Abstract
Background
Dysfunction of glutamate neurotransmission has been implicated in the pathology of schizophrenia and bipolar disorder, and one mechanism by which glutamate signalling can be altered is through RNA editing of ionotropic glutamate receptors (iGluRs). The objectives of the present study were to evaluate the editing status of iGluRs in the human prefrontal cortex, determine whether iGluR editing is associated with psychiatric disease or suicide and evaluate a potential association between editing and alternative splicing in the α-amino-3-hydroxy-5-methylisoxazole-4-propionate (AMPA) iGluR subunits’ pre-mRNA.
Methods
We studied specimens derived from patients with antemortem diagnoses of bipolar disorder (n = 31) or schizophrenia (n = 34) who died by suicide or other causes, and from psychiatrically healthy controls (n = 34) who died from causes other than suicide. The RNA editing at all 8 editing sites within AMPA (GluA2–4 subunits) and kainate (GluK1–2 subunits) iGluRs was analyzed using a novel real-time quantitative polymerase chain reaction assay.
Results
No differences in editing were detected among schizophrenia, bipolar or control groups or between suicide completers and patients who died from causes other than suicide. The editing efficiency was significantly higher in the flop than in the flip splicoforms of GluA3-4 AMPA subunits (all p < 0.001).
Limitations
The study is limited by the near absence of specimens from medication-naive psychiatric patients and considerable variation in medication regimens among individuals, both of which introduce considerable uncertainty into the analysis of potential medication effects.
Conclusion
We found that iGluR RNA editing status was not associated with bipolar disorder, schizophrenia or suicide. Differences in editing between flip and flop splicoforms suggest that glutamate sensitivity of receptors containing GluA3 and/or GluA4 flop subunits is moderated as a result of increased editing.
Introduction
Glutamate is the main neurotransmitter responsible for excitatory neurotransmission throughout the brain. Glutamate serves as a ligand for G-protein coupled metabotropic and ionotropic (iGluRs) receptors. The latter consists of N-methyl-d-aspartate, α-amino-3-hydroxy-5-methylisoxazole-4-propionate (AMPA) and kainate (KA) subtypes. Glutamate dysfunction has been linked to numerous diseases, including schizophrenia, bipolar disorder and major depressive disorder.1–6
The AMPA and KA receptors are ligand-gated nonselect-ive cation channels that mediate fast excitatory neurotransmission. Each receptor is a tetramer composed of a combination of subunits encoded by separate genes, GluA1–4 for AMPA, and GluK1–3 and KA1–2 for KA receptors. Individual subunit transcripts undergo posttranscriptional modifications that substantially alter the structure and function of the resulting receptors. Alternative splicing of GluA1–4 generates 2 different variants of each subunit (flip or flop), forming AMPA receptors with distinct kinetic properties.7–9 In addition, AMPA and KA receptor transcripts can be modified by RNA editing.10
The most frequent RNA editing process in eukaryotes involves conversion of adenosine to inosine catalyzed by the adenosine deaminases acting on RNA (ADAR) enzymes.11 As inosine is interpreted as a guanosine during splicing and translation, the editing event can result in recoding. A total of 8 specific adenosine nucleotides within iGluRs pre-mRNAs are subject to this type of editing, and in each case the editing event results in an amino acid change that impacts receptor function.
The most infamous editing event is the life-sustaining modification of the AMPA receptor subunit GluA2 at the glutamine-to-arginine (Q/R) site that results in the change of a glutamine to an arginine in the pore-forming region of the protein. The editing of GluA2 at the Q/R site occurs in vivo at a frequency of almost 100% and drastically reduces receptor calcium permeability. Mutant animals, in which this editing event does not occur, experience epileptic seizures and premature death, and the lack of GluA2 editing in motor neurons leads to neuronal death, both due to calcium toxicity.12–14 Other iGluR sites are edited to varying degrees, and these changes have less dramatic implications. Similar to the GluA2 Q/R site, editing of homologous Q/R sites within the KA receptor subunits GluK1 and GluK2 also influences calcium permeability.15 In addition, the GluK2 subunit has isoleucine-to-valine and tyrosine-to-cysteine sites. Editing at these sites limits the calcium permeability of the receptor in the absence of editing at the Q/R site.16,17 Another editing site, arginine-to-glycine (R/G), which is situated just upstream of the flip/flop splice site of GluA2–4 AMPA receptor subunits, influences the rate of receptor resensitization.18
Aberrant adenosine-to-inosine editing of iGluRs has been reported in association with a number of different neurologic and psychiatric diseases.19 Reduced efficiency of editing at the GluA2 Q/R site was observed in the prefrontal cortex (PFC) of patients with Alzheimer disease and patients with schizophrenia, in the striatum of patients with Huntington disease20 and in the spinal cord of patients with amyotrophic lateral sclerosis.5 Altered editing at the Q/R site of both GluA2 and GluK2 has also been implicated in epilepsy.21,22 In addition, editing of another neurotransmitter receptor, the serotonin 2C receptor (5-HT2CR), has been implicated in schizophrenia, depression and suicide.23–28 However, editing in iGluRs has never been comprehensively assessed in the human brain in the context of psychiatric conditions within the same brain region.
