Abstract
Human leukocyte antigen (HLA) molecules regulate the cellular immune system and may be determinants of infant susceptibility to human immunodeficiency virus type 1 (HIV-1) infection. Molecular HLA typing for class I alleles was performed on infants followed in a Kenyan perinatal cohort. Early HIV-1 infection status was defined as infection occurring at birth or month 1, while late infection via breast milk was defined as first detection of HIV-1 after 1 month of age. Likelihood ratio tests based on a proportional hazards model adjusting for maternal CD4 T cell count and HIV-1 viral load at 32 weeks of gestation were used to test associations between infant allelic variation and incident HIV-1 infection. Among 433 infants, 76 (18%) were HIV-1 infected during 12 months of follow-up. HLA B*18 was associated with a significantly lower risk of early HIV-1 transmission [relative risk (RR) = 0.26; 95% confidence interval (CI) 0.04–0.82], and none of the 24 breastfeeding infants expressing HLA B*18 who were uninfected at month 1 acquired HIV-1 late via breast milk. We observed a trend toward increased early HIV-1 acquisition for infants presenting HLA A*29 (RR = 2.0; 95% CI 1.0–3.8) and increased late HIV-1 acquisition via breast milk for both Cw*07 and Cw*08 (RR = 4.0; 95% CI 1.0–17.8 and RR = 7.2; 95% CI 1.2–37.3, respectively). HLA B*18 may protect breast-feeding infants against both early and late HIV-1 acquisition, a finding that could have implications for the design and monitoring of HIV-1 vaccines targeting cellular immune responses against HIV-1.
INTRODUCTION
Human leukocyte antigen (HLA) polymorphisms result in variability in antigen presentation to host lymphocytes and may be important determinants of susceptibility to infant HIV-1 acquisition and disease progression. Several associations between specific HLAs and HIV-1 mother-to-child transmission have been described, primarily in non-breast-feeding cohorts in Europe and North America.1–6 These studies focused on class II alleles in European and American children and found that HLAs DQB1*06 and DRB1*03 were associated with increased HIV-1 transmission, and several DQB1 alleles were associated with decreased transmission.1–5
More limited research has been conducted on HLA associations and vertical HIV-1 transmission among breast-feeding populations from developing countries where the majority of new pediatric HIV-1 infections occur. In an African breast-feeding cohort, the A2 supertype, a group of functionally related HLA subtypes, was associated with protection against infant HIV-1 infection during the first 6 months but not against later transmission through continued breast milk exposure.7 The relationship between the A2 supertype and risk of vertical transmission was independent of the protective effect of HLA discordance between mother and child.6,8
We examined associations between mother-to-child HIV-1 transmission and frequently expressed HLA alleles in a Kenyan perinatal cohort. Special attention was given to defining early and late transmission events to determine effects of HLA on HIV-1 acquisition among breast-feeding infants in sub-Saharan Africa.
MATERIALS AND METHODS
Study population and clinical procedures
Infants included in this study were born to HIV-1-infected mothers enrolled in an ongoing perinatal cohort in Nairobi evaluating infant cellular immune responses to HIV-1 exposure.9 Pregnant women had CD4 T cell counts and HIV-1 RNA viral loads measured at 32 weeks of pregnancy and were provided prophylactic zidovudine from 34–36 weeks gestation through delivery. Infants were fed according to maternal preference after being counseled regarding the risks and benefits associated with breast-feeding and formula feeding.10 Blood specimens were obtained from both breast-feeding and formula feeding infants within 48 hr of delivery for HLA typing and HIV-1 infection status, and at 1, 3, 6, 9, and 12 months of age to determine HIV-1 infection status. Infants with two consecutive positive filter paper specimens were considered HIV-1 infected and did not undergo additional HIV-1 testing.
