Abstract
Background
The left and right atria have different susceptibilities towards developing arrhythmias, with left atrial arrhythmias more commonly observed. To understand the molecular basis for such differences, we catalogued miRNA and mRNA expression differences by next generation sequencing.
Methods and Results
Four human left-right atrial pairs were subjected to whole-genome expression analyses via next generation sequencing of small RNAs, including microRNAs (miRNAs), and poly-A enriched mRNAs. Using a paired sample design, significant differences in the expression of 32 miRNAs were found in between the left and right atria at a p-value of <0.01. Hsa-miR-143 was the most highly expressed miRNA in the atria, as quantified by RNA-seq. There were 746 and 2292 differentially expressed mRNAs between the left and right atria at false discovery rates of <0.001 and <0.05, respectively. Transcription factor binding elements within 2 kb of RefSeq genes were determined and specific motifs were identified that were enriched in differentially expressed genes. Similarly, specific miRNA target sequences in 3' UTRs were also enriched in differentially expressed genes. In addition eleven novel non-coding RNAs of unknown function were found to be differentially expressed between the left and right atria.
Conclusions
There are significant differences in miRNA and mRNA expression profiles between the left and right atria, which may yield insight into increased the arrhythmogenesis of the left atria.
Keywords: arrhythmia, atrial fibrillation, atrium, gene expression
Introduction and Background
The electrophysiological properties of the heart are determined by the expression of ion channels, gap junctions and other accessory proteins, for example, the expression of hyperpolarization-activated, cyclic nucleotide-gated ion channels (HCN2, HCN4) which contribute to the pacemaker activity of the sinoatrial node (1). The molecular basis of some differences in action potentials between the cardiac ventricles, atria, and SA node have been previously elucidated (2); however, left-to-right differences, particularly in the atria are less well characterized. This is important, as the right and left atrium have different susceptibilities towards developing arrhythmias, such as atrial fibrillation (AF). AF is the most common chronic cardiac arrhythmia, and it increases the risk of stroke and mortality in both males and females (3). Left-to-right asymmetries in potassium channel expression (4) may promote reentry and have a role in the development of atrial fibrillation (AF) (5). Genome wide association studies have identified some of the genetic factors that are associated with an increased risk of developing AF (6); however, the mechanisms by which these genetic factors lead to AF are not currently known. Specifically, the strongest and most widely replicated single nucleotide polymorphisms (SNPs) associated with AF lie in an intergenic region of chromosome 4q25, which may play an as yet undiscovered role in regulating the expression of nearby genes, such as PITX2, a gene known to be involved in cardiac development and left/right patterning (7).
Expression of micro-RNAs (miRNAs) has also been shown to play a crucial role in cardiac development and disease, in part through the attenuation of ion channel expression (8). miRNAs are RNA polymerase II transcribed non-coding RNAs that bind to target transcripts in a sequence specific manner. This binding results in the repression of translation and can lead to transcript degradation (9) resulting in lower steady state mRNA levels. miRNAs have been shown to play an important role in determining the core transcriptional network and thus may play a role in cell type differentiation (10). miRNAs such as mir-1, modulate a wide-array of transcripts important in cardiac function (11). Previously, many significant left-right differences in mRNA expression were found in mouse atria using microarrays (12). In the current study we utilized next generation sequencing to establish a comprehensive catalogue of differentially expressed transcripts and miRNAs in the human left and right atria, which can serve as the basis for further investigations into the genetic etiology of AF.
Methods
Tissue Procurement
Surgically obtained left and right atrial appendages were obtained from human subjects at the time of cardiac surgery that involved excision of this tissue. These patients provided informed consent for use of discarded tissue for research. Atrial appendage tissue was also obtained from donor hearts that were not found to be suitable for transplantation in the intended recipient. The donors were expired, and the donor's families provided consent for use of the donor tissues for research. The study was performed in concordance with an approved IRB protocol. All samples were stored at −80°C prior to RNA extraction.
RNA-sequencing for left-right pairs
Total RNA was extracted using QIAGEN miRNeasy Mini Kits from four paired 20 mg atrial tissue samples processed simultaneously. mRNA sequencing libraries were made following the Illumina mRNA protocol. Briefly, RNA was purified by poly-A selection using oligo(dT) beads and chemically fragmented. First and second strand cDNA synthesis was followed by end-repair and 3' adenylation. 5' and 3' Illumina adapters were ligated and size-selected using gel-purification and PCR amplification. All eight samples were 50 bp paired-end sequenced on a single flow-cell using the Illumina Genome Analyzer IIx. Using the same total RNA preparations, a small RNA-sequencing library was constructed following Illumina's Alternative v1.5 small RNA-seq protocol. Briefly, 5' and 3' adapters were ligated onto the total RNA and amplified. The cDNA with adapters was size selected, by agarose gel electrophoresis, between 93–100 bp and 36 bp single-end sequencing was performed on all eight samples on a single flow-cell. RNA reads are deposited in the GEO database, accession GSE31999.
Paired-end read analysis
mRNA paired-end reads were aligned using TopHat (13) to the UCSC hg19 build using Reference Sequence (RefSeq) as a guide (-G option). Counts at known RefSeq genes were generated from the read alignments using custom Python scripts, where a read and its read pair were only counted once if the read-pairs were mapped to within 6 standard deviations of the average fragment size across reads. Genes with expression lower than 10 or lower reads summed across the samples were thrown out from further analysis. Count data was loaded into the R package, EdgeR (14) and were normalized between samples using the trimmed mean of M-values (TMM), which calculates the average library size after trimming the top and bottom 5% expressed transcripts, and by trimming the transcripts with the top and bottom 30% log-fold changes (15). To take into account the left-right paired experimental design, Cox-Reid conditional inference was used to estimate the tagwise dispersion for each of the genes (14). A modified Fisher's t-test was then used to determine differential expression between the left and right atria (14). P-values were then adjusted by the method of Benjamini-Hochberg to derive FDRs (16).
