Abstract
The ω-hydroxylase CYP4A11 catalyzes the transformation of epoxyeicosatrienoic acids to omega-hydroxylated-epoxyeicosatrienoic acids, endogenous peroxisome proliferator-activated receptor α (PPARα) agonists. PPARα activation increases high-density lipoprotein-cholesterol (HDL-C). A cytosine-for-thymidine (T8590C) variant of CYP4A11 encodes for a ω-hydroxylase with reduced activity. This study examined the relationship between CYP4A11 T8590C genotype and metabolic parameters in the Framingham Offspring Study and in a clinical practice-based biobank, BioVU. In women in the Framingham Offspring Study, the CYP4A11 8590C allele was associated with reduced HDL-C concentrations (54.2±0.9 mg/dL in CYP4A11 CC or CT genotype women versus 56.7±0.5 mg/dL in TT women, p=0.02), and with an increased prevalence of low HDL-C, defined categorically as ≤50mg/dL [odds ratio 1.39 (95% CI 1.02-1.90), p=0.04]. In the BioVU cohort, the CYP4A11 8590C allele was also associated with low HDL-C in women [odds ratio 1.69 (95% CI 1.03-2.77, p=0.04)]. There was no relationship between genotype and HDL-C in men in either cohort.
Introduction
The ω-hydroxylases (CYP4A and CYP4F) catalyze the NADPH-dependent oxidation of arachidonic acid to 19- and 20-hydroxyeicosatetraenoic acids (19- and 20-HETE), while the epoxygenases (CYP2C and CYP2J) catalyze the formation of four regioisomeric epoxyeicosatrienoic acids (EETs).[1, 2] The hydroxylase metabolite of arachidonic acid, 20-HETE can exert pro- or anti-hypertensive effects depending on its site-specific expression.[2] In the vasculature, 20-HETE causes vasoconstriction by inhibiting the calcium-dependent potassium channel and also enhances vasoconstriction induced by Ang II or endothelin. In contrast, 20-HETE causes natriuresis by inhibiting the tubular sodium channel. In addition, 20-HETE stimulates the expression of tubular epoxygenases and thereby increases the formation of EETs, which vasodilate and inhibit the epithelial sodium channel.
The ω-hydroxylases also catalyze the EETs to the potent peroxisome proliferator-activated receptor (PPAR) α agonists, the omega-hydroxylated-epoxyeicosatrienoic acids (HEETs).[3, 4] PPARα ligands modulate plasma triglycerides and elevate high-density lipoprotein-cholesterol (HDL-C) and exert anti-inflammatory effects.[5, 6] These observations suggest that monooxygenase products of arachidonic acid could modulate characteristics of the metabolic syndrome other than blood pressure (BP).
We have found that a cytosine-for-thymidine (T8590C) variant of CYP4A11, causing a phenylalanine-to-serine (F434S) protein polymorphism, encodes for a ω-hydroxylase with reduced 20-HETE synthase activity.[7] Several groups have reported an association of the loss-of-function CYP4A11 C allele with either hypertension or increased BP in a Tennessee cohort, among nondiabetic subjects in the Framingham Offspring Study, in the Monica Study, in the Mälmo Cancer study, in the African American Study of Kidney Disease, and in Evaluation of Nifedipine and Cerivastatin on Recovery of Coronary Endothelial function (ENCORE) trials, while others have reported association of different variants in CYP4A11 or CYP4F2 with BP or hypertension.[7-13]
Because HEETs are endogenous PPARα agonists, we tested the hypothesis that the CYP4A11 T8590C genotype is associated with additional characteristics of the metabolic syndrome, including HDL-C. We tested this hypothesis in two independent cohorts: the Framingham Offspring Study and BioVU, a large, clinical practice-based biobank at Vanderbilt University.
Methods
Framingham Offspring Study
The design and selection criteria of the Framingham Offspring Study have been described previously.[14] At each Framingham Heart Study examination, participants underwent routine medical history, physical examination, and laboratory assessment. BP was measured as the average of two readings in the left arm by a physician using a mercury column sphygmomanometer after the participant had sat for 5 minutes. Fasting plasma glucose, triglycerides, and HDL-C were measured using standardized assays. High-sensitivity C-reactive protein (CRP) was measured with a Dade Behring BN100 nephelometer. Smoking history and alcohol consumption were self-reported.
The components of the metabolic syndrome were defined using the modified definition of the National Cholesterol Education Program Adult Treatment Panel III guidelines: BP greater than or equal to 130 mm Hg systolic or 85 mm Hg diastolic or treatment for high BP, fasting glucose greater than or equal to 100 mg/dL (5.6mmol/L) or treatment with oral hypoglycemic agents or insulin, a waist circumference greater than or equal to 40.2 inches (102 cm) in men or 34.6 inches (88 cm) in women, fasting triglycerides greater than or equal to 150 mg/dL (1.7 mmol/L) or lipid-lowering treatment, and HDL-C less than 40 mg/dL (1.0 mmol/L) in men or 50 mg/dL (1.16 mmol/L) in women. Waist circumference was measured at visits 4 through 6 and CRP was measured at visit 5. All other traits were measured at visits 1 through 6. The presence of three or more NCEP-ATPIII components comprised the metabolic syndrome.