HLA typing
Molecular HLA typing was performed at the University of Nairobi using the first available infant blood specimen. HLA typing was not performed on maternal samples from this cohort. DNA was extracted from peripheral blood mononuclear cells using Gentra Purgene reagents. Class I HLA were determined using amplification refractory mutation system-polymerase chain reaction (ARMS-PCR)11 with primers specifically designed to characterize alleles expressed in an East African cohort.12
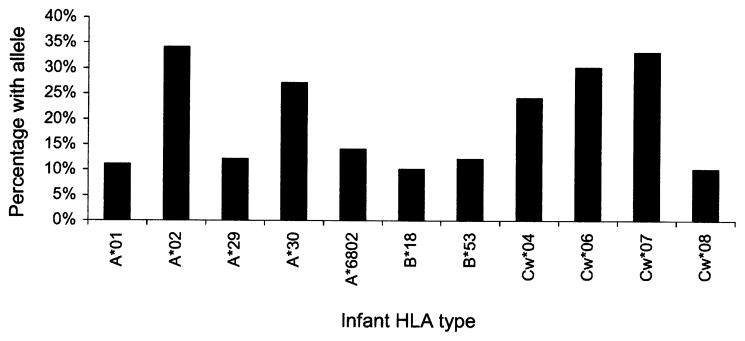
Infant class I HLA alleles were selected for analysis if the allele was represented at a frequency of 10% or greater in the cohort. We studied the following 11 alleles, many of which had been associated with HIV-1 transmission or disease progression in previous reports (Fig. 1): A*01,13–15 A*02,7,16 A*29,17 A*30, A*6802,7,17 B*18,18,19 B*53,20 Cw*04,21 Cw*06, Cw*07, Cw*08.
Fig. 1.
Frequency of infant HLA types (n = 433)
HIV-1 DNA filter paper assays
Polymerase chain reaction (PCR) was used to detect HIV-1 gag DNA in filter paper blood specimens as previously described.22 An infant was considered HIV-1 infected if two consecutive filter paper specimens were positive for HIV-1 DNA or if the last available filter paper specimen was positive for HIV-1 DNA. To determine precise timing of infant infection, HIV-1 RNA levels were measured on plasma samples obtained prior to the first positive filter paper using a Gen-Probe PCR assay sensitive for detection of Kenyan HIV-1 subtypes A, C, and D.23
Data analysis
Relative risk estimates and likelihood based 95% confidence intervals were computed for alleles expressed by at least 10% of infants using a proportional hazards model for interval-censored data. To address the multiplicity of alleles at each locus, we performed overall tests of association for these A, B, and C loci alleles.24,25 The overall test was also performed using all alleles expressed at these three loci regardless of frequency of expression within our cohort. P values based on the likelihood ratio test represent the probability that at least one allele in the specific locus is significantly associated with incident infant HIV-1 infection. We performed two analyses with outcomes (1) infant HIV-1 infection by 1 month of age or (2) infant HIV-1 infection between 1 and 12 months of age. The first analysis included all infants with maternal HIV-1 viral load data and the second analysis included all babies with maternal HIV-1 viral load data who were HIV-1 uninfected at month 1 and continued to breast-feed beyond 1 month of age. We performed the analysis adjusting for both maternal CD4 count and maternal HIV-1 viral load at 32 weeks of pregnancy. In the subset of women with maternal viral load measurements at delivery, the data were additionally analyzed using a model adjusting for this covariate.
RESULTS
Cohort characteristics and HLA allele frequencies
The study included 433 infants who participated in a perinatal HIV-1 transmission study between July 1999 and December 2002 (Table 1). Maternal CD4 T cell counts and maternal HIV-1 viral load results were available at 32 weeks of pregnancy for 433 and 332 mother–infant pairs, respectively. The mean maternal CD4 count was 474 cells/μl (range: 6–1628 cells/μl) and the mean maternal HIV-1 viral load was 4.6 log10 copies/ml (standard deviation 0.9 log10 copies/ml). At delivery, mean maternal viral load was lower than at 32 weeks of gestation (4.2 vs 4.6 log10 copies/ml) due to administration of perinatal zidovudine. Delivery HIV-1 viral load data, only available for a subset of 322 mother–infant pairs, were highly correlated with viral loads at 32 weeks of pregnancy (correlation coefficient = 0.68), indicating that the 32-week maternal viral load is a good marker for overall HIV-1 RNA levels in this cohort.
Table 1.