We have recently developed a novel real-time quantitative polymerase chain reaction (qPCR) assay to quantify the efficiency of editing at a single editing site.29 This assay enables exceptional discrimination between edited and unedited transcripts and is more precise and sensitive than 2 other common methods of editing analysis — sequencing of individual clones and restriction enzyme digestion — which were used in most of the previous iGluRs editing studies. The present study aimed to quantify editing efficiencies iGluRs in the human dorsolateral PFC (DLPFC) using our qPCR assay, determine whether this iGluR editing is associated with psychiatric disease or suicide and evaluate a potential association between editing and alternative splicing (flip v. flop) in AMPA receptors. We used a well-characterized cohort from the Stanley Medical Research Institute (SMRI), which consisted of specimens from patients with bipolar disorder or schizophrenia who died by suicide or from other causes and of psychiatrically heatlhy controls who died from causes other than suicide.
Methods
Specimens
We obtained specimens of total RNA extracted from the DLPFC from the SMRI Array Collection. The cohort consisted of individuals who had diagnosed bipolar disorder (n = 31) or schizophrenia (n = 34) and controls (n = 34; Table 1). Of the individuals with bipolar disorder or schizophrenia, 14 and 7, respectively, died by suicide. All controls died of causes other than suicide.
Table 1.
Antemortem demographic and clinical characteristics of the study cohort
| Group; mean (SEM)* | |||
|---|---|---|---|
|
|
|||
| Characteristic | Control | Bipolar disorder | Schizophrenia |
| No. of patients | 34 | 31 | 34 |
| Age at death, yr | 43.8 (1.3) | 44.9 (2.0) | 42.3 (1.4) |
| PMI, h | 29.5 (2.2) | 36.6 (3.3) | 31.9 (2.7) |
| Brain pH | 6.6 (0.05) | 6.5 (0.05) | 6.5 (0.04) |
| Sex, male:female | 25:9 | 15:16 | 25:9 |
| Cause of death | |||
| Suicide | 0 | 14 | 7 |
| Other | 34 | 17 | 27 |
| No. of patients with prescribed antidepressants† | 0 | 18 | 8 |
| No. of patients with prescribed antipsychotics | 0 | 19 | 34 |
| Lifetime antipsychotics (fluphenazine equivalents, g) | 0 | 10.3 (4.36) | 85.7 (17.45) |
| Lifetime drug abuse, no. | |||
| Little or none | 29 | 9 | 13 |
| Social | 4 | 4 | 4 |
| Moderate past | 0 | 6 | 3 |
| Moderate present | 1 | 3 | 3 |
| Heavy past | 0 | 4 | 3 |
| Heavy present | 0 | 5 | 6 |
| Unknown | 0 | 0 | 2 |
| Lifetime alcohol use, no. | |||
| Little or none | 18 | 4 | 10 |
| Social | 11 | 7 | 6 |
| Moderate past | 1 | 5 | 3 |
| Moderate present | 2 | 3 | 3 |
| Heavy past | 2 | 4 | 3 |
| Heavy present | 0 | 7 | 9 |
| Unknown | 0 | 1 | 0 |
PMI = postmortem interval; SEM = standard error of the mean.
Unless otherwise indicated.
Antidepressant drugs prescribed varied by patient and included fluoxetine, trazodone, doxepin, paroxetine, amitriptyline, mirtazapine, venlafaxine, sertraline, fluvoxamine and bupropion.
Assay validation
For each assay, unique PCR primers and TaqMan probes were designed according to the National Center for Biotechnology Information reference sequences, and the specificity of the resulting amplicons was confirmed by sequencing (Table 2). For each assay, amplicons identified to contain adenosine or guanosine at the editing sites were used to determine cross-hybridization and to generate a standard curve as described.29 The optimal probe concentrations were identified as described29 and were 312.5 nM for GluA3–4 flop assays and 250 nM for all other assays.
Table 2.
