can be toxic to the host cells. Titration may be necessary to determine the amount of template that is appropriate for the specific target or ZFN pair.
-
4)Some ZFN pairs do not efficiently cleave their genomic targets (See Anticipated Results). Possible explanations for poor activity include 1) insufficient specificity of the ZFN pair and 2) the context of the DNA target such as an inaccessible chromatin state. To maximize the change of success it is recommended to target multiple sites within the region of interest.
-
5)ZFNs may cleave at unintended off-target sites. Picking targets that are dissimilar to other genomic sequence may help to minimize potential off target cleavage. When possible in model organisms, outcrossing can be used to reduce the effects of potentially unwanted off-target effects.
-
1)
-
C)
Anticipated Results
Users can expect approximately 75% of CoDA ZFs will activate three-fold or more in the B2H reporter assay (Sander et al., 2011). Approximately 50% of ZFN pairs made from such ZF arrays are expected to induce NHEJ mutations with rates of 1% or higher in zebrafish and plants using the heterodimeric FokI nucleases (Curtin et al., 2011; Sander et al., 2011). Actual cleavage rates will be organism-/cell line-dependent. CoDA ZFs that exhibit B2H reporter assay fold-activation values between 1.57 and 3 fold activations may also show activity as ZFNs in zebrafish and plants. ZFs that activate transcription by 1.57-fold or less in the B2H reporter assay have been shown to have a high failure rate and are not expected to function well as nucleases (Ramirez et al., 2008).
- D) Time Considerations
-
1Construction of ZFN-expression vector:
- Cloning into the ZFN-expression vector takes 3–4 days with 2 overnight incubation steps and a few additional days for sequence verification.
-
2B2H Assay (Optional):The optional B2H protocol requires 8–10 days total. Construction of the zinc finger-Gal11P fusion protein plasmid takes 3–4 days with 2 overnight incubation steps and can be performed concurrently with construction of the B2H reporter plasmid. Construction of the B2H reporter plasmid and strain takes 5–7 days with 4 overnight incubation steps. The B2H assay takes 1 day for transformation, 1 day for starting overnights and the assay is run on the 3rd day. With practice, a full 96-well plate can easily be assayed at one time. For this reason, it is logical to construct 16 proteins and 16 reporter strains concurrently because picking 3 colonies of the protein and 3 of the basal control transformation for each strain will result in 96 samples.
-
1
Footnotes
Key references
Sander, J.D., Dahlborg, E.J., Goodwin, M.J., Cade, L., Zhang, F., Cifuentes, D., Curtin, S.J., Blackburn, J.S., Thibodeau-Beganny, S., Qi, Y., et al. (2011). Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA). Nat Methods 8, 67–69.
Internet Resources: ZiFiT (http://zifit.partners.org)
Literature Cited
- Beerli RR, Barbas CF., 3rd Engineering polydactyl zinc-finger transcription factors. Nat Biotechnol. 2002;20:135–141. doi: 10.1038/nbt0202-135. [DOI] [PubMed] [Google Scholar]
- Bibikova M, Beumer K, Trautman JK, Carroll D. Enhancing gene targeting with designed zinc finger nucleases. Science. 2003;300:764. doi: 10.1126/science.1079512. [DOI] [PubMed] [Google Scholar]
- Bibikova M, Golic M, Golic KG, Carroll D. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics. 2002;161:1169–1175. doi: 10.1093/genetics/161.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll D, Morton JJ, Beumer KJ, Segal DJ. Design, construction and in vitro testing of zinc finger nucleases. Nat Protoc. 2006;1:1329–1341. doi: 10.1038/nprot.2006.231. [DOI] [PubMed] [Google Scholar]
- Cornu TI, Thibodeau-Beganny S, Guhl E, Alwin S, Eichtinger M, Joung J, Cathomen T. DNA-binding Specificity Is a Major Determinant of the Activity and Toxicity of Zinc-finger Nucleases. Mol Ther. 2008;16:352–358. doi: 10.1038/sj.mt.6300357. [DOI] [PubMed] [Google Scholar]