Description of the Cohort
| Characteristic | n = 433 |
|---|---|
| Mean maternal CD4 T cell count at 32 weeks of gestation (standard deviation) | 474 cells/μl (240) |
| Mean HIV-1 viral load at 32 weeks of gestation (standard deviation)a | 4.6 log10 copies/ml (0.9) |
| Mean HIV-1 viral load at time of delivery (standard deviation)b | 4.2 log10 copies/ml (1.1) |
| Number HIV-1 infected | 76 (18%) |
| At birth, <48 hr postpartum | 29 (38%) |
| Between birth and month 1 | 36 (47%) |
| Between month 1 and month 12 | 11 (14%) |
| Number completing 12 months of follow-upc | 290 (67%) |
| Number lost to follow-up | 31 (7%) |
| Number of infant deaths | 49 (11%) |
| Number of breast-feeding infants | 322 (74%) |
| Number of breast-feeding infants ≥ 1 month of aged | 298 (73%) |
| HIV-1 uninfected and breast-feeding at 1 month | 253 (85%) |
| Median duration of breast-feeding in months (range) | 9 (1–12) |
Data available for 332 mother–infant pairs.
Data available for 322 mother–infant pairs.
Infants who died were not included in the denominator.
Among the 322 infants who ever breast-fed.
Among the 433 infants in the cohort, 322 (74%) were breast-fed at birth and 298 (73%) breast-fed after 1 month of age with a median duration of breast-feeding of 9 months (range: 1–12 months). Seventy-six (18%) infants were HIV-1 infected during 12 months of follow-up: 29 (38%) of the 76 were HIV-1 infected at birth, 36 (47%) between birth and the first month of life, and 11 (14%) between 1 and 12 months of age. The cumulative probability of HIV-1 infection was 7% at birth, 15% at 1 month, and 18% at 12 months. Infant mortality was 11% during follow-up with 49 deaths occurring among the 433 infants enrolled in the study, 26 (53%) of whom were HIV-1 infected and 23 (47%) of whom were HIV-1 uninfected at the last time of testing. Excluding infants who died, 257 (67%) infants completed 12 months of follow-up and 31 (7%) infants were lost to follow-up. Infants who were HIV-1 infected were not more likely to be lost to follow-up than infants who were HIV-1 uninfected (8% of 76 infected infants versus 7% of 357 uninfected infants; p = 0.8).
The prevalence of each of the 11 class I alleles evaluated in this cohort is depicted graphically in Figure 1. The most frequently expressed alleles were A*02 (34%), A*30 (27%), Cw*06 (30%), and Cw*07 (33%). An additional 51 alleles were typed in the cohort with 26 (51%) among these expressed by fewer than 5% of the 433 infants.
Infant HLAs associated with mother-to-child HIV-1 transmission
Early HIV-1 acquisition was defined as occurring at or before month 1 and late acquisition defined as occurring after month 1. While the 65 early infections may have occurred in utero, intrapartum, or through exposure to colostrum or breast milk, the 11 late infections resulted exclusively from exposure to HIV-1 in breast milk across oropharyngeal and gastrointestinal mucosa.
The B*18 haplotype was protective against early HIV-1 acquisition in this cohort. After adjusting for maternal CD4 T cell count and HIV-1 viral load at 32 weeks of pregnancy, infants expressing HLA B*18 were 74% less likely to become infected than infants not expressing this allele (RR = 0.26; 95% CI = 0.04–0.82) (Table 2). Two (5%) infants were HIV-1 infected at age 1 month among the 43 infants with B* 18 compared to 63 (16%) of 390 infants not expressing this haplotype. The overall test for the B locus was significant when HLA B*18 and HLA B*53 were included in the analysis (p = 0.02), but lacked power when all 28 B alleles were incorporated into the model (p = 0.72). These analyses were repeated adjusting for maternal HIV-1 viral load at the time of delivery, rather than at 32 weeks of pregnancy. In this analysis, the association between B*18 and protection persisted, the strength of the association was unchanged (RR = 0.28; 95% CI = 0.05–0.89), and the overall p value for the B locus remained significant (p = 0.03).
Table 2.