Gene set enrichment was done using the romer function in the R-package limma. Romer uses a model that is better suited for microarray data compared to edgeR, which estimates the expression and biological variation from a negative binomial as edgeR does. However romer was used nonetheless because it accounts for the correlation structure of genes and uses a novel rotation approach for calculating p-values that is applicable to small studies, unlike the permutation approach used in other gene-set methods. A pseudo count was first created by adding 0.5 counts to the counts of each RefSeq transcript. Counts were then converted to reads per kilobase exon model per million mapped reads (RPKM), multiplied by the TMM normalization factor and log base 2 transformed. Gene sets were obtained from Molecular Signature Databases from The Broad Institute (17). A parametric resampling method for generalized linear models (18) was used to obtain p-values (9999 iterations).
Short read analysis
Sequenced reads from the small RNA libraries were first collapsed down to redundant reads while preserving count information, followed by removal of adapter sequence at the 3' end using FastX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/). Reads were then aligned to mirBase release 17 using bowtie (19) with the –v option set to one, allowing for one mismatch per read. Reads were then collapsed to the mature sequence using a custom Python script. This was done because the mature sequence is too short to align to; in addition we could easily quantify cis-expression from the hairpin alignments. Data was loaded into edgeR, and normalized using the TMM method described above. Differential expression was calculated in a method similar to differential analysis for the mRNA-seq data.
RT-PCR
A custom designed Taqman-primer and probe set was used for qRT-qPCR of the PITX2c transcript (Supplemental Table 1), normalized to cardiac actin (ACTC1) expression using a primer limited probe set (assay number Hs00606316_m1 from Applied Biosystems). RT-PCR reactions were run in duplex and relative PITX2c expression was calculated by the ΔΔCT method.
Results
RNA-seq of left-right atrial appendages
Left-right atrial appendage pairs were obtained from four human subjects, three of whom underwent bilateral Maze surgery for the treatment of atrial fibrillation and valve disease, while the fourth pair of atrial appendages was obtained postmortem from an unused heart transplant donor. Supplemental Table 2 describes the characteristics of these subjects. Total RNA was extracted from all samples. To determine left-right atrial gene expression differences, whole-genome expression analysis of RNA samples was performed using RNA-sequencing for both the small RNA fraction containing miRNAs (36 bp single-end sequencing) and the poly-A enriched mRNA fraction (51 bp paired-end sequencing). miRNA-seq yielded between 7.7 and 17.1 million reads per sample that mapped to known miRNAs in miRBase(20). mRNA-seq yielded between 16.3 and 29.9 million reads per sample that mapped to known Reference Sequence (RefSeq) transcripts; all mapped reads were collapsed to their unique RefSeq transcript regardless of potential splice isoforms in order to obtain digital counts. The full summary statistics of the sequencing results are described in Supplemental Tables 3 and 4.
miRNA gene expression differences between the left and the right atria
After trimming the Illumina adapters from the miRNA reads, the majority of the reads were distributed around 22 bp (Figure 1). Of the total number of reads, 24.5 ± 5.7% (mean ± SD) mapped uniquely to known miRNAs (miRBase release 17). In addition, many sequences that mapped to more than one genomic locus represent valid miRNAs, as determined by alignment to the hairpin pre-miRNA which were collapsed into unique mature miRNAs, such that the majority of 22 bp reads that were multiple-mapped were in fact miRNAs. Overall for the 8 samples, 39 ± 8% of the total reads were mapped to miRNAs (Supplemental Table 3). Most of longer 36-bp reads that did not read into the Illumina adapters mapped to annotated tRNAs and rRNAs. The most highly expressed miRNA in the atria was mir-143, which represented, on average, 32.7 ± 1.5% and 26.7 ± 1.8% of all mapped reads in the left and right atria, respectively (Supplemental Table 5). The miRNA dataset was subjected to multidimension scaling (MDS) (Figure 2a) showing that left-right sidedness is measurably associated with the miRNA transcriptome. Using a generalized liner model (GLM) in edgeR software (14) to fit the pairwise data followed by removal of miRNAs whose average expression over the total library size in the left and right atria fell below 7.63×10−06, 32 miRNAs were differentially expressed between the left and right atria at a P-value ≤ 0.01 and a false discovery rate (FDR) <0.08 (Table 1). Of the top 32 differentially expressed miRNAs, 18 were expressed more so in the left atria than in the right. For example, hsa-mir-135b had 4.9-fold higher expression in the right vs. left side (FDR=4.02×10−5). In contrast, hsa-mir-100 had 3.2-fold higher expression in the left side (FDR=5.91×10−9).
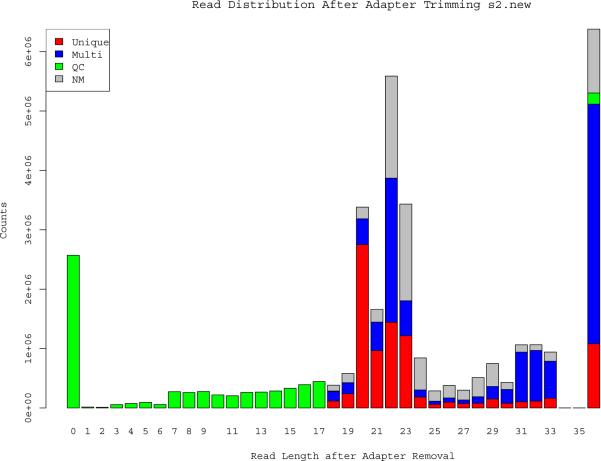
Figure 1.
Size-distribution of the small-RNA reads in one representative sample. The mode read length post adapter trimming occurs at 22bp which is expected from reads generated from miRNAs. The majority of sequences that did not read into the Illumina adapters aligned to tRNAs and rRNAs. Each read was classified as mapping uniquely to the human genome (Unique, red); having multiple alignments to the genome (Multi, blue); failing quality control (QC, green); or not mapping to the genome (NM, grey). The majority of the multiple aligned 20 – 23 bp reads correspond to miRNA families, which were collapsed into a single miRNA species for subsequent analyses.
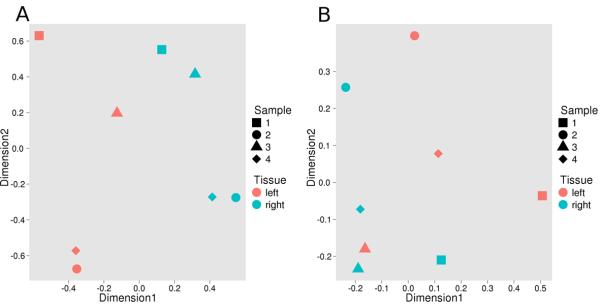
Figure 2.
