Abstract
In traditional morphology-based concepts many species of lichenized fungi have world-wide distributions. Molecular data have revolutionized the species delimitation in lichens and have demonstrated that we underestimated the diversity of these organisms. The aim of this study is to explore the phylogeography and the evolutionary patterns of the Xanthoparmelia pulla group, a widespread group of one of largest genera of macrolichens. We used a dated phylogeny based on nuITS and nuLSU rDNA sequences and performed an ancestral range reconstruction to understand the processes and explain their current distribution, dating the divergence of the major lineages in the group. An inferred age of radiation of parmelioid lichens and the age of a Parmelia fossil were used as the calibration points for the phylogeny. The results show that many species of the X. pulla group as currently delimited are polyphyletic and five major lineages correlate with their geographical distribution and the biosynthetic pathways of secondary metabolites. South Africa is the area where the X. pulla group radiated during the Miocene times, and currently is the region with the highest genetic, morphological and chemical diversity. From this center of radiation the different lineages migrated by long-distance dispersal to others areas, where secondary radiations developed. The ancestral range reconstruction also detected that a secondary lineage migrated from Australia to South America via long-distance dispersal and subsequent continental radiation.
Introduction
Methods for delimiting species, the fundamental taxonomic unit, have always fascinated evolutionary biologists [1]–[3]. Understanding the circumscription of species is important for biological and ecological studies and for conservation issues. However, the main challenge is to recognize species in organisms with relatively simple morphologies. In lichenized fungi traditional species circumscriptions are based on phenotypic characters, such as thallus and ascomatal morphology or chemical characters. However, there is a growing body of evidence from molecular studies that the traditional morphology-based species circumscriptions are insufficient to represent the diversity in lichenized ascomycetes [4]–[26]. A number of DNA sequence-based phylogenetic studies revealed the presence of distinct lineages within currently delimited species. Subsequent, detailed studies often revealed previously overlooked morphological subtleties or chemical differences among those clades and authors often refer to these as “semi-cryptic” species [8].
On a par with the phenotypic-based species circumscription, researchers often accepted wide distribution ranges for species occurring on different continents. This was at least partially due to a common belief by lichenologists in the “everything is everywhere” hypothesis [27], [28] applied to fungi, as discussed elsewhere [8], [29]. In several cases molecular data assisted in a better understanding of the biogeography of lichen-forming fungi where taxa were shown to represent different species on different continents, e.g. the Leptogium furfuraceum aggr. on different continents [18], Melanelixia glabra s. lat. in Europe and North America [14], Parmelina quercina s. lat. on different continents [4], Physcia aipolia aggr. in Europe and Australia [9], Xanthoparmelia spp. in North America and Australia [15], [16]. In the Leptogium furfuraceum aggr. complex sister-group relationships were found between populations from the same hemispheres which were incongruent with previous classifications based on morphological differences [18], and the dated phylogeny indicated that the species had migrated via transoceanic dispersal to different continents.
Here we report another case of a group of lichenized fungi where transoceanic dispersal to different continents is correlated with the phylogenetic lineages. The group studied here is the Xanthoparmelia pulla group which belongs to the family Parmeliaceae. This family represents one of the largest families of lichenized fungi [30], [31]. The main clade of the family is the parmelioid clade with almost 2000 species [32] currently classified in 27 genera, with Xanthoparmelia being the largest with over 800 accepted species [33]. The species in this genus characteristically occur on siliceous rocks or soil, predominantly in arid to subarid regions, with a center of distribution in the southern hemisphere. The genus is characterized by having cell wall polysaccharides of the Xanthoparmelia-type, small ascospores with an arachiform vacuolar body [34], and the presence of a pored epicortex [33], [35]. It has been hypothesized that the genus diversified in a rapid radiation following a shift towards drier habitats at the base of the Xanthoparmelia clade [36] leading to the high current diversity.
Previously, the Xanthoparmelia pulla group has been classified within the separate genus Neofuscelia based on the different cortical chemistry (having melanoid pigments and lacking usnic acid or atranorin, characteristic of the majority of Xanthoparmelia species) [37], [38]. A subsequent molecular study showed that the genus Neofuscelia was polyphyletic, with its clades scattered within Xanthoparmelia [33]. Consequently, the genus Neofuscelia was reduced to synonymy with Xanthoparmelia, as have other genera previously distinguished by cortical chemistry or growth form [33], [39]–[43]. The Xanthoparmelia pulla group is a monophyletic clade within the complete Xanthoparmelia clade, that includes the former Esslinger's Xanthoparmelia pulla species and other related species [44].
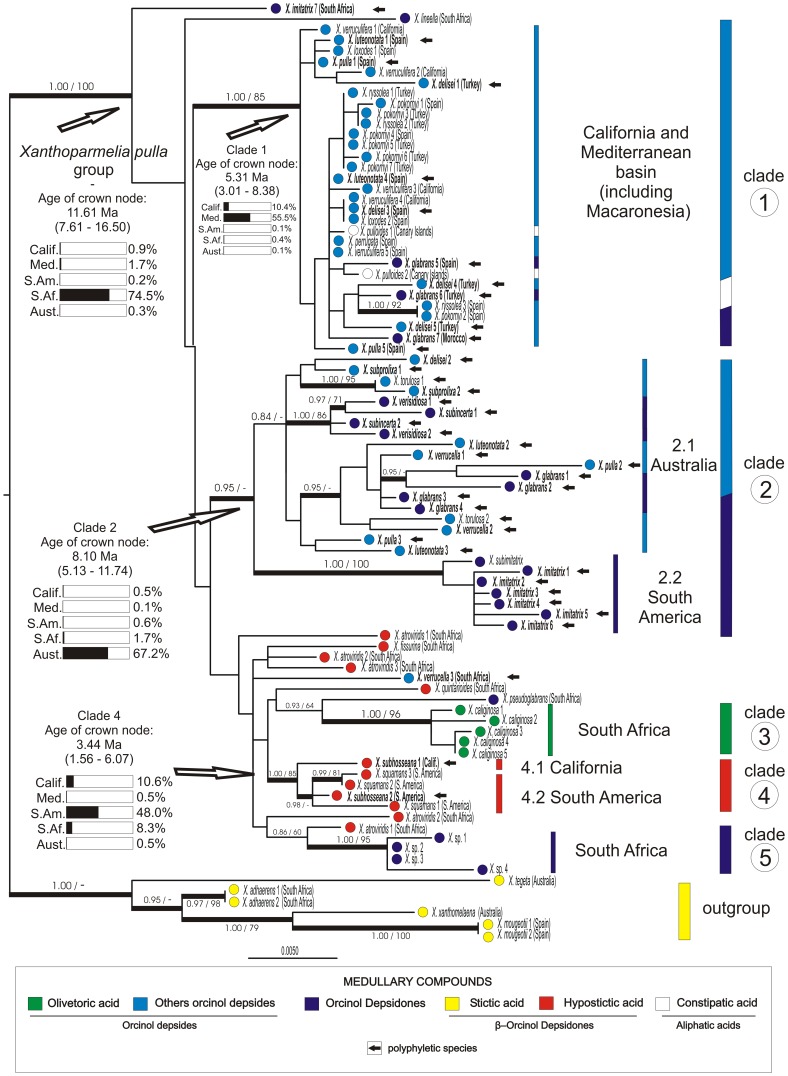
Although the clades were largely incongruent with the current species circumscription, we found a correlation of the main clades of X. pulla group with their geographical distribution and chemical profile (Fig. 1, 2). Clade 1 includes specimens from California, Macaronesia and areas around the Mediterranean basin, all of which contain depsides and depsidones derived from the orcinol pathway or with aliphatic acids; clade 2 includes specimens from Australia (subclade 2.1) and South America (subclade 2.2) with depsides and depsidones derived from the orcinol pathway; clade 3 derives from South African specimens containing olivetoric acid; clade 4 specimens with hypostictic acid from California (subclade 4.1) and South America (subclade 4.2); and clade 5 specimens with physodic acid (orcinol depsidones) from South Africa.
Figure 1. Phylogenetic relationships of the Xanthoparmelia pulla group based on nuITS and nuLSU rDNA sequences.
Topology based on maximum-likelihood (ML) analyses. Posterior probabilities and bootstrap values are indicated on each branch. Branches with posterior probabilities under Bayesian analysis equal or above 0.95 and/or bootstrap values equal or above 70% under MP are in bold. Medullary compounds, and results of the divergence time estimation and ancestral range reconstruction analyses are shown. Black arrow head indicates the polyphyletic species.