Risk of Infant HIV-1 Infection by 1 Month of Age
| Number (%) HIV-1 infecteda
|
Relative risk (95% confidence interval)b | ||
|---|---|---|---|
| With allele | Without allele | ||
| A locus | |||
| A*01 | 6 (13%) | 59 (15%) | 0.98 (0.37, 2.19) |
| A*02 | 15 (10%) | 50 (18%) | 0.73 (0.38, 1.36) |
| A*29 | 13 (25%) | 52 (14%) | 2.01 (1.01, 3.76) |
| A*30 | 19 (16%) | 46 (15%) | 1.09 (0.6, 1.93) |
| A*6802 | 10 (17%) | 55 (15%) | 1.18 (0.55, 2.29) |
| p valuec,d | 0.27 | ||
| B locus | |||
| B*18 | 2 (5%) | 63 (16%) | 0.26 (0.04, 0.82) |
| B*53 | 4 (8%) | 61 (16%) | 0.45 (0.14, 1.1) |
| p valuec,d | 0.02 | ||
| C locus | |||
| Cw*04 | 12 (11%) | 53 (16%) | 0.73 (0.36, 1.36) |
| Cw*06 | 18 (14%) | 47 (15%) | 0.92 (0.5, 1.63) |
| Cw*07 | 20 (14%) | 45 (15%) | 0.80 (0.44, 1.4) |
| Cw*08 | 5 (13%) | 60 (15%) | 0.98 (0.34, 2.31) |
| p valuec,d | 0.85 | ||
Includes all 433 infants in the cohort of whom 65 were HIV-1 infected by month 1.
Includes 332 infants with maternal CD4 T cell count and HIV-1 viral load data at 32 weeks gestation. Analysis adjusts for these correlates.
p values for test of overall locus association compared to a model without allele information, when including the alleles listed above.
p values for test of overall locus association equal 0.42, 0.72, and 0.99 for the A locus, B locus, and C locus, respectively, when including all 62 alleles expressed in the cohort.
While the overall test for the A locus was not statistically significant (p = 0.27), an association between early infection risk and HLA A*29 expression was observed in the adjusted analysis including both maternal CD4 T cell count and HIV-1 viral load at 32 weeks gestation (RR = 2.0; 95% CI = 1.0–3.8) (Table 2). Infants with A*29 had a 2-fold increased risk of infection before month 1, with 13 (25%) of the 52 infants expressing HLA A*29 becoming infected by month 1 compared to 52 (14%) of the 381 infants without this allele.
A trend toward protection was observed for HLA A*02 in an unadjusted analysis that did not control for maternal CD4 count and HIV-1 viral load. Infants expressing HLA A*02 were approximately 50% less likely to become HIV-1 infected before month 1 (RR = 0.55; 95% CI 0.3–0.9), an observation that is consistent with previous reports that also did not adjust for maternal viral load.7 We did not observe significant protection against HIV-1 acquisition for A*02 in the analysis that adjusted for the effect of maternal immunosuppression and viral load on vertical transmission risk (Table 2). Future studies focusing on HLAs A*02 and A*29 may clarify their contribution to mother-to-child HIV-1 transmission and disease progression.
In our analysis of late transmission events, risk of HIV-1 acquisition via breast milk after month 1 was not associated with the A, B, or C locus (Table 3). This may be because our evaluation of late HIV-1 infection was limited by the small number of infants (n = 11) who became HIV-1 infected between 1 and 12 months of age. Thus, our analysis may have lacked power to detect a significant difference in late transmission risk. However, we did observe that no infants expressing the B*18 allele acquired HIV-1 after month 1, suggesting that HLA B*18 may in fact protect against late breast milk infection (Table 3). We also observed that both Cw*07 and Cw*08 were associated with late HIV-1 acquisition (RR = 4.0; 95% CI = 1.0–18.0 and RR = 7.2; 95% CI 1.2–37.3, respectively). High relative risk estimates and wide confidence intervals for these two alleles reflect the fact that few infants were infected after month 1 and may not represent the true effect of these alleles on HIV-1 acquisition risk (Table 3).
Table 3.
Risk of Infant HIV-1 Infection After Month 1 for Breast-fed Infants
| Number (%) HIV-1 infecteda
|
Relative risk (95% confidence interval)b | ||
|---|---|---|---|
| With allele | Without allele | ||
| A locus | |||
| A*01 | 2 (6%) | 9 (4%) | 1.96 (0.28, 8.89) |
| A*02 | 5 (5%) | 6 (4%) | 1.48 (0.39, 5.59) |
| A*29 | 1 (3%) | 10 (5%) | 0.78 (0.04, 4.72) |
| A*30 | 2 (3%) | 9 (5%) | 0.76 (0.11, 3.14) |
| A*6802 | 1 (2%) | 10 (5%) | 0.49 (0.03, 2.87) |
| p valuec,d | 0.86 | ||
| B locus | |||
| B*18 | 0 | 11 (5%) | — |
| B*53 | 3 (8%) | 8 (4%) | 1.80 (0.39, 6.37) |
| p valuec,d | 0.42 | ||
| C locus | |||
| Cw*04 | 27% | 25% | 1.72 (0.34, 7.02) |
| Cw*06 | 27% | 30% | 1.62 (0.32, 6.73) |
| Cw*07 | 55% | 34% | 3.98 (1.01, 17.78) |
| Cw*08 | 27% | 8% | 7.19 (1.23, 37.3) |
| p valuec,d | 0.16 | ||
Includes all 253 infants in the cohort of whom 11 were HIV-1 infected between months 1 and 12.