Multidimension scaling (MDS) of gene expression differences between the left and right atria. A. MDS of the miRNA data reveals that the first dimension of miRNA expression imperfectly separated left (pink symbols) and right (blue symbols) atrial tissue, with the left atria tending to cluster on the left side of the plot. Different symbol shapes represent the four subjects, with sample 3 (triangle) representing the sinus rhythm donor. B. MDS of the mRNA data showing segregation of left and right atria in the first dimension.
Table 1.
Expression differences of miRNAs between the left and right atria at FDR <0.08 ranked by p-value.
| mirBase IDa | Atrial Concentrationb | Absolute Fold-Change | Expressed Higher | PValuec | FDRc |
|---|---|---|---|---|---|
| hsa-miR-10b | 6.05E-04 | 3.94 | Left | 2.13E-11 | 5.91E-09 |
| hsa-miR-100 | 1.38E-02 | 3.23 | Left | 3.50E-11 | 7.28E-09 |
| hsa-miR-135b | 8.58E-06 | 4.99 | Right | 5.79E-07 | 4.02E-05 |
| hsa-miR-487a | 1.38E-05 | 2.25 | Left | 8.77E-06 | 3.65E-04 |
| hsa-miR-4448 | 1.34E-05 | 2.28 | Left | 3.59E-05 | 1.30E-03 |
| hsa-miR-585 | 1.39E-05 | 2.27 | Right | 4.68E-05 | 1.56E-03 |
| hsa-miR-1275 | 1.58E-05 | 2.09 | Right | 7.83E-05 | 2.28E-03 |
| hsa-miR-483-5p | 2.43E-05 | 2.20 | Right | 8.08E-05 | 2.28E-03 |
| hsa-miR-4284 | 2.18E-05 | 2.45 | Left | 9.24E-05 | 2.39E-03 |
| hsa-miR-9 | 1.43E-04 | 1.92 | Left | 9.48E-05 | 2.39E-03 |
| hsa-miR-1973 | 1.89E-05 | 2.48 | Right | 1.05E-04 | 2.49E-03 |
| hsa-miR-125b-1* | 2.31E-05 | 1.90 | Left | 2.45E-04 | 4.75E-03 |
| hsa-miR-4497 | 3.63E-05 | 2.00 | Right | 2.88E-04 | 5.33E-03 |
| hsa-miR-425 | 9.38E-04 | 1.76 | Right | 6.25E-04 | 1.00E-02 |
| hsa-miR-125b | 6.58E-03 | 1.78 | Left | 6.27E-04 | 1.00E-02 |
| hsa-miR-92b | 8.89E-05 | 1.87 | Right | 8.18E-04 | 1.24E-02 |
| hsa-miR-150 | 1.25E-04 | 1.72 | Right | 1.16E-03 | 1.64E-02 |
| hsa-miR-708 | 5.80E-05 | 1.77 | Left | 1.18E-03 | 1.64E-02 |
| hsa-miR-495 | 1.51E-05 | 1.76 | Left | 1.64E-03 | 2.10E-02 |
| hsa-miR-3123 | 3.73E-05 | 2.04 | Left | 1.76E-03 | 2.22E-02 |
| hsa-miR-24-1* | 1.19E-04 | 1.78 | Left | 1.95E-03 | 2.39E-02 |
| hsa-miR-202* | 9.28E-06 | 1.86 | Left | 1.98E-03 | 2.39E-02 |
| hsa-miR-155 | 1.03E-04 | 1.72 | Right | 2.20E-03 | 2.48E-02 |
| hsa-miR-376c | 6.82E-05 | 1.67 | Right | 4.10E-03 | 4.17E-02 |
| hsa-miR-4792 | 2.89E-04 | 1.84 | Left | 4.75E-03 | 4.71E-02 |
| hsa-miR-766 | 1.01E-05 | 1.67 | Left | 5.05E-03 | 4.95E-02 |
| hsa-miR-675* | 1.94E-05 | 1.64 | Right | 7.15E-03 | 6.70E-02 |
| hsa-miR-378f | 1.06E-04 | 1.85 | Left | 7.61E-03 | 6.92E-02 |
| hsa-miR-133a | 1.11E-02 | 1.63 | Left | 8.95E-03 | 7.77E-02 |
| hsa-miR-423-5p | 1.10E-03 | 1.56 | Left | 9.22E-03 | 7.92E-02 |
| hsa-miR-146a | 6.24E-04 | 1.55 | Right | 9.40E-03 | 7.99E-02 |
miR-X* names represent the miR*or passenger strand of the primary miRNA stem-loop transcript.
Average fraction of total mapped miRNA reads in the left and right atria.
P values and FDRs are based on the EdgeR pairwise analysis.
