Abstract
Mitochondria, once viewed as functioning relatively autonomously in the cell, have increasingly been recognized to be involved in numerous signaling networks that impact on a wide range of cell biological processes. In addition to the many types of proteins that mediate these pathways, the importance of signaling functions regulated via lipids and lipid second messengers generated on the mitochondrial surface is also becoming well appreciated. We focus here on phosphatidic acid, a lipid second messenger produced via several different pathways that can in turn stimulate the formation of multiple other bioactive lipids. Taken together, fascinating roles for phosphatidic acid and the connected lipids in mitochondrial function and interaction with other organelles are being uncovered. These pathways present new opportunities for the development of therapeutic approaches relevant to reproduction, metabolism, and neurodegenerative disease.
Keywords: Mitochondria, phosphatidic acid, MitoPLD, lipid signaling, spermatogenesis
Introduction
Mitochondria, in addition to producing energy, participate in many other cell biological processes including propagation of calcium signaling and calcium storage, lipid synthesis and lipid transfer to and from other organelles, and apoptosis. The mitochondrial surface functions as a scaffold or platform upon which signaling events take place that regulate the above processes as well as mitochondrial morphology and subcellular trafficking. RNA processing events, including both translation and production of small RNAs (RNAi), also occur on the mitochondrial surface and require mitochondrial surface signaling or production of energy. Many of the signaling pathways involve the generation and degradation of specific lipids such as phosphatidic acid (PA), which can subsequently promote production of other signaling lipids. Disruption of these signaling pathways and the processes they affect cause a wide variety of diseases including neurodegenerative and metabolic disorders and infertility. In many cases, we do not have a full mechanistic understanding of how alterations in mitochondrial surface lipid signaling pathways link to the processes they control and subsequently to the diseases that ensue; thus numerous areas of investigation are intensively under exploration.
Organelle function
Phosphatidic acid biogenesis and signaling pathways
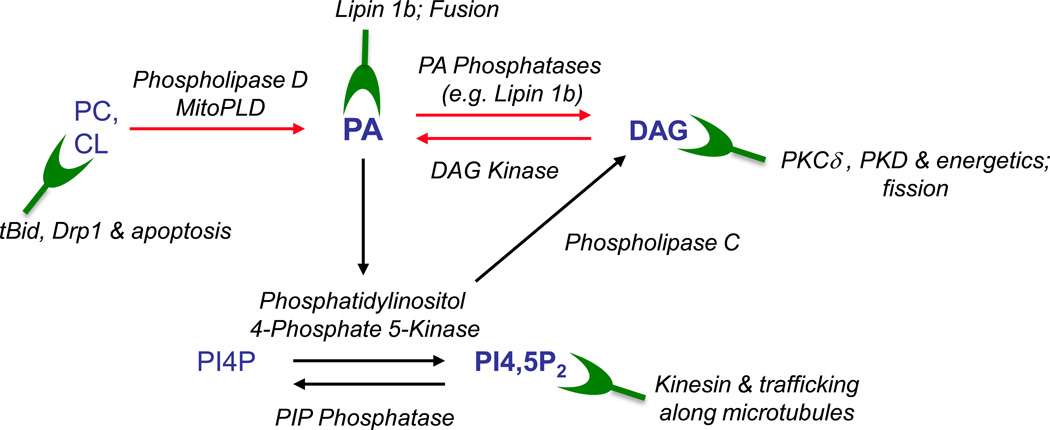
PA is a bioactive lipid that sits at the nexus of a number of lipid synthetic and signaling pathways (Fig. 1) (Jenkins and Frohman, 2005). PA can be generated via hydrolysis of the phospholipids phosphatidylcholine (PC) and cardiolipin (CL) by classical Phospholipase D (PLD) family members found on many cytoplasmic membrane surfaces and by MitoPLD, the PLD family member found on the mitochondrial surface (Choi et al., 2006), respectively. PA can also be generated by phosphorylation of the signaling lipid diacylglycerol (DAG) through the action of a large family of DAG kinases (DAGK), or via addition of a fatty acid side chain to lysoPA through the action of LysoPA acetyl transferases. In turn, PA can be reversibly be metabolized to LPA by Phospholipase A2, or dephosphorylated by PA phosphatases such as the enzyme Lipin 1 to form DAG (Han and Carman, 2010).
Fig. 1. Signaling Lipids.
Phosphatidylcholine (PC) and Cardiolipin (CL), which are major phospholipids in the plasma membrane and mitochondria, respectively, can be converted by members of the PLD superfamily into PA, which in turn can be used to generate DAG. PA also stimulates the enzyme Phosphatidylinositol 4-phosphate 5-Kinase to generate PIP2, which can be converted to DAG via the actions of Phospholipase C. Several of these actions are reversible, for example conversion of DAG back to PA by DAGK. Green shapes indicate proteins that bind to the lipids in the context of signaling on the mitochondrial surface and processes that the proteins and/or lipids are involved in.