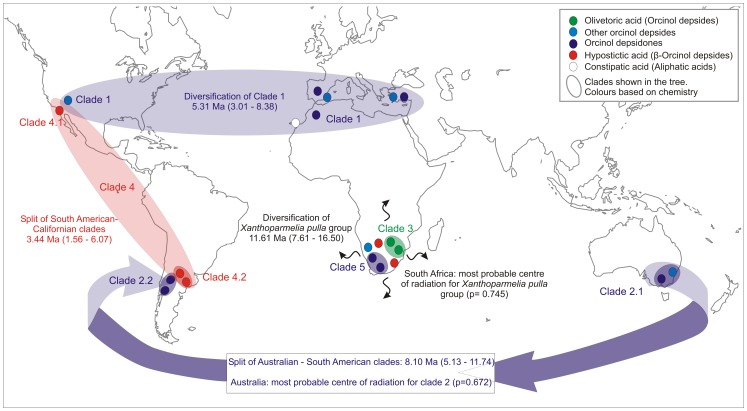
Figure 2. Schematic map showing the relationships between phylogeny, medullary compounds, ancestral range and divergence times estimation for the Xanthoparmelia pulla group.
The species delimitation within the Xanthoparmelia pulla group is currently based on a combination of morphological and chemical characters (Table 1). The morphological characters include the color of the lower surface, shape of the lobes, attachment to the substrate, and presence of vegetative propagules while the chemical differences pertain to upper cortical and medullary secondary metabolites. A number of the currently accepted species have a wide distribution spanning several continents. To address the species delimitation in this group and to test the hypothesis of widely distributed species we have generated a data set using two loci (nuITS rDNA, nuLSU rDNA) from specimens collected on different continents. The molecular data were used to perform phylogenetic reconstructions in a maximum likelihood (ML) and Bayesian (B/MCMC) framework. We have also estimated the timing of the diversification events leading to the main clades found in our study to discriminate between vicariance and long-distance dispersal as possible explanations for the current distribution patterns. A Bayesian-based approach of ancestral range reconstruction was used to identify potential areas in which the group and major clades within the group originated.
Table 1. Main differences of the species of Xanthoparmelia pulla group studied in this paper [38], [44], [47], [59], [60].
| Characters and distribution | |||||
| Species | Morphology of lobes | Isidia | Lower surface | Chemistry | Distribution |
| X. atroviridis | 1–2 mm broad, subirregular, contiguous to imbricate | Absent | Black, moderately rhizinate, rhizines concolorous, up to 0.4 mm long. | Medulla: Hypoconstictic, hypostictic, hyposalazinic acids; Cortex: HNO3 + violet | South Africa |
| X. caliginosa | 1–2.5 (−3.5) mm broad, subirregular, contiguous to imbricate | Sparse to crowded and areolate. Isidia erumpent, globose to cylindrical, 0.1–0.6 (−0.8) mm tall. | Dark brown to black, moderately rhizinate, rhizines more or less concolorous, up to 0.6 mm long. | Medulla: Olivetoric acid. Cortex: HNO3 + blue-green | South Africa |
| X. delisei | 1–4 mm broad, sublinear to irregular, flat to slightly concave or convex, becoming laciniate, often imbricate and entangled | Absent | Dark brown to black, often paler near the apices, moderately to densely rhizinate, the rhizines simple and concolorous with the lower surface, to 1 mm long | Medulla: glomelliferic, glomellic, perlatolic acids; ± gyrophoric acid. Cortex: HNO3 + blue-green | Europe, Asia, Africa, Australia, Macaronesia, South America |
| X. fissurina | 1–3 mm broad, contiguous to imbricate or entangled | Absent | Pale tan to pale brown, moderately rhizinate, rhizines concolorous, to 1 mm long | Medulla: hypostictic, hypoconstictic, hyposalazinic acids, unknown compounds. Cortex: HNO3 + blue-green | South Africa and South America |
| X. glabrans | 0.5–3.0 mm broad, sublinear to linear-elongate, imbricate to loosely entangled. | Absent | black, dull, slightly rugulose, moderately rhizinate; rhizines black, simple or fasciculate, to 1 mm long | Medulla: alectoronic acid; ± a-collatolic and gyrophoric acids. Cortex: HNO3 + blue-green | Australia, Europe, Africa, South America, New Zealand |
| X. imitatrix | 0.5–3.0 mm broad, sublinear to linear-elongate, imbricate to laciniate entangled, rarely developing subfruticose branches | Absent | Dark brown to black, sparsely to moderately rhizinate, rhizines simple, to 1.5 mm long | Medulla: physodic acid; ±4-O-methylphysodic and alectoronic acids. Cortex: HNO3 + blue-green | Australia, Africa, South America, New Zealand |
| X. lineella | 0.1–0.5mm broad, linear and dichotomously branched and entangled | Absent | Black, sparsely rhizinate, rhizines concolorous, to 1 mm long | Medulla: physodic acid; Cortex: HNO3 + blue-green | South Africa |
| X. loxodes | (0.5-)1–3(−5) mm broad, subirregular to sublinear, contiguous to entangled. | Sparsely to densely isidiate, isidia more or less spherical and distinctly pustular, erumpent | Dark brown to black, smooth to somewhat rugulose, moderately rhizinate, rhizines concolorous, to 1 mm long | Medulla: glomelliferic, glomellic and perlatolic acids; ±gyrophoric acid. Cortex: HNO3 + blue-green | Europe, North Africa, Asia, North America, Macaronesia |
| X. luteonotata | (0.5-)1–3 mm broad, sublinear to irregular, discrete to imbricate, rarely developing subfruticose branches | Absent | Pale tan to pale brown, moderately to densely rhizinate, rhizines simple, to 0.5 mm long | Medulla: ± divaricatic and stenosporic acids; ± gyrophoric acid. Cortex: HNO3 + blue-green | Australia, Europe, Africa, New Zealand |
| X. pokornyi | 1–2 mm broad, sublinear to linear, discrete to loosely imbricate or entangled | Absent | Pale tan to brown, moderately to sparsely rhizinate, rhizines concolorous or darkening, to 1 (−1.5) mm long | Medulla: stenosporic acid; ± gyrophoric and divaricatic acids. Cortex: HNO3 + blue-green | Europe, Asia |
| X. perrugata | 1–3 (−5) mm broad, sublinear to linear-elongate, discrete to imbricate or entangled. | Absent | Dark brown to black, moderately to densely rhizinate, rhizines simple, to 1.5 mm long. | Medulla: divaricatic acid; ± stenosporic, oxostenosporic, gyrophoric, lecanoric acids. Cortex: HNO3 + blue-green | Europe, North Africa, Australia, Asia |
| X. pseudoglabrans | 1–2.5 mm broad, subirregular to sublinear, imbricate to entangled | Absent | Black; moderately rhizinate or rhizines rather parse, concolorous with the lower surface | Medulla: alectoronic acid; ± a-collatolic acid. Cortex: HNO3 - | South Africa |
| X. pulla | 1–3 (−5) mm broad, sublinear to linear-elongate, discrete to imbricate or entangled | Absent | Dark brown to black, moderately to densely rhizinate, rhizines simple, to 1.5 mm long | Medulla: stenosporic acid; ± divaricatic, gyrophoric, perlatolic, 4-O-demethylstenosporic acid, oxostenosporic acids. Cortex: HNO3 + blue-green | Europe, Australia, New Zealand, Africa |
| X. pulloides | 1–2 mm broad, subirregular to sublinear, contiguous to subimbricate | Absent | Black, moderately rhizinate, rhizines concolorous, to 0.5 mm long | Medulla: constipatic and protoconstipatic acids; ± gyrophoric acid. Cortex: HNO3 + blue-green | Macaronesia, Asia |
| X. quintarioides | 1–2.5 (−3) mm broad, strongly convex and short-flabellate, discrete but close to more or less contiguous | Absent | Tan to pale brown, sparsely to moderately rhizinate, the rhizines short and hapterate | Medulla: hypostictic, hypoconstictic, cryptostictic acids; ± hyposalazinic acid. Cortex: HNO3 + blue-green | South Africa |
| X. ryssolea | 1–3 mm broad, linear elongate, subterete, convex. | Absent | Pale yellow-brown to red-brown, canaliculate, sparsely rhizinate, rhizines concolorous, to 0.6 mm long. | Medulla: stenosporic acid; ± gyrophoric, oxostenosporic, divaricatic acids. HNO3 + blue-green | Europe, Asia |
| X. squamans | 1–2 mm broad, sublinear, imbricate to contiguous | Absent | Dark brown to black, moderately to sparsely rhizinate, rhizines concolorous, to 1 mm long | Medulla: hypostictic, hypoconstictic, hyposalazinic acids. HNO3 + blue-green | South Africa, South America, New Zealand |
| X. subhosseana | 1–2 mm broad, subirregular, contiguous to slightly imbricate | Sparsely to densely isidiate. Isidia pustular, erumpent | Dark brown to black, moderately rhizinate, rhizines concolorous, to 0.6 mm long | Medulla: hypostictic, hyposalazinic, hypoconstictic acids. Cortex: HNO3 + blue-green | South Africa, North America, South America, New Zealand |
| X. subimitatrix | 0.5–2.0 mm broad, sublinear to subirregular, discrete to subimbricate | Absent | Pale tan to brown, moderately rhizinate, rhizines simple, brown or often blackened, to 0.8 mm long | Medulla: physodic acid and alectoronic acids. Cortex: HNO3 + blue-green | South Africa, Australia. |