mRNA gene expression differences between the left and the right atria
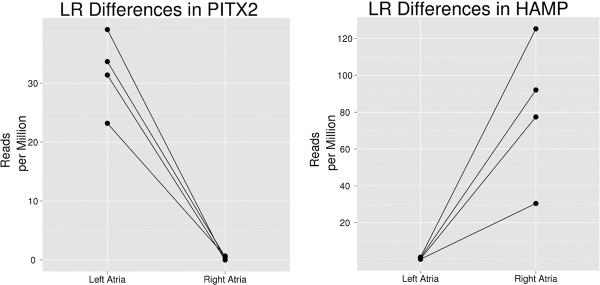
The majority of mRNA reads mapped to the human reference genome hg19 (85% ± 6.2) and more specifically to known RefSeq genes (67.7± 10.6 %). MDS plot of the transcripts showed separation between the left and right atria in dimension 1. (Figure 2b). 746 genes were called differentially expressed between the left and right atria at the stringent FDR of 0.001 with 305 genes over-expressed in the left atria and 441 genes over-expressed in the right atria. The top 20 differentially expressed genes ranked by p-value are shown in Table 2. PITX2 had 116-fold higher expression in the left vs. right atria with the 3rd most significant p-value (p = 8.72×10−68), accounting for 0.0032% of reads mapping to the transcriptome in the left atria, without appreciable expression in right atrial samples (Figure 3). Of the 6 RefSeq annotated PITX2 RNA isoforms, only those specific to PITX2c isoform were found (Supplemental Figure 1), although 3' library generation bias might under represent expression of other transcripts. To validate the PITX2 sequence results, we performed an internally-normalized quantitative RT-PCR using a TaqMan expression assay specific for PITX2c on 19 additional left right atrial pairs (17 surgical samples and 2 unused donor hearts). PITX2c was expressed 232 ± 165-fold (mean ± SD) higher in the left vs. right atria, confirming the RNA-seq results. In contrast, HAMP, encoding hepcidin antimicrobial protein, was expressed ~121-fold higher in the right vs. left atria (Figure 3). BMP10, a gene known to be down-regulated by PITX2c(21), had~282-fold higher expression in the right atria(Table 2). Two cardiomyocyte specific transcripts were differentially expressed among the top 25 significant genes: MYL2, a slow cardiac myosin regulatory light chain was expressed 10 fold higher in the left atria (p = 5.11×10−33), and HCN4, a pacemaker ion-channel was expressed more than 7 fold higher in the right atria (p = 1.28×10−24).
Table 2.
The top 20 left-right differentially expressed atrial genes ranked by p-value.
| Gene Symbol | Atrial Concentration* | Absolute Fold-Change | Expressed Higher | P-Value† | FDR† |
|---|---|---|---|---|---|
| HAMP | 6.42E-05 | 121.18 | Right | 1.55E-111 | 2.77E-107 |
| BMP10 | 5.51E-04 | 282.13 | Right | 1.39E-096 | 1.24E-092 |
| PITX2 | 9.19E-06 | 116.33 | Left | 8.72E-072 | 5.20E-068 |
| C2orf14 | 4.32E-06 | 126.97 | Right | 1.15E-048 | 5.13E-045 |
| C19orf33 | 4.83E-06 | 20.81 | Left | 2.70E-039 | 9.66E-036 |
| LOC100144602 | 1.60E-06 | 106.15 | Left | 1.32E-033 | 3.95E-030 |
| MYL2 | 2.08E-04 | 10.35 | Left | 5.11E-033 | 1.31E-029 |
| BDKRB1 | 3.61E-06 | 41.17 | Left | 1.10E-031 | 2.46E-028 |
| SALL1 | 2.57E-06 | 28.67 | Right | 6.53E-031 | 1.30E-027 |
| DNASE1L3 | 4.95E-06 | 12.47 | Right | 8.28E-031 | 1.48E-027 |
| KRT7 | 7.34E-06 | 14.19 | Left | 1.53E-030 | 2.48E-027 |
| FAM84A | 4.89E-06 | 17.27 | Right | 5.67E-030 | 8.46E-027 |
| IRX3 | 1.07E-05 | 8.39 | Right | 3.75E-029 | 5.16E-026 |
| THBS4 | 5.75E-05 | 9.54 | Left | 2.55E-027 | 3.27E-024 |
| ANKRD30BL | 2.26E-06 | > 1000 | Left | 1.97E-026 | 2.17E-023 |
| SYT4 | 7.43E-06 | 27.11 | Left | 2.05E-026 | 2.17E-023 |
| ALOX15 | 1.85E-05 | 15.03 | Left | 2.06E-026 | 2.17E-023 |
| CLDN18 | 8.92E-06 | 12.62 | Left | 5.19E-026 | 5.16E-023 |
| RBP4 | 1.50E-05 | 6.52 | Left | 1.10E-024 | 1.03E-021 |
| HCN4 | 1.99E-05 | 7.53 | Right | 1.28E-024 | 1.14E-021 |
Average fraction of total mapped miRNA reads in the left and right atria.
P values and FDRs are based on the EdgeR pairwise analysis.
Figure 3.
HAMP and PITX2 display inverse expression patterns between left and right atria. Data are normalized to one million reads per library. The lines connect each subject's paired samples.
To better characterize the differences between the left and right atria, gene set enrichment was performed using the molecular signatures database (MSigDB) from The Broad Institute (17) and testing was done with the competitive geneset test function romer from the limma R-package (18)(22). Various genes relating to the Gene Ontology (GO) term signal transduction and transcription were significantly up-regulated in the right atria compared to the left. For example, of the 1391 atrial-expressed RefSeq genes belonging to the signal transduction pathway, 205 had higher expression in the right atria in our prior pairwise edgeR analysis and 94 in the left, a significant enrichment (p= 0.0001 by re-sampling in limma, Supplemental Table 6). The mitochondrion is an example of a GO term geneset that was enriched in genes that were more highly expressed in the left atria (P= 0.0001). Various additional GO sets relating to the mitochondria were significantly enriched in the left atria, including components of the NADH dehydrogenase chain, mitochondrial matrix, and mitochondrial ribosomal complex (data not shown).
Using gene sets generated from shared conserved cis-regulatory motifs from the transcription factor targets (C3_TFT) database of MSigDB (23), genes containing or adjacent to several transcription factor binding motifs, or conserved motifs not yet associated with a specific transcription factor, were found to be enriched in right or left atria expression. Table 3 shows the list of 21 and 16 motifs that were associated with gene sets enriched in the right and left atria, respectively (at p=0.0001, with at least 5 genes enriched in one side). For example, 163 atrial-expressed RefSeq genes were close to a p300 binding element and this set of genes was expressed at significantly higher levels in the right vs. left atria (p=0.0001 by re-sampling analysis). We compared this atrial-expressed p300 motif containing geneset with our prior pairwise edgeR analysis of differentially expressed genes and found that out of these 163 genes, 25 genes were more highly expressed in the right atria, versus 7 genes in the left atria.
Table 3.