PA is a negatively-charged phospholipid that can effect changes in cell biological processes through several potential mechanisms in any specific setting. First, as a consequence of the small lipid head-group and negative charge, PA induces membrane bilayers to undergo negative curvature, which is thought to facilitate both fusion and fission of membrane organelles such as mitochondria (Choi et al., 2006) or secretory vesicles during trafficking to and from sites such as the plasma membrane (Huang et al., 2005) or Golgi complex (Yang et al., 2008). DAG can also facilitate fission events (Fernandez-Ulibarri et al., 2007). Second, PA can serve as a lipid anchor to recruit effectors such as fusogenic (Nakanishi et al., 2004), trafficking (Yang et al., 2008, Manifava et al., 2001), and lipid-modifying (Huang et al., 2011a) proteins to membrane surfaces and in some cases activate them.
Mitochondrial functions mediated by outer membrane signaling pathways
As opposed to being static organelles with the simple role of producing energy, mitochondria are now recognized to be dynamic in morphology, to require subcellular trafficking to perform their functions where needed, and to participate in a wide range of cell biological functions including cell death pathways and calcium homeostasis (Soubannier and McBride, 2009). Mitochondrial fusion facilitates mitochondrial DNA replication error checking and repair and has been proposed to be important for energy production, since fragmented mitochondria in cells lacking fusion machinery are less efficient at generating ATP (Chen et al., 2005). However, this remains an unsettled issue since it has not been demonstrated that mitochondria in the fragmented conformation are less able to produce ATP under physiological conditions. The heart, for example, relies strongly on oxidative phosphorylation to generate ATP and in this tissue, mitochondria are not fused. Fission is vital to permit damaged mitochondrial components to be recycled via autophagy (Twig et al., 2006) and to facilitate mitochondrial trafficking to sites where energy production is most required (Campello et al., 2006, Chada and Hollenbeck, 2004). Drp1, a dynamin-related protein that mediates mitochondrial fission by constricting and severing the mitochondrial membranes (Frank et al., 2001), is also tied to cell death pathways; fission is linked to apoptosis, resulting in release of cytochrome C and other mediators of cell death, via translocation of BCL-2 family members to the mitochondrial surface (Llambi et al., 2011).
Close approximation of mitochondria to the ER have long been noted at restricted regions of the ER known as mitochondrial-associated membranes (MAMs) and at restricted regions of the mitochondria newly recognized to constitute sites where fission will take place (Friedman et al., 2011). Functional interactions appear to take place in both directions; the ER has been shown to mediate the fission process possibly through recruiting Drp1; conversely, mitochondria take up calcium released by the ER during signaling events and propagate the calcium signaling wave through fusion of these mitochondria with ones further away from the ER and transfer of the calcium to them (Patergnani et al., 2011); calcium also regulates mitochondrial trafficking on microtubules (Saotome et al., 2008). MAMs are additionally critical for lipid transfer that needs to take place between mitochondria and the ER. Finally, as will be discussed subsequently, mitochondria play a poorly understood but fascinating signaling role in spermatogenesis at a stage earlier than which the fusion machinery is required (Huang et al., 2011a, Watanabe et al., 2011).
The coordination of these mitochondrial functions with cellular signaling pathways and requirements is currently understood to differing extents. Mitochondrial fusion is known to change in rate in response to extracellular stress and growth factor signaling and at specific points in the cell cycle (Soubannier and McBride, 2009), but the factors that regulate the rate of fusion have not been identified (Schauss et al., 2010); in this context, PA may have a role (Choi et al., 2006) as discussed below. Regulation of fission is better understood; a number of different types of post-transcriptional modifications of Drp1 facilitate its requirement to the mitochondrial surface. Interestingly, the mitochondrial-specific lipid CL, which is found mostly inside the mitochondria but also to a limited extent on the surface (Schlattner et al., 2009), functions to recruit and/or facilitate the action of both Drp1 and pro-apoptotic members of the BCL-2 protein family through a process that involves membrane remodeling and permeabilization (Montessuit et al., 2010) that PA and DAG might also affect. Finally, the lipid DAG recruits proteins involved in regulating energy production, such as PKCδ (Stahelin et al., 2004) and PKD1 (Cowell et al., 2009). DAG is also associated with membrane fission in the context of vesicle trafficking, and recent findings suggest that it may play a role in fission for mitochondria as well (Huang et al., 2011a), in particular at sites now also recognized to be MAMs (Friedman et al., 2011).