| X. subincerta | 0.5–1 mm broad, flat, sublinear, more or less imbricate | Isidia cylindrical, simple or densely branched, 0.08–0.5 mm tall. Apices syncorticate | Black, moderately rhizinate, rhizines simple, black, to 0.3 mm long | Medulla: glomelliferonic acid; ± loxodellonic and glomellonic acids. Cortex: HNO3 + blue-green | Australia, South Africa |
| X. subprolixa | 1–3 (−5) mm broad, sublinear to linear-elongate, discrete to imbricate or entangled. | Absent | Dark brown to black, often paler at apices, moderately to densely rhizinate, rhizines simple, to 1.5 mm long | Medulla: divaricatic acid; ± stenosporic, nordivaricatic acids. Cortex: HNO3 + blue-green | Australia, New Zealand |
| X. torulosa | 1.0–3.5 mm broad, sublinear to sublirregular, imbricate; laciniae at periphery and within thallus, ± subfruticose, sublinear to elongate, 0.3–1.0 mm broad. | Absent | Black, moderately to densely rhizinate; rhizines simple or occasionally tufted, slender. | Medulla: divaricatic acid; ± nordivaricatic, stenosporic acids. Cortex: HNO3 + blue-green | Australia |
| X. verisidiosa | 1–3 mm broad, irregular to sublinear, flat, short and rounded, contiguous to imbricate | Sparsely to densely isidiate. Isidia cylindrical, simple or becoming densely branched, 0.2–1 mm tall. Apices syncorticate | Black, moderately to sparsely rhizinate, rhizines simple, black to 0.4 mm long | Medulla: alectoronic and a-collatolic acids. Cortex: HNO3 + blue-green | Australia, New Zealand, South Africa |
| X. verrucella | 0.5–2 mm wide, irregular to sublinear, flat, imbricate to entangled. | Moderate to densely isidiate. Isidia cylindrical, simple or becoming branched, to 1 mm tall. Apices syncorticate | Black, sparsely to moderately rhizinate, rhizines simple, simple, black, to 0.4 mm long. | Medulla: divaricatic acid; ± stenosporic acid. Cortex: HNO3 + blue-green | Australia, New Zealand, South Africa |
| X. verruculifera | 1–2 mm broad, subirregular to sublinear, contiguous to imbricate | Sparcely to densely isidiate. Isidia pustular, erumpent. | Dark brown to black, moderately rhizinate, rhizines concolorous, to 0.8 mm long | Medulla: divaricatic acid; ± stenosporic and gyrophoric acids. Cortex: HNO3 + blue-green | North Africa, Europe, North America |
Results
Phylogenetic analyses
One hundred sixty-eight DNA sequences of ITS and nuLSU rDNA of 88 representative specimens of Xanthoparmelia were assembled. One hundred forty of these sequences were newly generated in this study. The specimens included 25 currently accepted species in the Xanthoparmelia pulla group, four unassigned specimens, and six samples of four species as outgroup. A data matrix of 1283 unambiguously aligned characters, with 454 characters in the ITS and 829 characters in the nuLSU rDNA was used for phylogenetic analyses. The data set included 1081 constant characters. The general time-reversible model with a gamma distribution and invariant model of rate heterogeneity (GTR+I+G) was employed for analyses of the single-loci and concatenated data sets. Since no strongly supported conflicts between the two single-locus ML phylogenetic trees were detected, a combined data set was analyzed. In the B/MCMC analysis of the combined data set, the likelihood parameters in the sample had the following averaged values for the partitioned data set (± standard deviation): base frequencies π(A) = 0.25 (±1.54E-4), π(C) = 0.24 (±1.42E-4), π(G) = 0.28 (±1.58E-4), π(T) = 0.23 (±1.51E-4); rate matrix r(AC) = 4.42 (±1.43E-4), r(AG) = 0.23 (±8.35E-4), r(AT) = 9.53 (±2.19E-4), r(CG) = 4.82 (±1.45E-4), r(CT) = 0.54 (±9.26E-4), r(GT) = 2.98 (±1.11E-4) and the gamma shape parameter α = 0.21 (±3.86E-4). The likelihood parameters in the sample had a mean likelihood of LnL = −4608.25 (±0.49), while the ML tree had a likelihood of LnL = −4163.64.
The phylogenetic estimates of the ML and B/MCMC analyses were congruent, hence only the ML tree (Fig. 1) is shown here. Specimens of the Xanthoparmelia pulla group form a strongly supported monophyletic group with five main, mostly well-supported, clades (Fig. 1). The clades do not agree with the current species circumscription, with 11 species being polyphyletic, five of them with specimens from different continents entering different clades. For example, all the Northern Hemisphere specimens identified as X. luteonotata, X. pulla, X. delisei or X. glabrans belong to clade 1 while all the Australian specimens of the same species belong to clade 2.1. Similarly, specimens of X. imitatrix from South America belong to clade 2.2 and are not directly related to the South African specimen.
Estimates of divergence times and ancestral range reconstructions
A Bayesian phylogenetic tree was dated to estimate the age of the X. pulla group and its main clades. The results of the divergence time analysis are summarized in Fig. 2, and the whole parmelioid tree is shown as supp. mat. (Fig. S1). The Xanthoparmelia pulla group started to diversify around 11.61 Ma (7.61 – 16.50 Ma), the age of the crown node of clade 1 was estimated at 5.31 Ma (3.01 – 8.38 Ma), the ancestor of clade 2 around 8.10 Ma (5.13 – 11.74 Ma), and the crown of clade 4 around 3.44 Ma (1.56 – 6.07 Ma).
The results of the ancestral range reconstruction analyses are summarized in Figs. 1 and 2. This established that South Africa was the most likely origin of the X. pulla group, with a marginal probability of 0.745, indicating localized uncertainty. The four other areas explored (South America, Australia, California and the Mediterranean basin) were rejected with probabilities below 0.05. For the base of clade 1, the Mediterranean basin was reconstructed as the most likely ancestral range with a marginal probability of 0.555, but California could not be rejected (probability of 0.104). For clade 2, Australia was recovered as the most likely ancestral area with marginal probability of 0.672; while South America, the other area from which specimens of this clade occur, was rejected as potential ancestral area (p<0.05); similarly, California, the Mediterranean basin and South Africa were also rejected as ancestral areas. South America was found to be the most likely origin for clade 4 (which also includes specimens from California and South America) with a marginal probability of 0.48, although neither California nor South Africa were rejected (probabilities of 0.106 and 0.083, respectively). Australia and the Mediterranean basin were rejected as ancestral ranges for clade 4.
Discussion
Understanding the diversity and delimiting species in lichenized fungi has been a long standing challenge and current studies using molecular data have dramatically changed our ability to distinguish species in this group [8], [23], [45]. The Xanthoparmelia pulla group is a good example for illustrating the difficulties in in distinguishing species by morphology due to the remarkable plasticity of morphological characters in this group. Consequently, secondary metabolites have played an important role in delimiting species in this group [44], [46], [47]. Following the current classification using a combination of vegetative morphology and secondary chemistry, a number of species have broad geographical distributions spanning several continents.
Here we have used molecular data to investigate the current classification within the group and attempt to explain their distribution. We used likelihood-based and Bayesian approaches to investigate the evolutionary origin of the group and timing of speciation events. Hopefully such data will reveal evolutionary patterns so we may develop a framework for their taxonomic classification which better reflects the phylogenetic relationships in the X. pulla group. Our results clearly indicate that the species as currently delimited are polyphyletic (Fig. 1). This is consistent with results from other studies of Xanthoparmelia species believed to occur on different continents which were subsequently found to represent distinct lineages [15], [16]. Further, similar patterns have been found in other groups of lichenized fungi [4], [14], [18], [48].