BioVU at Vanderbilt University
BioVU is currently the largest clinical practice-based biobank in the United States (106,464 samples from adults as of April 1, 2011). This Vanderbilt biobank accrues DNA samples extracted from blood drawn for routine clinical testing after the samples have been retained for 3 days and are scheduled to be discarded. These DNA samples are then linked to a “synthetic derivative” (or de-identified mirror image) of each individual’s electronic medical record at the Vanderbilt University Medical Center. New clinical data are added as they are generated, and each record in the synthetic derivative is labeled with the same unique research identifier as the DNA sample, maintaining the link between clinical data and DNA. BioVU has been validated as a robust resource for studies of disease onset and treatment outcome.[15, 16]
For the current study, a subset of 708 representative study subjects were initially chosen from BioVU, based upon predetermined criteria designed to maximize the density of longitudinal lipid data. These criteria included: (1) subjects using Vanderbilt Medical Center for their primary care, (2) subjects with clinical lipid panels (containing low density lipoprotein-C, HDL-C and triglycerides) obtained on at least three separate dates, (3) subjects with “European American” entered as their observer-reported race, (4) subjects age 44-68 years, based on the interquartile range in our previous validation work,[17] and (5) subjects across the full range of BMI distribution (i.e., matching BMI distribution for the entire cohort).
All clinical lipid data were electronically extracted from the electronic medical records linked to BioVU. Lipid traits were expressed as the median untreated lipid value for each individual. Age, gender, BMI and estrogen exposure data were extracted. BMI was calculated using weight and height data measured closest in time to each lipid panel. Because waist circumference measurements were not available in BioVU, BMI>30kg/M2 was used to define obesity.
Genetic analysis
Genotyping for SNP T8590C (rs1126742) in exon 10 of CYP4A11 was performed by amplification and sequencing of a DNA segment covering exon 10 through exon 11, as previously described.[7]
Statistical analysis
Two different methods were used to assess the relationship between CYP4A11 T8590C genotype and continuous variables in the Framingham Offspring Study. Multiple linear regression was first used to assess relationships in unrelated subjects (807 men and 818 women). Covariates used in the model were the exam-specific age, age2, BMI, number of cigarettes smoked per day, ounces of alcohol consumed per week and menopausal status and estrogen status in women. Gender was included as a covariate in analyses of men and women combined. Serum triglycerides and CRP were not normally distributed and were log-transformed for analysis. We also treated components of the metabolic syndrome as dichotomous variables and calculated the odds ratio of having a specific criterion for the metabolic syndrome such as a low HDL-C. For CRP, we calculated the odds ratio of having a concentration in the 5th quintile. Lastly, we repeated these analyses in both unrelated and related subjects (1107 men and 1238 women), using Generalized Estimating Equations (GEE) analysis to control for correlation of the outcome traits within families. In this generation, familial relationships were siblings or cousins.
For the BioVU cohort, HDL-C was also analyzed as a continuous variable using multiple linear regression and as a dichotomous variable using logistic regression. These analyses were performed using PLINK (http://pngu.mgh.harvard.edu/~purcell/plink/). Age, age2, BMI, and gender were included as covariates. Nine out of 360 female subjects had a history of taking estrogen and these were excluded from the final analyses.
All data are presented as means ± standard deviations unless otherwise stated and p<0.05 was considered significant.
Results
Discovery Cohort – Framingham Offspring Study
Tables 1 and 2 provide the baseline (visit 1) clinical characteristics for the unrelated sample and the combined unrelated and family sample, respectively. CYP4A11 T8590C genotype frequencies were in Hardy-Weinberg equilibrium in both samples. Tables S1 and S2 (supplement) provide the prevalence of each component of the metabolic syndrome at visit 1 and 5 for the unrelated sample and the combined unrelated and family sample, respectively. The frequency of obesity (BMI >30kg/M2) is provided because waist circumference was not measured at visit 1. The frequency of each trait was higher in men than in women. In addition, the frequency of metabolic syndrome traits increased from visit 1 to visit 5.
Table 1.
Characteristics of Unrelated Subjects in the Framingham Offspring Study at Visit 1
| Parameter | Men (807) | Women (816) | Combined (1623) |
|---|---|---|---|
| Genotype, N (%) CC:CT:TT | 7(0.9):190(23.5):610(75.6) | 14(1.7):172(21.1):630(77.2) | 21(1.3):362(22.3):1240(76.4) |
| Age, years | 36.6±9.5 | 35.7±9.3 | 36.1±9.4 |
| BMI, kg/M2 | 26.5±3.6 | 23.8±4.4 | 25.1±4.2 |
| SBP, mmHg | 124.4±13.6 | 117.0±14.9 | 120.7±14.7 |
| DBP, mmHg | 80.4±9.3 | 75.1±10.0 | 77.8±10.0 |
| Glucose, mg/dL | 104.7±11.5 | 98.7±10.1 | 101.7±11.2 |
| Triglycerides, mg/dL | 109.9±86.5 | 71.3±47.2 | 90.7±72.3 |
| HDL-cholesterol, mg/dL | 44.5±11.3 | 57.0±14.8 | 50.7±14.6 |
| Tobacco exposure, cigarettes/day | 16.4±16.1 | 10.1±12.1 | 13.2±14.6 |
| Alcohol intake, ounces/week | 5.0±5.2 | 2.2±2.8 | 3.6±4.4 |
BMI indicates body mass index, SBP indicates systolic blood pressure, DBP indicates diastolic blood pressure, HDL indicates high density lipoprotein
Table 2.