Cell Physiology
PA signaling roles on the mitochondrial surface
MitoPLD, a divergent member of the PLD superfamily, was found to localize to the mitochondrial surface via insertion of an N-terminal transmembrane and to produce PA there via the hydrolysis of CL (Choi et al., 2006). This was an unexpected finding since MitoPLD is most similar to a prokaryotic PLD that functions as an endonuclease, and after that most similar to bacterial cardiolipin synthase (CLS), which also functions via a PLD-like mechanism (CLS in animals is an unrelated gene with no sequence similarity to PLD family members). Nonetheless, nuclease and CLS activities have not been demonstrable for MitoPLD, whereas evidence for production of PA comes from several lines of approach.
Increasing levels of production of PA on the mitochondrial surface results in mitochondrial aggregation, a phenomena associated with exaggerated levels of proteins known to mediate fusion; conversely, acute elimination of MitoPLD-generation of PA using RNAi or expression of a dominant-negative allele that lacks catalytic activity results in mitochondria fragmentation. In principle, this type of alteration in the balance of fusion and fission could reflect either a role for MitoPLD / PA in facilitating fusion or in inhibiting fission. Assays that specifically measure rates of fusion, however, suggest that PA functions to facilitate the fusion process, albeit the mechanism through which it does so is unknown. Mouse embryo fibroblasts lacking MitoPLD have shortened mitochondria, although the phenotype is less dramatic than that seen with acute depletion of MitoPLD via RNAi targeting or expression of a dominant-negative allele (Huang et al., 2011a), suggesting compensatory mechanisms that could involve increased fusogenic activity by other components such as Mfn1, or decreased Drp1-mediated fission. Finally, Drosophila lacking MitoPLD (as achieved by RNAi-mediated knockdown) exhibit increased dorsal lamina closure during gastrulation, a process promoted by mitochondrial fragmentation (Muliyil et al., 2011).
As discussed earlier, PA can be metabolized to DAG by the PA phosphatase Lipin 1 (Han and Carman, 2010), mutation of which has been associated with metabolic and neurological disease. MitoPLD-produced PA recruits Lipin 1 to the mitochondrial surface, leading to DAG production there (Huang et al., 2011a). Interestingly, the Lipin 1 catalytic domain translocates to fission sites in a PA-independent manner and strongly promotes mitochondrial fission, suggesting a two-step mechanism wherein PA generated during the fusion process recruits Lipin1 to the mitochondrial surface, resulting in a conformational change in Lipin1 that exposes a second mitochondrial interaction mechanism that focuses Lipin on fission sites to produce the profission lipid DAG there and facilitate division. In some circumstances, fusion and fission events have been linked both temporarily and spatially (Liu et al., 2009, Twig et al., 2008), suggesting a collusion between the protein machinery that this lipid signaling scenario would fit well with. Since Lipin 1 is frequently recruited to the ER, another site of PA abundance, it is also possible that Lipin 1 localized at ER MAM sites might interact with mitochondrial fission sites in a trans-organelle manner. The DAG produced by Lipin may also serve to recruit PKD (Cowell et al., 2009) or PKCδ (Stahelin et al., 2004) in the context of extracellular or stress-signaled fusion events.
Organelle Pathology
Roles for PA early in spermatogenesis
Mice lacking MitoPLD are viable and grossly normal (Huang et al., 2011a, Watanabe et al., 2011), although phenotypes in brain and other tissue systems might yet be uncovered once provoked or formally assessed. A fully-penetrant phenotype, however, consists of male infertility involving lack of progression of spermatogenesis during meiosis beyond the early pachytene stage (Huang et al., 2011a, Watanabe et al., 2011). The direct cause of the meiotic block is that spermatocytes lacking MitoPLD fail to generate a specialized form of RNAi known as piRNA that is required to suppress transposon mobilization which otherwise causes lethal levels of genomic damage if not checked (Suh and Blelloch, 2011). A similar phenotype is present in Drosophila, in which MitoPLD was independently identified as Zucchini (Zuc) in a genetic screen for fertility (Hales and Fuller, 1997).
These findings raise the question of how generation of PA on the surface of the mitochondria could regulate biogenesis of piRNA. Intriguingly, the piRNA is generated in an organelle known as the nuage or intermitochondrial cement, an electron-dense amorphous structure that is juxtaposed to the mitochondria during this stage of meiosis (Chuma et al., 2009). The nuage contains the RNA precursors used to generate the piRNA small RNAs as well as many of the other proteins known to be required for piRNA generation. Strikingly, the nuage is not associated with the mitochondria in spermatocytes lacking MitoPLD, and as well, both the mitochondria and other piRNA-generating proteins normally found in the nuage are mislocalized to the peri-centrosomal region (Huang et al., 2011a, Watanabe et al., 2011).