Top left-right atria differentially regulated genesets by presence of transcription factor binding motifs ± 2 kb from the start site of transcription of atrial expressed genes.
| Motif | Right* | Left* | Hypothesis |
|---|---|---|---|
| V$AP4_01 | 19 | 10 | Right |
| AACTTT_UNKNOWN | 195 | 74 | Right |
| AACWWCAANK_UNKNOWN | 10 | 1 | Right |
| V$P300_01 | 25 | 7 | Right |
| V$CREBP1_01 | 14 | 6 | Right |
| V$MYOGNF1_01 | 6 | 3 | Right |
| V$IRF1_01 | 23 | 9 | Right |
| V$IRF2_01 | 10 | 5 | Right |
| V$TAL1BETAE47_01 | 24 | 12 | Right |
| V$TAL1ALPHAE47_01 | 24 | 11 | Right |
| V$HEN1_01 | 21 | 12 | Right |
| V$TAL1BETAITF2_01 | 26 | 14 | Right |
| V$GATA3_01 | 24 | 13 | Right |
| V$EVI1_04 | 34 | 6 | Right |
| V$EVI1_05 | 17 | 7 | Right |
| V$MZF1_02 | 19 | 6 | Right |
| V$ZID_01 | 21 | 8 | Right |
| V$IK2_01 | 28 | 7 | Right |
| V$CDP_01 | 7 | 4 | Right |
| ACCTGTTG_UNKNOWN | 15 | 5 | Right |
| V$ELK1_02 | 3 | 8 | Right |
| V$CREB_01 | 14 | 16 | Left |
| V$EVI1_02 | 7 | 7 | Left |
| V$NRF2_01 | 3 | 8 | Left |
| ACTAYRNNNCCCR_UNKNOWN | 9 | 10 | Left |
| V$USF_C | 12 | 14 | Left |
| V$PPARA_01 | 3 | 4 | Left |
| V$ATF_B | 11 | 13 | Left |
| V$GABP_B | 3 | 8 | Left |
| V$TEL2_Q6 | 14 | 4 | Left |
| V$SF1_Q6 | 19 | 13 | Left |
| V$COUP_DR1_Q6 | 13 | 11 | Left |
| GGAANCGGAANY_UNKNOWN | 0 | 9 | Left |
| V$CREB_Q4_01 | 11 | 12 | Left |
| V$AP4_Q6_01 | 16 | 23 | Left |
| V$ER_Q6_02 | 18 | 25 | Left |
Inclusion in this table was performed by filtering genesets that contained at least 5 genes that were significantly up regulated in the right or left atria, and by having the minimum p-value (10−4) under the model-free hypothesis including all genes near the specified DNA element.
Number of genes from the geneset that are contained within those found to be expressed significantly higher in the left or right atria via the edgeR analysis (FDR < 0.05), not including those with moderate expression bias.
We repeated this analysis using shared conserved miRNA binding motifs in the 3' UTR of atrial expressed genes. We found 16 and 1 miRNA motif genesets that were expressed higher in the right and left atria, respectively (p=0.0001, with at least 5 genes enriched in one side, Table 4). For example, 165 atrial-expressed genes contained a miR-133a binding site in their 3' UTRs, and this set of genes was expressed significantly higher in the right vs. left atria (p=0.0001 by re-sampling analysis). This corresponds well with the observed higher expression of mir133a in the left vs. right atria (1.63-fold difference, Table 1), leading to decreased expression of its mRNA target genes, resulting in a higher right-sided expression for these targets.
Table 4.
Top left-right atria differentially regulated genesets by presence of conserved miRNA motif in the 3' UTR of atrial expressed genes.
| Motif | Right* | Left* | Hypothesis |
|---|---|---|---|
| CACTGCC,MIR-34A,MIR-34C,MIR-449 | 20 | 13 | Right |
| GGGACCA,MIR-133A,MIR-133B | 11 | 1 | Right |
| ATGCTGC,MIR-103,MIR-107 | 18 | 7 | Right |
| AGGGCCA,MIR-328 | 8 | 1 | Right |
| CTGAGCC,MIR-24 | 17 | 7 | Right |
| CCTGTGA,MIR-513 | 11 | 5 | Right |
| GGCAGCT,MIR-22 | 24 | 13 | Right |
| CCTGCTG,MIR-214 | 19 | 3 | Right |
| GACAGGG,MIR-339 | 8 | 2 | Right |
| GAGCCAG,MIR-149 | 16 | 5 | Right |
| CCCAGAG,MIR-326 | 12 | 1 | Right |
| GCAAGGA,MIR-502 | 10 | 3 | Right |
| CAGTCAC,MIR-134 | 8 | 0 | Right |
| TCCAGAG,MIR-518C | 14 | 4 | Right |
| CACCAGC,MIR-138 | 17 | 5 | Right |
| CAGCCTC,MIR-485-5P | 16 | 3 | Right |
| CCATCCA,MIR-432 | 4 | 6 | Left |
Genesets were filtered to include only those that contained at least 5 that were significantly up regulated in the right or left atria, and by having the minimum p-value (10−4) under the model-free hypothesis.
Number of genes from the geneset that are contained within those found to be expressed higher in the left or right atria via the edgeR analysis (FDR < 0.05).
We examined AF and PR interval (the time difference between sinus atrial node firing and atrioventricular node firing measured by electrocardiogram) GWAS hits and found three adjacent coding genes, PITX2, SULF2, and WNT11, that had significant left-right atrial gene expression differences (Supplemental Table 7), suggesting that these genes may play a functional role in AF pathogenesis.
Left-right expression differences in poorly annotated transcripts
Cufflinks (24) was used to assemble transcripts from aligned sequencing reads in order to identify potentially novel transcripts. 521 transcripts were found to be differentially expressed that were either classified as novel or considered a novel processed transcript by the Ensembl database (release number 61). After manual curation to filter out alternative splice variants of known genes and very lowly expressed transcripts (i.e. that were expressed below 5 fragments per kilobase of the transcript per million mapped reads of the transcriptome), thirteen novel transcripts, ranging from 2 to 4 exons, were found to be differentially expressed that were not found in the Ensembl annotation, all of which appear to function as non-coding RNAs based upon in silico analysis. As a check to ensure that the transcripts did not arise due to misalignment of sequencing reads, eleven of the novel transcripts had 95–100% of the sequencing reads mapping uniquely to the genome in an exon contiguous fashion using BLAT (25) (Table 5), with the other two mapping to multiple sites. The most significantly differentially expressed novel transcript (p<10−16) is adjacent to the TBX5 gene, a heart-specific transcription factor.