Quantitative polymerase chain reaction editing assays used in the study*
| Target | Accession no. | Primers | Probes | % CH |
|---|---|---|---|---|
| GluA2 flip R/G | NM_000826.2 | F-ATGGCATCGCAACACCTAAAG | FAM-TCCTCATTAAGAACCCC | 3.1 |
| R-TCACTGAGTTTCAATACTGCAAGATTT | VIC-CCTCATTAGGAACCCCA | |||
| GluA2 flop R/G | NM_001083619.1 | F-ATGGCATCGCAACACCTAAAG | FAM-ATCCTCATTAAGAAATGCGG | 2.8 |
| R-CATTCAGTTTTAGTACTGCGAGGTTAA | VIC-ATCCTCATTAGGAAATGC | |||
| GluA2 Q/R | NM_000826.3 | F-GGAAGAGAAACACAAAGTAGTGAATCA | FAM-CATCCTTGCTGCATAA | 1.8 |
| R-AGAGAGGGATCTTGGCGAAATAT | VIC-CATCCTTGCCGCATAA | |||
| GluA3 flip R/G | NM_007325.4 | F-TGGTGTGGCAACCCCTAAAG | FAM-CAGCATTAAGAACGCCTG | 4.1 |
| R-CACTGAGTTTCAATACTGCAAGGTTT | VIC-CAGCATTAGGAACGCCT | |||
| GluA3 flop R/G | NM_000828.4 | F-TGGTGTGGCAACCCCTAAAG | FAM-CAGCATTAAGAAATG | 1.4 |
| R-GCCTTGCTCATTCAGTTTTAATACTG | VIC-CAGCATTAGGAAATG | |||
| GluA4 flip R/G | NM_000829.3 | F-CTATGGAGTAGCAACGCCCAA | FAM-ACAGGAGTTCTTAATGAGGA | 1.9 |
| R-TGCCTCACTGAGTTTCAAAACG | VIC-ACAGGAGTTCCTAATGAGGA | |||
| GluA4 flop R/G | NM_001077243.2 | F-CTATGGAGTAGCAACGCCCAA | FAM-CTCATTAAGAAATGCTG | 2.6 |
| R-CAAGAGGCCTTGTTCATTCAGTTT | VIC-CTCATTAGGAAATGCT | |||
| GluK1 Q/R | NM_000830 | F-GACGTGGTGGAAAACAATTTTACTT | FAM-CTCTCATGCAGCAAGGA | 1.0 |
| R-ACTATTCTGGTCGATAGAGCTTTGG | VIC-TCTCATGCGGCAAGGA | |||
| GluK2 Q/R | NM_021956 | F-CTAAATAGTTTCTGGTTTGGAGTTGGA | FAM-TCTCATGCAGCAAGGT | 3.3 |
| R-TCCTGGTGGACAGTGCTTTG | VIC-TCTCATGCGGCAAGGT | |||
| GluK2 I/V | NM_021956 | F-CCTGAATCCTCTCTCCCCTGAT | FAM-TGGATGTATATTCTGCTGGCT | 2.0 |
| R-ACTAAACCTGGCTATGACAAAGAGC | VIC-GATGTATGTTCTGCTGGCT | |||
| GluK2 Y/C | NM_021956 | F-CCTGAATCCTCTCTCCCCTGAT | FAM-TTCTGCTGGCTTACTTGGGTG | 8.0 |
| R-ACTAAACCTGGCTATGACAAAGAGC | VIC-TTCTGCTGGCTTGCTTGGGT |
CH = cross-hybridization; I/V = isoleucine-to-valine site; Q/R = glutamine-to-arginine; R/G = arginine-to-glycine site; Y/C = tyrosine-to-cysteine site.
Each assay consisted of a set of primers (forward [F] and reverse [R]) to amplify both edited and unedited transcripts, and 2 TaqMan probes. One probe was labelled with FAM and was designed to recognize the unedited, genomic base, A. Another probe was labelled with VIC and was designed to recognize the edited base, G, allowing for the edited and unedited transcripts to be measured simultaneously in the same reaction. We show sequences of primers and probes and percentages of cross-hybridization for each assay. Cross-hybridization is determined as the frequency with which a probe hybridizes to its mismatched template (for details, see Wong and colleagues29). Edited base is indicated in boldface type.
Editing analysis
We performed qPCR using a touchdown program that consisted of a 2 minute hold at 50°C and a 10 minute hold at 95°C, followed by 10 touchdown cycles (each consisting of 95°C for 15 seconds and 74°C for 1 minute with a Δ–1°C for each cycle) and 40 additional cycles with an annealing temperature of 64°C. For the GluA2 Q/R assay, the annealing temperatures were modified from 74°C to 75°C for the starting touchdown cycle and from 64°C to 65°C for the additional cycles. The editing efficiency at each site and in each specimen was calculated as a percentage: the quantity derived from the VIC assay (guanosine) divided by the total quantities from the VIC and FAM assays (guanosine plus adenosine).
Statistical analysis
We compared mean editing efficiencies among the 3 groups (schizophrenia, bipolar disorder and controls) using a separate analysis of covariance model for each site. Using separate t tests, we compared editing in specimens from those who died by suicide with specimens from those who died from other causes. Paired t tests were performed to compare editing in flip versus flop variants in the entire cohort (n = 99). We considered 8 possible covariates for inclusion: age, sex, postmortem interval, brain pH, lifetime alcohol use, lifetime drug use, prescription of antidepressant drugs (yes or no) and lifetime dosage of prescription antipsychotics (fluphenazine equivalents [mg]). The Mallow Cp model selected all these covariates, excluding sex, for the analyses.30 All hypothesis testing was conducted at the 5% level of significance using SAS, version 9.2.