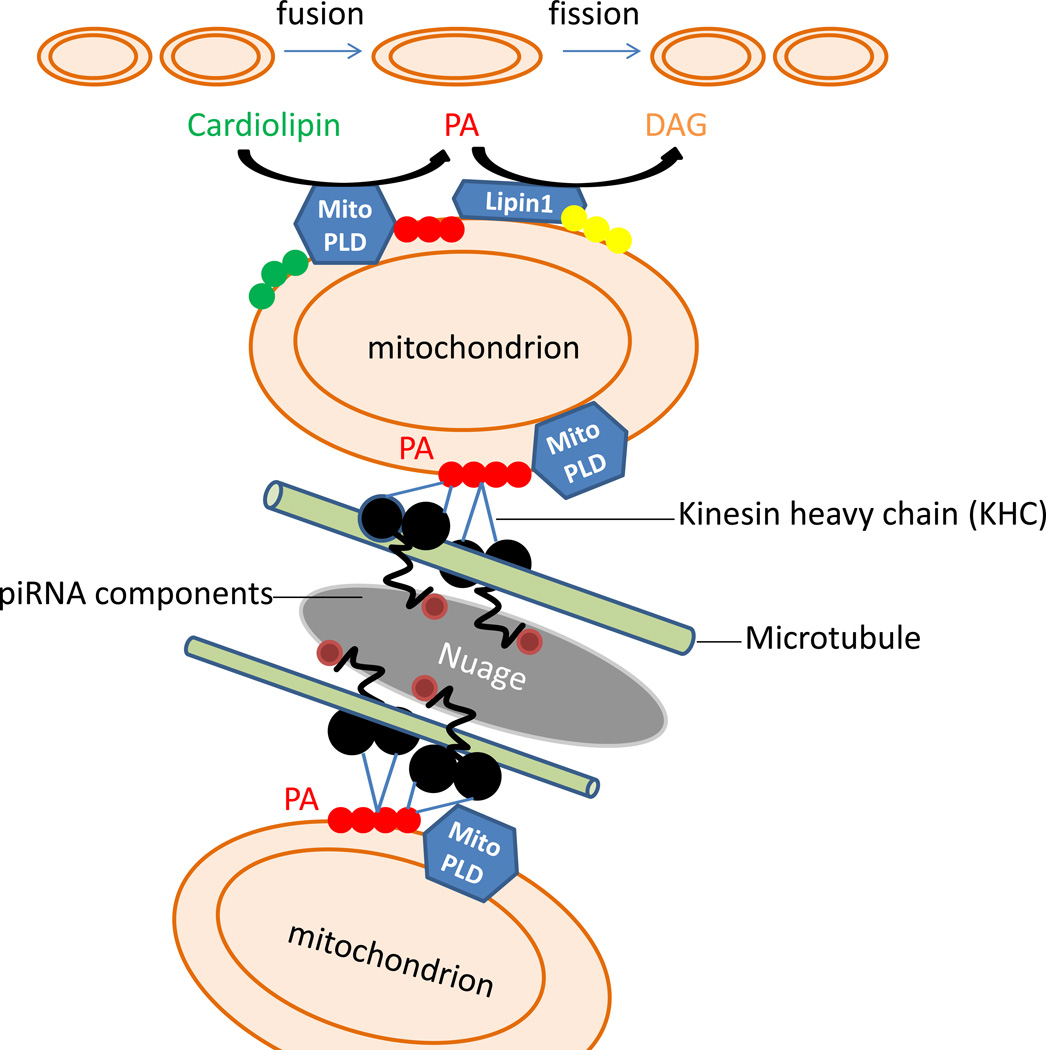
A number of possibilities exist at present. First, given its homology to the endonuclease branch of the PLD superfamily, it remains possible that MitoPLD/Zuc could be the actual RNA cleaving enzyme that generates the piRNA, even though this activity has not thus far been demonstrable, since the enzyme that performs the RNA processing has not yet been identified. Assuming, however, that the key event is dependent on the generation of PA, there are several other possibilities. First, the physical association between the nuage and mitochondria might be mediated by the production of PA, since it is a negatively-charged lipid and the nuage is highly abundant in basic proteins (Paniagua et al., 1985), creating a potential for electrostatic interaction. Second, as has been shown in many other systems, MitoPLD could recruit and /or activate cytoplasmic or nuage proteins via PA-binding domains to function in associating the nuage with the mitochondrial surface or more directly generating the piRNA or loading it onto the proteins that will perform the suppression of transposon mobilization. Finally, an intriguing possibility comes from the finding that the microtubule motor Kinesin heavy chain (KHC) is a PA-binding protein (Manifava et al., 2001). A significant fraction of KHC is found in association with mitochondria, and in cells lacking kinesin, the mitochondria mislocalize to the peri-centrosome (Tanaka et al., 1998). Developmental RNAs, in particular large ones like those used as templates for piRNA, are frequently transported along microtubules. Halting of the RNA movement in some cases has been shown to proceed from KHC or an adaptor protein binding to an immotile target (Zimyanin et al., 2008). As one possibility, piRNA template RNAs and/or nuage translocating on microtubules via KHC might halt at mitochondria with high levels of surface PA through a PA-KHC interaction (Fig.2). In the absence of KHC-PA interaction, mitochondria and the piRNA precursors would fail to interact, permitting centrally-directed dynein transport to predominate. In an elegant study, it was shown that mitochondrial fusion proceeds and resolves differently depending on whether the mitochondria impact on the same microtubule in a head-on collision as opposed to being on tangential microtubules and undergoing a glancing side-to-side impact (Liu et al., 2009). One could hypothesize that interaction of mitochondria and piRNA precursors translocating on different microtubules might be hard to resolve if KHC binds strongly to PA and the PA levels are high in that setting, leading to the formation of a “sink” of juxtaposed mitochondria and RNA processing granules, as is seen at this stage of spermatogenesis.
