Abstract
The role of the hydrophobic side chains of Ile-172 and Leu-232 in catalysis of the reversible isomerization of R-glyceraldehyde 3-phosphate (GAP) to dihydroxyacetone phosphate (DHAP) by triosephosphate isomerase (TIM) from Trypanosoma brucei brucei (Tbb) has been investigated. The I172A and L232A mutations result in 100- and 6-fold decreases in kcat/Km for the isomerization reaction, respectively. The effect of the mutations on the product distributions for the catalyzed reactions of GAP and of [1-13C]-glycolaldehyde ([1-13C]-GA) in D2O is reported. The 40% yield of DHAP from wildtype TbbTIM-catalyzed isomerization of GAP with intramolecular transfer of hydrogen is found to decrease to 13% and to 4%, respectively, for the reactions catalyzed by the I172A and L232A mutants. Likewise, the 13% yield of [2-13C]-GA from isomerization of [1-13C]-GA in D2O is found to decrease to 2% and to 1%, respectively, for the reactions catalyzed by the I172A and L232A mutants. The decrease in the yield of the product of intramolecular transfer of hydrogen is consistent with a repositioning of groups at the active site that favors transfer of the substrate-derived hydrogen to the protein or the oxygen anion of the bound intermediate. The I172A and L232A mutations result in: (a) A >10-fold decrease (I172A) and a 17-fold increase (L232A) in the second-order rate constant for TIM-catalyzed reaction of [1-13C]-GA in D2O. (b) A 170-fold decrease (I172A) and 25-fold increase (L232A) in the third-order rate constant for phosphite dianion (HPO32-) activation of TIM-catalyzed reaction of GA in D2O. (c) A 1.5-fold decrease (I172A) and a larger 16-fold decrease (L232A) in Kd for activation of TIM by HPO32- in D2O. The effects of the I172A mutation on the kinetic parameters for wildtype TIM-catalyzed reactions of the whole substrate and substrate pieces are consistent with a decrease in the basicity of the carboxylate side chain of Glu-167 for the mutant enzyme. The data provide striking evidence that the L232A mutation leads to a ca. 1.7 kcal/mol stabilization of a catalytically active loop-closed form of TIM (EC) relative to an inactive open form (EO).
Introduction
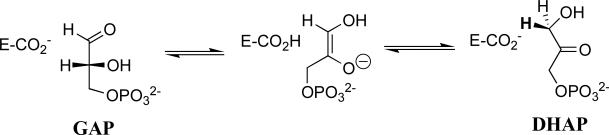
Triosephosphate isomerase (TIM) is the prototypical protein catalyst of proton transfer at α-carbonyl carbon.1-5 This small protein (dimer, 26 kDa/subunit) catalyzes the reversible 1,2-shift of the pro-R proton at dihydroxyacetone phosphate (DHAP) to give (R)-glyceraldehyde 3-phosphate (GAP) by a single-base proton transfer mechanism through an enzyme-bound cis-enediolate intermediate (Scheme 1).1,6 The most important metabolic role for TIM is to enable DHAP, a product of aldolase-catalyzed cleavage of fructose 1,6-bisphosphate, to enter glycolysis. The enzyme has the more general “housekeeping” function of maintaining rapid equilibrium between GAP and DHAP so that these compounds are available, as needed, for glycolysis, the glycerol-3-phosphate shuttle and the pentose phosphate pathway.7 Deficiencies in TIM are recognized as a recessive genetic disease associated with chronic hemolytic anemia and progressive neuromuscular dysfunction that often result in early childhood death.7,8
Scheme 1.
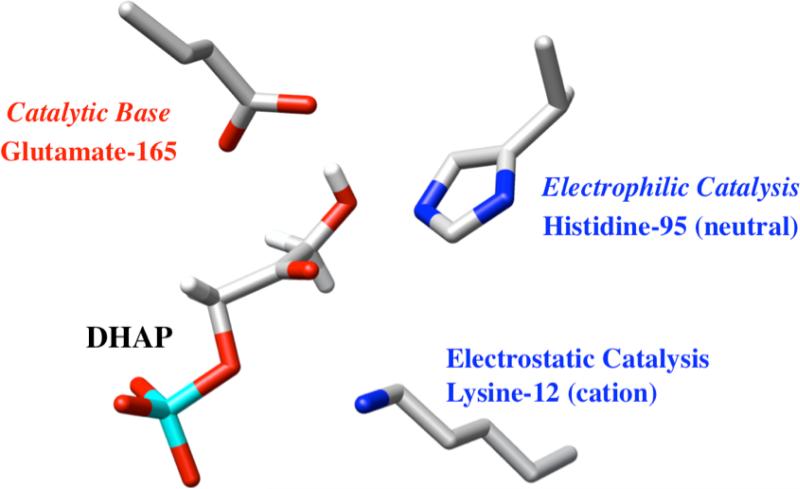
The chemical mechanism for TIM-catalyzed isomerization of triosephosphates was established by the early 1990s (Figure 1). Isomerization at the active site of TIM is similar to nonenzymatic isomerization in water.9 Catalysis proceeds by deprotonation of the bound substrate using the carboxylate side chain of Glu-165 10,11 and is assisted by the neutral electrophilic imidazole side chain of His-95.12,13 The alkylammonium side chain of Lys-12 plays an important electrostatic role in stabilizing the negatively charged bound substrate.14-17 We have proposed that the functional groups of these side chains are activated for catalysis compared to the corresponding groups in water.5,18 We present here the results of experiments designed to probe the mechanism for side-chain activation.
Figure 1.
The orientation of the catalytic side chains at the active site of TIM from yeast [PDB entry 1NEY]
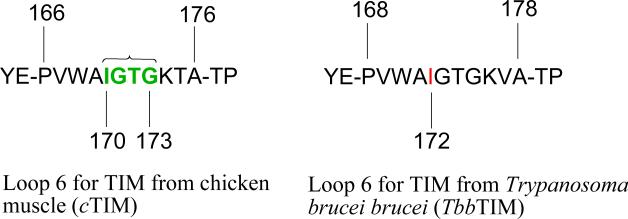
Binding interactions between TIM and the phosphodianion group of GAP account for 80% of the ca. 14 kcal/mol enzymatic stabilization of the transition state for deprotonation of carbon.19 Roughly one-half of this binding energy is recovered in phosphite dianion (HPO32-) activation of TIM for catalysis of deprotonation of the truncated substrate glycolaldehyde.20 High-resolution X-ray crystal structures of complexes between TIM and substrate DHAP 21 or between TIM and the tight-binding inhibitors phosphoglycolate (PGA)22 and phosphoglycolohydroxamate (PGH)23,24 show that the phosphodianion binding interactions are utilized to drive a large conformational change of the enzyme, the most prominent feature of which is closure of flexible loop 6 (residues 166 – 176 for TIM from chicken muscle, Scheme 2), over the enzyme active site.21,25-27 The deletion of residues 170-173 (green in Scheme 2), and the introduction of a peptide bond between Ala-169 and Lys-174 disrupt the loop-dianion interactions without significantly affecting the protein fold. This results in a substantial 105-fold decrease in kcat and a 2.3-fold increase in Km for isomerization of GAP,28 which serve as direct evidence that interactions between loop 6 and the substrate dianion activate TIM for catalysis.
Scheme 2.