Results
We assessed the editing efficiencies of all iGluR editing sites using site-specific multiplex qPCR assays.29 Each assay consisted of a single set of primers, which amplify both edited and unedited transcripts, and 2 probes, which were designed to recognize either the unedited or edited base and were labelled with different fluorophores (FAM or VIC). This enabled simultaneous measurement of edited and unedited transcripts in the same reaction. To assure that the probes accurately discriminated between edited and unedited transcripts that differ only by a single base pair, we assessed the cross-hybridization and found it to be less than 8% for all of the assays (Table 2).
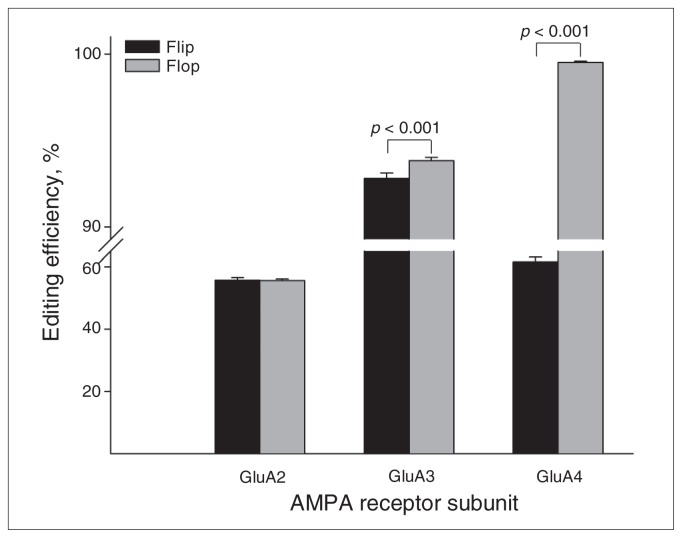
The mean editing efficiencies at each of the 8 editing sites are displayed in Table 3. We found no differences among the control, schizophrenia and bipolar disorder groups. Likewise, there were no differences between those who died by suicide and those who died from other causes (all p > 0.05). When the entire cohort (n = 99) was analyzed, paired t tests revealed a small but significant difference in editing efficiency between GluA3 flip and flop R/G sites (p < 0.001; Table 3, Fig. 1). In addition, we observed a striking difference in the R/G editing efficiency (Δ = 37.83%) between GluA4 flip and flop variants (p < 0.001; Table 3, Fig. 1). In both cases, flop receptors were edited to a greater extent than their alternatively spliced counterparts.
Table 3.
Editing efficiencies of the ionotropic glutamate receptors in the human dorsolateral prefrontal cortex, by diagnosis and manner of death
| Target | Entire cohort, n = 99 | Diagnostic group; mean (SEM)* | Manner of death; mean (SEM)* | |||
|---|---|---|---|---|---|---|
|
| ||||||
| Control, n = 34 | Bipolar disorder, n = 31 | Schizophrenia, n = 34 | Nonsuicide, n = 78 | Suicide, n = 21 | ||
| GluA2 flip R/G | 55.8 (0.8) | 56.7 (1.3) | 55.1 (1.2) | 55.4 (1.5) | 55.7 (0.9) | 56.5 (1.4) |
| GluA2 flop R/G | 55.7 (0.5) | 56.2 (0.6) | 55.9 (0.6) | 55.0 (1.0) | 55.5 (0.5) | 56.7 (0.6) |
| GluA2 Q/R | 96.1 (0.2) | 95.7 (0.3) | 96.2 (0.3) | 96.6 (0.2) | 96.2 (0.2) | 96.0 (0.3) |
| GluA3 flip R/G | 92.8 (0.3) | 92.5 (0.7) | 92.8 (0.5) | 93.1 (0.4) | 92.8 (0.4) | 92.8 (0.6) |
| GluA3 flop R/G | 93.8 (0.2) | 93.8 (0.4) | 94.1 (0.3) | 93.7 (0.3) | 93.8 (0.2) | 93.9 (0.4) |
| GluA4 flip R/G | 61.7 (1.5) | 61.4 (2.7) | 61.6 (2.4) | 62.0 (2.6) | 61.8 (1.7) | 60.4 (3.1) |
| GluA4 flop R/G | 99.5 (0.1) | 99.6 (0.1) | 99.6 (0.1) | 99.4 (0.1) | 99.5 (0.1) | 99.7 (0.1) |
| GluK1 Q/R | 59.8 (0.5) | 60.3 (0.6) | 59.0 (0.8) | 60.0 (1.0) | 59.8 (0.6) | 60.1 (1.0) |
| GluK2 Q/R | 81.2 (0.4) | 80.8 (0.7) | 80.8 (0.7) | 81.9 (0.5) | 81.2 (0.4) | 81.2 (0.8) |
| GluK2 I/V | 49.3 (0.7) | 50.7 (1.0) | 48.5 (1.1) | 48.6 (1.3) | 49.1 (0.8) | 49.9 (1.5) |
| GluK2 Y/C | 80.8 (0.6) | 80.4 (0.3) | 80.7 (0.5) | 80.3 (0.5) | 80.1 (0.5) | 80.2 (0.3) |
I/V = isoleucine-to-valine site; Q/R = glutamine-to-arginine; R/G = arginine-to-glycine site; SEM = standard error of the mean; Y/C = tyrosine-to-cysteine site.