Subject characteristics in Combined Unrelated and Family Sample of the Framingham Offspring Study at Visit 1
| Men (N=1107) | Women (N=1238) | Combined (2345) | |
|---|---|---|---|
| Genotype, N (%) CC:CT:TT | 8(0.7):252 (22.8):847 (76.5) | 20(1.6):271(21.9):947(76.5) | 28(1.2):523(22.3):1794(76.5) |
| Age, years | 35.1±9.9 | 34.7±9.7 | 34.9±9.8 |
| BMI, kg/M2 | 26.3±3.6 | 23.7±4.4 | 25.0±4.3 |
| SBP, mmHg | 124.6±13.8 | 116.3±14.5 | 120.2±14.7 |
| DBP, mmHg | 80.7±9.6 | 75.0±9.9 | 77.6±10.2 |
| Glucose, mg/dL | 104.2±11.2 | 98.1±9.8 | 101.0±10.9 |
| Triglycerides, mg/dL | 109.1±86.5 | 71.6±46.8 | 89.5±71.1 |
| HDL-cholesterol, mg/dL | 44.5±11.1 | 56.8±14.4 | 50.9±14.3 |
| Tobacco exposure, cigarettes/day | 15.5±15.8 | 9.9±12.0 | 12.6±14.2 |
| Alcohol intake, ounces/week | 4.9±5.3 | 2.2±2.8 | 3.5±4.4 |
BMI indicates body mass index, SBP indicates systolic blood pressure, DBP indicates diastolic blood pressure, HDL indicates high density lipoprotein
We first assessed the association of CYP4A11 T8590C genotype with individual metabolic traits as dichotomous variables at visits 1 through 6. Odds ratios were calculated with and without adjustment for age, age2, BMI, number of cigarettes smoked per day, ounces of alcohol consumed per week, and menopausal status and estrogen use in women. Gender was also included as a covariate in the analyses of both men and women. When men and women were analyzed together, there was no relationship between CYP4A11 T8590C genotype and the frequency of high BP, low HDL-C, hyperlipidemia, hyperglycemia, increased waist circumference (or increased BMI on those visits at which waist circumference was not measured), or the metabolic syndrome, at any visit in either the unrelated or the combined sample. In women in the combined sample, however, the CYP4A11 8590C allele was associated with an increased prevalence of low HDL-C (less than or equal to 50 mg/dL or 1.16 mmol/L) at the first 3 visits (Odds Ratio 1.39, 95% C.I. 1.02-1.90, p=0.02, Figure 1). There was no association in men. The odds ratio for having a low HDL-C in women who were carriers of the CYP4A11 8590 C versus TT homozygotes decreased progressively with each successive visit.
Figure 1. Framingham Offspring Study. Relationship between CYP4A11 T8590C genotype and the likelihood of having a low concentration of high density lipoprotein cholesterol (HDL-C), defined as less than 50mg/dL (1.16 mmol/L) in women.
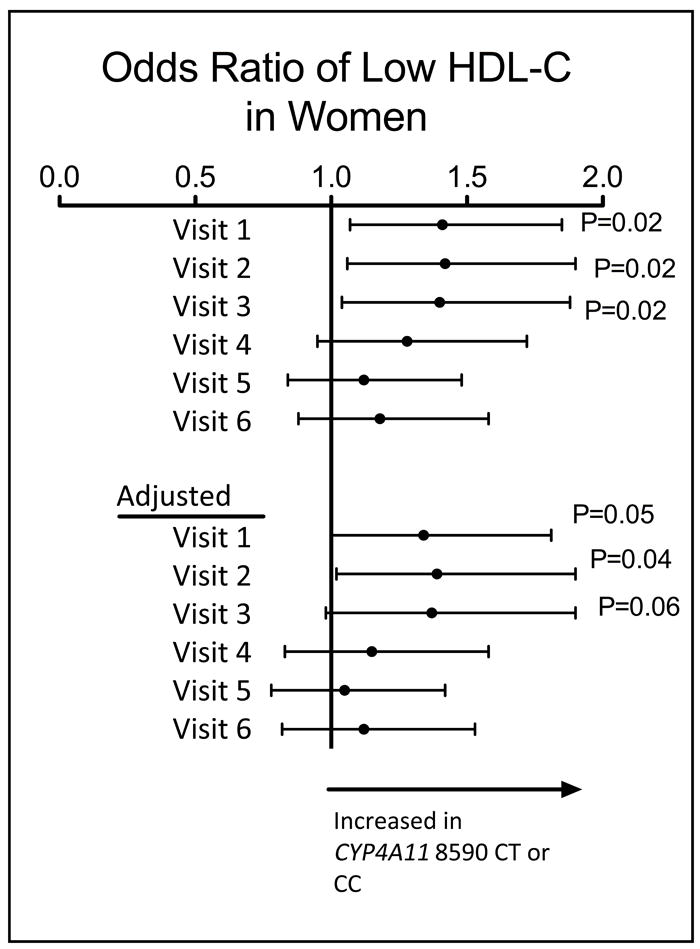
Data are from the combined unrelated and family sample. The analysis was adjusted for exam-specific age, age2, body mass index, number of cigarettes smoked per day, ounces of alcohol consumed per week and menopausal status and estrogen status in women.
When treated as a continuous variable, HDL-C was significantly lower in women in the unrelated sample who carried the CYP4A11 8590C allele compared to TT homozygotes at the first three visits and at visit 5 (Table 3). After adjustment for age, age2, BMI, number of cigarettes smoked per day, ounces of alcohol consumed per week, and menopausal status and estrogen use, this relationship between HDL-C and genotype was significant only at visit 5. In the combined sample, HDL-C was significantly lower in women who carried the C allele at all visits in the unadjusted analysis and at visits 2, 5, and 6 after adjustment (Table 4).
Table 3.