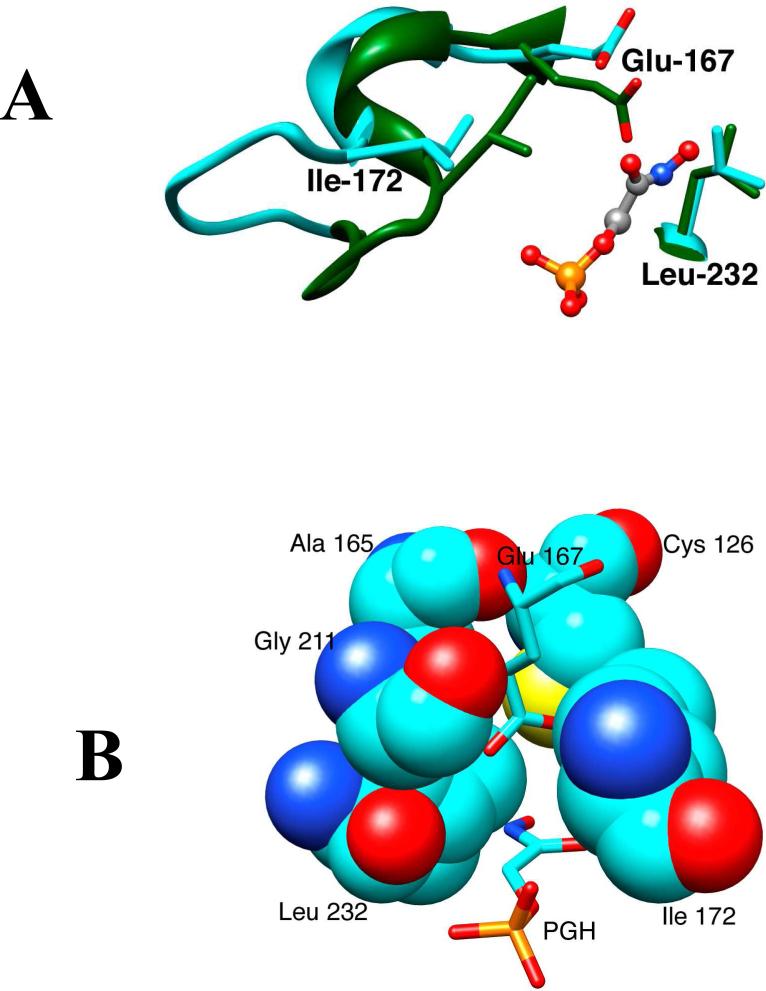
Wierenga has provided a lucid and detailed description of the many changes in protein structure observed to occur upon ligand binding to TIM.4,29 The closure of loop 6 of TIM from Trypanosoma brucei brucei (TbbTIM, residues 168 – 178 shown in Scheme 2) over bound PGA results in movement of the hydrophobic side chain of Ile-172 [Ile-170 for cTIM] toward the carboxylate side chain of Glu-167 and “drives” this anionic side chain toward the hydrophobic side chain of Leu-232 [Leu-230 for cTIM], which maintains a nearly fixed position (Figure 2A).22 The result is to sandwich the carboxylate anion between the hydrophobic side-chains of Ile-172 and Leu-232 and to shield the anion from interactions with the aqueous solvent (Figure 2B).22,23,30 At the same time, loop closure extrudes several water molecules to the bulk solvent. Desolvation of the active-site should result in an increase in the basicity of the carboxylate side chain that will activate the side chain for deprotonation of bound substrate.5,18 This analysis suggests that the greasy side chains of Ile-172 and Leu-232 of TbbTIM play a critical role in TIM-catalyzed deprotonation of carbon.
Figure 2.
(A) Models from X-ray crystal structures of the unliganded open (cyan, PDB entry 5TIM) and PGH-liganded closed (green, PDB entry 1TRD) forms of TIM from Trypanosoma brucei brucei in the region of the enzyme active site. Closure of loop 6 (residues 168 – 178, Scheme 2) over the bound phosphodianion ligand results in movement of the hydrophobic side chain of Ile-172 toward the carboxylate side chain of the catalytic base Glu-167. This is accompanied by movement of Glu-167 toward the hydrophobic side chain of Leu-232, that maintains a nearly fixed position. (B) A structure that shows the positions of Ile-172 and Leu-232 at the active site “hydrophobic cage” for wildtype Trypanosoma brucei TIM in complex with PGH (PDB entry 1TRD).
We recently reported the surprising result that the L232A mutation of TbbTIM, which we thought would be crippling, instead causes a 17-fold increase in the second-order rate constant for enzyme-catalyzed deprotonation of the truncated substrate glycolaldehyde!31 Additional kinetic data reported for the catalyzed reactions of the substrate pieces glycolaldehyde and HPO32- support the conclusion that the L232A mutation results in the stabilization of a high-energy, catalytically active loop-closed form of TIM by 1.7 kcal/mol relative to the inactive open form of TIM.31 We now report the results of an expanded study on the effect of I172A, I172V and L232A mutations on the kinetic parameters and the products of TIM-catalyzed reactions of the whole substrates GAP and DHAP, and the substrate pieces. We had envisioned similar functions for the hydrophobic side chains of Ile-172 and Leu-232 in the desolvation and “clamping” of the carboxylate side chain of Glu-167. However, we observe that the I172A and L232A mutations result in very different changes in the kinetic parameters of the wildtype TbbTIM, which show that the two clamping side-chains play different roles in catalysis.
EXPERIMENTAL
Materials
Rabbit muscle α−glycerol 3-phosphate dehydrogenase and glyceraldehyde 3-phosphate dehydrogenase were purchased from Sigma. These enzymes were exhaustively dialyzed against 20 mM triethanolamine buffer (pH 7.5) at 7 °C prior to their use in coupled enzyme assays. Bovine serum albumin (BSA) was from Roche. CM Sepharose Fast Flow was from GE Healthcare. d,l-Glyceraldehyde 3-phosphate diethyl acetal (barium salt), DHAP (lithium or magnesium salt), NADH (disodium salt), dithiothreitol (DTT), Dowex 50WX4-200R, triethanolamine hydrochloride (TEA•HCl) and imidazole were purchased from Sigma. NAD (free acid) was purchased from MP Biomedicals or Calzyme, and sodium phosphite (dibasic, pentahydrate) and hydrogen arsenate heptahydrate were from Fluka. Sodium phosphite was dried under vacuum prior to use.20 [1-13C]-Glycolaldehyde ([1-13C]-GA, 99% enriched with 13C at C-1, 0.09 M in water) was purchased from Omicron Biochemicals. Deuterium oxide (99% D) and deuterium chloride (35% w/w, 99.9% D) were from Cambridge Isotope Laboratories. The barium salt of d-glyceraldehyde 3-phosphate diethyl acetal was prepared according to a literature procedure.32 Imidazole was recrystallized from benzene. All other chemicals were reagent grade or better and were used without further purification.
Stock solutions of GAP and d,l-glyceraldehyde 3-phosphate (d,l-GAP) at pH 7.5 were prepared by hydrolysis of the corresponding diethyl acetals (barium salt) using Dowex 50WX4-200R (H+ form) in boiling H2O or D2O as described previously.33 The resulting solutions were stored at -20 °C and adjusted to the appropriate pH or pD by the addition of 1 M NaOH or 1 M NaOD before use in enzyme assays.
The pTIM plasmid that contains the gene for wildtype TIM from Trypanosoma brucei brucei (Tbb) was a generous gift from Professor Rik Wierenga. The L232A mutant of TIM was prepared as described in earlier work.31 Site-directed mutagenesis to introduce the I172A and I172V mutations was performed by following the Stratagene protocol and using Pfu Ultrahigh Fidelity DNA polymerase. The primers 5′-CCC-GTT-TGG-GCC-GCG-GGT-ACC-GGC-AAG-GTG-GCG-ACA-CC-3′ and 5′-CCC-GTT-TGG-GCC-GTC-GGT-ACC-GGC-AAG-GTG-GCG-ACA-CC-3′ were used, respectively, to introduce the I172A and I172V mutations. The product of the PCR reaction in a volume of 30 μL was treated with 20 units of the restriction enzyme DpnI at 37 °C for one hour in order to degrade the methylated DNA. The E. coli strain K802 was transformed with 1 μL of the DpnI-digested PCR product. Several colonies were selected, and the plasmid DNA was purified using the QIAprep Miniprep Kit from Qiagen. The DNA sequences of the genes for the I172A and I172V mutant enzymes were verified by sequencing at the Roswell Park Cancer Institute (Buffalo, NY).