Table 5.
Novel non-Ensembl annotated transcripts that were differentially expressed between the left and right atria ranked by p-value.
| Transcript boundary | Nearest gene | Exons | Absolute Fold Change | Expressed Higher | p-value |
|---|---|---|---|---|---|
| chr12:114883593-114885094 | TBX5 | 2 | 2.01 | Right | ≤10−16 |
| chr6:36810354-36812333 | CPNE5 | 2 | 1.84 | Left | ≤10−16 |
| chr12:58325277-58329330 | XRCC6BP1 | 2 | 6.69 | Right | 2.2E-16 |
| chr2:27939711-27961143 | SLC4A1 | 2 | 2.94 | Left | 1.4E-14 |
| chr2:50999236-51003553 | NRXN1 * | 2 | 1.77 | Left | 1.2E-12 |
| chr19:50989460-51003531 | JOSD2 | 3 | 4.95 | Right | 1.1E-04 |
| chr15:25243485-25247622 | SNOR108 | 4 | 2.32 | Right | 4.7E-04 |
| chr19:50990647-50999173 | C19orf63 | 2 | 1.75 | Right | 7.6E-04 |
| chr16:58467328-58496760 | NDRG4 | 3 | 1.58 | Left | 1.2E-03 |
| chr13:114054026-114066562 | ADPRHL1 | 2 | 1.39 | Left | 2.5E-03 |
| chr19:50991540-51005189 | JOSD2 | 2 | 3.13 | Left | 1.5E-02 |
Transcript is intronic
In order to determine if the one atrial donor sample (#3) significantly affected the results, we repeated the analysis after removing this sample. The top differentially expressed miRNAs, mRNAs and non-coding RNAs were largely conserved (Supplemental Tables 8–10).
Discussion
We found large differences in miRNA and mRNA expression levels between the left and the right atria in our genome-wide analysis of total RNA expression, including the well-known left-right patterning genes PITX2 and BMP10. Of the 17 named genes in our top 20 differentially expressed transcripts (Table 2), six have recently been validated to be differentially expressed by microarray and/or RT-PCR in the same direction in either mouse and/or human atria, including the top three transcripts HAMP, BMP10, PITX2 (26). Several novel non-coding messenger RNAs were asymmetrically expressed as well. These data suggest that the left and the right atria have significantly different gene expression profiles, which may have electrophysiological and pathophysiological consequences. Although a limitation of this study is the combination of diseased and non-diseased subjects, our results were largely conserved after elimination of the normal donor (Supplemental Tables 8–10). Differences in transcription between AF and non-AF in the left atria are of great interest, but we were not powered to detect these differences.
The top locus associated with AF maps to an intergenic region on chromosome 4q25, and PITX2 is the closest adjacent gene. Previous work in humans has shown that PITX2c is expressed primarily in the left atria (26)(27). We confirmed this result and found practically no PITX2c expression in the right atria, nor did we detect expression of any other PITX2 isoform in the left atria. Kirchhof et al. reported that Pitx2c was the only isoform expressed In mouse atria, and that it was expressed only in the left atria (12), agreeing with our data in humans. However, a prior study in humans detected multiple isoforms of PITX2 in both atria using RT-qPCR (28). RNA-seq has been reported to be more reliable for low-abundance transcripts compared to RT-qPCR(29); although, our finding of no other PITX2 isoforms may be due to insufficient read coverage to detect transcripts expressed at less than 10 mRNA copies per million mRNA molecules. In contrast to PITX2c, BMP10, a transcript known to be directly repressed by PITX2, was expressed only in the right atria agreeing with recently published data(26). PITX2 has been reported to repress the expression of hsa-mir-1-1(27), although our sample size is small we observed a trend for an inverse correlation between PITX2 expression and hsa-mir-1-1 expression in the left atria (r= −0.84, p= 0.099). However, SHOX2, TBX3 and NKX2-5, other atrial expressed genes known to be regulated by PITX2c (28)(30), showed no difference in expression levels between the left and the right atria (data not shown). PITX2 expression is high throughout the left atria just after birth in mice, but it is only expressed in a small subset of left atrial cells in adult mice (28). We speculate that developmental epigenetic influences or other overriding transcription factors might also regulate SHOX2, TBX3 and NKX2-5 expression in the adult atria diminishing the effect of PITX2. Although PITX2c is expressed only at low levels in these adult samples, the differential expression of PITX2c and BMP10 reported here suggests that in adults these factors may play a continuing role, potentially in the pathogenesis of AF.
In addition to genes with well-known roles in cardiac physiology, we found that the HAMP gene was expressed exclusively in the right atria. HAMP encodes hepcidin antimicrobial protein, a protein that is mainly produced by the liver and that controls iron absorption in the intestine. Mutations in HAMP cause hemochromatosis type 2B leading to iron overload in many organ systems. Furthermore, hemochromatosis type 2B often presents with cardiomyopathy, heart failure, and/or major arrhythmias which are a prominent cause of death in the absence of treatment, suggesting that HAMP may have a local role in iron regulation in the heart(31).
Hsa-miR-143 was the most abundantly expressed miRNA in human atria in our analysis, accounting for ~30% of all mapped miRNAs reads in the atria. However, it was previously reported, by sequencing a library of cloned miRNA cDNAs, that the most abundant miRNA expressed in the mouse heart is mir-1, accounting for 45% of miRNA expression(32). There are several potential explanations for this discrepancy. First, we specifically looked at miRNA expression in the atria and not the whole-heart, and it has been shown that the atria and ventricles have vastly different expression profiles(33). Second, miRNA expression in the heart may not be conserved across species; for example, cardiac p300 binding site enhancer regions are weakly conserved between mice and humans(34). The high expression level of miR-143 in the atria is concordant with its role in development in mice, where it has been shown to play a critical role in the formation of the outflow tract by repressing KLF4 and promoting differentiation(35). Recent work in zebrafish has shown that mir-143 expression during cardiogenesis is dependent on the beating of the heart; although, mir-143 is not expressed in zebrafish atria, but rather in the outflow tract and ventricles(36). mir-143 has also been shown to be critical for cardiac chamber formation through the direct repression of adducin3, an F-actin capping protein in zebrafish(37).