Relationship between CYP4A11 T8590C Genotype and HDL-C in Unrelated Sample from Framingham Offspring Study
| LS Mean HDL-C | |||||
|---|---|---|---|---|---|
| CC or CT | TT | Unadjusted P | Adjusted P* | ||
| Visit 1 | Women | 54.9±1.1 | 57.6±0.6 | 0.03 | 0.12 |
| Men | 44.6±0.8 | 44.5±0.5 | 0.84 | 0.74 | |
| Combined | 49.6±0.8 | 51.1±0.4 | 0.09 | 0.15 | |
| Visit 2 | Women | 52.3±1.1 | 54.9±0.6 | 0.04 | 0.13 |
| Men | 43.0±0.8 | 43.0±0.5 | 0.92 | 0.85 | |
| Combined | 47.3±0.8 | 48.9±0.4 | 0.06 | 0.17 | |
| Visit 3 | Women | 55.3±1.2 | 57.9±0.7 | 0.05 | 0.27 |
| Men | 44.3±0.9 | 44.9±0.5 | 0.55 | 0.96 | |
| Combined | 49.5±0.8 | 51.3±0.5 | 0.06 | 0.43 | |
| Visit 4 | Women | 54.5±1.2 | 56.6±0.6 | 0.12 | 0.31 |
| Men | 43.6±0.8 | 43.1±0.5 | 0.61 | 0.67 | |
| Combined | 48.8±0.8 | 49.8 | 0.24 | 0.51 | |
| Visit 5 | Women | 53.4±1.2 | 56.7±0.6 | 0.01 | 0.02 |
| Men | 43.0±0.8 | 42.9±0.5 | 0.86 | 0.88 | |
| Combined | 48.0±0.8 | 49.9±0.4 | 0.03 | 0.06 | |
| Visit 6 | Women | 55.8±1.2 | 58.2±0.7 | 0.09 | 0.28 |
| Men | 43.4±0.9 | 43.5±0.5 | 0.92 | 0.88 | |
| Combined | 49.4±0.8 | 50.9±0.5 | 0.10 | 0.26 | |
HDL-C indicates high density lipoprotein cholesterol. Data are presented as estimated means ± standard error.
Adjusted for exam-specific age, age2, body mass index, number of cigarettes smoked per day, ounces of alcohol consumed per week and menopausal status and estrogen status in women.
Table 4.
Relationship between CYP4A11 T8590C Genotype and HDL-C in Combined Unrelated and Family Sample from Framingham Offspring Study
| LS Mean HDL-C | |||||
|---|---|---|---|---|---|
| CC or CT | TT | Unadjusted P | Adjusted P* | ||
| Visit 1 | Women | 54.9±0.9 | 57.4±0.5 | 0.01 | 0.06 |
| Men | 44.4±0.7 | 44.4±0.4 | 0.95 | 1.00 | |
| Combined | 49.9±0.6 | 51.2±0.4 | 0.07 | 0.1 | |
| Visit 2 | Women | 52.1±0.8 | 54.8±0.5 | 0.004 | 0.02 |
| Men | 42.8±0.7 | 43.0±0.4 | 0.78 | 0.76 | |
| Combined | 47.6±0.6 | 49.1±0.4 | 0.02 | 0.04 | |
| Visit 3 | Women | 55.5±0.9 | 57.9±0.5 | 0.02 | 0.07 |
| Men | 44.3±0.7 | 45.0±0.5 | 0.34 | 0.68 | |
| Combined | 50.1±0.6 | 51.6±0.4 | 0.047 | 0.09 | |
| Visit 4 | Women | 54.2±0.9 | 57.9±0.5 | 0.02 | 0.07 |
| Men | 42.9±0.7 | 43.2±0.4 | 0.62 | 0.88 | |
| Combined | 48.7±0.6 | 50.2±0.4 | 0.048 | 0.11 | |
| Visit 5 | Women | 54.4±0.9 | 56.7±0.5 | 0.02 | 0.03 |
| Men | 42.6±0.7 | 43.0±0.4 | 0.563 | 0.42 | |
| Combined | 48.8±0.6 | 50.3±0.4 | 0.05 | 0.04 | |
| Visit 6 | Women | 55.6±0.9 | 58.6±0.6 | 0.005 | 0.02 |
| Men | 42.8±0.7 | 43.6±0.4 | 0.324 | 0.33 | |
| Combined | 49.5±0.7 | 51.5±0.4 | 0.01 | 0.01 | |
HDL-C indicates high density lipoprotein cholesterol. Data are presented as estimated means ± standard error.
Adjusted for exam-specific age, age2, body mass index, number of cigarettes smoked per day, ounces of alcohol consumed per week and menopausal status and estrogen status in women.
C-reactive protein was measured at visit 5 as a marker of inflammation. Because many inflammatory biomarkers are contained within the proteome of the HDL particle, we further tested the hypothesis that CYP4A11 T8590C genotype was associated with CRP concentrations in the Framingham Offspring Study. There was no relationship between CYP4A11 T8590C genotype and CRP concentrations in men in either the unrelated or the combined sample. In women in the combined sample, CYP4A11 CC genotype was significantly associated with having a CRP in the uppermost quintile (odds ratio 4.1, 95% CI 1.6-10.1). In linear regression models, logCRP+1 was significantly increased in CYP4A11 CC homozygotes compared to in those with the TT genotype in the unrelated sample (P=0.02, P=0.06 after adjustment) and the combined unrelated and family sample (P=0.04, P=0.31 after adjustment), but not after adjustment for covariates.