The I172A, I172V and L232 mutant enzymes were expressed using the E. coli BL21 pLysS strain grown in LB medium at 18 °C and the proteins were purified by a published protocol.34 The enzymes obtained from chromatography over a CM Sepharose column were judged to be homogenous by gel electrophoresis. The concentration of the protein was determined from the absorbance at 280 nm using the extinction coefficient of 3.5 × 104 M-1cm-1 calculated using the ProtParam tool available on the Expasy server.35,36
Preparation of Solutions
Solution pH or pD was determined at 25 °C using an Orion model 720A pH meter equipped with a radiometer pHC4006-9 combination electrode that was standardized at pH 7.0 and 4.0 or pH 7.0 and 10.0 at 25 °C. Values of pD were obtained by adding 0.40 to the observed reading of the pH meter.37 Imidazole and phosphite buffers were prepared as described in previous work.20,38 Triethanolamine buffers at pH 7.5 were prepared by neutralization of the hydrochloride salt with 1 M NaOH. Solutions of [1-13C]-GA were prepared and the concentration of this compound determined as described in earlier work.20
Enzyme Assays
All enzyme assays were carried out at 25 °C. One unit of enzyme activity is defined as the amount of enzyme that converts one mol of substrate to product in one minute under the specified reaction conditions. The change in the concentration of NADH was calculated from the change in absorbance at 340 nm using an extinction coefficient of 6220 M-1 cm-1. α-Glycerol 3-phosphate dehydrogenase was assayed by monitoring the oxidation of NADH by DHAP, as described in earlier work.33 Glyceraldehyde 3-phosphate dehydrogenase was assayed by monitoring the enzyme-catalyzed reduction of NAD+ by GAP. The assay solution (1.0 mL) contained 30 mM TEA (pH 7.5, I = 0.1, NaCl), 1 mM NAD+, 2 mM GAP, 5 mM disodium hydrogen arsenate, 3 mM DTT and 0.01% BSA.
The TIM-catalyzed isomerization of GAP was monitored by coupling the formation of DHAP to the oxidation of NADH catalyzed by α-glycerol 3-phosphate dehydrogenase.39 The TIM-catalyzed isomerization of DHAP was monitored by coupling the formation of GAP to the reduction of NAD+ catalyzed by glyceraldehyde 3-phosphate dehydrogenase.40 The assay mixtures (1.0 mL) contained 30 mM TEA (pH 7.5, I = 0.1, NaCl), DHAP (0.15 – 5.0 mM), 1 mM NAD, sodium arsenate (2 – 15 mM), 3 mM DTT, 0.01% BSA, 1 unit of GAPDH, and ca. 0.8 nM wildtype TbbTIM.
The values of kcat and Km for mutant TbbTIM-catalyzed isomerization of GAP or DHAP were determined from the fit to the Michaelis-Menten equation of initial velocities (vi) determined at varying concentrations of substrate. The arsenate dianion used with the glyceraldehyde 3-phosphate dehydrogenase coupling enzyme in the assay for TIM-catalyzed isomerization of DHAP is a competitive inhibitor of wildtype TbbTIM with Ki = 4.6 mM.41 Values for kcat, Km and Ki for mutant TbbTIM-catalyzed reaction of DHAP were determined from the nonlinear least squares fit to eq 1 of initial velocities determined at varying concentrations of arsenate dianion and DHAP.
| (1) |
1H NMR Analyses
1H NMR spectra at 500 MHz were recorded in D2O at 25 °C using a Varian Unity Inova 500 spectrometer that was shimmed to give a line width ≤ 0.7 Hz for each peak of the doublet due to the C-1 proton of GAP hydrate, or ≤ 0.5 Hz for the most downfield peak of the double triplet due to the C-1 proton of [1-13C]-GA hydrate. Spectra (16-64 transients) were obtained using a sweep width of 6000 Hz, a pulse angle of 90°, an acquisition time of 6 s and a relaxation delay of 60 s (>5T1) for experiments on the TIM-catalyzed isomerization of GAP in D2O or 120 s (>8T1) for experiments on the TIM-catalyzed reaction of [1-13C]-GA in D2O.33,38 It was shown in a control experiment with GAP that identical relative peak areas for substrate and product protons were observed for spectra determined using the long 120 s and the shorter 60 s delay time. Baselines were subjected to a first-order drift correction before determination of peak areas. Chemical shifts are reported relative to HOD at 4.67 ppm.
Isomerization of GAP in D2O
The yields of the products of I172A and L232A mutant TbbTIM-catalyzed reaction of GAP in D2O at 25 °C were determined by 1H NMR analysis, as described in studies on wildtype TIM.33,39 The enzyme was exhaustively dialyzed at 7 °C against 40 mM imidazole (70% free base) in D2O at pD 7.9 and I = 0.1 (NaCl). The reaction was initiated by adding enzyme to give a final solution that contained 5 or 10 mM GAP, 20 mM imidazole (pD 7.9 at I = 0.1, NaCl), and 3.4 nM L232A TbbTIM or 65 nM I172A TbbTIM in a volume of 750 μL. The reactions at 25 °C were monitored by 1H NMR spectroscopy. Spectra (16 transients) were recorded continuously during the reaction until the conversion of 80 – 90% of GAP to products. The peak areas were normalized using the signal for the C-(4,5) protons of the imidazole buffer as an internal standard. The fraction of the remaining substrate GAP (fGAP), and the fractional product yields were determined from the integrated areas of the appropriate 1H NMR signals, as described in earlier work.33
Reactions of [1-13C]-GA in D2O
The yields of the I172V, I172A and L232A mutant TbbTIM-catalyzed reactions of [1-13C]-GA in the presence or absence of HPO32- in D2O at 25 °C were determined by 1H NMR analyses as described in earlier work.38,39 The enzyme was exhaustively dialyzed at 7 °C against 30 mM imidazole (20% free base, pD 7.0) in D2O (I = 0.024 or 0.1 (NaCl)). The reactions in the absence of phosphite were initiated by adding enzyme to give a final solution that contained 20 mM [1-13C]-GA, 20 mM imidazole (20% free base, pD 7.0, I = 0.1 (NaCl)) and enzyme [0.16 – 0.34 mM L232A TbbTIM; 0.4 mM I172V TbbTIM; 0.3 – 0.7 mM I172A TbbTIM] in a volume of 850 μL. The reactions in the presence of HPO32- were initiated by adding enzyme to give a final solution that contained 20 mM [1-13C]-GA, 20 mM imidazole (20% free base, pD 7.0), 1 – 40 mM HPO32- (50% dianion, pD 7.0) and enzyme [1 - 50 μM L232A or I172V TbbTIM; 150 – 700 μM I172A TbbTIM] in a volume of 850 μL (I = 0.1). In every case, 750 μL of the reaction mixture was transferred to an NMR tube, the 1H NMR spectrum was recorded immediately and spectra were then recorded at regular intervals. The remaining solution was incubated at 25 °C and the activity of TIM towards catalysis of isomerization of GAP was monitored. No significant loss in activity of TIM was observed during any of these reactions. At the end of each NMR experiment, the protein was removed by ultrafiltration and the solution pD was determined. There was no significant change in pD during any of these reactions.
The observed 1H NMR peak areas for the reaction products were normalized, as described in previous work, using the signal due to the C-(4,5) protons of imidazole or the upfield peak of the doublet due to the P – H proton of phosphite as an internal standard.38 The fraction of the substrate [1-13C]-GA remaining and the fractional yields of the identifiable reaction products [2-13C]-GA, [2-13C, 2-2H]-GA, [1-13C, 2-2H]-GA and [1-13C, 2,2-di-2H]-GA (Chart 1), were determined from the integrated areas of the relevant 1H NMR signals for these compounds as described in earlier work.38,39 The disappearance of 30 – 70% [1-13C]-GA was monitored, and product yields were determined over the first ca. 20 – 30% of the reaction
Chart 1.