- Curtin SJ, Zhang F, Sander JD, Haun WJ, Starker C, Baltes NJ, Reyon D, Dahlborg EJ, Goodwin MJ, Coffman AP, et al. Targeted mutagenesis of duplicated genes in soybean with zinc-finger nucleases breakthrough technologies. Plant Physiol. 2011;156:466–473. doi: 10.1104/pp.111.172981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley JE, Yeh J-RJ, Maeder ML, Reyon D, Sander JD, Peterson RT, Joung JK. Rapid Mutation of Endogenous Zebrafish Genes Using Zinc Finger Nucleases Made by Oligomerized Pool ENgineering. PLoS ONE. 2009;4:e4348. doi: 10.1371/journal.pone.0004348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez B, Schwimmer LJ, Fuller RP, Ye Y, Asawapornmongkol L, Barbas CF., 3rd Modular system for the construction of zinc-finger libraries and proteins. Nat Protoc. 2010;5:791–810. doi: 10.1038/nprot.2010.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handel EM, Alwin S, Cathomen T. Expanding or restricting the target site repertoire of zinc-finger nucleases: the inter-domain linker as a major determinant of target site selectivity. Mol Ther. 2009;17:104–111. doi: 10.1038/mt.2008.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt JA, Thibodeau SA, Hirsh AS, Pabo CO, Joung JK. Highly specific zinc finger proteins obtained by directed domain shuffling and cell-based selection. Proc Natl Acad Sci U S A. 2003;100:12271–12276. doi: 10.1073/pnas.2135381100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Lee HJ, Kim H, Cho SW, Kim JS. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res. 2009;19:1279–1288. doi: 10.1101/gr.089417.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci U S A. 1996;93:1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder ML, Thibodeau-Beganny S, Osiak A, Wright DA, Anthony RM, Eichtinger M, Jiang T, Foley JE, Winfrey RJ, Townsend JA, et al. Rapid "open-source" engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol Cell. 2008;31:294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder ML, Thibodeau-Beganny S, Sander JD, Voytas DF, Joung JK. Oligomerized pool engineering (OPEN): an 'open-source' protocol for making customized zinc-finger arrays. Nat Protoc. 2009;4:1471–1501. doi: 10.1038/nprot.2009.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Holmes MC, Wang J, Guschin DY, Lee YL, Rupniewski I, Beausejour CM, Waite AJ, Wang NS, Kim KA, et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- Rahman SH, Maeder ML, Joung JK, Cathomen T. Zinc-finger nucleases for somatic gene therapy: the next frontier. Hum Gene Ther. 2011 doi: 10.1089/hum.2011.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez CL, Foley JE, Wright DA, Muller-Lerch F, Rahman SH, Cornu TI, Winfrey RJ, Sander JD, Fu F, Townsend JA, et al. Unexpected failure rates for modular assembly of engineered zinc-fingers. Nat Methods. 2008;5:374–375. doi: 10.1038/nmeth0508-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander JD, Dahlborg EJ, Goodwin MJ, Cade L, Zhang F, Cifuentes D, Curtin SJ, Blackburn JS, Thibodeau-Beganny S, Qi Y, et al. Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA) Nat Methods. 2011;8:67–69. doi: 10.1038/nmeth.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago Y, Chan E, Liu PQ, Orlando S, Zhang L, Urnov FD, Holmes MC, Guschin D, Waite A, Miller JC, et al. Targeted gene knockout in mammalian cells by using engineered zinc-finger nucleases. Proc Natl Acad Sci U S A. 2008;105:5809–5814. doi: 10.1073/pnas.0800940105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend JA, Wright DA, Winfrey RJ, Fu F, Maeder ML, Joung JK, Voytas DF. High-frequency modification of plant genes using engineered zinc-finger nucleases. Nature. 2009;459:442–445. doi: 10.1038/nature07845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- Wright DA, Thibodeau-Beganny S, Sander JD, Winfrey RJ, Hirsh AS, Eichtinger M, Fu F, Porteus MH, Dobbs D, Voytas DF, et al. Standardized reagents and protocols for engineering zinc finger nucleases by modular assembly. Nat Protoc. 2006;1:1637–1652. doi: 10.1038/nprot.2006.259. [DOI] [PubMed] [Google Scholar]
- Zhang F, Maeder ML, Unger-Wallace E, Hoshaw JP, Reyon D, Christian M, Li X, Pierick CJ, Dobbs D, Peterson T, et al. High frequency targeted mutagenesis in Arabidopsis thaliana using zinc finger nucleases. Proc Natl Acad Sci U S A. 2010;107:12028–12033. doi: 10.1073/pnas.0914991107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Maeder ML, Mali P, Pruett-Miller SM, Thibodeau-Beganny S, Chou BK, Chen G, Ye Z, Park IH, Daley GQ, et al. Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells. Cell Stem Cell. 2009;5:97–110. doi: 10.1016/j.stem.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]