Means represent % edited.
Fig. 1.
Editing efficiencies at the R/G site of different splicoforms (flip and flop) of the the α-amino-3-hydroxy-5-methylisoxazole-4-propionate (AMPA) receptor subunits GluA2–4. Shown are means and standard errors of the mean for the entire sample (n = 99). Significant editing differences were observed between flip and flop in GluA3 and GluA4 (all p < 0.001).
Discussion
Considerable evidence supports the involvement of the glutamatergic system in the pathology of schizophrenia and bipolar disorder.3,6 Thus, it is important that we investigate potential functional alterations that may occur in iGluRs in the context of these disorders. The function of iGluRs is influenced by numerous factors, including editing of their pre-mRNA. Although editing at the GluA2 Q/R site, which is crucial for limiting calcium permeability in GluA2-containing AMPA receptors, has been extensively studied,5,13,20 the editing status of all other iGluR sites, which have more subtle effects on receptor properties, has not been examined in detail in the context of psychiatric disease or suicide. In the present study we determined the editing efficiencies for all iGluR sites within the DLPFC in a well-characterized cohort of individuals who had schizophrenia or bipolar disorder and controls. Our novel methodological approach allowed for a significant improvement in precision and sensitivity of measurements compared with previously used methods. As reflected by low variability in our data, we observed highly consistent levels of editing at all iGluR sites across individuals, which is in line with the results of recent studies that assessed editing in different molecules.31,32 These results confirm that, as was reported for other targets, RNA editing of iGluRs is a tightly regulated process, emphasizing the importance of this posttranscriptional modification for the functionality of the glutamatergic system in the brain.32 However, we did not detect any diagnosis-or suicide-specific alterations.
A previous study of a well-established model of schizophrenia (phencyclidine [PCP]-treated rats) showed decreased editing at the R/G site of GluA2–3 flop subunits and increased editing of the Q/R site of GluK2 in the PFC, suggesting decreased glutamate signalling in the PFC of those animals.33 However, that study used a different methodology. In addition, chronic PCP treatment of mice only mimics the main behavioural and neurobiological defects associated with schizophrenia.33 In contrast, our study was designed to probe the intrinsic editing differences in humans with distinct psychiatric diagnoses. Our findings did not support the existence of editing differences in humans with schizophrenia compared with controls. This lack of significant editing alterations extends to the GluA2 Q/R site, which was reported to have lower editing efficiency in schizophrenia compared with controls.20 That study, however, compared only 6 schizophrenia and 8 control specimens and used a restriction enzyme digestion, which is substantially less accurate than the qPCR assay used in our study.29 For all specimens, we also observed slightly lower editing efficiency at the Q/R site than previously reported (96% v. 99%).5,20 This discrepancy is likely due to an overestimation of editing efficiency in previous publications as a result of the bias associated with analysis by enzymatic restriction (for details, see Wong and colleagues29) and/or to an existence of a small cross-hybridization (about 1.8%) between the unedited probe and mismatched edited template in our GluA2 Q/R assay.
Changes in editing of the 5-HT2CR have previously been reported in individuals with schizophrenia;28 however, replication attempts have been unsuccessful.24,27 Later studies demonstrated an association between 5-HT2CR editing and suicide.23–26 In particular, one study conducted in our laboratory, which used samples from the same cohort and brain region as the present study, showed that the specimens from individuals who had committed suicide displayed greater 5-HT2CR editing than those from individuals who died from other causes, regardless of psychiatric diagnosis.26 One could hypothesize that if these differences in editing were the result of greater ADAR activity in suicide completers, the editing efficiency of other ADAR substrates would also be elevated in these individuals. However, we detected no association between suicide and iGluR editing. This discrepancy may be a result of cell type–specific editing alterations, since iGluRs and ADARs are ubiquitously expressed, whereas 5-HT2CRs are mostly confined to GABAergic interneurons within layer 5 of the PFC.34,35 Therefore, an increase in ADAR activity in suicide completers could be confined to γ-aminobutyric acid (GABA) cells, with a consequential increase in editing only in this cell population, precluding the detection of iGluRs editing changes in the homogenate cortical preparations used in the study. Alternatively, 5-HT2CR editing differences in suicide completers may stem from causes other than ADAR activity that are specific to the 5-HT2CR.36,37 To investigate these possibilities, editing analysis in cells coexpressing iGluRs and 5-HT2CRs would be essential. Unfortunately, such studies are not currently feasible since our preliminary experiments have demonstrated that the required immunohistochemical treatment severely compromises the quality of RNA, precluding the necessary quantitative analysis.