Observed first-order rate constants, kobs (s-1), for the reactions of [1-13C]-GA were determined from the slopes of linear semi-logarithmic plots of reaction progress against time (eq 2), where fS is the fraction of [1-13C]-GA remaining at time t. Observed second-order rate constants, (kcat/Km)obs (M-1 s-1), for the TIM-catalyzed reaction of [1-13C]-GA were determined from the values of kobs using eq 3, where fhyd = 0.94 is the fraction of [1-13C]-GA present as the hydrate.20
| (2) |
| (3) |
RESULTS
The kinetic parameters for wildtype and I172A, I172V and L232A mutant TIM-catalyzed isomerization of GAP and DHAP, determined at pH 7.5 and 25 °C (I = 0.1, NaCl), are reported in Table 1. Arsenate is a required activator of glyceraldehyde 3-phosphate dehydrogenase, that was used as the coupling enzyme in assays of TIM-catalyzed isomerization of DHAP. Table 1 also reports the values of Ki for arsenate inhibition of this TIM-catalyzed reaction. The values of kcat/Km were calculated using the total concentration of GAP in the enzyme assay. However, GAP exists largely (96%) in the hydrated form, and TIM is specific for the carbonyl form of this substrate (4%).42 The values for kcat/Km calculated for catalysis of the reactive carbonyl form of GAP are given in parenthesis in Table 1.
Table 1.
Kinetic Parameters for the Isomerization Reactions of GAP and DHAP Catalyzed by TIM from Chicken Muscle, from Trypanosoma brucei brucei, and by Mutant Enzymes.a
| TIM | GAP | DHAP | |||||
|---|---|---|---|---|---|---|---|
| kcatb (s-1) | Kmb (M) | kcat/Kmb,c (M-1 s-1) | kcatd (s-1) | Kmd (mM) | kcat/Kmd (M-1 s-1) | Ki (mM)d (arsenate) | |
| WT Tbbe | 2100 | 2.5 × 10-4 | 8.4 × 106 (1.7 × 108) | 300 | 7.0 × 10-4 | 4.3 × 105 | 4.6f |
| I172A Tbb | 12 | 1.5 × 10-4 | 8.0 × 104 (1.6 × 106) | 17 | 3.7 × 10-3 | 4.6 × 103 | 8.6 |
| L232A Tbbg | 220 | 1.4 × 10-4 | 1.5 × 106 (3.0 × 107) | 4.7 | 7.7 × 10-5 | 6.1 × 104 | 4.6 |
| I172V Tbb | 225 | 5.8 × 10-5 | 3.9 × 106 (7.8 × 107) | 65 | 2.0 × 10-4 | 3.2 × 105 | 4.5 |
| WT chickenh | 8300 | 4.2 × 10-4 | 2.0 × 107 | 600 | 6.5 × 10-4 | 9.2 × 105 | 11 |
| E165D chickenh | 4.2 | 7.8 × 10-5 | 5.4 × 104 | 4.1 | 1.2 × 10-3 | 3.4 × 103 | 12 |
| E165D/S96P chickenh | 68 | 6.6 × 10-5 | 1.0 × 106 | 3.4 | 5.3 × 10-5 | 6.4 × 104 | 2.1 |
Under standard assay conditions: 30 mM TEA, pH 7.5 and 25 °C (I = 0.1, NaCl). In most cases, the variation in the values of kcat and Km determined in different experiments is less than ± 15%.
Determined from the fit to the Michaelis-Menten equation of initial velocities (vi) determined at ≥ 8 concentrations of GAP.
The values in parenthesis are calculated for catalysis of the reactive carbonyl form of GAP, that is present as 4% of the total substrate.42
Determined from the fit of the initial velocities (vi) to eq 1. The concentration of arsenate was held constant as values of vi were determined at eight different [DHAP]. This was repeated for a total of three fixed arsenate concentrations between 2 and 15 mM.
Data from ref 39, unless noted otherwise.
Data from ref 39.
Data from ref 31.
Data from ref 43.
TIM-Catalyzed Isomerization of GAP in D2O
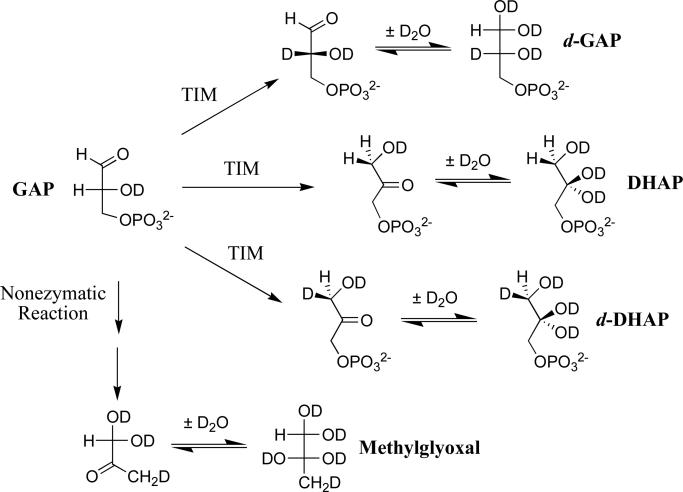
The disappearance of GAP and the appearance of the products of its nonenzymatic and TIM-catalyzed reactions in D2O (Scheme 3) at pD 7.9 (20 mM imidazole) and 25 °C were monitored by 1H NMR spectroscopy.17,33,39 Methylglyoxal is the product of the nonenzymatic elimination reaction of GAP (Scheme 3).9 The observed fractional product yields, (fP)obs, were calculated as the ratio AP/ ΣAP, where AP is the normalized 1H NMR peak area of a single product proton and ΣAP, is the sum of the normalized peak areas of single protons for every reaction product (eq 4). Values of (fP)obs were determined at 4 – 6 different times over the first three hours of the reaction, during which time > 50% of GAP was consumed. The thermodynamically favored TIM-catalyzed conversion of d-GAP to d-DHAP results in <10% decreases and increases, respectively, in the apparent yields of these products.33,39 Values of fT (Table 2) were determined as the y-intercept of linear plots (not shown) of (fP)obs against the reaction time.33,39 The values of fE (eq 5-7) reported in Table 2 are the yields calculated as a fraction of the three products of the TIM-catalyzed reactions of GAP (Scheme 3).
| (4) |
| (5) |
| (6) |
| (7) |
Scheme 3.
Table 2.
Product Yields, Expressed as Mole Fractions, and the Derived Product Rate Constant Ratios for Reaction of GAP in D2O Catalyzed by Wildtype and Mutant Forms of TbbTIM at 25 °C.a
| TIM | Fractional Product Yieldb | ||||
|---|---|---|---|---|---|
| DHAP | d-DHAP | d-GAP | MG | ||
| L232A TIMc | f T d | 0.036 | 0.75 | 0.086 | 0.12 |
| f E e | 0.041 | 0.86 | 0.098 | ||
| I172A TIMf | f T d | 0.12 | 0.33 | 0.45 | 0.10 |
| f E e | 0.13 | 0.37 | 0.50 | ||
| Wildtype TbbTIMg | f E | 0.40 ± 0.02 | 0.37 ± 0.02 | 0.22 ± 0.002 | |
| Rate Constant Ratioh | kex/(kC1)H | (kC1)D/(kC2)D |
|---|---|---|
| L232A TIM | 23 | 8.8 |
| I172A TIM | 6.7 | 0.74 |
| Wildtype TbbTIM | 1.5 | 1.7 |
For the reaction of GAP at pD 7.9 (20 mM imidazole) and I = 0.1 (NaCl).
The product yields (Scheme 3) were determined at 4 – 6 different times over a 6-8 hour reaction time.
For the reaction of 5 mM GAP catalyzed by 3.4 nM Tbb L232A.
The yields, fT, reported in this table were determined as the y-intercept of linear plots of (fP)obs (eq 4) against time.33 The product yields determined in different experiments carried out under the same reaction conditions are typically reproducible to better than 5%.17,33,39
For the reaction of 5 mM GAP catalyzed by 65 nM Tbb I172A TIM.
Data from ref 39.
The ratios of rate constants, defined in Figure 4, calculated from the ratios of product yields as described in the text.
