Abstract
Biotin synthase (BS) catalyzes the oxidative addition of a sulfur atom to dethiobiotin (DTB) to generate the biotin thiophane ring. This enzyme is an S-adenosylmethionine (AdoMet) radical enzyme that catalyzes the reductive cleavage of AdoMet, generating methionine and a transient 5´-deoxyadenosyl radical. In our working mechanism, the 5´-deoxyadenosyl radical oxidizes DTB by abstracting a hydrogen from C6 or C9, generating a dethiobiotinyl carbon radical that is quenched by sulfide from a [2Fe-2S]2+ cluster. A similar reaction sequence directed at the other position generates the second C-S bond in the thiophane ring. Since the BS active site holds only one AdoMet and one DTB, it follows that dissociation of methionine and 5´-deoxyadenosine and binding of a second equivalent of AdoMet must be intermediate steps in the formation of biotin. During these dissociation/association steps, a discrete DTB-derived intermediate must remain bound to the enzyme. In the present work, we confirm that the conversion of DTB to biotin is accompanied by the reductive cleavage of 2 equivalents of AdoMet. A discrepancy between DTB consumption and biotin formation suggests the presence of an intermediate, and we use LCMS to demonstrate that this intermediate is indeed 9-mercaptodethiobiotin, generated at ~10% of the total enzyme concentration. The amount of intermediate observed is increased when the reaction is run with substoichiometric levels of AdoMet or with defective enzyme containing the mutation Asn153Ser. The retention of 9-mercaptodethiobiotin as a tightly-bound intermediate is consistent with a mechanism involving the stepwise radical-mediated oxidative abstraction of sulfide from an iron-sulfur cluster.
Biotin synthase (BioB)1 is an AdoMet radical enzyme that catalyzes formation of the thiophane ring of biotin (1, 2). Similar to other AdoMet radical enzymes, BioB utilizes a reduced [4Fe-4S]+ cluster to catalyze the one electron reduction of the AdoMet sulfonium, generating methionine and a transient 5´-deoxyadenosyl radical. This high energy radical then abstracts hydrogen atoms from the C6 and C9 positions of the substrate (3), dethiobiotin (DTB), facilitating the formation of two new C-S bonds in the biotin thiophane ring. These two bonds could be formed in either a concerted or a stepwise reaction sequence; these mechanisms can be distinguished based upon the reaction stoichiometry and the presence or absence of a stable chemical intermediate.
The structure of BioB from Escherichia coli solved at 3.4 Å resolution shows a dimeric protein in which each monomer contains an active site encapsulated within an (αβ)8 barrel motif (4). Electron density maps indicate that each active site contains one equivalent each of AdoMet and DTB sandwiched between a [4Fe-4S]2+ cluster and a [2Fe-2S]2+ cluster. As modeled within this density, the C5´ position of AdoMet, the initial site of radical generation, is in van der Waals contact with both the C6 methylene and the C9 methyl positions of DTB (dC-C = 4.1 and 3.9 Å, respectively). Based upon this structural data alone, a high-energy 5´-deoxyadenosyl radical could potentially abstract a hydrogen atom from either position, initially resulting in the formation of a DTB-centered radical at C6 or C9. Either dethiobiotinyl carbon radical could then be quenched by reacting with a μ-sulfide from the nearby [2Fe-2S]2+ cluster (5), resulting in formation of one new C-S bond at either the C6 or C9 position.
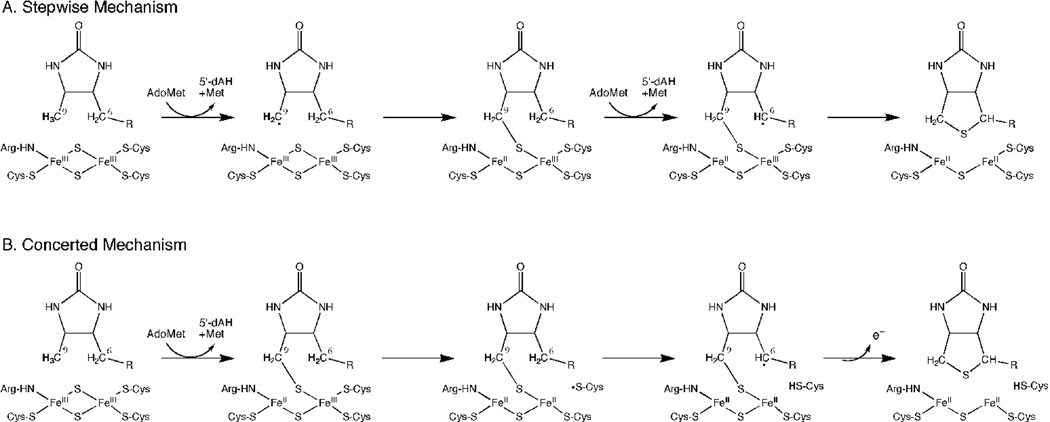
At this point, two plausible reaction sequences could be proposed. A concerted reaction (Figure 1B) could occur in which the second hydrogen atom abstraction is catalyzed by a cysteine thiyl radical from the partially degraded [2Fe-2S] cluster, followed rapidly by formation of the second C-S bond. A second electron derived from the sulfur atom must also be eliminated, possibly by electron transfer back to the [4Fe-4S]2+ cluster. This mechanism would be consistent with a product stoichiometry of 1:1 5´-deoxyadenosine:biotin and predicts that any DTB-derived intermediate would be very short-lived. Alternatively, a stepwise mechanism (Figure 1A) could occur in which a thiolated intermediate remains bound while 5´-deoxyadenosine and methionine dissociate and a second equivalent of AdoMet binds. This mechanism would be consistent with a product stoichiometry of 2:1 5´-deoxyadenosine:biotin and with the transient formation of either 6- or 9-mercaptodethiobiotin (6- or 9-MDTB) as a long-lived tightly-bound intermediate.
Figure 1.
Stepwise and concerted mechanisms for thiophane ring formation via a 9-MDTB intermediate. (A) In a stepwise mechanism, reductive cleavage of AdoMet is coupled to abstraction of a hydrogen atom from C9 and formation of a new C-S bond. Reductive cleavage of second equivalent of AdoMet is coupled to abstraction of a hydrogen from the C6 position, leading to completion of the thiophane ring and destruction of the [2Fe-2S]2+ cluster. 9-MDTB is formed as a stable intermediate. (B) In a concerted mechanism, initial C-S bond formation requires reductive cleavage of AdoMet and proceeds in the same manner, but a cysteine thiyl radical generated during degradation of the [2Fe-2S]2+ cluster abstracts a hydrogen atom from C6, completing the biotin thiophane ring without any additional requirement for AdoMet. One excess electron derived from sulfur oxidation could be transferred back to the catalytic [4Fe-4S]2+ cluster.
Several prior studies have provided evidence supporting 9-MDTB as an intermediate in the enzymatic reaction. In early studies, Marquet and coworkers fed 3H-DTB to E. coli grown on 35S-sulfate and isolated a novel DTB-derived compound that contained labeled sulfur and could be further converted to biotin (6). Shaw and coworkers isolated a DTB-derived sulfur-containing intermediate after adding 14C-DTB to E. coli extracts that contained overexpressed E. coli BioB (7). Synthetic 35S-labeled 6- and 9-MDTB were fed to E. coli and B. sphaericus, and both strains could convert 9-MDTB to biotin with ~75% retention of the labeled sulfur. Surprisingly, E. coli could also convert 6-MDTB to biotin, but with complete loss of the labeled sulfur (8). 9-MDTB has also been isolated as a biotin precursor from extracts of plant cells (Lavandula vera L.) (9). More recently, Marquet and coworkers have demonstrated that synthetic 9-MDTB is converted to biotin by purified His6-BioB from E. coli (10), although the Michaelis constant for 9-MDTB (Km ≈ 200 µM) is much higher than had been previously reported for DTB (Km = 2 µM (11)).