We also examined the relation between editing and alternative splicing and found no differences in the frequency of editing at the R/G site in flip- versus flop-containing GluA2 transcripts. However, in GluA3 and GluA4, flop variants were edited at a significantly higher frequency than flip variants at this site. This is mostly in agreement with data obtained from the adult rodent brain.18,38 Previous studies indicated that inclusion of flop over flip subunits increases the rate of desensitization, whereas editing at the R/G site increases the rate of resensitization of the receptor.39 Thus, our studies suggest that, in the context of GluA3 and GluA4, flop variants are more likely to be edited than unedited, thus increasing the glutamate sensitivity of the flop-containing receptors. Although the significance of these findings is not immediately evident, they provide insight into the functional regulation of the iGluRs that is achieved by a combination of editing and splicing mechanisms. Of note, most AMPA receptors in the human DLPFC contain GluA2 and GluA3 subunits,40 and, although the expression of GluA4 is low in this region, GluA4-containing receptors may be of importance for specific subpopulations of cortical neurons.41,42
Limitations
Common to most postmortem studies of individuals with psychiatric diseases, the main shortcoming of the present study was the potential influence of treatment. We may have failed to observe a diagnosis-associated difference in iGluR editing because this inherent difference had been corrected by antipsychotic or antidepressant exposure. To our knowledge, no studies have been conducted to evaluate the potential effects of antipsychotics on iGluR editing, and studies of the effect of antidepressants on iGluR editing have yielded inconsistent results.38,43 The antidepressants prescribed to the patients with schizophrenia and bipolar disorder whose specimens were included in this study were too variable, and there were no antipsychotic-naive patients with schizophrenia for comparison to enable meaningful subgroup analyses (Table 1). However, although we cannot rule out a potential influence of medications, our statistical analysis revealed no significant effect of the medication history on our findings.
Conclusion
We have used a novel sensitive assay to assess editing of the iGluRs in the DLPFC of controls and of patients with bipolar disorder or schizophrenia. We detected no differences in editing among the groups or between suicide completers and those who died from other causes. Editing differences have been previously reported for 5-HT2CR in suicide completers and for GluA2 Q/R in individuals with schizophrenia. The apparent inconsistency with the previously reported suicide-specific 5-HT2CR editing alterations in this cohort may stem from cellular specificity of editing abnormalities in individuals with psychiatric conditions, which will require further investigation using cell-specific assays. We also detected that GluA3–4 flop variants were edited at a significantly higher frequency than their flip counterparts, suggesting an association between editing and splicing that influences the sensitivity of the receptors to glutamate.
Acknowledgments
This study was supported by the Department of Veterans Affairs (VA), Veterans Health Administration, Office of Research and Development, Biomedical Laboratory Research and Development, by a VA Merit award (S. Dracheva), NIMH/NIH grant MH090352 (S. Dracheva), a grant from the American Foundation for Suicide Prevention (S. Dracheva) and by the VISN3 Mental Illness Research and Education Clinical Center (S. Dracheva). Postmortem brain tissue was donated by The Stanley Medical Research Institute.
Footnotes
Competing interests: None declared for R. Lyddon and S. Navarrett. As above for S. Dracheva.
Contributors: R. Lyddon and S. Dracheva designed the study, analyzed the data and wrote the article. R. Lyddon and S. Navarrett acquired the data. All authors reviewed the article and approved its publication.