TIM-Catalyzed Reaction of [1-13C]-GA in D2O
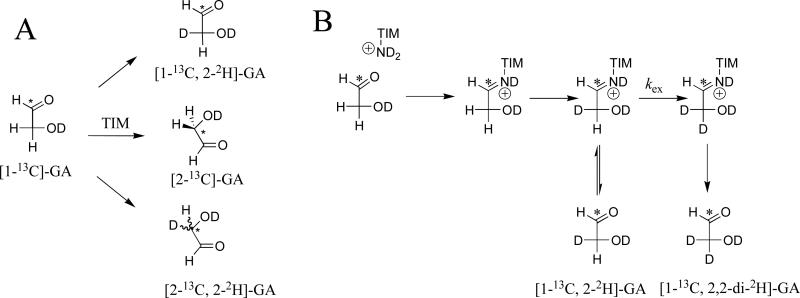
The mutant TbbTIM-catalyzed reactions of [1-13C]-GA in D2O were monitored by 1H NMR spectroscopy.38 The experimental results provide both the kinetic parameters for the TIM-catalyzed reaction of [1-13C]-GA and the yield for the reaction products (Chart 1). The product yields were determined at four different times during the first 20-30% of the reaction of [1-13C]-GA and are invariant (± 5%) over this time. The product yields reported in Table 3 were calculated as the average of 3 or 4 separate determinations. The disappearance of [1-13C]-GA was monitored for 30 – 70% of the reaction. The observed second-order rate constants (kcat/Km)obs (M-1 s-1) for the TIM-catalyzed reaction of [1-13C]-GA were determined from the values of kobs using eq 3.20,38 Values of (kcat/Km) for reactions at the active site of TIM were determined using eq 8, where fE is the sum of the fractional yields of [1-13C, 2-2H]-GA, [2-13C]-GA and [2-13C, 2-2H]-GA from TIM-catalyzed reactions of [1-13C]-GA (Scheme 4A).
Table 3.
Product Yields, Expressed as Mole Fractions, from the Reactions of [1-13C]-GA Catalyzed by Mutant TbbTIMs in the Absence and Presence of Phosphite Dianion in D2O at 25 °C.a
| TIM-mutant | [HPO32-]/mM | [2-13C]-GA | [2-13C, 2-2H]-GA | [1-13C, 2-2H]-GA | [1-13C, 2,2-di-2H]-GA |
|---|---|---|---|---|---|
| 0 | 0.01 | 0.45 ± 0.02 | 0.47 ± 0.02 | 0 | |
| 2.4 | 0.01 ± 0.001 | 0.50 ± 0.05 | 0.50 ± 0.03 | 0 | |
| 5 | 0.01 ± 0.001 | 0.50 ± 0.01 | 0.49 ± 0.02 | 0 | |
| L232Ab | 10 | 0.01 ± 0.001 | 0.47 ± 0.01 | 0.52 ± 0.02 | 0 |
| 15 | 0.01 ± 0.001 | 0.47 ± 0.02 | 0.52 ± 0.01 | 0 | |
| 20 | 0.01 ± 0.001 | 0.48 ± 0.02 | 0.51 ± 0.02 | 0 | |
| Averagec | 0.01 ± 0.001 | 0.48 ± 0.02 | 0.51 ± 0.02 | ||
| [HPO32-]/mM | [2-13C]-GA | [2-13C, 2-2H]-GA | [1-13C, 2-2H]-GA | [1-13C, 2,2-di-2H]-GA | |
| 0 | 0.02 (0.04)e | 0.18 (0.40)e | 0.25 (0.55)e | 0.25 | |
| 5 | 0.12 | 0.62 | 0.26 | 0 | |
| I172Vd | 10 | 0.13 | 0.63 | 0.24 | 0 |
| 15 | 0.12 | 0.62 | 0.26 | 0 | |
| 20 | 0.14 | 0.65 | 0.21 | 0 | |
| Averagec | 0.13 ± 0.01 | 0.63 ± 0.01 | 0.24 ± 0.02 | ||
| TIM | [HPO32-]/mM | [2-13C]-GA | [2-13C, 2-2H]-GA | [1-13C, 2-2H]-GA | [1-13C, 2,2-di-2H]-GA |
| 0g | 0 (n.d.) | 0 (n.d.) | 0.15 ± 0.02 | 0.38 ± 0.08 | |
| 5 | 0.02 ± 0.001 | 0.26 ± 0.03 | 0.30 ± 0.05 | 0.14 ± 0.04 | |
| 10 | 0.02 ± 0.001 | 0.23 ± 0.02 | 0.29 ± 0.05 | 0.14 ± 0.06 | |
| I172Af | 15 | 0.02 ± 0.001 | 0.27 ± 0.03 | 0.32 ± 0.02 | 0.18 ± 0.06 |
| 20 | 0.02 ± 0.001 | 0.25 ± 0.04 | 0.29 ± 0.07 | 0.17 ± 0.02 | |
| Averagec | 0.02 ± 0.001 | 0.25 ± 0.02 | 0.30 ± 0.02 | ||
| Wildtype TbbTIM HIPO32- Activated Reactionsh | 0.13 | 0.63 | 0.24 | ||
For reactions of 20 mM [1-13C]-GA at pD 7.0 and I = 0.1 (NaCl).
Fractional yields of the products of L232A TbbTIM-catalyzed reactions of [1-13C]-GA. The quoted errors are the average of product yields from two different experiments.
Average product yields for the phosphite dianion-activated reaction of [1-13C]-GA.
Fractional yields of the products of I172V TbbTIM-catalyzed reactions of [1-13C]-GA.
Fractional yields of [2-13C]-GA, [2-13C, 2-2H]-GA, and [1-13C, 2-2H]-GA from reaction at the enzyme active site (Scheme 4A).
Fractional yields of the products of I172A TbbTIM-catalyzed reactions of [1-13C]-GA. The quoted errors are the average of product yields from at least two different experiments
n.d. = not detected.
Data from ref 39.
Scheme 4.
L232A mutant
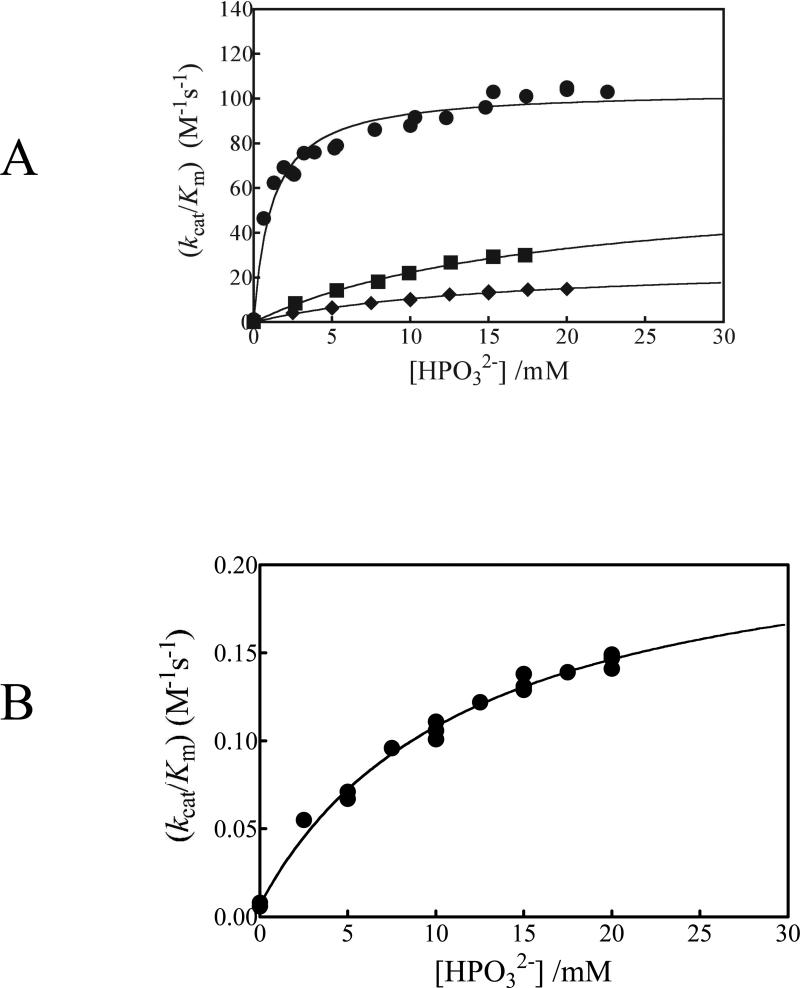
The yield of the products of the L232A mutant TbbTIM-catalyzed reactions of [1-13C]-GA in D2O at pD 7.0 (20 mM imidazole, I = 0.1, NaCl) and 25 °C in the absence or the presence of HPO32- are reported in Table 3. Only [1-13C, 2-2H]-GA, [2-13C]-GA and [2-13C, 2-2H]-GA from reactions at the enzyme active site (Scheme 4A) were observed (fE = 1.0, eq 8). Figure 3A (●) shows a plot of the increase, with increasing [HPO32-], in the second order-rate constant (kcat/Km) for L232A mutant TIM-catalyzed reaction of the carbonyl form of [1-13C]-GA that was reported in an earlier preliminary communication.31
| (8) |
Figure 3.