Validation Cohort – BioVU
Table 5 provides the baseline characteristics of patients in the replication data set from BioVU. CYP4A11 T8590C genotypes were in Hardy-Weinberg equilibrium. Table S3 provides the prevalence of components of the metabolic syndrome for this cohort. As observed in the Framingham Offspring study, the loss-of-function CYP4A11 8590C allele was associated with increased risk of having a low HDL-C concentration in women in BioVU, but not in men (Table 6 and Figure 2). The odds ratio, level of significance, and effect size were similar to those observed in women in the Framingham Offspring cohort. None of the other variables related to the metabolic syndrome were associated with the CYP4A11 T8590C variant.
Table 5.
Characteristics of Unrelated Subjects in the Vanderbilt BioVU Replication Cohort
| Parameter | Men (348) | Women (360) | Combined (708) |
|---|---|---|---|
| Genotype, N (%) CC:CT:TT | 5(1.4):93(26.7):248(71.3) | 8(2.2):83(23.1):269(74.7) | 13(1.8):176(24.9):517(73.0) |
| Age, years | 59.3±6.4 | 58.8±6.6 | 59.0±6.5 |
| BMI, kg/M2 | 29.3±5.3 | 29.1±7.4 | 29.2±6.4 |
| SBP, mmHg | 128.1±10.4 | 125.9±11.3 | 127.0±10.9 |
| DBP, mmHg | 79.3±6.5 | 76.4±6.6 | 77.8±6.7 |
| Glucose, mg/dL | 103.5±34.3 | 99.1±31.7 | 101.3±33.1 |
| Triglycerides, mg/dL | 164.6±84.7 | 132.6±76.2 | 148.3±82.0 |
| HDL-cholesterol, mg/dL | 45.7±11.4 | 59.8±17.5 | 52.9±16.4 |
BMI indicates body mass index, SBP indicates systolic blood pressure, DBP diastolic blood pressure, HDL high density lipoprotein
Table 6.
Relationship between CYP4A11 T8590C and components of the metabolic syndrome in the Vanderbilt BioVU Cohort
| Obesity | Hypertriglyceridemia | High Blood Pressure | Impaired Fasting Glucose | Low HDL-C | Metabolic Syndrome | |
|---|---|---|---|---|---|---|
| Unadjusted P | ||||||
| All (N=708) | 0.31 | 0.54 | 0.83 | 0.28 | 0.14 | 0.92 |
| Female (N=360) | 0.32 | 0.75 | 0.93 | 0.12 | 0.03 | 0.53 |
| Male (N=348) | 0.67 | 0.73 | 0.58 | 0.82 | 0.97 | 0.37 |
| Adjusted P* | ||||||
| All (708) | 0.56 | 0.91 | 0.25 | 0.15 | 0.83 | |
| Female (N=360) | 0.91 | 0.90 | 0.06 | 0.04 | 0.76 | |
| Male (N=348) | 0.61 | 0.69 | 0.96 | 0.96 | 0.48 |
Adjusted for age, age2 and body mass index. Subjects taking estrogen were excluded. HDL-C indicates high density lipid cholesterol
Figure 2. BioVU. Relationship between CYP4A11 T8590C genotype and the likelihood of having traits related to the metabolic syndrome.
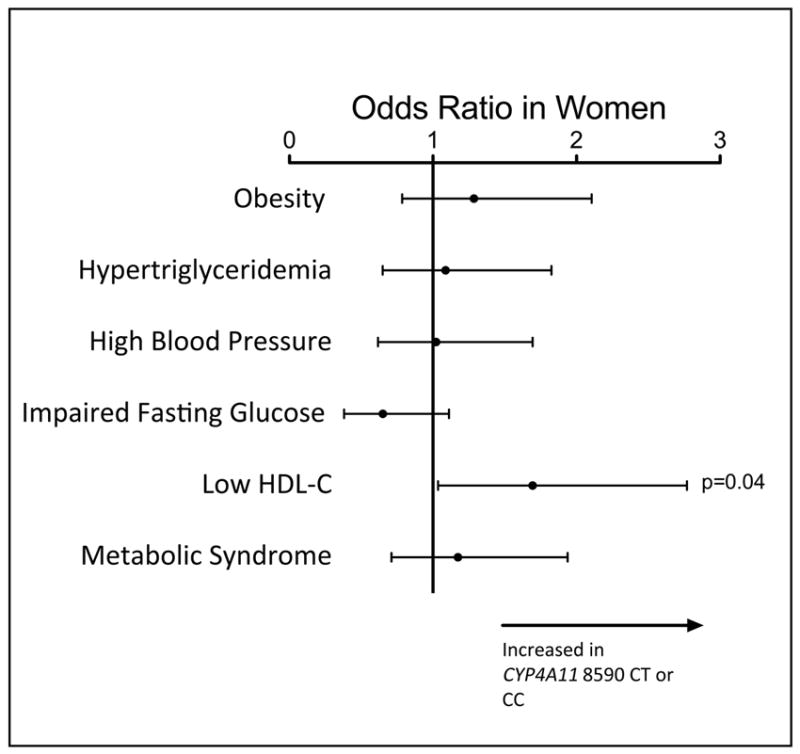
Continuous variables have been converted to binary traits based upon the criteria published by NCEP-ATPIII. Low high density lipoprotein (HDL) cholesterol was defined as <50mg/dL in women. From this clinic practice-based biobank, body mass index (BMI) >30kg/M2 was used to define obesity, rather than gender-specific waist circumference. The analysis was adjusted for age, age2, and BMI.