In the present study, we demonstrate that biotin synthase catalyzes the stepwise addition of sulfur to DTB. First, we examine the stoichiometry of substrate consumption and product formation, and find a 2:1 stoichiometry most consistent with formation of a discrete intermediate. We then use electrospray ionization mass spectrometry to search for trace intermediates and products related to DTB, and find only one compound whose mass and HPLC elution are consistent with 9-MDTB. With WT BioB, this compound is formed and decays on a time-scale that is kinetically competent to be an intermediate in biotin formation. In contrast, an Asn153Ser mutant of BioB generates 9-MDTB as a stoichiometric product due to an inability to retain the bound intermediate during 5´-deoxyadenosine/AdoMet exchange. Finally, we used partially deuterated (2H-9-methyl)-DTB to demonstrate that the intermediate is formed through replacement of a hydrogen/deuterium at C9 with sulfur. We propose a stepwise mechanism for biotin formation in which reductive cleavage of AdoMet and intermediate dissociation of 5´-deoxyadenosine are the most likely rate-limiting steps.
Materials and Methods
Materials
All reagents were purchased from commercial sources and used without further purification. AdoMet was purchased as the p-toluenesulfonate salt and contains <5% 5’-methylthioadenosine and <1% S-adenosyl-L-homocysteine contamination. Synthetic 9-MDTB disulfide and (2H-9-methyl)-DTB were gifts from Prof. Andrée Marquet (Université Paris) and were synthesized according to published procedures (3, 8). His6-BioB was prepared as previously described (12) and the hexahistidine tag was not removed. The concentration of WT and Asn153Ser BioB was determined based on the UV/visible absorbance of the [2Fe-2S]2+ cluster (ε452 = 8400 M−1 cm−1). The Asn153Ser BioB mutant was constructed by the Quickchange method (Stratagene) using the WT plasmid pJJ15-4A as a template (12), and PCR primers (5’-GGATTACTACAACCACAGCCTGGACACCTCGCCGG-3’, 5’-CCGGCGAGGTGTCCAGGCTGTGGTTGTAGTAATCC-3’, Integrated DNA Technologies) that overlap the Asn153 codon. The mutated plasmid was isolated from DH5α and the sequence of the entire BioB gene confirmed. The isolated plasmids were transformed into BL21(DE3)pLysS (Novagen) and the Asn153Ser mutant protein was expressed and purified as previously described for WT enzyme (12).
Activity Assays
Biotin synthase (25–100 µM in 200 µl final volume) was added to septum-covered vials containing 50 mM Tris HCl, 10 mM KCl, 5 mM DTT, pH 8.0 and degassed under argon. The [4Fe-4S]2+ cluster was reconstituted by incubation with 4 equiv. each Na2S and FeCl3 for 5 min. Flavodoxin (20 µM), ferredoxin (flavodoxin): NADP+ oxidoreductase (2 µM), NADPH (1 mM), and AdoMet (1 – 5 equiv.) were added while the vials were continuously degassed. Turnover was initiated by addition of DTB (1 – 5 equiv.), and the vials were incubated in a 37°C water bath for 5 – 240 min. Acetic acid/sodium acetate, pH 4.5 (20 µl of 4.5 M stock) was added to quench the reaction and precipitate the protein. The vials were incubated on ice to ensure full precipitation, the precipitate removed by centrifugation for 10 min at 18,000 × g, and the supernatant transferred to an HPLC autosampler vial. Samples were injected on a Waters Atlantis dC18 reversed-phase column (3.0 × 150 mm, 5 µm) and eluted with a 25 min linear gradient from 2 – 25% acetonitrile in H2O containing 0.1 % H3PO4 at 35 °C. DTB (tR = 21 min), 9-MDTB (tR = 23.6 min), and biotin (tR = 18 min) were detected at 210 nm, while 5´-deoxyadenosine (tR = 7.5 min) was detected at 260 nm. Fresh external standards containing biotin, DTB, and 5’-deoxyadenosine (10–250 µM) were prepared and analyzed prior to and following each set of samples to ensure accurate identification and quantitation. 9-MDTB was quantified by comparison to the integration of an external biotin standard; this analysis assumes 9-MDTB has an identical extinction coefficient as biotin. The substrate and product stoichiometry (equivalents) is reported based on the concentration of BioB monomer in the assay solution.
Mass Spectrometry
The mass of BioB reaction products was determined by high-pressure liquid chromatography with electrospray-ionization mass spectrometry detection (HPLC ESI-MS). Chromatographic separation was performed on an Agilent 1100 series liquid chromatography system (Agilent Technologies) equipped with an Eclipse XDB-C18 column (4.6 mm × 150 mm, 5µm, Agilent) using a 25 min linear gradient from 95:5 to 70:30 H2O:acetonitrile (both containing 0.1% v/v formic acid) at a flow rate of 0.7 ml/min. The LC system was coupled to an Agilent 6210 time-of-flight mass spectrometer equipped with an electrospray ionization source operating in positive ion mode. Nitrogen was used as the nebulizer drying gas (pressure, 30 psi; flow rate, 7 L/min; temperature, 350 °C). Additional ESI-MS parameters were as follows: spray capillary voltage, 3500 V; skimmer voltage, 65 V; fragmenter voltage, 175 V; OctRFV voltage, 250V. Mass spectra were acquired over the range m/z = 150–1000 at a scan speed of 0.99 scan/s. Agilent MassHunter Workstation Software (v B.01.03, Agilent) was used for instrument control, data acquisition, and data analysis.
Results
Conversion of Dethiobiotin to Biotin Produces Two Equivalents of 5´-Deoxyadenosine
Formation of the biotin thiophane ring requires that hydrogen atoms are abstracted from both the C6 and C9 positions of DTB. The reductive cleavage of the AdoMet sulfonium generates a transient 5´-deoxyadenosyl radical that initiates the chemistry by abstraction of a hydrogen atom from one of these positions (initial abstraction from C9 is shown in Figure 1). Although the nearby [2Fe-2S]2+ cluster has been identified as the most likely sulfur donor, the subsequent reaction sequence has not been fully elucidated. A stepwise mechanism (Figure 1A) would require reductive cleavage of a second equivalent of AdoMet to facilitate abstraction of a hydrogen atom from the second position (5), while a concerted mechanism (Figure 1B) could be postulated in which this second hydrogen atom abstraction is coupled to FeS cluster degradation (13). These mechanisms are distinguished by the stoichiometry of product formation.