Dependence of the second-order rate constants (kcat/Km) for the TIM-catalyzed turnover of the carbonyl form of [1-13C]-GA in D2O on the concentration of HPO32- at pD 7.0 and 25 °C (I = 0.1, NaCl). The data were fit to eq 9 derived for the model shown in Scheme 5. (A) (●) L232A TbbTIM, (■) wildtype TbbTIM,39 (◆) I172V TbbTIM. (B) I172A TbbTIM.
I172V mutant
The yield of the products of the I172V mutant TbbTIM-catalyzed reactions of [1-13C]-GA in D2O at pD 7.0 (20 mM imidazole, I = 0.1, NaCl) and 25 °C in the absence or the presence of HPO32- are reported in Table 3. No [1-13C, 2,2-di-2H]-GA from a nonspecific protein-catalyzed reaction (Scheme 4B) was detected for the relatively fast reactions in the presence of HPO32- (fE = 1.0). A 25% yield of [1-13C, 2,2-di-2H]-GA was observed for the slow reaction in the absence of HPO32-. The total yield of all of the reaction products from Chart 1 was 70%, and the yield of the products for Scheme 4A was 45% (fE = 0.45). These product yields are similar to those determined for unactivated wildtype TbbTIM-catalyzed reactions of [1-13C]-GA.38,39 Figure 3A (◆) shows a plot of the increase, with increasing [HPO32-], in the second-order rate constant (kcat/Km) for I172V mutant TIM-catalyzed reaction of the carbonyl form of [1-13C]-GA.
I172A mutant
The yield of the products of the I172A mutant TIM-catalyzed reactions of [1-13C]-GA in D2O at pD 7.0 (20 mM imidazole, I = 0.1, NaCl) and 25 °C in the absence or the presence of HPO32- (Chart 1) are reported in Table 3. No [2-13C]-GA or [2-13C, 2-2H]-GA from the isomerization of [1-13C]-GA were detected for the reaction in the absence of phosphite, but these products are observed for the phosphite dianion-activated enzyme-catalyzed reactions. The dianion activation is much weaker than for wildtype TbbTIM, and a significant yield of [1-13C, 2,2-di-2H]-GA from the nonspecific protein-catalyzed reaction was observed in all cases. The data in Table 3 were used to calculate a value of fE = (0.02 + 0.25 + 0.30) = 0.57 ± 0.04 for the sum of the yields of [2-13C]-GA, [2-13C, 2-2H]-GA and [1-13C, 2-2H]-GA from reactions at the enzyme active site (Scheme 4A). Table 4 reports values of kobs (s-1, eq 1), (kcat/Km)obs (M-1 s-1, eq 2) and (kcat/Km) (M-1 s-1, eq 8) for reactions in the presence of HPO32-. Most or all of the 15% yield of [1-13C, 2-2H]-GA from the unactivated mutant TIM-catalyzed reaction is expected to form by the nonspecific pathway shown in Scheme 4B that gives an even larger 38% yield of [1-13C, 2,2-di-2H]-GA. The yield of [1-13C, 2-2H]-GA is observed to increase by two-fold (15% to 30%) for the phosphite activated reactions, as the yield of [1-13C, 2,2-di-2H]-GA from the protein catalyzed reaction decreases from 38% to 14% (Table 3). This shows that most or all of the [1-13C, 2-2H]-GA observed in these HPO32--activated reactions forms by Scheme 4A. We cannot exclude the possibility of a ca. 5% yield of [1-13C, 2-2H]-GA by Scheme 4B for reactions in the presence of the dianion activator. However, this uncertainty will not affect any conclusions from our discussion of the mechanism for dianion activation of the I172A mutant. Finally, we note that the initial 14% yield of [1-13C, 2,2-di-2H]-GA observed for the reaction in the presence of 5 mM HPO32- remains nearly constant as the concentration of the activator is increased to 20 mM. This shows that phosphite dianion acts both as an activator (Scheme 4A) and as a Brønsted-base catalyst of the protein-catalyzed reaction (Scheme 4B), as was proposed in an earlier study of reactions catalyzed by cTIM.38
Table 4.
Kinetic Data for the Reaction of [1-13C]-GA Catalyzed by the I172A mutant of TbbTIM in D2O in the Absence and Presence of Phosphite Dianion in D2O at 25 °C.a
| [HPO32-], mM | [TIM], M | kobs, (s-1)b | (kcat/Km)obs (M-1s-1)c | (kcat/Km) (M-1s-1)d |
|---|---|---|---|---|
| 0 | 3.1 × 10-4 | 7.5 × 10-7 | 0.040 | |
| 0 | 3.4 × 10-4 | 1.1 × 10-6 | 0.055 | |
| 0 | 7.3 × 10-4 | 1.8 × 10-6 | 0.041 | |
| 2.5 | 4.0 × 10-4 | 2.3 × 10-6 | 0.096 | 0.055 |
| 5 | 3.4 × 10-4 | 2.5 × 10-6 | 0.12 | 0.071 |
| 5 | 6.9 × 10-4 | 5.3 × 10-6 | 0.13 | 0.071 |
| 5 | 3.3 × 10-4 | 2.4 × 10-6 | 0.12 | 0.067 |
| 7.5 | 4.0 × 10-4 | 4.1 × 10-6 | 0.17 | 0.096 |
| 10 | 1.7 × 10-4 | 2.0 × 10-6 | 0.19 | 0.11 |
| 10 | 3.4 × 10-4 | 3.6 × 10-6 | 0.18 | 0.10 |
| 10 | 4.0 × 10-4 | 4.6 × 10-6 | 0.19 | 0.11 |
| 12.5 | 4.0 × 10-4 | 5.3 × 10-6 | 0.22 | 0.12 |
| 15 | 1.7 × 10-4 | 2.4 × 10-6 | 0.23 | 0.13 |
| 15 | 2.1 × 10-4 | 3.1 × 10-6 | 0.24 | 0.14 |
| 15 | 4.0 × 10-4 | 5.6 × 10-6 | 0.23 | 0.13 |
| 17.5 | 4.0 × 10-4 | 5.9 × 10-6 | 0.24 | 0.14 |
| 20 | 1.5 × 10-4 | 2.40 × 10-6 | 0.26 | 0.15 |
| 20 | 1.7 × 10-4 | 2.7 × 10-6 | 0.26 | 0.15 |
| 20 | 1.7 × 10-4 | 2.5 × 10-6 | 0.25 | 0.14 |
| 20 | 4.0 × 10-4 | 6.4 × 10-6 | 0.26 | 0.15 |
DISCUSSION
The conservative I172V mutation results in only a 2-fold decrease in kcat/Km for isomerization of GAP that masks larger 4-fold and 9-fold decreases, respectively, in the individual kinetic parameters Km and kcat. The L232A and I172A mutations of TbbTIM result in 6-fold and 100-fold decreases, respectively, compared to wildtype TIM in kcat/Km for isomerization of GAP and of DHAP (Table 1),31 but a 9-fold decrease and 5-fold increase in Km for isomerization of DHAP. A larger enzyme activity is therefore observed for the L232A mutant, when the [DHAP] is low and the relative velocity is determined by the 13-fold greater value of kcat/Km for the L232A mutant, while a larger enzyme activity is observed for the I172A mutant when the [DHAP] is saturating and the relative velocity is determined by the 3.6-fold greater value of kcat for the I172A mutant.