The ancestral range reconstruction points to South Africa as the most likely origin of the X. pulla group. Although there is a certain degree of uncertainty in the reconstruction (marginal probability of 0.745), the analysis rejected other areas as potential ancestral areas for the group. Interestingly, South Africa has the highest morphological and chemical diversity within the group and the specimens studied here belong to different, unrelated lineages (Fig. 1). South African specimens containing olivetoric acid cluster in clade 3 and those with physodic acid in clade 5. The phylogenetic relationships of other specimens from South Africa with hypostictic acid, physodic acid or other orcinol depsides and depsidones are still unresolved. The South African specimens show remarkable morphological variability, including subcrustose and foliose species. Further, many Xanthoparmelia species occur in arid climates and the diversification of X. pulla group occurred around 11.61 Ma (7.61 – 16.50). At this time the Cape Region underwent a major aridification [49], which may be responsible for the rapid radiation and current richness of the Cape flora. Thus, it is likely that the X. pulla group originated in South Africa around the same time. Unlike most Cape region elements in flowering plants, the species of the X. pulla group subsequently extended their distribution by transoceanic dispersal.
Within the X. pulla group, the five lineages identified are characterized by the presence of different substance classes and in some cases they diverged secondarily in different geographical areas. The correlation between chemical pathways and the lineages found in molecular studies has also been found in Pertusariaceae among lichenized fungi [50]–[52].
Clade 1 includes specimens containing orcinol depsides and depsidones that occur in California and the Mediterranean basin. Neither internally supported subclades nor a geographical pattern was found within this clade, and specimens with different phenotypical characters from different geographical areas are intermingled. In fact, shifting between orcinol depsides and depsidones can occur by one-step transformations [46]. By contrast, in other genera (e.g. Melanelixia, Parmelina, Leptogium) with similar disjunct distributions (North America and the Mediterranean basin), the geographical distribution correlates with the clades found in the molecular study. In the X. pulla group this pattern was not found, possibly due to insufficient sampling or absence of a phylogenetic signal in the markers used. This might be due to the slower evolutionary rates of lichenized fungi from arid and subarid regions compared to oceanic parmelioid lichens [36]. The diversification age of clade 1 was estimated at 5.31 Ma (3.01 – 8.38 Ma), at the end of the Miocene, which was a geological period when numerous groups radiated in arid conditions. The Mediterranean region was suggested as the ancestral area of this clade by the ancestral range analysis, but the result was poorly supported.
Clade 2 also contains species with orcinol depsides and depsidones and comprises two disjunct lineages, one occurring in Australia (clade 2.1) and the other in South America (2.2). The estimated age for clade 2 (8.10 Ma; 5.13 – 11.74 Ma) rules out the possibility of vicariance, since the breakup of Australia, Antarctica and South America occurred between 35–52 Ma ago [53]. The ancestral range reconstruction points to Australia as the ancestral area of clade 2. This would be consistent with long distance dispersal from Australia to South America, a phenomenon frequently found in many plants groups [54]. Within the Australian clade several strongly supported lineages are not consistent with the current species delimitation of the group, indicating that the phenotypical characters used to distinguish species in the group have limited phylogenetic validity. Similar disparities between phylogenetic relationships and current species delimitations were found within the yellow Xanthoparmelia species from western North American [55], [56].
Clade 4 includes specimens containing hypostictic acid from California and South America. Here again all the South American specimens form a monophyletic group. Specimens of X. subhosseana occurring in different continents are not closely related. The most likely ancestral origin of clade 4 is South America (marginal probability of 0.48), although neither California nor South Africa could be rejected.
Our study indicates that the X. pulla group started to radiate during the Miocene in South Africa, where the highest diversity of this group is found. From this region, different lineages with distinct secondary metabolites belonging to different chemical pathways were dispersed to other regions, where they experienced rapid and more recent radiations. In some cases our results showed that the sympatric species of the X. pulla group in an area belong to distantly related groups. For example, the Californian X. pulla flora includes species from clade 1 and clade 4.1, the latter most probably having migrated from South America. Indeed our study indicated that the current taxonomic circumscription of species in the group does not agree with the evolutionary hypotheses inferred by molecular markers. The incongruence of phenotype-based classification and molecular phylogeny is a challenge for the classification of these fungi. Additional studies will be needed to determine whether the lineages found here represent cryptic species or whether new phenotypical characters can be found to distinguish these distinct lineages (as has been found in some other Parmeliaceae [10], [57]). Future research should address how such parallel evolution of phenotypical characters in lichenized fungi could be explained in order to provide a better framework to test the adaptive value of these characters [58]. Our results here have important implications for conservation and ecological issues, since species were found to have much more restricted distribution than previously thought.
Materials and Methods
Taxon sampling
Eighty two specimens of the Xanthoparmelia pulla group from California, the Mediterranean basin, Macaronesia, South America, Australia and South Africa were used for the phylogenetic study. The specimens were identified following the current species delimitations [38], [44], [47], [59], [60]. Chemical constituents were identified using thin layer chromatography (TLC) [61]–[64], and gradient-elution high performance liquid chromatography (HPLC) [65]. The major medullary compounds were classified into four major groups based on their chemical structure: 1) Orcinol depsides: olivetoric, divaricatic, stenosporic and glomelliferic acids. 2) Orcinol depsidones, physodic, alectoronic and glomelliferonic acids. 3) β-Orcinol depsidones: stictic acid (only present in the outgroup) and hypostictic acid. 4) aliphatic acids: constipatic acid. All necessary permits were obtained for the described field studies. Collecting permits in Australia were all obtained by J.A. Elix (ca. 50 permit numbers for each states and over several years) and in Chile by W. Quilhot. For European locations specific permission was not required, since the locations were neither in privately-owned or protected areas. The field studies did not involve endangered or protected species.
Molecular study
Total DNA was extracted from frozen lobes of thalli crushed with sterile glass pestles, using the DNeasy Plant Mini Kit (Qiagen) following the manufacturer's instructions and modifications of Crespo et al. [66]. The following primers were used: ITS1-LM [67] and ITS2-KL [68] for nuITS rDNA, and LR0R and LR5 [69] for nuLSU rDNA.
For each amplification we used a reaction mixture of 25 μL, containing: 2.5 μL of 10x DNA polymerase buffer (including MgCl2 2mM) (Biotools), 1.25 μL of each primer, 0.75 μL of DNA polymerase (1U/μL), 0.5 μL of dNTPs containing 10 mM of each base (Biotools), 5 μL of DNA (third elution of DNA extraction) and 13.5 μL dH2O. Amplifications were carried out in an automatic thermocycler (Techne Progene 3000) with the following steps: an initial denaturation at 94°C for 5 min; 35 cycles of 94°C for 1 min, 58°C (nuITS rDNA) or 56°C (nuLSU rDNA) for 1 min, and 72°C for 1.5 min; a final extension at 72°C for 5 min. PCR products were cleaned with DNA Purification Kit (Flavorgen) and sequenced with the same primers using the ABI Prism Dye Terminator Cycle Sequencing Ready reaction kit (Applied Biosystems) with the following program: initial denaturation at 94°C for 3 min, 25 cycles at 96°C for 10s, 50°C for 5s and 60°C for 4 min. Sequencing reactions were electrophoresed on a 3730 DNA analyzer (Applied Biosystems). The sequence fragments were assembled with Bioedit v. 7.0 [70] and manually adjusted.
Sequence alignment and selection of the substitution model
We used a dataset of 2 loci of 82 specimens representing 25 species of the Xanthoparmelia pulla group and 6 specimens as outgroup. The sequences were mainly generated in this study (140 sequences) and the others taken from our previous studies [33], [41]. The outgroup selection was based on previous phylogenetic studies [41]. GenBank accession numbers and sources of the specimens are listed in Table 2.
Table 2. Specimens used in this study with country of collection, voucher information and GenBank accession numbers.