Includes 190 infants with maternal CD4 T cell count and HIV-1 viral load data at 32 weeks gestation. The analysis adjusts for these correlates.
p values for test of overall locus association compared to a model without allele information, when including the alleles listed above.
p values for test of overall locus association equal 0.63, 0.25, and 0.99 for the A locus, B locus, and C locus, respectively, when including all 62 alleles expressed in the cohort.
DISCUSSION
In this breast-feeding cohort in Nairobi, HLA B*18 was associated with protection against mother-to-child HIV-1 transmission before 1 month of age and between months 1 and 12 through breast milk exposure. These data support a role for HLA regulation of the infant immune response to HIV-1 infection and highlight the importance of genetic markers in the development and evaluation of antiviral interventions such as HIV-1 vaccines and pharmaceutical agents.
Several studies among highly exposed persistently seronegative adults in both Kenya and Thailand suggest that HLA B*18 is associated with decreased risk of heterosexual HIV-1 acquisition and that HIV-specific cytotoxic T lymphocyte (CTL) responses regulated by B*18 are responsible for its protective effect.18,19,26 Regional differences in HIV-1 subtypes and sub-sequent variability in important CTL epitopes may explain why other studies did not find an association between the B*18 haplotype and HIV-1 transmission.6,20 In Nairobi and Thailand, predominant subtypes are A and E, respectively, whereas in North America and Europe, subtype B is more common.6,20 In this study, we hypothesize that infants expressing HLA B*18 may be capable of mounting an effective early and late immune response against HIV-1 through presentation of HIV-1 antigens to infant CTLs. Other immune mechanisms, such as noncyto-toxic CD8+ T cells and natural killer (NK) cells, may also contribute to an HLA-mediated antiviral effect and control of HIV-1 infection.27,28
We observed trends for increased risk of early HIV-1 transmission for infants presenting HLA A*29 and increased risk of late transmission with Cw*07 and Cw*08. Increased HIV-1 acquisition risk may result from the inability of infants expressing these particular alleles to mount a protective response or to sustain effective immunity against HIV-1. Viral escape has been proposed as a mechanism for the decreased protection that has been observed in association with some HLA types.29 This occurs when infants express alleles associated with immune escape mutations and may be the case for HLA A*29. Adults with the A*29 haplotype have been found to progress rapidly to AIDS and death,17 a phenomenon associated with failure to maintain effective cellular immune control. Resistant HIV-1 variants may also be selected as a result of an effective maternal immune response and these may evade infant responses when infants mount the same immune responses due to shared haplotype. This explanation would be more likely for common alleles such as Cw*07, an allele with a prevalence of more than 30% in this cohort. Such alleles have a higher probability of being expressed as both maternal and paternal alleles, increasing the chance that the infant will share alleles with the mother.
In conclusion, specific HLA class I alleles in the A, B, and C loci influenced risk of infant HIV-1 acquisition before and after 1 month of age. These results may stimulate additional investigations into the mechanisms through which HLA polymorphisms affect mother-to-child HIV-1 transmission and may contribute to the development of effective epitope-based vaccines for this population.
Acknowledgments
Supported by the U.S. National Institutes of Health through Grants RO1 HD-23412 and K23 HD-41879. C. Farquhar, B. Lohman, J. Slyker, P. Otieno, and E. Obimbo were scholars in the AIDS International Training and Research Program supported by the National Institutes of Health/Fogarty International Center (D43-TW00007).