Discussion
The ω-hydroxylases such as CYP4A11 catalyze the metabolism of EETs to HEETs, potent endogenous PPARα agonists.[3, 4] PPARα activation can increase HDL-C by increasing concentration of apo A-I and A-II and by stimulating the reverse cholesterol transport pathway.[5] The capacity of PPARα agonists to improve dyslipidemia appears to be modulated by estradiol.[18] PPARα activation can also exert anti-inflammatory effects, suppressing the acute phase response.[6, 19] We tested the hypothesis in two independent cohorts that individuals who carry the loss-of-function CYP4A11 8590C allele have phenotypic characteristics consistent with decreased production of an endogenous PPARα agonist. We found that in women, but not in men, the CYP4A11 8590C allele was associated with lower HDL-C. Genetic variability in CYP4A11 8590C was also associated with increased inflammation, as determined by elevated CRP concentrations, in women. The magnitude of the effect of carrying the CYP4A11 8590C allele was comparable to the effect of pharmacological PPARα agonists on HDL-C and CRP.[20, 21]
Previous studies have characterized the effect of genetic variation in genes encoding for Apo A-I, the major apolipoprotein of HDL; apolipoprotein receptor ligands such as apo E, and CETP, involved in the transfer of lipids between triglyceride rich lipoproteins and HDL particles.[22] The transcription factor PPARα regulates many of the proteins involved in lipid homeostasis and rare variants in the gene encoding PPARα are associated with decreased HDL-C.[23, 24] In contrast to our findings, Hermann et al recently reported higher concentrations of HDL-C in 15 homozygotes for the CYP4A11 8590C allele compared to heterozygotes and homozygotes for the T allele in the ENCORE trials, although the authors did not conduct a multivariable analysis controlling for potentially confounding differences in age. The reason for the divergence of findings in the two populations studied here and the ENCORE trial is not immediately evident. ENCORE subjects were predominantly male and carriers of the CYP4A11 8590C allele were significantly younger than subjects in the CYP4A11 8590TT genotype group. Regardless, this and the present study provide the first evidence that variation in a gene encoding an enzyme involved in the formation of an endogenous PPARα agonist affects HDL-C concentrations.
We found that CYP4A11 T8590C genotype was associated with HDL-C only in women. HDL-C concentrations are higher in women than in men, even after menopause.[25] At the same time, the relationship between decreases in HDL-C and cardiovascular risk is more pronounced in women compared to men,[25] emphasizing the importance of understanding gene × gender interactions.
Gender-specific effects have been reported for polymorphisms in other genes affecting HDL-C. Several variants in genes encoding for apo B, apo A-I, apo A-V, CETP and scavenger receptor class B type 1 (SRB1) have been associated with HDL-C in women but not in men, whereas a -75A/G variant in apo A-I has been reported to associate with apo A concentrations in men and not women.[9, 26-30] Gender differences in PPARα expression and action are also well-established. For example, dietary fatty acid intake alters the expression of hepatic PPARα in female rodents, but not in male rodents.[31, 32] Moreover, while CYP4A11 catalyzes the formation of endogenous PPARα ligands, PPARα also regulates CYP4A11 expression and this effect is greater in female mice than in male mice.[33] Hence, it is possible that the effect of the loss-of-function CYP4A11 8590C allele on decreased formation of PPARα agonists is magnified in women.
We analyzed the relationship between CYP4A11 T8590C genotype and HDL-C as a continuous variable, as well as dichotomous variable. Although HDL-C was consistently 2-3 mg/dL lower among CYP4A11 8590C allele carriers, the association of CYP4A11 genotype with the dichotomous variable of HDL-C less than 50mg/dL was not seen after visit 3 in women of the Framingham Offspring study. This may reflect the increased prevalence of low HDL-C among women over time as well as changes in the prevalence of confounding factors with aging of the population, even though we controlled for many of these factors. Numerous environmental factors impact on HDL-C including alcohol intake, estrogen replacement, dietary fat intake, use of statins or other medications, and activity level. In the BioVU cohort, we excluded women taking estrogen and analyzed untreated lipid concentrations. Of note, Roberts et al reported an age-dependent association of variants in the PPARα target SRB1 gene (SCARB1) and HDL-C in Amish women, noting an association in women younger than 50 years but not in women aged 50 years or older.[34]
In summary, CYP4A11 T8590C genotype is associated with HDL-C and CRP in women in the Framingham Offspring study and in a biobank-based validation cohort. These data support the hypothesis that CYP4A11 catalyzes the EETs to potent PPARα agonists. Given recent data from the ENCORE trials suggesting that CYP4A11 8590CC genotype is associated with increased HDL-C concentration, additional studies are warranted to further define this relationship.
Supplementary Material
Acknowledgments
This work was funded by Vanderbilt CTSA grant 1 UL1 RR024975, and by NIH Grants R01 HL060906, R01 DK080007, and P01 DK038226. The Framingham studies are also supported by a contract from the National Heart, Lung and Blood Institute (contract no. N01-HC-25195).