We found increased expression of mir-133 in the left atria and a corresponding decrease in gene expression of mir-133 targets in the left atria. mir-133 specifies a cardiac progenitor lineage by preventing the expression of non-muscle genes, but it also inhibits further differentiation into cardiomyocytes (38)(39), which may suggest a more progenitor like state of the left atria when compared to the right atria.
Polyadenlyated long intergenic non-coding RNAs (lincRNAs) have been shown to regulate gene expression in trans by acting as scaffolds for chromatin modifying proteins such as the polycomb repressor complex 2 (PRC2), which regulate gene expression via histone modification(40). siRNA mediated down regulation of specific PRC2 associated lincRNAs, such as HOTAIR, leads to the up regulation of a specific set of ~100 to 300 PRC2 repressed genes. We found a conserved lincRNA that was differentially expressed between the left and right atria located 47 kb up-stream the gene encoding transcription factor TBX5. There are 2 SNPs near TBX5 associated with PR interval(41), which may be relevant to AF pathogenesis. We speculate that the lincRNA adjacent to TBX5 may also be regulated by these SNPs, altering expression of downstream genes through chromatin remodeling. We found many lincRNAs were expressed in the heart, suggesting the potential for widespread cardiac regulation of gene expression by these non-coding RNAs.
Conclusions
RNA-sequencing provides a comprehensive assessment of gene expression, using a method that is more sensitive for lowly expressed transcripts than microarrays(42). Using RNA-sequencing, we demonstrated signatures of miRNA and mRNA gene expression that distinguish human left atria from right atria. In addition we identified expression differences for transcripts that have not been annotated and are not present on microarrays. This catalog of left and right atrial gene expression should be useful for the functional determination of how SNPs associated with AF actually increase AF susceptibility.
Supplementary Material
AF is the most common chronic cardiac arrhythmia, and AF is associated with increased risk of stroke and mortality. While the heart beat originates in the right atrium, ectopic electrical activity that initiates AF frequently comes from the pulmonary veins that enter the left atrium. Altered function is often related to changes in gene expression. Here we show differences in expression of many transcripts by RNA-sequencing of both small RNAs and mRNAs between paired left and right atrial samples. We confirm higher expression in the left atria of the PITX2C mRNA, which is adjacent to the locus most highly associated with AF in genome-wide association studies (GWAS). PITX2 is a transcription factor that is critical in left-right patterning. In addition, we find many long intergenic noncoding RNA differences between the left and right atria that may lead to functional differences between the two chambers, including a novel transcript adjacent to TBX5, a locus also implicated by GWAS. Our study provides a catalog of molecular differences between these two chambers and serves as a useful resource for investigators. In future studies, it will be of interest to evaluate the impact of age, disease and genetics on the gene expression differences between the left and right atria.
Acknowledgments
Funding Sources: This work was supported by NIH National Heart, Lung and Blood Institute grant HL090620 (Chung, Barnard, Smith, and Van Wagoner). It was also supported in part by NIH National Center for Research Resources (NCRR) Case Western Reserve University/Cleveland Clinic grant CTSA UL1-RR024989 (Chung, Van Wagoner); the Heart and Vascular Institute, Department of Cardiovascular Medicine, Cleveland Clinic (Chung); Leducq Foundation grant 07-CVD 03 (Van Wagoner, Chung); and the Atrial Fibrillation Innovation Center, State of Ohio (Van Wagoner, Chung). Jeffrey Hsu was supported by the Howard Hughes Med Into Grad Scholar program, and by the Molecular Medicine training grant from NIH National Institute of General Medical Sciences T32GM088088.
Footnotes
Journal Subject Codes: [5] Arrhythmias, clinical electrophysiology, drugs; [155] Physiological and pathological control of gene expression
Conflict of Interest Disclosures: None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ludwig A, Zong X, Stieber J, Hullin R, Hofmann F, Biel M. Two pacemaker channels from human heart with profoundly different activation kinetics. EMBO J. 1999;18:2323–2329. doi: 10.1093/emboj/18.9.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schram G, Pourrier M, Melnyk P, Nattel S. Differential Distribution of Cardiac Ion Channel Expression as a Basis for Regional Specialization in Electrical Function. Circ Res. 2002;90:939–950. doi: 10.1161/01.res.0000018627.89528.6f. [DOI] [PubMed] [Google Scholar]
- 3.Benjamin EJ, Wolf PA, D'Agostino RB, Silbershatz H, Kannel WB, Levy D. Impact of atrial fibrillation on the risk of death: the Framingham Heart Study. Circulation. 1998;98:946–952. doi: 10.1161/01.cir.98.10.946. [DOI] [PubMed] [Google Scholar]
- 4.Voigt N, Trausch A, Knaut M, Matschke K, Varró A, Van Wagoner DR, et al. Left-to-right atrial inward rectifier potassium current gradients in patients with paroxysmal versus chronic atrial fibrillation. Circ Arrhythm Electrophysiol. 2010;3:472–80. doi: 10.1161/CIRCEP.110.954636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Atienza F, Almendral J, Moreno J, Vaidyanathan R, Talkachou A, Kalifa J, et al. Activation of inward rectifier potassium channels accelerates atrial fibrillation in humans: evidence for a reentrant mechanism. Circulation. 2006;114:2434–42. doi: 10.1161/CIRCULATIONAHA.106.633735. [DOI] [PubMed] [Google Scholar]
- 6.Gudbjartsson DF, Arnar DO, Helgadottir A, Gretarsdottir S, Holm H, Sigurdsson A, et al. Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature. 2007;448:353–357. doi: 10.1038/nature06007. [DOI] [PubMed] [Google Scholar]
- 7.Galli D, Domínguez JN, Zaffran S, Munk A, Brown N a, Buckingham ME. Atrial myocardium derives from the posterior region of the second heart field, which acquires left-right identity as Pitx2c is expressed. Development. 2008;135:1157–1167. doi: 10.1242/dev.014563. [DOI] [PubMed] [Google Scholar]