By comparison, a 12-fold decrease and 2-fold increase, respectively, compared to wildtype TIM has been reported in the values of Km for isomerization of DHAP catalyzed by the E165D/S96P and E165D mutants of TIM from chicken muscle (Table 1).43 Consequently, the E165D mutant exhibits a 19-fold smaller kcat/Km and a 1.2-fold larger kcat compared with the E165D/S96P double mutant. These data suggest that the interactions between wildtype TIM and functionality at C-1 and C-2 are balanced in the binding of DHAP and GAP, but that this balance is upset by L232A or E165D/S96P mutations that result in the stabilization of the complex to DHAP, while the I172A or E165D mutations result in a destabilization of this complex. These variations in Km may reflect changes in the stabilization of the Michaelis complexes by hydrogen bond(s) to the substrate hydroxyl group. X-ray crystal structures determined for the E165D44 and E165D/S96P45 mutant enzymes show subtle changes in the positioning of catalytic residues at the enzyme active site, but the structures of the I172A and L232A mutants were not determined.
Reactions of GAP in D2O
Deprotonation of enzyme-bound GAP by the basic side chain of Glu-167 gives an intermediate and the protonated side chain. In a solvent of D2O the –H derived from substrate partitions between intramolecular transfer to form DHAP, and essentially irreversible exchange with –D derived from solvent to form the D-labeled carboxylic acid. This -D then partitions between transfer to C-1 and to C-2 of the intermediate to form d-DHAP and d-GAP, respectively. Table 2 compares the yields of DHAP, d-DHAP and d-GAP for wildtype TIM-catalyzed reactions of GAP in D2O,39 with the product yields for the reactions catalyzed by L232A and I172A mutant TIMs, and reports values of the following rate constant ratios calculated from these product yields: (1) kex/(kC1)H, the ratio of the sum of the yields of products that form after irreversible deuterium exchange (d-DHAP + d-GAP), and the product that forms by intramolecular transfer of –H (DHAP); (2) (kC1)D/(kC2)D, the ratio of the yields of d-DHAP and d-GAP that form by deuteration of the intermediate at C-1 and C-2, respectively.
If hydron transfer were directly to solvent D2O, then the exchange between enzyme and solvent should be accelerated by general bases such as imidazole,46 as has been documented for carbonic anhydrase.47 However, there is no detectable imidazole catalysis of the exchange reaction with D2O that results in an increase in the yields of d-DHAP and d-GAP.48 This provides strong evidence that hydron transfer reactions of the reaction intermediate cannot involve the bulk aqueous solvent, and must therefore be with -D at the enzyme active site.48 The experimental results are consistent with X-ray crystal structures of complexes between TIM and intermediate analogs, which show that these analogs are sequestered at an enzyme active site that is shielded from interaction with bulk solvent.21-23,27,49
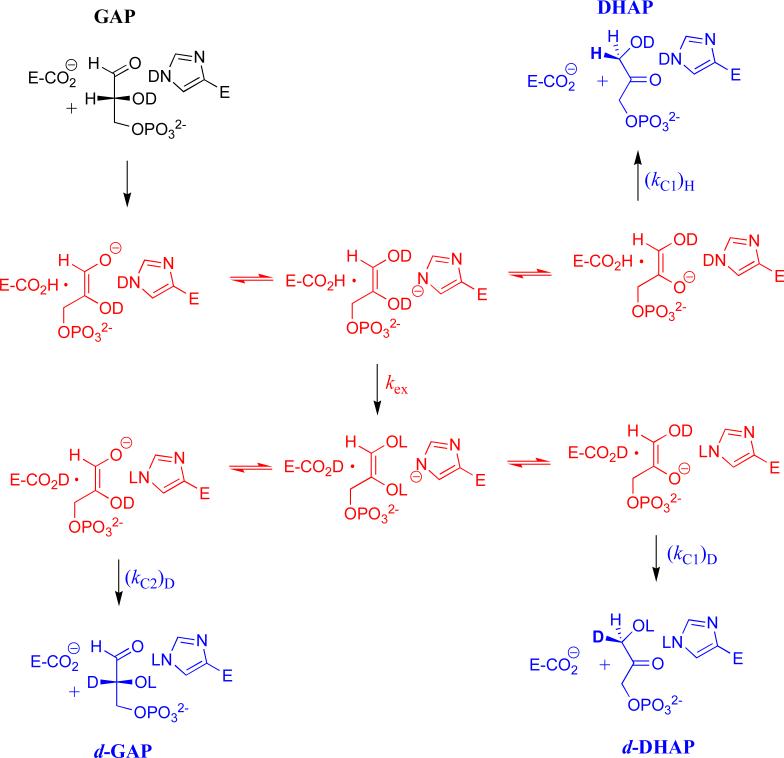
We propose the Scheme shown in Figure 4 as a working model to rationalize the effects of the L232A, I172A and other mutations on the product yields for reactions of GAP in D2O. The exchange reaction (kex) is proposed to involve the –H from substrate and a pool with a minimal size of two -D, one derived from the substrate -OD and the second from the imidazole side chain of His-95. The presence of this side chain distinguishes this Scheme from the simpler criss-cross mechanism that was proposed to explain the surprising effects of the H95Q mutation on the product yields for a reaction in tritiated water.12
Figure 4.
A minimal mechanism to rationalize the yields of the products of wildtype and mutant TIM-catalyzed reactions of GAP in D2O. The -H derived from substrate is exchanged with a pool of two -D at the enzyme, the intermediate –OD and the -D at N-3 of the imidazole side chain of His-95. Fast transfer between four basic sites; the carboxylate of Glu-165, the two oxygen anions of the isomeric intermediates, and the N-3 imidazolate of His-95 scrambles a total of three hydrons. The steps that result in scrambling are not shown, but are subsumed in the macroscopic rate constant kex. A mechanism, referred to as the criss-cross mechanism, has been proposed to explain the effects of the H95Q mutation on the product yields for reactions of GAP in 3H-labeled water.12 In this mechanism (not shown) the -H derived from substrate exchanges with a single hydron at the intermediate –OD, and there is no scrambling of hydrons at the C-1 and C-2 hydroxyl groups of the intermediate. In cases where a >2/3 yield of deuterium labeled products is obtained, the substrate-derived hydron either undergoes fast irreversible exchange with deuterium at TIM, or reversible exchange that scrambles a pool of >2 -D at the enzyme active site.50
The increase in kex/(kC1)H from 1.5 for wildtype TbbTIM to 6.7 for the I172A mutant to 23 for L232A mutant (Table 2) shows that these mutations favor the D-exchange reaction compared with intramolecular transfer of –H. The -CH2CH2CO2-/-CH2CH2CO2H side chain of Glu-167 is presumably aligned optimally at the wildtype enzyme TIM to react at both C-1 and C-2 of the bound substrate/intermediate.21 We suggest that substitution of the bulky hydrophobic side chain of Ile-172 or Leu-232 by a smaller methyl group results in movement of the glutamate side chain away from optimal alignment, and favors proton transfer to the imidazolate anion of His-95 compared to carbon-2 of the reaction intermediate. The large 96% yield of d-DHAP and d-GAP observed for the L232A mutant TbbTIM-catalyzed reaction requires either: (a) that the exchange step kex is nearly irreversible, as was proposed for the criss-cross mechanism;12 (2) the rapid scrambling of substrate –H with a large pool of enzyme-bound –D, as proposed by Irwin Rose;50 or (c) that the mutations favor direct exchange between the protonated side chain of E167 and deuterium from bulk solvent. These different explanations will be difficult to distinguish.
The increase in (kC1)D/(kC2)D (Figure 4) from 1.7 for wildtype TbbTIM to 8.8 for the L232A mutant shows that this mutation also causes an increase in the reactivity of the intermediate for protonation at carbon-1, that is masked in kex/(kC1)H by the even larger relative increase in kex. It is interesting that the L232A mutation causes both a decrease in Km for DHAP compared with GAP that is consistent with an increased binding affinity for DHAP, and in the relative reactivity of the enediolate phosphate for protonation at carbon-1 to form DHAP, compared with protonation at carbon-2 to form GAP. We propose that the interactions at the L232A mutant enzyme that stabilize the Michaelis complex to DHAP [perhaps a hydrogen bond to the carbon-1 hydroxyl] are also expressed at, and stabilize, the transition state for protonation of the intermediate to form DHAP.