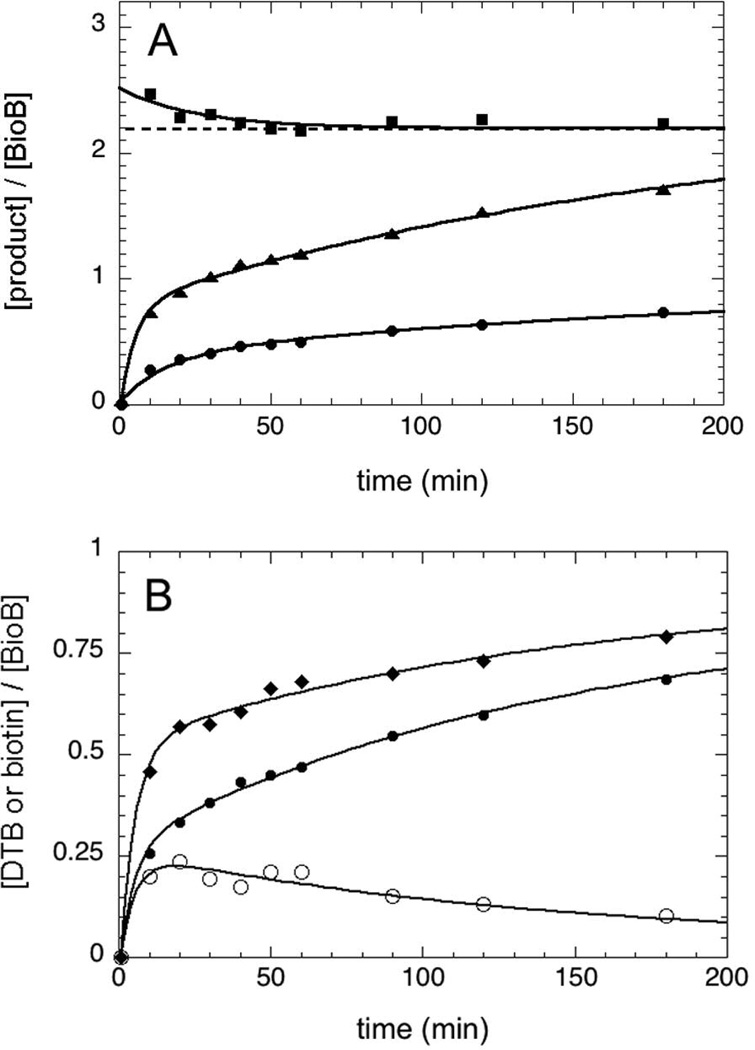
The concentration of biotin and 5´-deoxyadenosine formed during turnover of BioB was determined by HPLC analysis with UV detection (Figure 2A). As is typical for BioB assays in which the air-sensitive [4Fe-4S]2+ cluster is reconstituted in situ (5), biotin is formed more rapidly over the first 30–60 min (k1obs = 0.07 min−1), and then more slowly over the next 4 h (k2obs = 0.005 min−1), yielding a maximum of 0.85 equiv biotin per BioB monomer after 4 h. 5´-Deoxyadenosine is formed with similar kinetics over the entire time course. The relative stoichiometric ratio of these two products is initially ~2.5 but falls over the first 1–2 h to a limiting value of 2.20 ± 0.06. This stoichiometry is consistent with a stepwise mechanism for thiophane ring formation (Figure 1A).
Figure 2.
Stoichiometry of substrate consumption and product formation. (A) The concentration of biotin (●) and 5´-deoxyadenosine (▲) produced in an assay containing BioB (50 µM monomer), AdoMet (200 µM), and DTB (100 µM) was determined by HPLC analysis. The 5´-deoxyadenosine:biotin ratio (■) decreases to a limiting value of 2.20 (± 0.05). (B) The concentration of DTB consumed (♦) is compared to the concentration of biotin produced (●) under the same conditions. An initial burst of DTB consumption is consistent with production of an intermediate (○) with a maximal concentration of ~20% of the enzyme concentration. Assays were conducted at 37 °C under argon as described in the Materials and Methods. All concentration curves are fit by a two-exponential function with k1obs = 0.07 min−1 and k2obs = 0.005 min−1.
The initial decrease in the product ratio suggests that slightly more 5´-deoxyadenosine is initially being produced than can be explained based upon the amount of biotin produced. Control experiments in which DTB was not added to the assay failed to produce any detectable 5´-deoxyadenosine, and therefore BioB does not catalyze uncoupled reductive cleavage of AdoMet under our assay conditions. Instead, we suspected that an intermediate or an alternate product was being formed. We determined the concentration of DTB remaining throughout the assay, and subtracted the initial concentration to obtain the amount of DTB that had been consumed (Figure 2B, diamonds). There is a clear discrepancy between DTB consumed and biotin produced (Figure 2B, filled circles), reaching a maximum of ~0.2 equiv. after 10–20 min (Figure 2B, open circles). The most likely cause of this discrepancy is the initial formation of a DTB-derived intermediate.
Identification of 9-Mercaptodethiobiotin Formed During Turnover of WT BioB
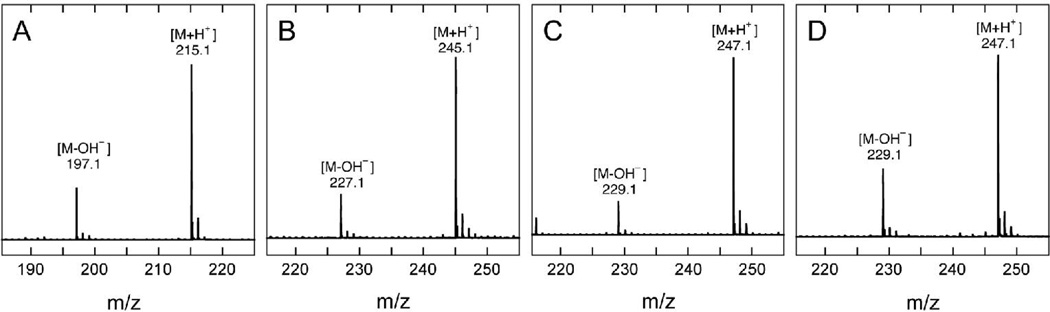
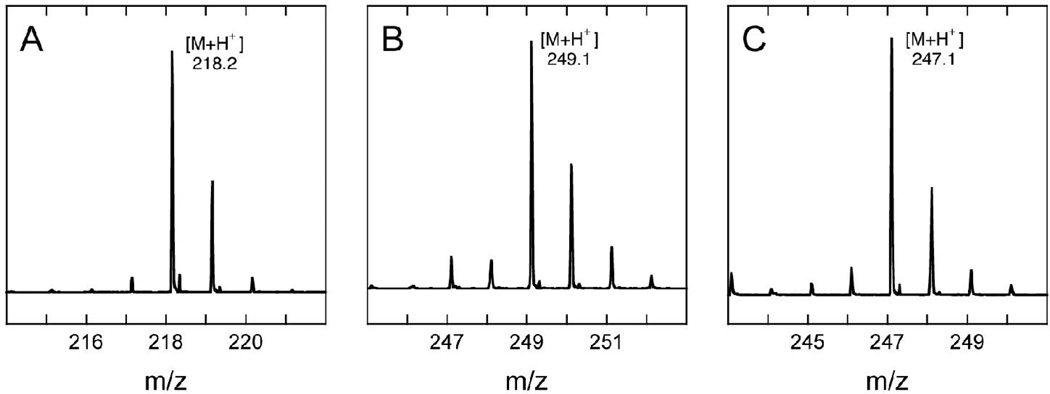
The most likely DTB-derived intermediates are the monothiolated 6- or 9-mercaptodethiobiotin, which are predicted to have monoisotopic masses (M = 246.10) that are higher than either DTB (M = 214.13) or biotin (M = 244.09). We carried out an assay with wild-type BioB for 30 min and quenched with acetic acid to stop the reaction and denature and precipitate the protein. The soluble fraction was analyzed using HPLC electrospray-ionization mass spectrometry, initially focusing on a mass window (246 – 249) that would only include a thiolated intermediate. We observe formation of a new compound that elutes ~2 min after DTB with [M+H+] = 247.1 (Figure 3C), which corresponds to addition of one sulfur to form either 6- or 9-MDTB. The incorporation of a sulfur atom into this new product is clearly demonstrated by the presence of a monoisotopic peak at [M+H+] = 249.1 with ~5% abundance, which corresponds to the natural abundance of 34S (4.2 %) plus a minor fraction of molecules with two 13C atoms (1.2 %). This isotopic distribution is similar to that observed for biotin (Figure 3B), but clearly different than observed for DTB (Figure 3A). To determine which of the two possible MDTB isomers was formed, we compared these results to a sample of synthetic 9-MDTB obtained from Dr. Andreé Marquet (10). We observed a similar elution time (Figure 4) and mass spectrum (Figure 3D) as was observed for the intermediate generated by WT BioB. We did not observe any other peaks corresponding to monothiolated products or intermediates (ie. 6-MDTB) by LCMS. Although we did not have a sample of synthetic 6-MDTB for comparison and the reversed-phase HPLC elution profile of 6-MDTB has not been reported (8), the absence of a second peak with the appropriate monothiolated mass suggests that 6-MDTB is not formed in the enzymatic reaction Further evidence that the thiol is incorporated at C9 is provided by studies with 2H-labeled DTB described below.