Reference List
- 1.Carroll MA, McGiff JC. A new class of lipid mediators: cytochrome P450 arachidonate metabolites. Thorax. 2000;55(Suppl 2):S13–S16. doi: 10.1136/thorax.55.suppl_2.S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sarkis A, Lopez B, Roman RJ. Role of 20-hydroxyeicosatetraenoic acid and epoxyeicosatrienoic acids in hypertension. Curr Opin Nephrol Hypertens. 2004;13:205–214. doi: 10.1097/00041552-200403000-00009. [DOI] [PubMed] [Google Scholar]
- 3.Cowart LA, Wei S, Hsu MH, Johnson EF, Krishna MU, Falck JR, et al. The CYP4A isoforms hydroxylate epoxyeicosatrienoic acids to form high affinity peroxisome proliferator-activated receptor ligands. J Biol Chem. 2002;277:35105–35112. doi: 10.1074/jbc.M201575200. [DOI] [PubMed] [Google Scholar]
- 4.Gatica A, Aguilera MC, Contador D, Loyola G, Pinto CO, Amigo L, et al. P450 CYP2C epoxygenase and CYP4A omega-hydroxylase mediate ciprofibrate-induced PPARalpha-dependent peroxisomal proliferation. J Lipid Res. 2007;48:924–934. doi: 10.1194/jlr.M700002-JLR200. [DOI] [PubMed] [Google Scholar]
- 5.Nissen SE, Nicholls SJ, Wolski K, Howey DC, McErlean E, Wang MD, et al. Effects of a potent and selective PPAR-alpha agonist in patients with atherogenic dyslipidemia or hypercholesterolemia: two randomized controlled trials. JAMA. 2007;297:1362–1373. doi: 10.1001/jama.297.12.1362. [DOI] [PubMed] [Google Scholar]
- 6.Cuzzocrea S, Mazzon E, Di PR, Peli A, Bonato A, Britti D, et al. The role of the peroxisome proliferator-activated receptor-alpha (PPAR-alpha) in the regulation of acute inflammation. J Leukoc Biol. 2006;79:999–1010. doi: 10.1189/jlb.0605341. [DOI] [PubMed] [Google Scholar]
- 7.Gainer JV, Bellamine A, Dawson EP, Womble KE, Grant SW, Wang Y, et al. Functional variant of CYP4A11 20-hydroxyeicosatetraenoic acid synthase is associated with essential hypertension. Circulation. 2005;111:63–69. doi: 10.1161/01.CIR.0000151309.82473.59. [DOI] [PubMed] [Google Scholar]
- 8.Mayer B, Lieb W, Gotz A, Konig IR, Aherrahrou Z, Thiemig A, et al. Association of the T8590C polymorphism of CYP4A11 with hypertension in the MONICA Augsburg echocardiographic substudy. Hypertension. 2005;46:766–771. doi: 10.1161/01.HYP.0000182658.04299.15. [DOI] [PubMed] [Google Scholar]
- 9.Hermann M, Hellermann JP, Quitzau K, Hoffmann MM, Gasser T, Meinertz T, et al. CYP4A11 polymorphism correlates with coronary endothelial dysfunction in patients with coronary artery disease-The ENCORE Trials. Atherosclerosis. 2009 doi: 10.1016/j.atherosclerosis.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 10.Fava C, Montagnana M, Almgren P, Rosberg L, Lippi G, Hedblad B, et al. The V433M variant of the CYP4F2 is associated with ischemic stroke in male Swedes beyond its effect on blood pressure. Hypertension. 2008;52:373–380. doi: 10.1161/HYPERTENSIONAHA.108.114199. [DOI] [PubMed] [Google Scholar]
- 11.Ward NC, Tsai IJ, Barden A, van Bockxmeer FM, Puddey IB, Hodgson JM, et al. A single nucleotide polymorphism in the CYP4F2 but not CYP4A11 gene is associated with increased 20-HETE excretion and blood pressure. Hypertension. 2008;51:1393–1398. doi: 10.1161/HYPERTENSIONAHA.107.104463. [DOI] [PubMed] [Google Scholar]
- 12.Sugimoto K, Akasaka H, Katsuya T, Node K, Fujisawa T, Shimaoka I, et al. A polymorphism regulates CYP4A11 transcriptional activity and is associated with hypertension in a Japanese population. Hypertension. 2008;52:1142–1148. doi: 10.1161/HYPERTENSIONAHA.108.114082. [DOI] [PubMed] [Google Scholar]
- 13.Gainer JV, Lipkowitz MS, Yu C, Waterman MR, Dawson EP, Capdevila JH, et al. Association of a CYP4A11 variant and blood pressure in black men. J Am Soc Nephrol. 2008;19:1606–1612. doi: 10.1681/ASN.2008010063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kannel WB, Feinleib M, McNamara PM, Garrison RJ, Castelli WP. An investigation of coronary heart disease in families. The Framingham offspring study. Am J Epidemiol. 1979;110:281–290. doi: 10.1093/oxfordjournals.aje.a112813. [DOI] [PubMed] [Google Scholar]
- 15.Ritchie MD, Denny JC, Crawford DC, Ramirez AH, Weiner JB, Pulley JM, et al. Robust replication of genotype-phenotype associations across multiple diseases in an electronic medical record. Am J Hum Genet. 2010;86:560–572. doi: 10.1016/j.ajhg.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilke RA. High-density lipoprotein (HDL) cholesterol: leveraging practice-based biobank cohorts to characterize clinical and genetic predictors of treatment outcome. Pharmacogenomics J. 2010 doi: 10.1038/tpj.2010.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Denny JC, Ritchie MD, Basford MA, Pulley JM, Bastarache L, Brown-Gentry K, et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics. 2010;26:1205–1210. doi: 10.1093/bioinformatics/btq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoon M. The role of PPARalpha in lipid metabolism and obesity: focusing on the effects of estrogen on PPARalpha actions. Pharmacol Res. 2009;60:151–159. doi: 10.1016/j.phrs.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 19.Kleemann R, Gervois PP, Verschuren L, Staels B, Princen HM, Kooistra T. Fibrates down-regulate IL-1-stimulated C-reactive protein gene expression in hepatocytes by reducing nuclear p50-NFkappa B-C/EBP-beta complex formation. Blood. 2003;101:545–551. doi: 10.1182/blood-2002-06-1762. [DOI] [PubMed] [Google Scholar]