- 8.Saxena A, Tabin CJ. miRNA-processing enzyme Dicer is necessary for cardiac outflow tract alignment and chamber septation. Proc Natl Acad Sci USA. 2010;107:87–91. doi: 10.1073/pnas.0912870107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, et al. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science (80-) 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 10.Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Z, Lu Y, Yang B. MicroRNAs and atrial fibrillation: new fundamentals. Cardiovasc Res. 2011;89:710–721. doi: 10.1093/cvr/cvq350. [DOI] [PubMed] [Google Scholar]
- 12.Kirchhof P, Kahr PC, Kaese S, Piccini I, Vokshi I, Scheld H-H, et al. PITX2c is Expressed in the Adult Left Atrium, and Reducing Pitx2c Expression Promotes Atrial Fibrillation Inducibility and Complex Changes in Gene Expression. Circ Cardiovasc Genet. 2011;4:123–133. doi: 10.1161/CIRCGENETICS.110.958058. [DOI] [PubMed] [Google Scholar]
- 13.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57:289–300. [Google Scholar]
- 17.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette M a, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Majewski IJ, Ritchie ME, Phipson B, Corbin J, Pakusch M, Ebert A, et al. Opposing roles of polycomb repressive complexes in hematopoietic stem and progenitor cells. Blood. 2010;116:731–739. doi: 10.1182/blood-2009-12-260760. [DOI] [PubMed] [Google Scholar]
- 19.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32:D109–11. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tessari A, Pietrobon M, Notte A, Cifelli G, Gage PJ, Schneider MD, et al. Myocardial Pitx2 differentially regulates the left atrial identity and ventricular asymmetric remodeling programs. Circ Res. 2008;102:813–22. doi: 10.1161/CIRCRESAHA.107.163188. [DOI] [PubMed] [Google Scholar]
- 22.Wu D, Lim E, Vaillant F, Asselin-Labat M-L, Visvader JE, Smyth GK. ROAST: rotation gene set tests for complex microarray experiments. Bioinformatics. 2010;26:2176–2182. doi: 10.1093/bioinformatics/btq401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, et al. Systematic discovery of regulatory motifs in human promoters and 3' UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trapnell C, Williams B a, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:516–520. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kent WJ. BLAT — The BLAST-Like Alignment Tool. Genome Res. 2002;12:656. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kahr PC, Piccini I, Fabritz L, Greber B, Schöler H, Scheld HH, et al. Systematic Analysis of Gene Expression Differences between Left and Right Atria in Different Mouse Strains and in Human Atrial Tissue. PLoS ONE. 2011;6:e26389. doi: 10.1371/journal.pone.0026389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chinchilla A, Daimi H, Lozano-Velasco E, Dominguez JN, Caballero R, et al. PITX2 Insufficiency Leads to Atrial Electrical and Structural Remodeling Linked to Arrhythmogenesis. Circ Cardiovasc Genet. 2011;4:269–279. doi: 10.1161/CIRCGENETICS.110.958116. [DOI] [PubMed] [Google Scholar]
- 28.Wang J, Klysik E, Sood S, Johnson RL, Wehrens XHT, Martin JF. Pitx2 prevents susceptibility to atrial arrhythmias by inhibiting left-sided pacemaker specification. Proc Natl Acad Sci USA. 2010;107:9753–9758. doi: 10.1073/pnas.0912585107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matkovich SJ, Zhang Y, Van Booven DJ, Dorn GW. Deep mRNA sequencing for in vivo functional analysis of cardiac transcriptional regulators: application to Galphaq. Circ Res. 2010;106:1459–1467. doi: 10.1161/CIRCRESAHA.110.217513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Faucourt M, Houliston E, Besnardeau L, Kimelman D, Lepage T. The pitx2 homeobox protein is required early for endoderm formation and nodal signaling. Dev Biol. 2001;229:287–306. doi: 10.1006/dbio.2000.9950. [DOI] [PubMed] [Google Scholar]
- 31.Roetto A, Papanikolaou G, Politou M, Alberti F, Girelli D, Christakis J, et al. Mutant antimicrobial peptide hepcidin is associated with severe juvenile hemochromatosis. Nat Genet. 2003;33:21–22. doi: 10.1038/ng1053. [DOI] [PubMed] [Google Scholar]
- 32.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 33.Barth AS, Merk S, Arnoldi E, Zwermann L, Kloos P, Gebauer M, et al. Functional profiling of human atrial and ventricular gene expression. Pflugers Arch. 2005;450:201–208. doi: 10.1007/s00424-005-1404-8. [DOI] [PubMed] [Google Scholar]
- 34.Blow MJ, McCulley DJ, Li Z, Zhang T, Akiyama JA, Holt A, et al. ChIP-Seq identification of weakly conserved heart enhancers. Nat Genet. 2010;42:818–822. doi: 10.1038/ng.650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, et al. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:705–710. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miyasaka KY, Kida YS, Banjo T, Ueki Y, Nagayama K, Matsumoto T, et al. Heartbeat regulates cardiogenesis by suppressing retinoic acid signaling via expression of miR-143. Mech Dev. 2010;128:18–28. doi: 10.1016/j.mod.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 37.Deacon DC, Nevis KR, Cashman TJ, Zhou Y, Zhao L, Washko D, et al. The miR-143-adducin3 pathway is essential for cardiac chamber morphogenesis. Development. 2010;137:1887–1896. doi: 10.1242/dev.050526. [DOI] [PubMed] [Google Scholar]
- 38.Ivey KN, Muth A, Arnold J, King FW, Yeh R-F, Fish JE, et al. MicroRNA regulation of cell lineages in mouse and human embryonic stem cells. Cell Stem Cell. 2008;2:219–229. doi: 10.1016/j.stem.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kato Y, Miyaki S, Yokoyama S, Omori S, Inoue A, Horiuchi M, Asahara H. Real-time functional imaging for monitoring miR-133 during myogenic differentiation. Int J Biochem Cell Biol. 2009;41:2225–2231. doi: 10.1016/j.biocel.2009.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holm H, Gudbjartsson DF, Arnar DO, Thorleifsson G, Thorgeirsson G, Stefansdottir H, et al. Several common variants modulate heart rate, PR interval and QRS duration. Nat Genet. 2010;42:117–122. doi: 10.1038/ng.511. [DOI] [PubMed] [Google Scholar]
- 42.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.