| GenBank accession no. | ||||
| Species | Country | Herbarium acc. no. | nuITS | nuLSU |
| Xanthoparmelia adhaerens 1 | South Africa | MAF-Lich 16212 | HM125744 | HM125766 |
| X. adhaerens 2 | South Africa | MAF-Lich 16213 | HM125746 | HM125768 |
| X. atroviridis 1 | South Africa | MAF-Lich 17163 | JQ912329 | JQ912425 |
| X. atroviridis 1 | South Africa | MAF-Lich 17168 | JQ912351 | JQ912448 |
| X. atroviridis 2 | South Africa | MAF-Lich 17158 | JQ912314 | JQ912415 |
| X. atroviridis 2 | South Africa | MAF-Lich 17154 | JQ912320 | JQ912419 |
| X. atroviridis 3 | South Africa | MAF-Lich 17153 | JQ912349 | JQ912446 |
| X. caliginosa 1 | South Africa | MAF-Lich 17157 | JQ912350 | JQ912447 |
| X. caliginosa 2 | South Africa | MAF-Lich 17150 | JQ912315 | - |
| X. caliginosa 3 | South Africa | MAF-Lich 17156 | JQ912333 | JQ912430 |
| X. caliginosa 4 | South Africa | MAF-Lich 17152 | JQ912317 | - |
| X. caliginosa 5 | South Africa | MAF-Lich 17186 | JQ912348 | JQ912445 |
| X. delisei 1 | Turkey | MAF-Lich 17139 | JQ912307 | JQ912408 |
| X. delisei 2 | Australia | MAF-Lich 7432 | AY581067 | AY578930 |
| X. delisei 3 | Spain | MAF-Lich 7659 | AY581068 | AY578931 |
| X. delisei 4 | Turkey | MAF-Lich 17134 | JQ912308 | JQ912409 |
| X. delisei 5 | Turkey | MAF-Lich 17135 | JQ912305 | JQ912406 |
| X. fissurina | South Africa | MAF-Lich 17162 | JQ912353 | JQ912450 |
| X. glabrans 1 | Australia | CANB 746334 | JQ912291 | JQ912393 |
| X. glabrans 2 | Australia | CANB 746340 | JQ912289 | JQ912391 |
| X. glabrans 3 | Australia | MAF-Lich 7665 | AY581069 | AY578932 |
| X. glabrans 4 | Australia | CANB 681875.1 | JQ912290 | JQ912392 |
| X. glabrans 5 | Spain | MAF-Lich 9912 | AY581072 | AY578935 |
| X. glabrans 6 | Turkey | MAF-Lich 17137 | JQ912306 | JQ912407 |
| X. glabrans 7 | Morocco | MAF-Lich 17144 | JQ912286 | JQ912388 |
| X. imitatrix 1 | Chile | MAF-Lich 17132 | JQ912344 | JQ912441 |
| X. imitatrix 2 | Chile | MAF-Lich 17126 | JQ912288 | JQ912390 |
| X. imitatrix 3 | Chile | MAF-Lich 17123 | JQ912287 | JQ912389 |
| X. imitatrix 4 | Chile | MAF-Lich 17127 | JQ912342 | JQ912439 |
| X. imitatrix 5 | Chile | MAF-Lich 17122 | JQ912285 | JQ912387 |
| X. imitatrix 6 | Chile | MAF-Lich 17124 | JQ912326 | JQ912422 |
| X. imitatrix 7 | South Africa | MAF-Lich 17155 | JQ912352 | JQ912449 |
| X. lineella | South Africa | MAF-Lich 17160 | JQ912319 | JQ912418 |
| X. loxodes 1 | Spain | MAF-Lich 7072 | AY581076 | AY578940 |
| X. loxodes 2 | Spain | MAF-Lich 6206 | AY581070 | AY578933 |
| X. luteonotata 1 | Spain | MAF-Lich 17120 | JQ912341 | JQ912438 |
| X. luteonotata 2 | Australia | CANB 746358 | JQ912293 | - |
| X. luteonotata 3 | Australia | CANB 746366.1 | JQ912292 | JQ912394 |
| X. luteonotata 4 | Spain | MAF-Lich 17119 | JQ912340 | JQ912437 |
| X. mougeotii 1 | Spain | MAF-Lich 6802 | AY37006 | AY578966 |
| X. mougeotii 2 | Spain | MAF-Lich 9916 | AY581100 | AY578967 |
| X. perrugata | Spain | MAF-Lich 17118 | JQ912324 | - |
| X. pokornyi 1 | Spain | MAF-Lich 6052 | AY037005 | AY578934 |
| X. pokornyi 2 | Spain | MAF-Lich 9908 | AY581075 | AY578939 |
| X. pokornyi 3 | Turkey | MAF-Lich 17140 | JQ912310 | JQ912411 |
| X. pokornyi 4 | Spain | MAF-Lich 17117 | JQ912323 | - |
| X. pokornyi 5 | Turkey | MAF-Lich 17143 | JQ912313 | JQ912414 |
| X. pokornyi 6 | Turkey | MAF-Lich 17142 | JQ912312 | JQ912413 |
| X. pokornyi 7 | Turkey | MAF-Lich 17136 | JQ912304 | JQ912405 |
| X. pseudoglabrans | South Africa | MAF-Lich 17161 | JQ912316 | JQ912416 |
| X. pulla 1 | Spain | MAF-Lich 17115 | - | JQ912420 |
| X. pulla 2 | Australia | CANB 739130.1 | JQ912294 | JQ912395 |
| X. pulla 3 | Australia | CBG 9810185 | JQ912295 | JQ912396 |
| X. pulla 5 | Spain | MAF-Lich 6794 | AY581071 | AJ 421433 |
| X. pulloides 1 | Spain | MAF-Lich 17121 | JQ912347 | JQ912444 |
| X. pulloides 2 | Spain | MAF-Lich 6784 | AY037004 | AY578936 |
| X. quintarioides | South Africa | MAF-Lich 17159 | JQ912318 | JQ912417 |
| X. ryssolea 1 | Turkey | MAF-Lich 17141 | JQ912311 | JQ912412 |
| X. ryssolea 2 | Turkey | MAF-Lich 17138 | JQ912309 | JQ912410 |
| X. ryssolea 3 | Spain | MAF-Lich 17116 | JQ912322 | - |
| X. sp. 1 | South Africa | MAF-Lich 17166 | JQ912330 | JQ912426 |
| X. sp. 2 | South Africa | MAF-Lich 17167 | JQ912331 | JQ912427 |
| X. sp. 3 | South Africa | MAF-Lich 17165 | - | JQ912429 |
| X. sp. 4 | South Africa | MAF-Lich 17164 | JQ912339 | JQ912436 |
| X. squamans 1 | Chile | MAF-Lich 17128 | JQ912325 | JQ912421 |
| X. squamans 2 | Chile | MAF-Lich 17129 | JQ912327 | JQ912423 |
| X. squamans 3 | Chile | MAF-Lich 17131 | JQ912343 | JQ912440 |
| X. subhosseana 1 | USA | MAF-Lich 17149 | JQ912337 | JQ912434 |
| X. subhosseana 2 | Chile | MAF-Lich 17133 | JQ912345 | JQ912442 |
| X. subimitatrix | Chile | MAF-Lich 17130 | JQ912328 | JQ912424 |
| X. subincerta 1 | Australia | CANB 746346.1 | JQ912296 | JQ912397 |
| X. subincerta 2 | Australia | MAF-Lich 7494 | AY581073 | AY578937 |
| X. subprolixa 1 | Australia | MAF-Lich 7667 | AY581074 | AY578938 |
| X. subprolixa 2 | Australia | CANB 746355 | JQ912297 | JQ912398 |
| X. tegeta | Australia | MAF-Lich 7523 | AY581107 | AY578975 |
| X. torulosa 1 | Australia | CANB 746363.1 | JQ912299 | JQ912400 |
| X. torulosa 2 | Australia | CANB 746351 | JQ912298 | JQ912399 |
| X. verisidiosa 1 | Australia | CANB 746341.1 | JQ912301 | JQ912402 |
| X. verisidiosa 2 | Australia | CANB 746345.1 | JQ912300 | JQ912401 |
| X. verrucella 1 | Australia | CANB 746353 | JQ912303 | JQ912404 |
| X. verrucella 2 | Australia | CANB 746349 | JQ912302 | JQ912403 |
| X. verrucella 3 | South Africa | MAF-Lich 17151 | JQ912332 | JQ912428 |
| X. verruculifera 1 | USA | MAF-Lich 17146 | JQ912334 | JQ912431 |
| X. verruculifera 2 | USA | MAF-Lich 17147 | JQ912336 | JQ912433 |
| X. verruculifera 3 | USA | MAF-Lich 17145 | JQ912338 | JQ912435 |
| X. verruculifera 4 | USA | MAF-Lich 17148 | JQ912335 | JQ912432 |
| X. verruculifera 5 | Spain | MAF-Lich 17114 | JQ912321 | - |
| X. xanthomelaena | Australia | MAF-Lich 16447 | HM125740 | HM125761 |
New sequences are in bold.
The two loci were aligned separately with Muscle 3.6 [71] and the ambiguous positions were removed manually. The general time reversible model including estimation of invariant sites (GTR+I+G) was selected by jModelTest v 0.1.1 [72] as the most appropriate nucleotide substitution model for both loci.
Phylogenetic Analyses
Potential conflict between the two loci was assessed by comparison of the ML analyses obtained with Garli 0.96 [73] for each locus, using 100 pseudoreplicates for the bootstrap analyses. The phylogenetic analyses of the combined matrix were done using maximum likelihood (ML) and a Bayesian approach. ML analysis was performed using Garli 0.96 [73] with default settings and 100 replicates for the bootstrap analyses. The Bayesian analysis was performed using MrBayes 3.1.1 [74] using the GTR+I+G model, and the data set partitioned into nu ITS and nu LSU. Each partition was allowed to have its own parameter values [75]. Heating of chains was set to 0.2, with 5 million generations sampled every 500th tree. The first 1000 trees were discarded as burn in. We used AWTY [76] to compare splits frequencies in the different runs and to plot cumulative split frequencies to insure that stationarity was reached. Of the remaining 18000 trees (9000 from each of the parallel runs) a majority rule consensus tree with average branch lengths and posterior probabilities was calculated using the sumt option of MrBayes. Clades with bootstrap support equal or above 70 % under ML and/or posterior probabilities ≥0.95 in the Bayesian analysis were considered as strongly supported. Phylogenetic trees were visualized using the program FigTree [77].