References
- 1.Kilpatrick DC, Hague RA, Yap PL, Mok JY. HLA antigen frequencies in children born to HIV-infected mothers. Dis Markers. 1991;9:21–26. [PubMed] [Google Scholar]
- 2.Just JJ, Abrams E, Louie LG, et al. Influence of host genotype on progression to acquired immunodeficiency syndrome among children infected with human immunodeficiency virus type 1. J Pediatr. 1995;127:544–549. doi: 10.1016/s0022-3476(95)70110-9. [DOI] [PubMed] [Google Scholar]
- 3.Just JJ, Louie LL, Abrams E, et al. Genetic risk factors for perinatally acquired HIV-1 infection. Paediatr Perinat Epidemiol. 1992;6:215–224. doi: 10.1111/j.1365-3016.1992.tb00762.x. [DOI] [PubMed] [Google Scholar]
- 4.Greggio NA, Cameran M, Giaquinto C, Zacchello F, Koroliuk D, Colizzi V. DNA HLA-DRB1 analysis in children of positive mothers and estimated risk of vertical HIV transmission. Dis Markers. 1993;11:21–26. doi: 10.1155/1993/292684. [DOI] [PubMed] [Google Scholar]
- 5.Winchester R, Chen Y, Rose S, Selby J, Borkowsky W. Major histocompatibility complex class II DR alleles DRB1*1501 and those encoding HLA-DR13 are preferentially associated with a diminution in maternally transmitted human immunodeficiency virus 1 infection in different ethnic groups: Determination by an automated sequence-based typing method. Proc Natl Acad Sci USA. 2001;92:12374–12378. doi: 10.1073/pnas.92.26.12374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Polycarpou A, Ntais C, Korber BT, et al. Association between maternal and infant class I and II HLA alleles and of their concordance with the risk of perinatal HIV type I transmission. AIDS Res Hum Retroviruses. 2002;18(11):741–746. doi: 10.1089/08892220260139477. [DOI] [PubMed] [Google Scholar]
- 7.MacDonald KS, Embree JE, Nagelkerke NJ, et al. The HLA A2/6802 supertype is associated with reduced risk of perinatal human immunodeficiency virus type 1 transmission. J Infect Dis. 2001;183:503–506. doi: 10.1086/318092. [DOI] [PubMed] [Google Scholar]
- 8.MacDonald KS, Embree J, Njenga S, et al. Mother-child class I HLA concordance increases perinatal human immunodeficiency virus type 1 transmission. J Infect Dis. 1998;177:551–556. doi: 10.1086/514243. [DOI] [PubMed] [Google Scholar]
- 9.Farquhar C, VanCott TC, Mbori-Ngacha DA, et al. Salivary secretory leukocyte protease inhibitor is associated with reduced transmission of human immunodeficiency virus type 1 through breast milk. J Infect Dis. 2002;186(8):1173–1176. doi: 10.1086/343805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nduati R, John G, Mbori-Ngacha D, et al. Effect of breastfeeding and formula feeding on transmission of HIV-1: A randomized clinical trial. JAMA. 2000;283(9):1167–1174. doi: 10.1001/jama.283.9.1167. [DOI] [PubMed] [Google Scholar]
- 11.Bunce M, O’Neill CM, Barnardo MC, et al. Phototyping: Comprehensive DNA typing for HLA-A, B, C, DRB1, DRB3, DRB4, DRB5 & DQB1 by PCR with 144 primer mixes utilizing sequence-specific primers (PCR-SSP) Tissue Antigens. 1995;46:355–367. doi: 10.1111/j.1399-0039.1995.tb03127.x. [DOI] [PubMed] [Google Scholar]
- 12.Bird TG, Kaul R, Rostron T, et al. HLA typing in a Kenyan cohort identifies novel class I alleles that restrict cytotoxic T-cell responses to local HIV-1 clades. AIDS. 2002;16(14):1899–1904. doi: 10.1097/00002030-200209270-00006. [DOI] [PubMed] [Google Scholar]
- 13.Steel CM, Ludlam CA, Beatson D, et al. HLA haplotype A1 B8 DR3 as a risk factor for HIV-related disease. Lancet. 1988;1(8596):1185–1188. doi: 10.1016/s0140-6736(88)92009-0. [DOI] [PubMed] [Google Scholar]
- 14.Brettle RP, McNeil AJ, Burns S, et al. Progression of HIV: Follow-up of Edinburgh injecting drug users with narrow serocon-version intervals in 1983–1985. AIDS. 1996;10(4):419–430. [PubMed] [Google Scholar]