Figure 3.
Electrospray ionization mass spectra of (A) DTB prior to assay, (B) biotin produced by WT BioB, (C) 9-MDTB produced by WT BioB, and (D) synthetic 9-MDTB. Experimentally determined masses for the most abundant ions are listed.
Figure 4.
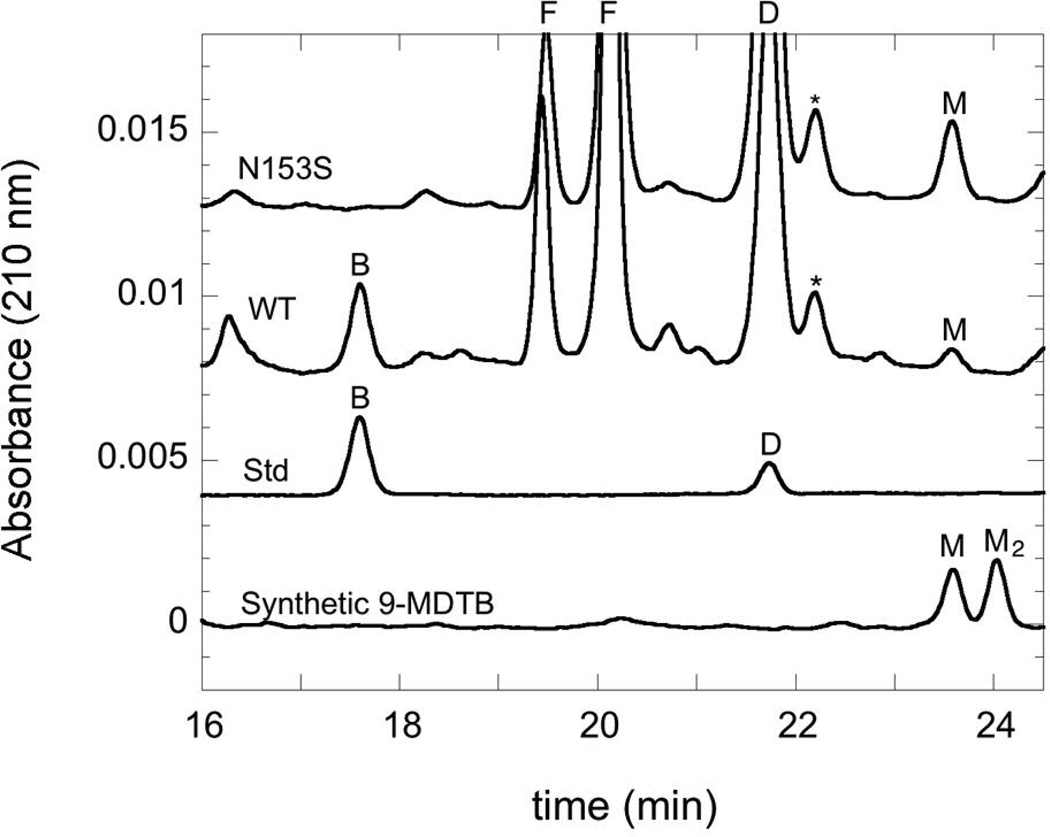
HPLC chromatograms demonstrating 9-MDTB and biotin production in assays of WT and Asn153Ser BioB, monitored by absorbance at 210 nm. A mixed standard containing biotin and DTB (100 µM each) is shown for comparison. A sample of synthetic 9-MDTB disulfide dimer (~100 µM) reduced with dithiothreitol (10 mM) shows two major peaks, identified by LCMS as monomer (tR = 23.6 min) and dimer (tR = 24.0 min). Chromatograms have been subjected to baseline flattening to improve the overlay. Abbreviations: B, biotin, D, DTB, M, 9-MDTB, F, flavin mononucleotide and riboflavin (from flavodoxin), *, contaminant found in DTB.
9-Mercaptodethiobiotin is a Kinetically Competent Intermediate
Although 9-MDTB, as observed using LCMS, was a likely candidate for an intermediate formed on the path to biotin formation, it was also possible that the small amount we observed was instead an aborted product that dissociated prematurely from the enzyme. One test of the competence of this intermediate is whether it is formed and consumed within a time-frame consistent with the observed rate of biotin formation (kinit ≈ 0.03–0.07 min−1 (5)). Using synthetic 9-MDTB as a standard, we were able to assign a minor compound that appears during the assay of WT enzyme as this putative intermediate (Figure 4). The synthetic sample also contains 9-MDTB disulfide dimer, which elutes slightly later from the HPLC and was readily identified by LCMS, but this compound was not observed in enzyme-generated samples. Both of these compounds absorb at 210 nm, but not at 260 nm, and elute from C18 reversed-phase HPLC columns slightly later than DTB.
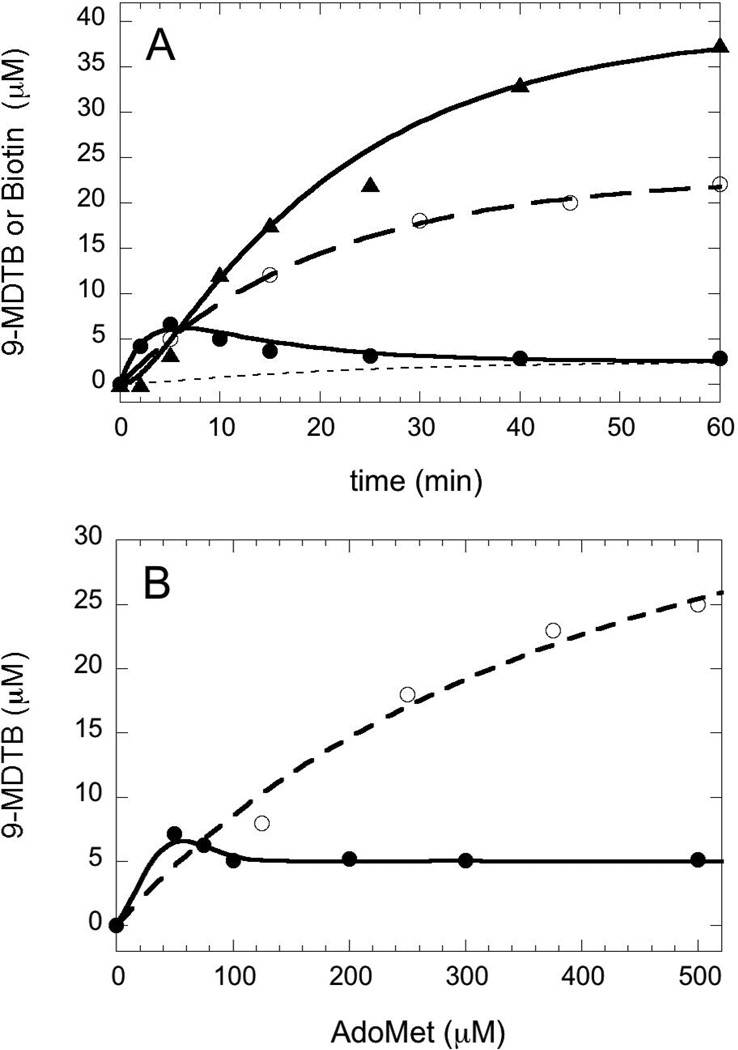
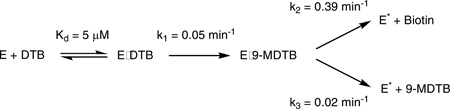
We examined the rate of 9-MDTB and biotin formation under our standard assay conditions, which in particular use a 10-fold excess of AdoMet and therefore should minimize formation of the intermediate by favoring binding of a second equiv of AdoMet. We find that 9-MDTB is formed rapidly and reaches a maximum concentration of 6.6 µM or 0.13 equiv (based on BioB monomer concentration) at 5 min, and then is slowly consumed over the following 60 min (Figure 5A, solid circles). In contrast, biotin is formed following a noticeable lag time of ~3 min and accumulates slowly to a concentration of 37 µM or 0.74 equiv (Figure 5A, triangles). We can model this data with a simple kinetic scheme involving irreversible formation of the intermediate at k1 = 0.05 ± 0.02 min−1, followed by irreversible conversion of the intermediate to biotin and inactive enzyme at k2 = 0.39 ± 0.05 min−1 (Figure 5A, solid curves). However, the intermediate is not completely consumed by the reaction, reaching a limiting concentration of ~2.6 µM after 1–2 hrs. One possible explanation is that the intermediate escapes from the enzyme at a slow background rate of ~0.02 min−1 (Figure 5A, dotted line). In principle this free 9-MDTB should be recaptured by additional active enzyme and converted to biotin, but this is unlikely in the presence of excess DTB, which competes for the reaction with BioB (KmDTB = 2 µM, Km9-MDTB ≈ 200 µM). The kinetic mechanism used to model the data is summarized in the scheme below and predicts that biotin should be formed at an overall initial rate of 0.044 ± 0.016 min−1, while recent reports place the rate of biotin production at 0.002 – 0.2 min−1 (5, 10, 13, 14). E* indicates the generation of inactive enzyme missing the [2Fe-2S]2+ cluster following dissociation of either 9-MDTB or biotin.