Calibration of nodes and dating analysis
The ages of the X. pulla group and its major clades were estimated by a divergence time analysis based on a calibrated phylogeny of the parmelioid lichens [78]. We used a matrix of two loci (nu ITS and LSU) with a proportional number of samples of each parmelioid clade to have a representative tree and trend in speciation through time [79]. The matrix included 299 specimens of parmelioid lichens and 3 specimens of the genus Usnea (as outgroup); 62 new sequences of Xanthoparmelia species outside the X. pulla group were included. GenBank accession numbers with the specimens of the dating analysis are listed in Table S1. The sequence alignment, selection of the nucleotide substitution model, and phylogenetic analyses were done using the same procedures used for the Xanthoparmelia pulla dataset (see above).
The divergence time analyses were performed using BEAST v.1.6.1 [80]. We used a starting tree obtained from a ML analysis using Garli 0. 96 [73] of the concatenated dataset, calculated an ultrametric tree using nonparametric rate smoothing (NPRS) implemented in TreeEdit v.10a10 [81]. The age of the crown node of the parmelioid lichens was calibrated at 60 Ma, following Amo de Paz et al. [78]. The starting tree was topologically congruent with the parmelioid phylogeny presented in Crespo et al. [32].
For the divergence time analyses we used two points of calibration: the age of the crown node of the parmelioid lichens set at 60.28 Ma (49.81 – 73.55 Ma) [78], and the age of the crown node of the genus Parmelia (dated from the fossil Parmelia ambra from the Dominican amber, 15–45 Ma, [82] as discussed previously) [78].
The BEAST analysis was performed using the GTR+I+G substitution model, a Birth-Death process tree prior, and a relaxed clock model (uncorrelated lognormal) for the concatenated dataset. Calibration points were defined as prior distribution, minimal ages and calibrated with a lognormal distributions: 1) the parmelioid crown node at uniform distribution between 49 – 73 Ma; 2) the Parmelia crown node at log-normal mean = 2.77, offset = 14, lognormal standard deviation = 0.5. The analysis was run for 10 million generations, with parameter values sampled every 1000 generation. We checked the stationary plateau with Tracer v. 1.5 [83]. We discarded 10% of the initial trees as burn in and the consensus tree was calculated using Tree Annotator v 1.6.1 [80]. The results were visualized with FigTree v. 1.3.1 [84]. Ages of the X. pulla clades were estimated for nodes with more than 0.95 of posterior probability in the BEAST runs and in the previous Bayesian analysis.
Ancestral range reconstructions
The biogeographical analysis to reconstruct the ancestral area was performed using an indirect Bayesian approach to character state reconstruction [85] implemented in SIMMAP v1.5 [86] following [87]. This analysis integrates the combination of the uncertainty in the tree, branch lengths and the substitution models using Markov chain Monte Carlo. We treated the biogeographic regions as discrete characters. The major areas in which X. pulla species are distributed were categorised broadly into five areas: California, Mediterranean basin (including Macaronesia), South America, South Africa, and Australia. Presence/absence was coded as binary states and each area was given equal probability. We performed the ancestral state reconstruction analysis on a sub-sample of 1000 trees derived from the MrBayes tree sampling of the Xanthoparmelia pulla group.
We also performed ancestral range reconstruction analysis using dispersal-extinction-cladogenesis (DEC) implemented in Lagrange program [88]. The results were inconclusive due to the lack of confidence in parts of the X. pulla phylogeny and hence the results are not included in this paper.
Supporting Information
Chronogram of parmelioid lichens focusing in Xanthoparmelia pulla group. Calibration points: A, inferred age of radiation of parmelioid lichens and B, age of the Parmelia fossil. The Xanthoparmelia pulla group is highlighted by a box and the dated clades are indicated by a branch in bold.
(TIF)
GenBank accession numbers of parmelioid lichens (except X. pulla group, see Table 2 ) used for divergence time analysis. New sequences are in bold.
(DOC)
Acknowledgments
We thank L. Mucina (Perth) for his help in the organization of the field work in South Africa. GAP thanks W. Quilot (Valparaiso) and C. Rubio (Valparaiso) and C. Cargill (Canberra) who kindly facilitated the visits to the Universidad de Valparaiso (Chile) and ANU and CANBR (Australia).
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This study was supported by a grant from the Spanish Ministerio de Ciencia e Innovación (CGL2010-21646) and a grant by the National Science Foundation (USA) (DEB-0949147). The Universidad Complutense de Madrid is thanked for providing all complementary facilities and funds (SYSTEMOL research group UCM-BSCH GR35/10A). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Wiens JJ, Penkrot TA. Delimiting species using DNA and morphological variation and discordant species limits in spiny lizards (Sceloporus). Systematic Biology. 2002;51:69–91. doi: 10.1080/106351502753475880. [DOI] [PubMed] [Google Scholar]
- 2.Cracraft J. Species concepts and speciation analysis. Current Ornithology. 1983;1:159–187. [Google Scholar]
- 3.Mayr E. Animal species and evolution. Cambridge, MA: Harvard University Press. 1963.
- 4.Arguello A, Del Prado R, Cubas P, Crespo A. Parmelina quercina (Parmeliaceae, Lecanorales) includes four phylogenetically supported morphospecies. Biological Journal of the Linnean Society. 2007;91:455–467. [Google Scholar]
- 5.Leavitt SD, Fankhauser JD, Leavitt DH, Porter LD, Johnson LA, et al. Complex patterns of speciation in cosmopolitan “rock posy” lichens – Discovering and delimiting cryptic fungal species in the lichen-forming Rhizoplaca melanophthalma species-complex (Lecanoraceae, Ascomycota). Molecular Phylogenetics and Evolution. 2011;59:587–602. doi: 10.1016/j.ympev.2011.03.020. [DOI] [PubMed] [Google Scholar]
- 6.Leavitt SD, Johnson L, St Clair LL. Species delimitation and evolution in morphologially and chmically diverse communities of the lichen-forming genus Xanthoparmelia (Parmeliaceae, Ascomycota) in western North America. American Journal of Botany. 2011;98:175–188. doi: 10.3732/ajb.1000230. [DOI] [PubMed] [Google Scholar]
- 7.Leavitt SD, Johnson LA, Goward T, St Clair LL. Species delimitation in taxonomically difficult lichen-forming fungi: An example from morphologically and chemically diverse Xanthoparmelia (Parmeliaceae) in North America. Molecular Phylogenetics and Evolution. 2011;60:317–332. doi: 10.1016/j.ympev.2011.05.012. [DOI] [PubMed] [Google Scholar]
- 8.Lumbsch HT, Leavitt SD. Goodbye morphology? A paradigm shift in the delimitation of species in lichenized fungi Fungal Diversity. 2011;50:59–72. [Google Scholar]
- 9.Elix JA, Corush J, Lumbsch HT. Triterpene chemosyndromes and subtle morphological characters characterise lineages in the Physcia aipolia group in Australia (Ascomycota). Systematics and Biodiversity. 2009;7:479–487. [Google Scholar]
- 10.Wirtz N, Printzen C, Lumbsch HT. The delimitation of Antarctic and bipolar species of neuropogonoid Usnea (Ascomycota, Lecanorales): a cohesion approach of species recognition for the Usnea perpusilla complex. Mycological Research. 2008;112:472–484. doi: 10.1016/j.mycres.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 11.Cubero OF, Crespo A, Esslinger TL, Lumbsch HT. Molecular phylogeny of the genus Physconia (Ascomycota, Lecanorales) inferred from a Bayesian analysis of nuclear ITS rDNA sequences. Mycological Research. 2004;108:498–505. doi: 10.1017/s095375620400975x. [DOI] [PubMed] [Google Scholar]
- 12.Molina MD, Crespo A, Blanco O, Lumbsch HT, Hawksworth DL. Phylogenetic relationships and species concepts in Parmelia s.str. (Parmeliaceae) inferred from nuclear ITS rDNA and beta-tubulin sequences. Lichenologist. 2004;36:37–54. [Google Scholar]
- 13.Crespo A, Molina MC, Blanco O, Schroeter B, Sancho LG, et al. rDNA ITS and beta-tubulin gene sequence analyses reveal two monophyletic groups within the cosmopolitan lichen Parmelia saxatilis. Mycological Research. 2002;106:788–795. [Google Scholar]
- 14.Divakar PK, Figueras G, Hladun NL, Crespo A. Molecular phylogenetic studies reveal an undescribed species within the North American concept of Melanelixia glabra (Parmeliaceae). Fungal Diversity. 2010;42:47–55. [Google Scholar]
- 15.Hodkinson BP, Lendemer JC. Molecular analyses reveal semi-cryptic species in Xanthoparmelia tasmanica. Bibliotheca Lichenologica. 2011;106:108–119. [Google Scholar]
- 16.Thell A, Elix JA, Søchting U. Xanthoparmelia lineola s. l. in Australia and North America. Bibliotheca Lichenologica. 2009;99:393–404. [Google Scholar]
- 17.Velmala S, Myllys L, Halonen P, Goward T, Ahti T. Molecular data show that Bryoria fremontii and B. tortuosa (Parmeliaceae) are conspecific. Lichenologist. 2009;41:231–242. [Google Scholar]
- 18.Otálora MAG, Martínez I, Aragón G, Molina MC. Phylogeography and divergence date estimates of a lichen species complex with a disjunct distribution pattern. American Journal of Botany. 2010;97:216–223. doi: 10.3732/ajb.0900064. [DOI] [PubMed] [Google Scholar]
- 19.Otalora MAG, Martinez I, Molina MC, Aragon G, Lutzoni F. Phylogenetic relationships and taxonomy of the Leptogium lichenoides group (Collemataceae, Ascomycota) in Europe. Taxon. 2008;57:907–921. [Google Scholar]
- 20.Sérusiaux E, Villarreal A JC, Wheeler T, Goffinet B. Recent origin, active speciation and dispersal for the lichen genus Nephroma (Peltigerales) in Macaronesia. Journal of Biogeography: no–no. 2011.