Fig. 2. Mitochondrial surface lipid signaling roles in mitochondria dynamics and piRNA biogenesis.
Upper part of schema: mitochondrial fusion is facilitated by MitoPLD-generated PA. The fusion event is terminated through the conversion of PA to DAG by Lipin1, leading to fission. Disruption of these pathways may have neurological or metabolic consequences through decreasing the rate of fusion or via effects on CL and Drp1- or DAG and Lipin1-mediated fission events or signaling to other enzymes such as PKCδ. Lower part of schema: piRNA template RNAs and/or nuage translocating on microtubules via KHC in spermatocytes are proposed to halt via a PA-KHC interaction at mitochondria exhibiting high levels of surface MitoPLD-generated PA. In the absence of MitoPLD-generated PA, piRNA biogenesis does not occur, leading to transposon activation, DNA damage, triggering of meiotic checkpoints, spermatocyte apoptosis, and infertility.
Future Outlook
Many directions are now timely to undertake to address questions that have arisen from these findings. As one possibility, many of the piRNA generating proteins are expressed in somatic tissues (Yan et al., 2011), including MitoPLD (Choi et al., 2006), and the nuage-like P-body RNA granules in which siRNAs and miRNAs are generated have been shown to associate physically with mitochondria (Huang et al., 2011b). Strikingly, the ability of the mitochondria to generate ATP has a substantial effect on the ability of P-bodies to generate the siRNAs and miRNAs, and key nuage proteins such as Ago2 mislocalize away from the P-body when the mitochondria are energetically compromised (Huang et al., 2011b). Whether MitoPLD and PA function in the role mitochondria play in facilitating generation of siRNAs and/or miRNAs in somatic tissues remains unknown.
Small molecule inhibitors have been generated for other members of the PLD superfamily (Scott et al., 2009, Su et al., 2009b). The first inhibitor, FIPI, was identified in a large screen for PLD2 inhibitors and then shown to inhibit both PLD1 and PLD2 but not MitoPLD (Su et al., 2009a); combinatorial chemistry on the initial lead was employed in parallel to generate isoform-selective inhibitors (Scott et al., 2009). Although MitoPLD generates the same product, PA, as PLD1 and PLD2, it uses a different substrate, CL, and it is evolutionarily quite divergent from PLD1 and PLD2, which are 50% identical to each other. Hence it is not surprising that FIPI does not inhibit MitoPLD. At present, the in vitro CL hydrolysis assay for MitoPLD is labor-intensive (Choi et al., 2006); however, it would be straightforward to identify leads by performing high through-put cell-based screens for compounds capable of blocking MitoPLD-induced mitochondrial aggregation. Given the absence at present for other phenotypes in mice lacking MitoPLD, a MitoPLD small molecule inhibitor presents an intriguing opportunity for a reversible and fully-effective male contraceptive agent.
Finally, therapeutic gain might also be achieved through manipulation of the mitochondrial fusion and fission machinery. mDiviA, a small molecule inhibitor of Drp1-mediated fission, has been reported to confer benefit via suppression of apoptosis in the setting of kidney transplants (Brooks et al., 2009) and cardiac (Ong et al., 2010) and brain (Grohm et al., 2012) ischemia reperfusion injuries. On the other hand, ablation of Mfn1, which promotes fusion, triggers mitochondrial fragmentation in cardiomyocytes and has been reported to increase tolerance to stress-induced mitochondrial dysfunction and cell death (Papanicolaou et al., 2012). Whether other functions for Drp1 and Mfn1 underlie these findings or whether manipulating mitochondrial morphology in itself offers therapeutic possibilities requires further exploration.