Figure 5.
Formation of 9-MDTB and biotin by WT and Asn153Ser BioB under standard assay conditions. (A) WT BioB (50 µM) produces a small burst of 9-MDTB (●) in the first 5–10 min that then dissipates over 1–2 h. Biotin (▲) is produced following 3 min lag, reaching 0.74 equiv after 60 min. These data are fit to a two-step kinetic scheme (k1 = 0.05 min−1 and k2 = 0.39 min−1), as depicted in the text, that includes slow dissociation of 9-MDTB from the enzyme (dotted curve, k3 = 0.02 min−1). Asn153Ser BioB (○) produces 9-MDTB at the same rate as WT BioB (kobs = 0.05 min−1) (B) The AdoMet concentration dependence of 9-MDTB formation. WT BioB (●) produces a maximum of 0.15 equiv 9-MDTB at in the presence of 1 equiv AdoMet in a 45 min assay. A further increase in AdoMet concentration decreases the amount of 9-MDTB detected to 0.1 equiv. Asn153Ser BioB (○) produces up to 0.9 equiv of 9-MDTB in a 3 hr assay with KmAdoMet = 480 µM.
Substoichiometric AdoMet Results in a Modest Increase in 9-MDTB Formation
Following generation of enzyme-bound 9-MDTB in the first half of the reaction (k1 in the scheme above), 5´-deoxyadenosine and methionine must dissociate and a second equivalent of AdoMet must bind prior to conversion of 9-MDTB to biotin. In the absence of the second equivalent of AdoMet, the intermediate would be predicted to accumulate at significantly higher concentrations. We do observe a slight increase in the concentration of 9-MDTB in assays that have less than two equivalents of AdoMet (Figure 5B), but the maximal level of 9-MDTB (7.7 µM) does not approach saturation of the enzyme (50 µM). This observation could be explained by a model in which the second equivalent of AdoMet binds with much higher affinity than the first equivalent. Once the BioB•9-MDTB intermediate state is formed, a conformational change could increase the affinity of this form of the enzyme for AdoMet, allowing the BioB•9-MDTB complex to absorb AdoMet that transiently dissociates from other unreacted BioB molecules. In addition, we observe that the concentration of 9-MDTB formed is only slightly decreased (4.8 µM) at two equivalents of AdoMet, and is not further decreased at higher concentrations. The failure to further suppress the intermediate at higher AdoMet concentrations indicates that AdoMet binding is easily saturated (low Kd) and is not rate-limiting (fast kon) for the second half of the reaction scheme, formation of the C6-S bond.
An Asn153Ser Mutant Enzyme Generates 9-Mercaptodethiobiotin as a Stoichiometric Product
As a part of ongoing efforts to understand the role of conserved residues in controlling the radical chemistry catalyzed by BioB, we have generated mutations at asparagine 153, a residue the forms bidentate hydrogen-bonds that bridge the AdoMet ribose to the DTB ureido ring (4). All mutations of this residue result in enzyme that is inactive for biotin production (C. Farrar and J. Jarrett, unpublished data, see also (15)). However, mutation of Asn153 to Ser results in an enzyme that produces only one equiv of 5´-deoxyadenosine (not shown), and that produces 9-MDTB as a stoichiometric product (Figure 4, top trace). As might be expected, this mutant appears to be somewhat defective in binding AdoMet (Km = 480 µM vs. 5 µM for WT (16)) and DTB (Km = 300 µM vs. 2 µM for WT (11)), but at high concentrations of both substrates (>1 mM) produces ~0.9 equiv of 9-MDTB after a 3 h assay at 37 °C (Figure 5B). In the presence of saturating substrate concentrations, 9-MDTB is formed at an initial rate (kinit = 0.05 min−1) that is identical to the modeled rate of formation of 9-MDTB by the WT enzyme (Figure 5A).
The observation that the Asn153Ser mutant can make 9-MDTB but cannot complete the biotin thiophane ring indicates that the mutant enzyme cannot carry out the second half-reaction in the stepwise mechanism. In principle, this could be due to improper positioning of the C6 position of 9-MDTB with respect to the second equiv of AdoMet, resulting in a protein that retains bound 9-MDTB but in an unreactive conformation. We examined whether 9-MDTB remains bound to WT and Asn153Ser proteins following a 2 h assay by passing the assay mixture through a 10 KDa centrifugal concentrator (Millipore) under anaerobic conditions. The concentration of products in the eluted small molecule fraction was compared to the total concentration using our standard assay HPLC protocol, and the difference in concentration is presumed to be due to tight binding to the enzyme. Control samples with no protein demonstrated that neither substrate binds to the filter alone. After a 2 h assay, WT enzyme produced ~0.1 equiv 9-MDTB, and we find that ~50% is tightly bound to the protein, while the remainder is found in solution due to slow dissociation during the assay (similar to Figure 5A). In contrast, the Asn153Ser mutant enzyme produced ~0.5 equiv 9-MDTB, and we found that <10% is bound to the protein, while the remainder is found free in solution. We conclude that the Asn153Ser mutation increases the rate at which 9-MDTB dissociates from the protein, such that binding of the second equiv of AdoMet and formation of the C6-S bond cannot kinetically compete and do not occur.