- 21.Wedin M, Westberg M, Crewe AT, Tehler A, Purvis OW. Species delimitation and evolution of metal bioaccumulation in the lichenized Acarospora smaragdula (Ascomycota, Fungi) complex. Cladistics. 2009;25:161–172. doi: 10.1111/j.1096-0031.2009.00240.x. [DOI] [PubMed] [Google Scholar]
- 22.Kroken S, Taylor JW. A gene genealogical approach to recognize phylogenetic species boundaries in the lichenized fungus Letharia. Mycologia. 2001;93:38–53. [Google Scholar]
- 23.Crespo A, Lumbsch HT. Cryptic species in lichen-forming fungi. IMA Fungus. 2010;1:167–170. doi: 10.5598/imafungus.2010.01.02.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Divakar PK, Blanco O, Hawksworth DL, Crespo A. Molecular phylogenetic studies on the Parmotrema reticulatum (syn. Rimelia reticulata) complex, including the confirmation of P. pseudoreticulatum as a distinct species. Lichenologist. 2005;37:55–65. [Google Scholar]
- 25.Baloch E, Grube M. Pronounced genetic diversity in tropical epiphyllous lichen fungi. Molecular Ecology. 2009;18:2185–2197. doi: 10.1111/j.1365-294X.2009.04183.x. [DOI] [PubMed] [Google Scholar]
- 26.Högnabba F, Wedin M. Molecular phylogeny of the Sphaerophorus globosus species complex. Cladistics. 2003;19:224–232. [Google Scholar]
- 27.Fenchel T, Finlay BJ. The ubiquity of small species: Patterns of local and global diversity. Bioscience. 2004;54:777–784. [Google Scholar]
- 28.Finlay BJ. Global dispersal of free-living microbial eukaryote species. Science. 2002;296:1061–1063. doi: 10.1126/science.1070710. [DOI] [PubMed] [Google Scholar]
- 29.Lumbsch HT, Buchanan PK, May TW, Mueller GM. Phylogeography and biogeography of fungi. Mycological Research. 2008;112:423–424. doi: 10.1016/j.mycres.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 30.Crespo A, Lumbsch HT, Mattsson JE, Blanco O, Divakar PK, et al. Testing morphology-based hypotheses of phylogenetic relationships in Parmeliaceae (Ascomycota) using three ribosomal markers and the nuclear RPB1 gene. Molecular Phylogenetics and Evolution. 2007;44:812–824. doi: 10.1016/j.ympev.2006.11.029. [DOI] [PubMed] [Google Scholar]
- 31.Hale BW, DePriest PT. Mason E. Hale's list of epithets in the parmelioid genera. Bryologist. 1999;102:462–544. [Google Scholar]
- 32.Crespo A, Kauff F, Divakar PK, Amo G, Arguello A, et al. Phylogenetic generic classification of parmelioid lichens (Parmeliaceae, Ascomycota) based on molecular, morphological and chemical evidence. Taxon. 2010;59:1735–1753. [Google Scholar]
- 33.Blanco O, Crespo A, Elix JA, Hawksworth DL, Lumbsch HT. A molecular phylogeny and a new classification of parmelioid lichens containing Xanthoparmelia-type lichenan (Ascomycota: Lecanorales). Taxon. 2004;53:959–975. [Google Scholar]
- 34.Del Prado R, Ferencova Z, Armas-Crespo V, De Paz GA, Cubas P, et al. The arachiform vacuolar body: an overlooked shared character in the ascospores of a large monophyletic group within Parmeliaceae (Xanthoparmelia clade, Lecanorales). Mycological Research. 2007;111:685–692. doi: 10.1016/j.mycres.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 35.Blanco O, Crespo A, Ree RH, Lumbsch HT. Major clades of parmeliold lichens (Parmeliaceae, Ascomycota) and the evolution of their morphological and chemical diversity. Molecular Phylogenetics and Evolution. 2006;39:52–69. doi: 10.1016/j.ympev.2005.12.015. [DOI] [PubMed] [Google Scholar]
- 36.Lumbsch HT, Hipp AL, Divakar PK, Blanco O, Crespo A. Accelerated evolutionary rates in tropical and oceanic parmelioid lichens (Ascomycota). BMC Evolutionary Biology 8. 2008. [DOI] [PMC free article] [PubMed]
- 37.Esslinger TL. A new status for the brown Parmeliae. Mycotaxon. 1978;7:45–54. [Google Scholar]
- 38.Esslinger TL. Notes on the brown-colored species of Parmeliaceae (lichenized Ascomycota) in southern Africa. Bryologist. 2000;103:568–591. [Google Scholar]
- 39.Elix JA. The lichen genus Paraparmelia, a synonym of Xanthoparmelia (Ascomycota, Parmeliaceae). Mycotaxon. 2003;87:395–403. [Google Scholar]
- 40.Amo de Paz G, Lumbsch HT, Cubas P, Elix JA, Crespo A. The morphologically deviating genera Omphalodiella and Placoparmelia belong to Xanthoparmelia (Parmeliaceae). The Bryologist. 2010;113:376–386. [Google Scholar]
- 41.Amo de Paz G, Lumbsch HT, Cubas P, Elix JA, Crespo A. The genus Karoowia (Parmeliaceae, Ascomycota) includes unrelated clades nested within Xanthoparmelia. Australian Systematic Botany. 2010;23:173–184. [Google Scholar]
- 42.Thell A, Feuerer T, Elix JA, Kärnefelt I. A contribution to the phylogeny and taxonomy of Xanthoparmelia (Ascomycota, Parmeliaceae). Journal of the Hattori Botanical Laboratory. 2006;100:797–807. [Google Scholar]
- 43.Hawksworth DL, Crespo A. Proposal to conserve the name Xanthoparmelia against Chondropsis nom. cons. (Parmeliaceae). Taxon. 2002;51:807. [Google Scholar]
- 44.Esslinger TL. A chemosystematic revision of the brown Parmeliae. Jour Hattori Bot Lab. 1977;42:1–211. [Google Scholar]
- 45.Crespo A, Perez-Ortega S. Cryptic species and species pairs in lichens: A discussion on the relationship between molecular phylogenies and morphological characters. Anales del Jardín Botánico de Madrid. 2009;66:71–81. [Google Scholar]
- 46.Culberson CF, Culberson WL, Esslinger TL. Chemosyndromic variation in the Parmelia pulla group. Bryologist. 1977;80:125–135. [Google Scholar]
- 47.Elix JA. Chemical variations of the lichen Neofuscelia pulla (Ascomycotina: Parmeliaceae) sensu Esslinger. Australasian Lichenology. 2002;51:7–9. [Google Scholar]
- 48.Elix JA, Corush J, Lumbsch HT. Triterpene chemosyndromes and subtle morphological characters characterise lineages in the Physcia aipolia group in Australia (Ascomycota). Systematics and Biodiversity. 2009;7:479–487. [Google Scholar]
- 49.Richardson JE, Weitz FM, Fay MF, Cronk QCB, Linder HP, et al. Rapid and recent origin of species richness in the Cape flora of South Africa. Nature. 2001;412:181–183. doi: 10.1038/35084067. [DOI] [PubMed] [Google Scholar]
- 50.Lumbsch HT, Schmitt I, Barker D, Pagel M. Evolution of micromorphological and chemical characters in the lichen-forming fungal family Pertusariaceae. Biological Journal of the Linnean Society. 2006;89:615–626. [Google Scholar]
- 51.Schmitt I, Lumbsch HT. Molecular phylogeny of the Pertusariaceae supports secondary chemistry as an important systematic character set in lichen-forming ascomycetes. Molecular Phylogenetics and Evolution. 2004;33:43–55. doi: 10.1016/j.ympev.2004.04.014. [DOI] [PubMed] [Google Scholar]
- 52.Lumbsch HT, Schmitt I. Molecular data shake the Pertusariaceae tree into order. Lichenology. 2002;1:37–43. [Google Scholar]
- 53.Sanmartin I, Ronquist F. Southern Hemisphere biogeography inferred by event-based models: Plant versus animal patterns. Systematic Biology. 2004;53:216–243. doi: 10.1080/10635150490423430. [DOI] [PubMed] [Google Scholar]