References
- 1.Ure J, Baudry M, Perassolo M. Metabotropic glutamate receptors and epilepsy. J Neurol Sci. 2006;247:1–9. doi: 10.1016/j.jns.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 2.Zarate C, Jr, Machado-Vieira R, Henter I, et al. Glutamatergic modulators: The future of treating mood disorders? Harv Rev Psychiatry. 2010;18:293–303. doi: 10.3109/10673229.2010.511059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Javitt DC. Glutamatergic theories of schizophrenia. Isr J Psychiatry Relat Sci. 2010;47:4–16. [PubMed] [Google Scholar]
- 4.Cortese BM, Phan KL. The role of glutamate in anxiety and related disorders. CNS Spectr. 2005;10:820–30. doi: 10.1017/s1092852900010427. [DOI] [PubMed] [Google Scholar]
- 5.Kawahara Y, Ito K, Sun H, et al. Glutamate receptors: RNA editing and death of motor neurons. Nature. 2004;427:801. doi: 10.1038/427801a. [DOI] [PubMed] [Google Scholar]
- 6.Mitchell ND, Baker GB. An update on the role of glutamate in the pathophysiology of depression. Acta Psychiatr Scand. 2010;122:192–210. doi: 10.1111/j.1600-0447.2009.01529.x. [DOI] [PubMed] [Google Scholar]
- 7.Mosbacher J, Schoepfer R, Monyer H, et al. A molecular determinant for submillisecond desensitization in glutamate receptors. Science. 1994;266:1059–62. doi: 10.1126/science.7973663. [DOI] [PubMed] [Google Scholar]
- 8.Koike M, Tsukada S, Tsuzuki K, et al. Regulation of kinetic properties of GluR2 AMPA receptor channels by alternative splicing. J Neurosci. 2000;20:2166–74. doi: 10.1523/JNEUROSCI.20-06-02166.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pei W, Huang Z, Wang C, et al. Flip and flop: a molecular determinant for AMPA receptor channel opening. Biochemistry. 2009;48:3767–77. doi: 10.1021/bi8015907. [DOI] [PubMed] [Google Scholar]
- 10.Gott JM, Emeson RB. Functions and mechanisms of RNA editing. Annu Rev Genet. 2000;34:499–531. doi: 10.1146/annurev.genet.34.1.499. [DOI] [PubMed] [Google Scholar]
- 11.Bass BL. RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem. 2002;71:817–46. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brusa R, Zimmermann F, Koh DS, et al. Early-onset epilepsy and postnatal lethality associated with an editing-deficient Glur-B allele in mice. Science. 1995;270:1677–80. doi: 10.1126/science.270.5242.1677. [DOI] [PubMed] [Google Scholar]
- 13.Higuchi M, Maas S, Single FN, et al. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 2000;406:78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- 14.Hideyama T, Yamashita T, Suzuki T, et al. Induced loss of ADAR2 engenders slow death of motor neurons from Q/R site-unedited GluR2. J Neurosci. 2010;30:11917–25. doi: 10.1523/JNEUROSCI.2021-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swanson GT, Feldmeyer D, Kaneda M, et al. Effect of RNA editing and subunit co-assembly single-channel properties of recombinant kainate receptors. J Physiol. 1996;492(Pt 1):129–42. doi: 10.1113/jphysiol.1996.sp021295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burnashev N, Zhou Z, Neher, et al. Fractional calcium currents through recombinant GluR channels of the NMDA, AMPA and kainate receptor subtypes. J Physiol. 1995;485(Pt 2):403–18. doi: 10.1113/jphysiol.1995.sp020738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Köhler M, Burnashev N, Sakmann B, et al. Determinants of Ca2+ permeability in both TM1 and TM2 of high affinity kainate receptor channels: diversity by RNA editing. Neuron. 1993;10:491–500. doi: 10.1016/0896-6273(93)90336-p. [DOI] [PubMed] [Google Scholar]
- 18.Lomeli H, Mosbacher J, Melcher T, et al. Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science. 1994;266:1709–13. doi: 10.1126/science.7992055. [DOI] [PubMed] [Google Scholar]
- 19.Morabito MV, Emeson RB. RNA editing as a therapeutic target for CNS disorders. Neuropsychopharmacology. 2009;34:246. doi: 10.1038/npp.2008.159. [DOI] [PubMed] [Google Scholar]
- 20.Akbarian S, Smith MA, Jones EG. Editing for an AMPA receptor subunit RNA in prefrontal cortex and striatum in Alzheimer’s disease, Huntington’s disease and schizophrenia. Brain Res. 1995;699:297–304. doi: 10.1016/0006-8993(95)00922-d. [DOI] [PubMed] [Google Scholar]
- 21.Vollmar W, Gloger J, Berger E, et al. RNA editing (R/G site) and flip-flop splicing of the AMPA receptor subunit GluR2 in nervous tissue of epilepsy patients. Neurobiol Dis. 2004;15:371–9. doi: 10.1016/j.nbd.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 22.Vissel B, Royle GA, Christie BR, et al. The role of RNA editing of kainate receptors in synaptic plasticity and seizures. Neuron. 2001;29:217–27. doi: 10.1016/s0896-6273(01)00192-1. [DOI] [PubMed] [Google Scholar]
- 23.Gurevich I, Tamir H, Arango V, et al. Altered editing of serotonin 2C receptor pre-mRNA in the prefrontal cortex of depressed suicide victims. Neuron. 2002;34:349–56. doi: 10.1016/s0896-6273(02)00660-8. [DOI] [PubMed] [Google Scholar]