(2H-9-Methyl)-Dethiobiotin Demonstrates Initial Thiolation at C9
To further confirm that the intermediate formed by WT BioB is 9-MDTB, and not 6-MDTB, we carried out an assay using synthetic DTB that had been predominantly deuterated at the C9 position. The synthesis of this sample of (2H-9-methyl)-DTB by Marquet and coworkers has been previously described (3), and using ESI-MS we have independently determined that it contains (2H3)-DTB (71 %), (2H2)-DTB (4 %), and (2H4)-DTB (24 %) (Figure 6A and Table 1), with the latter partially deuterated at C6 (A. Marquet, personal communication). The major peak observed in the mass spectrum at [M+H+] = 218.2 (Figure 6A), is shifted +3 mass units from unlabeled DTB ([M+H+] = 215.1, Figure 3A). If the first step in biotin formation involves abstraction of a hydrogen/deuterium atom from C9, then one deuterium should be transferred to 5´-deoxyadenosine and the intermediate formed from labeled DTB should retain only two deuterium atoms. We observed that the major peak in the mass spectrum of the intermediate ([M+H+] = 249.1, Figure 6B) is indeed shifted by only +2 mass units from the unlabeled sample ([M+H+] = 247.1, Figure 3C), indicating that the thiol has been incorporated at the C9 position. A similar isotope shift is also observed for biotin formed in the same assay, with the major peak in the mass spectrum ([M+H+] = 247.1, Figure 6C) shifted by +2 mass units from the unlabeled sample ([M+H+] = 245.1, Figure 3B).
Figure 6.
Mass spectra of (A) synthetic trideuterated (2H-9-methyl)-DTB and (B) deuterated 9-MDTB and (C) deuterated biotin obtained from incubation of WT BioB with (2H-9-methyl)-DTB (200 µM) for 45 min at 37 °C under standard assay conditions. Isotopic peak integration was used to calculate the deuterium incorporation in each sample (Table 1).
Table 1.
Deuterium incorporation in 9-MDTB and Biotin Derived From (2H-9-methyl)-DTB
| Number of Deuterium Atomsa | |||||
|---|---|---|---|---|---|
| Sample | 0 | 1 | 2 | 3 | 4 |
| DTB | 0.6% | 0.8% | 4% | 71% | 24% |
| MDTB | 9% | 7% | 56% | 21% | 6% |
| Biotin | 3% | 7% | 64% | 22% | 2% |
The percentage of each sample with the indicated number of deuterium atoms, after correcting for a natural abundance distribution of 13C (1.11 %) and 34S (4.2 %) at each position. The location of deuterium within each sample has not been determined.
If the reaction catalyzed by BioB involves a clean abstraction of hydrogen/deuterium and addition of sulfur, with no isotope effect or isotopic equilibration, then the distribution of the isotope envelope should remain unchanged, except for a shift due to the removal of one deuterium atom. As is apparent from comparison of Figure 6A with 6B or 6C, the isotopic distribution found in the original (2H-9-methyl)-DTB was not retained in 9-MDTB or biotin (Table 1). After accounting for the minor contributions due to the natural abundance of 13C and 34S, we unexpectedly found that a significant fraction of intermediate and product retain 4 deuterium atoms. Since the C6 and C9 positions of biotin have only 3 possible deuterium positions, this indicates that a small amount of deuterium (2–6 %) has been transferred to other sites within 9-MDTB and biotin. In addition, we find that 10–15 % of each sample has 0 or 1 deuterium remaining, suggesting that multiple deuterium atoms have been removed from DTB. The reproducibility of the isotopic distribution for each sample is ±1 % for multiple analyses of the same sample, and ±2 % for multiple assays, and therefore any minor changes should not be overinterpreted. However, the broadening of the isotope envelope is consistently observed and is even more dramatic in several partially active mutant enzymes (data not shown).
Excess loss or gain of deuterium in a fraction of the product can be explained if we assume that hydrogen atom abstraction is a reversible process, and in addition, that BioB does not always perfectly discriminate the correct C6 and C9 positions. If a 5´-deoxyadenosyl radical abstracts a deuterium atom from C9, the 5´-methyl group of 5´-deoxyadenosine is free to rotate within the active site. If the dethiobiotinyl radical then reabstracts a hydrogen from 5´-deoxyadenosine, there is no net chemical reaction, but a deuterium has now been transferred from DTB to the 5´-deoxyadenosyl radical. This labeled 5´-deoxyadenosyl radical could then react with methionine and generate 2H-labeled AdoMet. A similar reversible reaction scheme has been proposed to explain the transfer of a tritium label from AdoMet to L-lysine catalyzed by the AdoMet radical enzyme lysine 2,3-aminomutase (17). Once 2H-AdoMet has been formed through this process, a similar reversible hydrogen atom abstraction from other sites in the substrate could result in minor amounts of deuterium being incorporated at sites other than C9 or C6. In general, these unproductive hydrogen atom transfer reactions would go unnoticed in unlabeled samples, as long as subsequent C-S bond formation proceeds with very high fidelity. A complete elucidation of this process would require a more cleanly labeled dethiobiotin sample, as well as a more precise analysis of substrates and products prior to and after catalysis, and is beyond the scope of the current work. Qualitatively, however, the broadening of the isotope envelope observed upon conversion of DTB to 9-MDTB and biotin suggests that only ~80–90 % of the dethiobiotinyl radical formed reacts with sulfur on the first pass, while the remainder reabstracts a hydrogen from 5´-deoxyadenosine.
Discussion
The incorporation of sulfur into biomolecules is a relatively rare reaction in biochemistry. Whenever the biosynthetic pathway and neighboring functional groups allow, this process is typically carried out through a nucleophilic process that takes advantage of the nucleophilicity of sulfur in thiocarboxylate, thiolate, or bisulfide moieties (18). Only three known enzymes incorporate sulfur by substituting directly for a hydrogen atom. In addition to the formation of the thiophane ring catalyzed by biotin synthase, lipoyl synthase (LipA) catalyzes the addition of two sulfur atoms to octanoyl-E2 protein or H protein (19), while MiaB catalyzes the addition of sulfur and a methyl group to N-6-isopentenyl adenosine at position 37 within most tRNA molecules (20). In each of the latter cases, the sulfur atoms incorporated into the thiolated product are proposed to derive from an iron-sulfur cluster bound within the enzyme (21).
The mechanism by which BioB catalyzes incorporation of sulfur into DTB to form the biotin thiophane ring is not well understood. We have proposed a stepwise mechanism (Figure 1A) in which reductive cleavage of AdoMet is coupled to oxidation of dethiobiotin via hydrogen atom abstraction at C9 and C6, generating substrate radicals that are quenched by a sulfur atom derived from a nearby [2Fe-2S]2+ cluster (1, 5). This mechanism is consistent with several prior studies. The amount of 5´-deoxyadenosine formed per biotin produced has been reported as 2.7 – 3.0 equivalents (7, 22), suggesting that 5´-deoxyadenosine is involved in abstraction of at least two hydrogen atoms from DTB, although some abortive AdoMet cleavage not coupled to biotin formation apparently also occurs. Marquet and coworkers found that synthetic DTB preferentially deuterated at C9 or C6 transfers a fraction of this deuterium to 5´-deoxyadenosine, albeit with only ~18% efficiency (3). Finally, both heavy atom labeling with 34S, 35S, or Se (23–26) and spectroscopic studies (5, 14, 27) implicate the [2Fe-2S]2+ cluster as the likely source of sulfur. However, the specific details of the reaction sequence have not been delineated. In a recent study, Huynh and coworkers demonstrated using Mössbauer spectroscopy and HPLC analysis that degradation of the [2Fe-2S]2+ cluster may precede biotin formation (14), suggesting that a sulfur atom could be transferred to a protein residue for subsequent incorporation into biotin. Based on the data presented herein, we would suggest that degradation of the [2Fe-2S]2+ cluster might instead be coupled to 9-MDTB formation (not biotin formation), and therefore 9-MDTB might be a long-lived intermediate present in the spectroscopic samples generated by Huynh and coworkers.