- 15.Kaslow RA, Duquesnoy R, VanRaden M, et al. A1, Cw7, B8, DR3 HLA antigen combination associated with rapid decline of T-helper lymphocytes in HIV-1 infection. A report from the Multi-center AIDS Cohort Study. Lancet. 1990;335(8695):927–930. doi: 10.1016/0140-6736(90)90995-h. [DOI] [PubMed] [Google Scholar]
- 16.Grene E, Pinto LA, Cohen SS, et al. Generation of alloantigen-stimulated anti-human immunodeficiency virus activity is associated with HLA-A*02 expression. J Infect Dis. 2001;183:409–416. doi: 10.1086/318085. [DOI] [PubMed] [Google Scholar]
- 17.Hendel H, Caillat-Zucman S, Lebuanec H, et al. New class I and II HLA alleles strongly associated with opposite patterns of progression to AIDS. J Immunol. 1999;162:6942–6946. [PubMed] [Google Scholar]
- 18.Beyrer C, Artenstein A, Rugpao S, et al. Epidemiologic and biologic characterization of a cohort human immunodeficiency virus type 1 highly exposed, persistently seronegative female sex workers in northern Thailand. J Infect Dis. 1999;179:59–67. doi: 10.1086/314556. [DOI] [PubMed] [Google Scholar]
- 19.Rowland-Jones SL, Dong T, Fowke KR, et al. Cytotoxic T cell responses to multiple conserved HIV epitopes in HIV-resistant prostitutes in Nairobi. J Clin Invest. 1998;102(9):1758–1765. doi: 10.1172/JCI4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rohowsky-Kochan C, Skurnick J, Molinaro D, Louria D. HLA antigens associated with susceptibility/resistance to HIV-1 infection. Hum Immunol. 1998;59(12):802–815. doi: 10.1016/s0198-8859(98)00086-x. [DOI] [PubMed] [Google Scholar]
- 21.Carrington M, Nelson GW, Martin MP, et al. HLA and HIV-1: Heterozygote advantage and B*35-Cw*04 disadvantage. Science. 1999;283:1748–1752. doi: 10.1126/science.283.5408.1748. [DOI] [PubMed] [Google Scholar]
- 22.Panteleeff DD, John G, Nduati R, et al. Rapid method for screening dried blood samples on filter paper for human immunodeficiency virus type 1 DNA. J Clin Microbiol. 1999;37(2):350–353. doi: 10.1128/jcm.37.2.350-353.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Emery S, Bodrug S, Richardson BA, et al. Evaluation of performance of the GenProbe human immunodeficiency virus type 1 viral load assay using primary subtype A, C, and D isolates from Kenya. J Clin Microbiol. 2000;38(7):2688–2695. doi: 10.1128/jcm.38.7.2688-2695.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prentice RL, Storb R, Brown KS, Mason MV. HLA and disease: Relative-risk regression methods and multiple testing considerations. Biometrics. 1984;40(3):653–661. [PubMed] [Google Scholar]
- 25.Farewell VT, Dahlberg S. Some statistical methodology for the analysis of HLA data. Biometrics. 1984;40(2):547–560. [PubMed] [Google Scholar]
- 26.MacDonald KS, Fowke KR, Kimani J, et al. Influence of HLA supertypes on susceptibility and resistance to human immunodeficiency virus type 1 infection. J Infect Dis. 2000;181:1581–1589. doi: 10.1086/315472. [DOI] [PubMed] [Google Scholar]
- 27.Mackewicz CE, Garovoy MR, Levy JA. HLA compatibility requirements for CD8+-T-cell-mediated suppression of human immunodeficiency virus replication. J Virol. 1998;72(12):10165–10170. doi: 10.1128/jvi.72.12.10165-10170.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trinchiri G. Recognition of major histocompatibility complex class I antigens by natural killer cells. J Exp Med. 1994;180(2):417–421. doi: 10.1084/jem.180.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goulder PJ, Pasquier C, Holmes EC, et al. Mother-to-child transmission of HIV infection and CTL escape through HLA-A2-SLYNVATL epitope sequence variation. Immunol Lett. 2001;79(1–2):109–116. doi: 10.1016/s0165-2478(01)00272-3. [DOI] [PubMed] [Google Scholar]