- 24.Niswender CM, Herrick-Davis K, Dilley GE, et al. RNA editing of the human serotonin 5-HT2C receptor. alterations in suicide and implications for serotonergic pharmacotherapy. Neuropsycho-pharmacology. 2001;24:478–91. doi: 10.1016/S0893-133X(00)00223-2. [DOI] [PubMed] [Google Scholar]
- 25.Iwamoto K, Kato T. RNA editing of serotonin 2C receptor in human postmortem brains of major mental disorders. Neurosci Lett. 2003;346:169–72. doi: 10.1016/s0304-3940(03)00608-6. [DOI] [PubMed] [Google Scholar]
- 26.Dracheva S, Patel N, Woo DA, et al. Increased serotonin 2C receptor mRNA editing: a possible risk factor for suicide. Mol Psychiatry. 2008;13:1001–10. doi: 10.1038/sj.mp.4002081. [DOI] [PubMed] [Google Scholar]
- 27.Dracheva S, Elhakem SL, Marcus SM, et al. RNA editing and alternative splicing of human serotonin 2C receptor in schizophrenia. J Neurochem. 2003;87:1402–12. doi: 10.1046/j.1471-4159.2003.02115.x. [DOI] [PubMed] [Google Scholar]
- 28.Sodhi MS, Burnet PW, Makoff AJ, et al. RNA editing of the 5-HT(2C) receptor is reduced in schizophrenia. Mol Psychiatry. 2001;6:373–9. doi: 10.1038/sj.mp.4000920. [DOI] [PubMed] [Google Scholar]
- 29.Wong K, Lyddon R, Dracheva S. TaqMan-based real-time quantitative PCR method for RNA editing analysis. Anal Biochem. 2009;390:173–80. doi: 10.1016/j.ab.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 30.Lance CE. Mallows’ Cp statistics. In: Everitt BS, Howell DC, editors. Encyclopedia of statistics in behavioral science. Chichester: John Wiley & Sons; 2005. pp. 1119–20. [Google Scholar]
- 31.Wahlstedt H, Daniel C, Enstero M, et al. Large-scale mRNA sequencing determines global regulation of RNA editing during brain development. Genome Res. 2009;19:978–86. doi: 10.1101/gr.089409.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Greenberger S, Levanon EY, Paz-Yaacov N, et al. Consistent levels of A-to-I RNA editing across individuals in coding sequences and non-conserved Alu repeats. BMC Genomics. 2010;11:608. doi: 10.1186/1471-2164-11-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barbon A, Fumagalli F, La VL, et al. Chronic phencyclidine administration reduces the expression and editing of specific glutamate receptors in rat prefrontal cortex. Exp Neurol. 2007;208:54–62. doi: 10.1016/j.expneurol.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 34.Pasqualetti M, Ori M, Castagna M, et al. Distribution and cellular localization of the serotonin type 2C receptor messenger RNA in human brain. Neuroscience. 1999;92:601–11. doi: 10.1016/s0306-4522(99)00011-1. [DOI] [PubMed] [Google Scholar]
- 35.Liu S, Bubar MJ, Lanfranco MF, et al. Serotonin2C receptor localization in GABA neurons of the rat medial prefrontal cortex: implications for understanding the neurobiology of addiction. Neuroscience. 2007;146:1677–88. doi: 10.1016/j.neuroscience.2007.02.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang W, Wang Q, Kanes SJ, et al. Altered RNA editing of serotonin 5-HT2C receptor induced by interferon: implications for depression associated with cytokine therapy. Brain Res Mol Brain Res. 2004;124:70–8. doi: 10.1016/j.molbrainres.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 37.Vitali P, Basyuk E, Le Meur E, et al. ADAR2-mediated editing of RNA substrates in the nucleolus is inhibited by C/D small nucleolar RNAs. J Cell Biol. 2005;169:745–53. doi: 10.1083/jcb.200411129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barbon A, Popoli M, La Via L, et al. Regulation of editing and expression of glutamate alpha-amino-propionic-acid (AMPA)/kainate receptors by antidepressant drugs. Biol Psychiatry. 2006;59:713–20. doi: 10.1016/j.biopsych.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 39.Penn AC, Greger IH. Sculpting AMPA receptor formation and function by alternative RNA processing. RNA Biol. 2009;6:517–21. doi: 10.4161/rna.6.5.9552. [DOI] [PubMed] [Google Scholar]
- 40.Dracheva S, McGurk SR, Haroutunian V. mRNA expression of AMPA receptors and AMPA receptor binding proteins in the cerebral cortex of elderly schizophrenics. J Neurosci Res. 2005;79:868–78. doi: 10.1002/jnr.20423. [DOI] [PubMed] [Google Scholar]
- 41.Paz JT, Bryant AS, Peng K, et al. A new mode of corticothalamic transmission revealed in the Gria4(−/−) model of absence epilepsy. Nat Neurosci. 2011;14:1167–73. doi: 10.1038/nn.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sagata N, Iwaki A, Aramaki T, et al. Comprehensive behavioural study of GluR4 knockout mice: implication in cognitive function. Genes Brain Behav. 2010;9:899–909. doi: 10.1111/j.1601-183X.2010.00629.x. [DOI] [PubMed] [Google Scholar]
- 43.Sawada J, Yamashita T, Aizawa H, et al. Effects of antidepressants on GluR2 Q/R site-RNA editing in modified HeLa cell line. Neurosci Res. 2009;64:251–8. doi: 10.1016/j.neures.2009.03.009. [DOI] [PubMed] [Google Scholar]