Organelle facts.
Mitochondria are dynamic organelles, frequently undergoing fusion and fission and trafficking along the cytoskeleton.
The mitochondrial surface serves as a scaffold for multiple cell biological processes.
Interaction of the mitochondria with the endoplasmic reticulum is important in the maintenance of calcium homeostasis and for exchange of phospholipids between the organelles. Interaction of mitochondria with nuage, an RNA granule and processing organelle, is critical for biogenesis of piRNA, a specialized form of RNAi, during spermatogenesis.
Lipid signals generated on the surface of the mitochondria help regulate mitochondrial trafficking, fusion, fission, and interaction with the endoplasmic reticulum and nuage. Key lipids on the mitochondrial surface also play important roles in apoptosis, mitophagy, and ROS generation.
Mitochondrial dysfunction resulting from altered lipid signaling of phosphatidic acid and connected lipids causes infertility, the major focus of this review, but may also contribute to neurodegenerative disease and metabolic disorders.
Acknowledgments
Supported by NIH GM071520 and GM084251 to MAF.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Brooks C, Wei Q, Cho SG, Dong Z. Regulation of mitochondrial dynamics in acute kidney injury in cell culture and rodent models. J Clin Invest. 2009;119:1275–1285. doi: 10.1172/JCI37829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campello S, Lacalle RA, Bettella M, Manes S, Scorrano L, Viola A. Orchestration of lymphocyte chemotaxis by mitochondrial dynamics. J Exp Med. 2006;203:2879–2886. doi: 10.1084/jem.20061877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chada SR, Hollenbeck PJ. Nerve growth factor signaling regulates motility and docking of axonal mitochondria. Curr Biol. 2004;14:1272–1276. doi: 10.1016/j.cub.2004.07.027. [DOI] [PubMed] [Google Scholar]
- Chen H, Chomyn A, Chan DC. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J Biol Chem. 2005;280:26185–26192. doi: 10.1074/jbc.M503062200. [DOI] [PubMed] [Google Scholar]
- Choi SY, Huang P, Jenkins GM, Chan DC, Schiller J, Frohman MA. A common lipid links Mfn-mediated mitochondrial fusion and SNARE-regulated exocytosis. Nat Cell Biol. 2006;8:1255–1262. doi: 10.1038/ncb1487. [DOI] [PubMed] [Google Scholar]
- Chuma S, Hosokawa M, Tanaka T, Nakatsuji N. Ultrastructural characterization of spermatogenesis and its evolutionary conservation in the germline: germinal granules in mammals. Mol Cell Endocrinol. 2009;306:17–23. doi: 10.1016/j.mce.2008.11.009. [DOI] [PubMed] [Google Scholar]
- Cowell CF, Doppler H, Yan IK, Hausser A, Umezawa Y, Storz P. Mitochondrial diacylglycerol initiates protein-kinase D1-mediated ROS signaling. J Cell Sci. 2009;122:919–928. doi: 10.1242/jcs.041061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Ulibarri I, Vilella M, Lazaro-Dieguez F, Sarri E, Martinez SE, Jimenez N, Claro E, Merida I, Burger KN, Egea G. Diacylglycerol is required for the formation of COPI vesicles in the Golgi-to-ER transport pathway. Mol Biol Cell. 2007;18:3250–3263. doi: 10.1091/mbc.E07-04-0334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank S, Gaume B, Bergmann-Leitner ES, Leitner WW, Robert EG, Catez F, Smith CL, Youle RJ. The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev Cell. 2001;1:515–525. doi: 10.1016/s1534-5807(01)00055-7. [DOI] [PubMed] [Google Scholar]
- Friedman JR, Lackner LL, West M, DiBenedetto JR, Nunnari J, Voeltz GK. ER tubules mark sites of mitochondrial division. Science. 2011;334:358–362. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grohm J, Kim SW, Mamrak U, Tobaben S, Cassidy-Stone A, Nunnari J, Plesnila N, Culmsee C. Inhibition of Drp1 provides neuroprotection in vitro and in vivo. Cell Death Differ. 2012 doi: 10.1038/cdd.2012.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hales KG, Fuller MT. Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell. 1997;90:121–129. doi: 10.1016/s0092-8674(00)80319-0. [DOI] [PubMed] [Google Scholar]