In an effort towards unraveling the specific reaction sequence that leads to thiophane ring formation, we have attempted to examine whether formation of the C6-S and C9-S bonds occurs in a stepwise or concerted manner. Recent studies of a similar C-S bond forming reaction catalyzed by the AdoMet radical enzyme lipoyl synthase (LipA) suggest that the two C-S bonds in lipoic acid are formed in a tightly-coupled stepwise mechanism involving attack of substrate radicals on a [4Fe-4S]2+ cluster (21). LipA from Sulfolobus solfataricus P2 converts octanoyl-E2 protein to lipoyl-E2 via an apparent 6-mercaptooctanoyl-E2 intermediate, as detected by mass spectrometry (28) In LipA from E. coli, a monothiolated mercaptooctanoyl-H protein has been detected as a minor reaction product by mass spectrometry, but becomes the major reaction product when perdeuterated octanoyl-H protein is used as a substrate (29). However, if a monothiolated intermediate is formed, it must remain tightly bound to LipA, since a mixture of 32S- and 34S-LipA produced only homogenous 32S- and 34S-lipoyl-H protein (30). However, despite the appeal of a stepwise mechanism involving a monothiolated intermediate, neither laboratory has provided kinetic evidence supporting an intermediate as opposed to an alternate abortive product (21).
In the case of BioB, isotopic labeling had indicated that a monothiolated intermediate or alternate product is formed from DTB (6, 7, 9), but failed to provide a positive identification of this species. However, synthetic 9-MDTB (but not 6-MDTB) was shown to be a chemically competent substrate for BioB, lending support to a stepwise mechanism with 9-MDTB as a putative intermediate (8, 10). Consistent with a stepwise mechanism, we determined the stoichiometry of 5´-deoxyadenosine to biotin formation of 2.2:1, and since the active site can only hold 1 equiv of AdoMet, this strongly suggests that a DTB-derived intermediate must remain bound during 5´-deoxyadenosine/AdoMet exchange. Using HPLC and ESI-MS analysis, we identified a minor component of the BioB reaction mixture as 9-mercaptodethiobiotin. In assays of WT BioB, 9-MDTB is formed within 10 min at 37 °C and is then converted to biotin over the following 1–2 hrs. A kinetic analysis of 9-MDTB formation and conversion to biotin implicates the first half-reaction, formation of the C9-S bond, as rate-limiting for the overall formation of biotin (k1 = 0.05 min−1). As a consequence, 9-MDTB is normally not observed at a stoichiometry greater than ~0.1 equiv per BioB monomer.
We do observe that a small fraction of the 9-MDTB is formed and not converted to biotin, and we have modeled this process as a slow escape of the intermediate from the enzyme at ~0.02 min−1. Several residues are involved in binding DTB, and presumably also 9-MDTB, with perhaps the most critical residue Asn153, which forms hydrogen bonds to both DTB and AdoMet. We find that the mutation Asn153Ser results in an “active” enzyme that can bind AdoMet and DTB and can convert DTB to 9-MDTB with a stoichiometry of ~1:1 9-MDTB:BioB monomer. However, the majority of this 9-MDTB is released into the buffer and not bound to the enzyme, suggesting that dissociation of 9-MDTB is faster than the second half-reaction for biotin formation (k2 = 0.39 min−1). We conclude that Asn153 is critical for retaining a tightly-bound intermediate, and could potentially also be important in aligning the C6 position of 9-MDTB with AdoMet for the second hydrogen atom abstraction.
The specific role of the [2Fe-2S]2+ cluster in generating 9-MDTB and ultimately biotin may be more complex than serving as a simple sulfur donor. Formation of a C-S bond from a dethiobiotinyl carbon radical and sulfide requires that the sulfide is oxidized by one electron, a process that can be accomplished within an iron-sulfur cluster via inner-sphere electron transfer to a Fe3+ ion. In BioB, this electron transfer should generate an EPR-detectable [2Fe-2S]+ cluster in a stoichiometry equivalent to the concentration of 9-MDTB. Huynh and coworkers report an EPR signal attributed to a transient [2Fe-2S]+ cluster, formed during an assay with a maximum of 0.31 spins/BioB monomer after 15 min (14). Subsequent formation of the C6-S bond would also require further oxidation of the sulfur atom, resulting in destruction of the FeS cluster and loss of this EPR signal. In addition to a role in sulfur oxidation, the [2Fe-2S]2+ cluster may also function as a protecting group for the 9-MDTB thiol. If the free thiol or thiolate were formed as an intermediate, a 5´-deoxyadenosyl radical formed in the second half-reaction of the enzyme would be rapidly quenched by hydrogen atom or electron transfer from the thiol or thiolate, resulting in abortive AdoMet cleavage and possibly also 9-MDTB decomposition. Coordination of the thiolate to an Fe3+ ion derived from the [2Fe-2S]+ cluster could prevent these undesired side reactions and promote completion of the biotin thiophane ring.
Supplementary Material
Acknowledgements
We would like to thank Dr. Andrée Marquet for the kind gift of synthetic 9-mercaptodethiobiotin and (2H-9-methyl)-dethiobiotin and for helpful discussions regarding interpretation of the isotope data.
Footnotes
This research has been supported by the NIH (R01 GM59175 to J.T.J.).
Abbreviations: AdoMet, S-adenosyl-L-methionine; BioB, biotin synthase; DTB, dethiobiotin; DTT, dithiothreitol; EPR, electron paramagnetic resonance; ESI-MS, electrospray ionization mass spectrometry; 9-MDTB, 9-mercaptodethiobiotin; Tris, tris(hydroxymethyl)aminomethane.