- 20.Scott R, Best J, Forder P, Taskinen MR, Simes J, Barter P, et al. Fenofibrate Intervention and Event Lowering in Diabetes (FIELD) study: baseline characteristics and short-term effects of fenofibrate [ISRCTN64783481] Cardiovasc Diabetol. 2005;4:13. doi: 10.1186/1475-2840-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keech A, Simes RJ, Barter P, Best J, Scott R, Taskinen MR, et al. Effects of long-term fenofibrate therapy on cardiovascular events in 9795 people with type 2 diabetes mellitus (the FIELD study): randomised controlled trial. Lancet. 2005;366:1849–1861. doi: 10.1016/S0140-6736(05)67667-2. [DOI] [PubMed] [Google Scholar]
- 22.Do HQ, Nazih H, Luc G, Arveiler D, Ferrieres J, Evans A, et al. Influence of cholesteryl ester transfer protein, peroxisome proliferator-activated receptor alpha, apolipoprotein E, and apolipoprotein A-I polymorphisms on high-density lipoprotein cholesterol, apolipoprotein A-I, lipoprotein A-I, and lipoprotein A-I:A-II concentrations: the Prospective Epidemiological Study of Myocardial Infarction study. Metabolism. 2009;58:283–289. doi: 10.1016/j.metabol.2008.09.026. [DOI] [PubMed] [Google Scholar]
- 23.Flavell DM, Pineda TI, Jamshidi Y, Evans D, Diamond JR, Elkeles RS, et al. Variation in the PPARalpha gene is associated with altered function in vitro and plasma lipid concentrations in Type II diabetic subjects. Diabetologia. 2000;43:673–680. doi: 10.1007/s001250051357. [DOI] [PubMed] [Google Scholar]
- 24.Chan E, Tan CS, urenberg-Yap M, Chia KS, Chew SK, Tai ES. The V227A polymorphism at the PPARA locus is associated with serum lipid concentrations and modulates the association between dietary polyunsaturated fatty acid intake and serum high density lipoprotein concentrations in Chinese women. Atherosclerosis. 2006;187:309–315. doi: 10.1016/j.atherosclerosis.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 25.Abbott RD, Wilson PW, Kannel WB, Castelli WP. High density lipoprotein cholesterol, total cholesterol screening, and myocardial infarction. The Framingham Study. Arteriosclerosis. 1988;8:207–211. doi: 10.1161/01.atv.8.3.207. [DOI] [PubMed] [Google Scholar]
- 26.Kessling A, Ouellette S, Bouffard O, Chamberland A, Betard C, Selinger E, et al. Patterns of association between genetic variability in apolipoprotein (apo) B, apo AI-CIII-AIV, and cholesterol ester transfer protein gene regions and quantitative variation in lipid and lipoprotein traits: influence of gender and exogenous hormones. Am J Hum Genet. 1992;50:92–106. [PMC free article] [PubMed] [Google Scholar]
- 27.Komurcu-Bayrak E, Onat A, Poda M, Humphries SE, Palmen J, Guclu F, et al. Gender-modulated impact of apolipoprotein A5 gene (APOA5) -1131T>C and c.56C>G polymorphisms on lipids, dyslipidemia and metabolic syndrome in Turkish adults. Clin Chem Lab Med. 2008;46:778–784. doi: 10.1515/CCLM.2008.161. [DOI] [PubMed] [Google Scholar]
- 28.Ordovas JM. HDL genetics: candidate genes, genome wide scans and gene-environment interactions. Cardiovasc Drugs Ther. 2002;16:273–281. doi: 10.1023/a:1021769523568. [DOI] [PubMed] [Google Scholar]
- 29.Saha N, Tay JS, Low PS, Humphries SE. Guanidine to adenine (G/A) substitution in the promoter region of the apolipoprotein AI gene is associated with elevated serum apolipoprotein AI levels in Chinese non-smokers. Genet Epidemiol. 1994;11:255–264. doi: 10.1002/gepi.1370110304. [DOI] [PubMed] [Google Scholar]
- 30.Sigurdsson G, Jr, Gudnason V, Sigurdsson G, Humphries SE. Interaction between a polymorphism of the apo A-I promoter region and smoking determines plasma levels of HDL and apo A-I. Arterioscler Thromb. 1992;12:1017–1022. doi: 10.1161/01.atv.12.9.1017. [DOI] [PubMed] [Google Scholar]
- 31.Morise A, Mourot J, Boue C, Combe N, Amsler G, Gripois D, et al. Gender-related response of lipid metabolism to dietary fatty acids in the hamster. Br J Nutr. 2006;95:709–720. doi: 10.1079/bjn20051721. [DOI] [PubMed] [Google Scholar]
- 32.Morise A, Thomas C, Landrier JF, Besnard P, Hermier D. Hepatic lipid metabolism response to dietary fatty acids is differently modulated by PPARalpha in male and female mice. Eur J Nutr. 2009;48:465–473. doi: 10.1007/s00394-009-0037-7. [DOI] [PubMed] [Google Scholar]
- 33.Savas U, Machemer DE, Hsu MH, Gaynor P, Lasker JM, Tukey RH, et al. Opposing roles of peroxisome proliferator-activated receptor alpha and growth hormone in the regulation of CYP4A11 expression in a transgenic mouse model. J Biol Chem. 2009;284:16541–16552. doi: 10.1074/jbc.M902074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roberts CG, Shen H, Mitchell BD, Damcott CM, Shuldiner AR, Rodriguez A. Variants in scavenger receptor class B type I gene are associated with HDL cholesterol levels in younger women. Hum Hered. 2007;64:107–113. doi: 10.1159/000101962. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