- Han GS, Carman GM. Characterization of the human LPIN1-encoded phosphatidate phosphatase isoforms. J Biol Chem. 2010;285:14628–14638. doi: 10.1074/jbc.M110.117747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Gao Q, Peng X, Choi SY, Sarma K, Ren H, Morris AJ, Frohman MA. piRNA-associated germline nuage formation and spermatogenesis require MitoPLD profusogenic mitochondrial-surface lipid signaling. Dev Cell. 2011a;20:376–387. doi: 10.1016/j.devcel.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Mollet S, Souquere S, Le Roy F, Ernoult-Lange M, Pierron G, Dautry F, Weil D. Mitochondria associate with P-bodies and modulate microRNA-mediated RNA interference. J Biol Chem. 2011b;286:24219–24230. doi: 10.1074/jbc.M111.240259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, Altshuller YM, Hou JC, Pessin JE, Frohman MA. Insulin-stimulated plasma membrane fusion of Glut4 glucose transporter-containing vesicles is regulated by phospholipase D1. Mol Biol Cell. 2005;16:2614–2623. doi: 10.1091/mbc.E04-12-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins GM, Frohman MA. Phospholipase D: a lipid centric review. Cell Mol Life Sci. 2005;62:2305–2316. doi: 10.1007/s00018-005-5195-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Weaver D, Shirihai O, Hajnoczky G. Mitochondrial 'kiss-and-run': interplay between mitochondrial motility and fusion-fission dynamics. Embo J. 2009;28:3074–3089. doi: 10.1038/emboj.2009.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llambi F, Moldoveanu T, Tait SW, Bouchier-Hayes L, Temirov J, McCormick LL, Dillon CP, Green DR. A unified model of mammalian BCL-2 protein family interactions at the mitochondria. Mol Cell. 2011;44:517–531. doi: 10.1016/j.molcel.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manifava M, Thuring JW, Lim ZY, Packman L, Holmes AB, Ktistakis NT. Differential binding of traffic-related proteins to phosphatidic acid- or phosphatidylinositol (4,5)- bisphosphate-coupled affinity reagents. J Biol Chem. 2001;276:8987–8994. doi: 10.1074/jbc.M010308200. [DOI] [PubMed] [Google Scholar]
- Montessuit S, Somasekharan SP, Terrones O, Lucken-Ardjomande S, Herzig S, Schwarzenbacher R, Manstein DJ, Bossy-Wetzel E, Basanez G, Meda P, Martinou JC. Membrane remodeling induced by the dynamin-related protein Drp1 stimulates Bax oligomerization. Cell. 2010;142:889–901. doi: 10.1016/j.cell.2010.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muliyil S, Krishnakumar P, Narasimha M. Spatial, temporal and molecular hierarchies in the link between death, delamination and dorsal closure. Development. 2011;138:3043–3054. doi: 10.1242/dev.060731. [DOI] [PubMed] [Google Scholar]
- Nakanishi H, de los Santos P, Neiman AM. Positive and negative regulation of a SNARE protein by control of intracellular localization. Mol Biol Cell. 2004;15:1802–1815. doi: 10.1091/mbc.E03-11-0798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong SB, Subrayan S, Lim SY, Yellon DM, Davidson SM, Hausenloy DJ. Inhibiting mitochondrial fission protects the heart against ischemia/reperfusion injury. Circulation. 2010;121:2012–2022. doi: 10.1161/CIRCULATIONAHA.109.906610. [DOI] [PubMed] [Google Scholar]
- Paniagua R, Nistal M, Amat P, Rodriguez MC. Presence of ribonucleoproteins and basic proteins in the nuage and intermitochondrial bars of human spermatogonia. J Anat. 1985;143:201–206. [PMC free article] [PubMed] [Google Scholar]
- Papanicolaou KN, Ngoh GA, Dabkowski ER, O'Connell KA, Ribeiro RF, Jr., Stanley WC, Walsh K. Cardiomyocyte deletion of mitofusin-1 leads to mitochondrial fragmentation and improves tolerance to ROS-induced mitochondrial dysfunction and cell death. Am J Physiol Heart Circ Physiol. 2012;302:H167–H179. doi: 10.1152/ajpheart.00833.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patergnani S, Suski JM, Agnoletto C, Bononi A, Bonora M, De Marchi E, Giorgi C, Marchi S, Missiroli S, Poletti F, Rimessi A, Duszynski J, Wieckowski MR, Pinton P. Calcium signaling around Mitochondria Associated Membranes (MAMs) Cell Commun Signal. 2011;9:19. doi: 10.1186/1478-811X-9-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saotome M, Safiulina D, Szabadkai G, Das S, Fransson A, Aspenstrom P, Rizzuto R, Hajnoczky G. Bidirectional Ca2+-dependent control of mitochondrial dynamics by the Miro GTPase. Proc Natl Acad Sci U S A. 2008;105:20728–20733. doi: 10.1073/pnas.0808953105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauss AC, Huang H, Choi SY, Xu L, Soubeyrand S, Bilodeau P, Zunino R, Rippstein P, Frohman MA, McBride HM. A novel cell-free mitochondrial fusion assay amenable for high-throughput screenings of fusion modulators. BMC Biol. 2010;8:100. doi: 10.1186/1741-7007-8-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlattner U, Tokarska-Schlattner M, Ramirez S, Bruckner A, Kay L, Polge C, Epand RF, Lee RM, Lacombe ML, Epand RM. Mitochondrial kinases and their molecular interaction with cardiolipin. Biochim Biophys Acta. 2009;1788:2032–2047. doi: 10.1016/j.bbamem.2009.04.018. [DOI] [PubMed] [Google Scholar]