- 54.Sanmartin I, Wanntorp L, Winkworth RC. West Wind Drift revisited: testing for directional dispersal in the Southern Hemisphere using event-based tree fitting. Journal of Biogeography. 2007;34:398–416. [Google Scholar]
- 55.Leavitt SD, Johnson L, St Clair LL. Species delimitation and evolution in morphologically and chemically diverse communities of the lichen-forming genus Xanthoparmelia (Parmeliaceae, Ascomycota) in western North America. American Journal of Botany. 2011;98:175–188. doi: 10.3732/ajb.1000230. [DOI] [PubMed] [Google Scholar]
- 56.Leavitt SD, Johnson LA, Goward T, St. Clair LL. Species delimitation in taxonomically difficult lichen-forming fungi: an example from morphologically and chemically diverse Xanthoparmelia (Parmeliaceae) in North America. Molecular Phylogenetics and Evolution in press: xx–xx. 2011. [DOI] [PubMed]
- 57.Wirtz N, Printzen C, Lumbsch HT. Using haplotype networks, estimation of gene flow and phenotypic characters to understand species delimitation in fungi of a predominantly Antarctic Usnea group (Ascomycota, Parmeliaceae). Organisms Diversity & Evolution. 2012;12:17–37. [Google Scholar]
- 58.Rivas Plata E, Lumbsch HT. Parallel evolution and phenotypic divergence in lichenized fungi: a case study in the lichen-forming fungal family Graphidaceae (Ascomycota: Lecanoromycetes: Ostropales). Molecular Phylogenetics and Evolution. 2011;61:45–63. doi: 10.1016/j.ympev.2011.04.025. [DOI] [PubMed] [Google Scholar]
- 59.Elix JA. New species of Neofuscelia (Lichenized Ascomycotina, Parmeliaceae) from the Southern Hemisphere. Mycotaxon. 1999;71:431–456. [Google Scholar]
- 60.Elix JA. Neofuscelia. Flora of Australia. 1994;55:68–85. [Google Scholar]
- 61.Culberson CF. Improved conditions and new data for the identification of lichen products by a standardized thin-layer chromatographic method. Journal of Chromatography. 1972;72:113–125. doi: 10.1016/0021-9673(72)80013-x. [DOI] [PubMed] [Google Scholar]
- 62.Culberson CF, Culberson WL, Johnson A. A standardized TLC analysis of ß-orcinol depsidones. Bryologist. 1981;84:16–29. [Google Scholar]
- 63.Culberson CF, Johnson A. Substitution of methyl tert.-butyl ether for diethyl ether in standardized thin-layer chromatographic method for lichen products. Journal of Chromatography. 1982;238:438–487. [Google Scholar]
- 64.Orange A, James PW, White FJ. Microchemical Methods for the Identification of Lichens: British Lichen Society. 101 p. 2001.
- 65.Feige GB, Lumbsch HT, Huneck S, Elix JA. Identification of lichen substance by a standardized high-performance liquid-chromatographic method. Journal of Chromatography. 1993;646:417–427. [Google Scholar]
- 66.Crespo A, Blanco O, Hawksworth DL. The potential of mitochondrial DNA for establishing phylogeny and stabilising generic concepts in the parmelioid lichens. Taxon. 2001;50:807–819. [Google Scholar]
- 67.Myllys L, Lohtander K, Källersjö M, Tehler A. Sequence insertions and ITS data provide congruent information on Roccella canariensis and R. tuberculata (Arthoniales, Euascomycetes) phylogeny. Molecular Phylogenetics and Evolution. 1999;12:295–309. doi: 10.1006/mpev.1999.0620. [DOI] [PubMed] [Google Scholar]
- 68.Lohtander K, Myllys L, Sundin R, Kallersjo M, Tehler A. The species pair concept in the lichen Dendrographa leucophaea (Arthoniales): Analyses based on ITS sequences. Bryologist. 1998;101:404–411. [Google Scholar]
- 69.Vilgalys R, Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology. 1990;172:4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- 71.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Posada D. jModelTest: Phylogenetic model averaging. Molecular Biology and Evolution. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 73.Zwickl DJ. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. Austin: The University of Texas at Austin. 2006.
- 74.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 75.Nylander JAA, Ronquist F, Huelsenbeck JP, Nieves-Aldrey JL. Bayesian phylogenetic analysis of combined data. Systematic Biology. 2004;53:47–67. doi: 10.1080/10635150490264699. [DOI] [PubMed] [Google Scholar]
- 76.Nylander JAA, Wilgenbusch JC, Warren DL, Swofford DL. AWTY (Are We There Yet?): a system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics. 2007;24:581–583. doi: 10.1093/bioinformatics/btm388. [DOI] [PubMed] [Google Scholar]
- 77.Page RDM. Treeview: an application to display phylogenetic trees on personal computers. Computer Applied Biosciences. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 78.Amo de Paz G, Cubas P, Divakar PK, Lumbsch HT, Crespo A. Origin and diversification of major clades in parmelioid lichens (Parmeliaceae, Ascomycota) during the Paleogene inferred by Bayesian analysis. Plos One 6. 2011. [DOI] [PMC free article] [PubMed]
- 79.Pybus OG, Harvey PH. Testing macro-evolutionary models using incomplete molecular phylogenies. Proceedings of the Royal Society of London Series B-Biological Sciences. 2000;267:2267–2272. doi: 10.1098/rspb.2000.1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Drummond AJ, Rambaut A. Beast: Bayesian evoluionary analysis by sampling trees. BMC Evolutionary Biology. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rambaut A. TreeEdit v.1.0a10. 2007. http://tree.bio.ed.ac.uk/software/treeedit/
- 82.Poinar GO, Jr, Peterson EB, Platt JL. Fossil Parmelia in new world amber. Lichenologist. 2000;32:263–269. [Google Scholar]
- 83.Rambaut A, Drummond AJ. Tracer v1.4, Available from. 2007. http://beast.bio.ed.ac.uk/Tracer.
- 84.Rambaut A. FigTree 1.2.2. 2009. http://tree.bio.ed.ac.uk/software/figtree/
- 85.Huelsenbeck JP, Bollback JP. Empirical and hierarchical Bayesian estimation of ancestral states. Systematic Biology. 2001;50:351–366. [PubMed] [Google Scholar]
- 86.Bollback JP. SIMMAP: Stochastic character mapping of discrete traits on phylogenies. BMC Bioinformatics 7. 2006. [DOI] [PMC free article] [PubMed]
- 87.Lopez-Vaamonde C, Wikstrom N, Kjer KM, Weiblen GD, Rasplus JY, et al. Molecular dating and biogeography of fig-pollinating wasps. Molecular Phylogenetics and Evolution. 2009;52:715–726. doi: 10.1016/j.ympev.2009.05.028. [DOI] [PubMed] [Google Scholar]
- 88.Ree RH, Smith SA. Maximum likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis. Systematic Biology. 2008;57:4–14. doi: 10.1080/10635150701883881. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Chronogram of parmelioid lichens focusing in Xanthoparmelia pulla group. Calibration points: A, inferred age of radiation of parmelioid lichens and B, age of the Parmelia fossil. The Xanthoparmelia pulla group is highlighted by a box and the dated clades are indicated by a branch in bold.
(TIF)
GenBank accession numbers of parmelioid lichens (except X. pulla group, see Table 2 ) used for divergence time analysis. New sequences are in bold.
(DOC)