Supporting Information Available
Additional LCMS data including total and extracted ion chromatograms, as well as spectra of selected unidentified peaks in the chromatogram. A detailed description of the filtration binding assay, including raw peak integration and processed data. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Jarrett JT. The novel structure and chemistry of iron-sulfur clusters in the adenosylmethionine-dependent radical enzyme biotin synthase. Arch. Biochem. Biophys. 2005;433:312–321. doi: 10.1016/j.abb.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 2.Marquet A, Tse Sum Bui B, Florentin D. Biosynthesis of biotin and lipoic acid. Vitam. Horm. 2001;61:51–101. doi: 10.1016/s0083-6729(01)61002-1. [DOI] [PubMed] [Google Scholar]
- 3.Escalletes F, Florentin D, Tse Sum Bui B, Lesage D, Marquet A. Biotin Synthase Mechanism: Evidence for Hydrogen Transfer from the Substrate into Deoxyadenosine. J. Am. Chem. Soc. 1999;121:3571–3578. [Google Scholar]
- 4.Berkovitch F, Nicolet Y, Wan JT, Jarrett JT, Drennan CL. Crystal structure of biotin synthase, an S-adenosylmethionine-dependent radical enzyme. Science. 2004;303:76–79. doi: 10.1126/science.1088493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ugulava NB, Sacanell CJ, Jarrett JT. Spectroscopic Changes during a Single Turnover of Biotin Synthase: Destruction of a [2Fe-2S] Sluster Accompanies Sulfur Insertion. Biochemistry. 2001;40:8352–8358. doi: 10.1021/bi010463x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salib AG, Frappier F, Guillerm G, Marquet A. On the mechanism of conversion of dethiobiotin to biotin in Escherichia coli. III. Isolation of an intermediate in the biosynthesis of biotin from dethiobiotin. Biochem Biophys Res Commun. 1979;88:312–319. doi: 10.1016/0006-291x(79)91731-5. [DOI] [PubMed] [Google Scholar]
- 7.Shaw NM, Birch OM, Tinschert A, Venetz V, Dietrich R, Savoy LA. Biotin synthase from Escherichia coli: isolation of an enzyme-generated intermediate and stoichiometry of S-adenosylmethionine use. Biochem J. 1998;330:1079–1085. doi: 10.1042/bj3301079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marquet A, Frappier F, Guillerm G, Azoulay M, Florentin D, Tabet J-C. Biotin Biosynthesis: Synthesis and Biological Evaluation of the Putative Intermediate Thiols. J. Am. Chem. Soc. 1993;115:2139–2145. [Google Scholar]
- 9.Baldet P, Gerbling H, Axiotis S, Douce R. Biotin biosynthesis in higher plant cells. Identification of intermediates. Eur. J. Biochem. 1993;217:479–485. doi: 10.1111/j.1432-1033.1993.tb18267.x. [DOI] [PubMed] [Google Scholar]
- 10.Tse Sum Bui B, Lotierzo M, Escalettes F, Florentin D, Marquet A. Further investigation on the turnover of Escherichia coli biotin synthase with dethiobiotin and 9-mercaptodethiobiotin as substrates. Biochemistry. 2004;43:16432–16441. doi: 10.1021/bi048040t. [DOI] [PubMed] [Google Scholar]
- 11.Sanyal I, Cohen G, Flint DH. Biotin synthase: purification, characterization as a [2Fe-2S] cluster protein, and in vitro activity of the Escherichia coli bioB gene product. Biochemistry. 1994;33:3625–3631. doi: 10.1021/bi00178a020. [DOI] [PubMed] [Google Scholar]
- 12.Ugulava NB, Gibney BR, Jarrett JT. Iron-Sulfur cluster Interconversion in Biotin Synthase: Dissociation and Reassociation of Iron during Conversion of [2Fe-2S] to [4Fe-4S] Clusters. Biochemistry. 2000;39:5206–5214. doi: 10.1021/bi9926227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ollagnier-de Choudens S, Mulliez E, Fontecave M. The PLP-dependent biotin synthase from Escherichia coli: mechanistic studies. FEBS Lett. 2002;532:465–468. doi: 10.1016/s0014-5793(02)03733-x. [DOI] [PubMed] [Google Scholar]
- 14.Jameson GN, Cosper MM, Hernandez HL, Johnson MK, Huynh BH. Role of the [2Fe-2S] cluster in recombinant Escherichia coli biotin synthase. Biochemistry. 2004;43:2022–2031. doi: 10.1021/bi035666v. [DOI] [PubMed] [Google Scholar]
- 15.Lotierzo M, Raux E, Tse Sum Bui B, Goasdoue N, Libot F, Florentin D, Warren MJ, Marquet A. Biotin synthase mechanism: mutagenesis of the YNHNLD conserved motif. Biochemistry. 2006;45:12274–12281. doi: 10.1021/bi060662m. [DOI] [PubMed] [Google Scholar]
- 16.Sanyal I, Gibson KJ, Flint DH. Escherichia coli Biotin Synthase: An Investigation into the Factors Required for Its Activity and Its Sulfur Donor. Arch. Biochem. Biophys. 1996;326:48–56. doi: 10.1006/abbi.1996.0045. [DOI] [PubMed] [Google Scholar]
- 17.Moss M, Frey PA. The Role of S-Adenosylmethionine in the Lysine 2,3-Aminomutase Reaction. J. Biol. Chem. 1987;262:14859–14862. [PubMed] [Google Scholar]
- 18.Mueller EG. Trafficking in persulfides: delivering sulfur in biosynthetic pathways. Nat Chem Biol. 2006;2:185–194. doi: 10.1038/nchembio779. [DOI] [PubMed] [Google Scholar]
- 19.Cicchillo RM, Lee KH, Baleanu-Gogonea C, Nesbitt NM, Krebs C, Booker SJ. Escherichia coli lipoyl synthase binds two distinct [4Fe-4S] clusters per polypeptide. Biochemistry. 2004;43:11770–11781. doi: 10.1021/bi0488505. [DOI] [PubMed] [Google Scholar]
- 20.Hernandez HL, Pierrel F, Elleingand E, Garcia-Serres R, Huynh BH, Johnson MK, Fontecave M, Atta M. MiaB, a bifunctional radical-S-adenosylmethionine enzyme involved in the thiolation and methylation of tRNA, contains two essential [4Fe-4S] clusters. Biochemistry. 2007;46:5140–5147. doi: 10.1021/bi7000449. [DOI] [PubMed] [Google Scholar]
- 21.Booker SJ, Cicchillo RM, Grove TL. Self-sacrifice in radical S-adenosylmethionine proteins. Curr Opin Chem Biol. 2007;11:543–552. doi: 10.1016/j.cbpa.2007.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guianvarc'h D, Florentin D, Tse Sum Bui B, Nunzi F, Marquet A. Biotin synthase, a new member of the family of enzymes which uses S-adenosylmethionine as a source of deoxyadenosyl radical. Biochem. Biophys. Res. Commun. 1997;236:402–406. doi: 10.1006/bbrc.1997.6952. [DOI] [PubMed] [Google Scholar]
- 23.Gibson KJ, Pelletier DA, Turner IM., Sr Transfer of sulfur to biotin from biotin synthase (BioB protein) Biochem. Biophys. Res. Commun. 1999;254:632–635. doi: 10.1006/bbrc.1998.9991. [DOI] [PubMed] [Google Scholar]
- 24.Tse Sum Bui B, Escalletes F, Chottard G, Florentin D, Marquet A. Enzyme-mediated sulfide production for the reconstitution of [2Fe-2S] clusters into apo-biotin synthase of Escherichia coli. Eur. J. Biochem. 2000;267:2688–2694. doi: 10.1046/j.1432-1327.2000.01284.x. [DOI] [PubMed] [Google Scholar]
- 25.Tse Sum Bui B, Florentin D, Fournier F, Ploux O, Mejean A, Marquet A. Biotin synthase mechanism: on the origin of sulphur. FEBS Lett. 1998;440:226–230. doi: 10.1016/s0014-5793(98)01464-1. [DOI] [PubMed] [Google Scholar]
- 26.Tse Sum Bui B, Mattioli TA, Florentin D, Bolbach G, Marquet A. Escherichia coli biotin synthase produces selenobiotin. Further evidence of the involvement of the [2Fe-2S]2+ cluster in the sulfur insertion step. Biochemistry. 2006;45:3824–3834. doi: 10.1021/bi052388m. [DOI] [PubMed] [Google Scholar]
- 27.Tse Sum Bui B, Benda R, Schunemann V, Florentin D, Trautwein AX, Marquet A. Fate of the (2Fe-2S)2+ cluster of Escherichia coli biotin synthase during reaction: a Mössbauer characterization. Biochemistry. 2003;42:8791–8798. doi: 10.1021/bi034426c. [DOI] [PubMed] [Google Scholar]
- 28.Douglas P, Kriek M, Bryant P, Roach PL. Lipoyl synthase inserts sulfur atoms into an octanoyl substrate in a stepwise manner. Angew Chem Int Ed Engl. 2006;45:5197–5199. doi: 10.1002/anie.200601910. [DOI] [PubMed] [Google Scholar]
- 29.Cicchillo RM, Iwig DF, Jones AD, Nesbitt NM, Baleanu-Gogonea C, Souder MG, Tu L, Booker SJ. Lipoyl synthase requires two equivalents of S-adenosyl-l-methionine to synthesize one equivalent of lipoic acid. Biochemistry. 2004;43:6378–6386. doi: 10.1021/bi049528x. [DOI] [PubMed] [Google Scholar]
- 30.Cicchillo RM, Booker SJ. Mechanistic investigations of lipoic acid biosynthesis in Escherichia coli: both sulfur atoms in lipoic acid are contributed by the same lipoyl synthase polypeptide. J Am Chem Soc. 2005;127:2860–2861. doi: 10.1021/ja042428u. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.