- Scott SA, Selvy PE, Buck JR, Cho HP, Criswell TL, Thomas AL, Armstrong MD, Arteaga CL, Lindsley CW, Brown HA. Design of isoform-selective phospholipase D inhibitors that modulate cancer cell invasiveness. Nat Chem Biol. 2009;5:108–117. doi: 10.1038/nchembio.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soubannier V, McBride HM. Positioning mitochondrial plasticity within cellular signaling cascades. Biochim Biophys Acta. 2009;1793:154–170. doi: 10.1016/j.bbamcr.2008.07.008. [DOI] [PubMed] [Google Scholar]
- Stahelin RV, Digman MA, Medkova M, Ananthanarayanan B, Rafter JD, Melowic HR, Cho W. Mechanism of diacylglycerol-induced membrane targeting and activation of protein kinase Cdelta. J Biol Chem. 2004;279:29501–29512. doi: 10.1074/jbc.M403191200. [DOI] [PubMed] [Google Scholar]
- Su W, Chen Q, Frohman MA. Targeting phospholipase D with small-molecule inhibitors as a potential therapeutic approach for cancer metastasis. Future Oncol. 2009a;5:1477–1486. doi: 10.2217/fon.09.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su W, Yeku O, Olepu S, Genna A, Park JS, Ren H, Du G, Gelb MH, Morris AJ, Frohman MA. 5-Fluoro-2-indolyl des-chlorohalopemide (FIPI), a phospholipase D pharmacological inhibitor that alters cell spreading and inhibits chemotaxis. Mol Pharmacol. 2009b;75:437–446. doi: 10.1124/mol.108.053298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh N, Blelloch R. Small RNAs in early mammalian development: from gametes to gastrulation. Development. 2011;138:1653–1661. doi: 10.1242/dev.056234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Kanai Y, Okada Y, Nonaka S, Takeda S, Harada A, Hirokawa N. Targeted disruption of mouse conventional kinesin heavy chain, kif5B, results in abnormal perinuclear clustering of mitochondria. Cell. 1998;93:1147–1158. doi: 10.1016/s0092-8674(00)81459-2. [DOI] [PubMed] [Google Scholar]
- Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, Alroy J, Wu M, Py BF, Yuan J, Deeney JT, Corkey BE, Shirihai OS. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. Embo J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twig G, Graf SA, Wikstrom JD, Mohamed H, Haigh SE, Elorza A, Deutsch M, Zurgil N, Reynolds N, Shirihai OS. Tagging and tracking individual networks within a complex mitochondrial web with photoactivatable GFP. Am J Physiol Cell Physiol. 2006;291:C176–C184. doi: 10.1152/ajpcell.00348.2005. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Chuma S, Yamamoto Y, Kuramochi-Miyagawa S, Totoki Y, Toyoda A, Hoki Y, Fujiyama A, Shibata T, Sado T, Noce T, Nakano T, Nakatsuji N, Lin H, Sasaki H. MITOPLD is a mitochondrial protein essential for nuage formation and piRNA biogenesis in the mouse germline. Dev Cell. 2011;20:364–375. doi: 10.1016/j.devcel.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, Hu HY, Jiang X, Maierhofer V, Neb E, He L, Hu Y, Hu H, Li N, Chen W, Khaitovich P. Widespread expression of piRNA-like molecules in somatic tissues. Nucleic Acids Res. 2011;39:6596–6607. doi: 10.1093/nar/gkr298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JS, Gad H, Lee SY, Mironov A, Zhang L, Beznoussenko GV, Valente C, Turacchio G, Bonsra AN, Du G, Baldanzi G, Graziani A, Bourgoin S, Frohman MA, Luini A, Hsu VW. A role for phosphatidic acid in COPI vesicle fission yields insights into Golgi maintenance. Nat Cell Biol. 2008;10:1146–1153. doi: 10.1038/ncb1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimyanin VL, Belaya K, Pecreaux J, Gilchrist MJ, Clark A, Davis I, St Johnston D. In vivo imaging of oskar mRNA transport reveals the mechanism of posterior localization. Cell. 2008;134:843–853. doi: 10.1016/j.cell.2008.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]