Product Yields from the Reactions of [1-13C]-GA in D2O
The products of the reaction of [1-13C]-GA in D2O form by essentially the same mechanism as shown in Figure 4 for the reactions of GAP. Deprotonation of [1-13C]-GA gives an enediolate and the protonated side chain,17 that partitions between intramolecular transfer of -H to form [2-13C]-GA, and essentially irreversible exchange with D2O to give [2-13C, 2-2H]-GA and [1-13C, 2-2H]-GA as the final reaction products (Scheme 4A). The conservative I172V mutation has no effect on the yields of the products of the phosphite dianion-activated reactions of [1-13C]-GA in D2O (Table 3), while the yield of [2-13C]-GA from phosphite-activated reactions with intramolecular transfer of –H decreases from 13% (wildtype TbbTIM) to 2% (I172A mutant) or 1% (L232A mutant). This mirrors the change in the yield of product DHAP (Table 2) of the reaction in D2O with intramolecular transfer of –H, from 40% (wildtype TbbTIM) to 13% (I172A mutant) to 4% (L232A mutant). The similar effects of these mutations on the product yields from reaction of the whole substrate GAP and from the reaction of substrate pieces [1-13C]-GA + HPO32- in D2O suggests a common explanation for these changes that was discussed above for the reaction of GAP.
An interesting question is whether the binding of HPO32- to TIM affects the products of the TIM-catalyzed reactions of [1-13C]-GA. The same yields of [2-13C]-GA, [2-13C, 2-2H]-GA and [1-13C, 2-2H]-GA (Scheme 4A) are observed from the wildtype TbbTIM-catalyzed reactions of [1-13C]-GA in either the presence or the absence of phosphite dianion.38,39 In this case, the interactions between HPO32- and TIM affect the rate of the reactions of [1-13C]-GA (Figure 3), but not the relative yields of the reaction products. Similarly, the same yield of products (Scheme 4A) is observed for the robust L232A TIM-catalyzed reactions of [1-13C]-GA in either the presence or the absence of phosphite dianion (Table 3).
The yields of products for the I172V TIM-catalyzed reactions of [1-13C]-GA in the absence of HPO32- is different from the yields observed for the phosphite-activated reactions (Table 3). In particular, enzyme-bound HPO32- causes the ratio of the yields of isomerization reaction products [2-13C]-GA and [2-13C, 2-2H]-GA to change from 1/9 to 1/5 for reasons that we do not understand. The binding of HPO32- to TIM results in an increase in the rate of the reactions shown in Scheme 4A, relative to the rate of the protein-catalyzed reactions (Scheme 4B). Therefore, the decrease in the yield of [1-13C, 2-2H]-GA from reactions in the absence of phosphite dianion (fP = 0.55) compared to its presence (fP = 0.24) reflects the change in the fraction of the reaction that proceeds by the pathway shown in Figure 4B when the enzyme is strongly activated by phosphite dianion.20,38,51
No [2-13C]-GA or [2-13C, 2-2H]-GA is observed for the unactivated I172A TIM-catalyzed reactions of [1-13C]-GA. This strongly suggests that the observed products [1-13C, 2-2H]-GA and [1-13C, 2,2-di-2H]-GA form by nonspecific protein-catalyzed reactions (Scheme 4B). The binding of phosphite dianion to I172A TIM activates the enzyme for catalysis of the reactions of [1-13C]-GA to form the products shown in Scheme 4A. This might cause the partitioning of the reaction intermediate to change from the exclusive formation of [1-13C, 2-2H]-GA in the unactivated reaction, to the formation of a full complement of reaction products (Scheme 4A) for the phosphite-activated reaction. We consider this unlikely, because of the negligible or small effects of HPO32- on the yields of the products of the wildtype, L232A and I172V mutant TbbTIM-catalyzed reactions.
TbbTIM-Catalyzed Reactions of [1-13C]-GA in D2O
Table 5 reports second-order rate constants (kcat/Km)E (Scheme 5) for wildtype and mutant TbbTIM-catalyzed reactions of [1-13C]-GA in the absence of phosphite dianion. These rate constants were calculated from the observed second-order rate constant using eq 8, where fE is the yield of the products from reaction at the enzyme active site (Scheme 4A). Only the products from Scheme 4A were detected for the robust L232A mutant enzyme-catalyzed reaction (fE = 1.0), for which a value of (kcat/Km)E = 1.2 M-1 s-1 was determined. The value of (kcat/Km)E = 0.03 M-1 s-1 for the I172V mutant TIM-catalyzed reaction was calculated from (kcat/Km)obs = 0.063 M-1 s-1 and fE = 0.45 (eq 8, Table 3). Finally, the I172A mutant shows no detectable [2-13C]-GA or [2-13C, 2-2H]-GA from isomerization of [1-13C]-GA. The upper limit of (kcat/Km)E < 0.003 M-1 s-1 was calculated from (kcat/Km)obs = 0.045 M-1 s-1 (Table 4) assuming that no more than 1/2 of the observed 15% yield of [1-13C, 2-2H]-GA forms by a reaction at the enzyme active site (fE < 0.075, eq 8).
| (9) |
Table 5.
Kinetic Parameters for the Phosphite-Activated and Unactivated Reactions of [1-13C]-GA Catalyzed by Wildtype Tbb and Mutant TIMs in D2O at 25 °C.a,b
Scheme 5.
Phosphite Activation of TbbTIM-Catalyzed Reactions of [1-13C]-GA in D2O
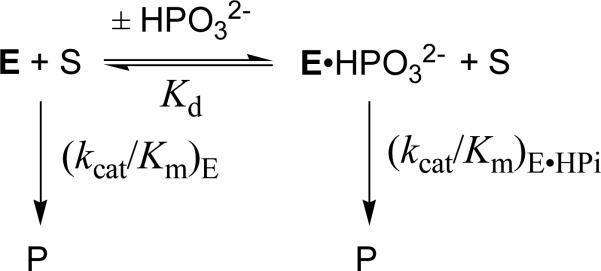
The effect of increasing [HPO32-] on (kcat/Km) for the reactions of [1-13C]-GA catalyzed by wildtype (■), I172V (◆) and L232A (●) TbbTIM is shown in Figure 3A. Figure 3B shows the effect of increasing [HPO32-] on (kcat/Km) for reactions catalyzed by the I172A mutant. These data were fit to eq 9 derived for Scheme 5.20 In Scheme 5, [1-13C]-GA undergoes a TIM-catalyzed reaction (E) with the second-order rate constant (kcat/Km)E. TIM also undergoes conversion to the activated complex E•HPO32- (Kd) that catalyzes the reaction of [1-13C]-GA with the second-order rate constant (kcat/Km)E•HPi.20 The values of (kcat/Km)E from Table 5 were used in nonlinear least squares fitting of the data to eq 9 to obtain the values for (kcat/Km)E•HPi and Kd reported in Table 5.
I172V Mutant
This conservative mutation results in only a two-fold decrease in (kcat/Km)E and (kcat/Km)E•HPi (Scheme 5) for deprotonation of [1-13C]-GA by E and by E•HPO32-, respectively, and no change in the 900-fold enzyme activation by phosphite dianion (Table 5). We conclude that the I172V mutation has a similar small effect on the kinetic parameters for the TbbTIM-catalyzed reactions of the substrate pieces and on kcat/Km for catalysis of isomerization of the whole substrates GAP and DHAP (Table 1). It is interesting that the corresponding I170V mutation has in one case been noted as the cause of a TIM-deficiency in humans that gives rise to a genetic disease.52 Evidently, there are drastic physiological consequences associated with the minor effects of this mutation on the kinetic parameters for TIM.
I172A Mutant
The I172A mutation results in a >23-fold decrease in (kcat/Km)E, a 280-fold decrease in (kcat/Km)E•HPi and a 20% decrease in Kd compared to the wildtype enzyme-catalyzed reactions of [1-13C]-GA (Table 5). A comparison of the limit for (kcat/Km)E with (kcat/Km)E•HPi shows that the binding of HPO32- to the I172A mutant results in at least 70-fold activation of the mutant TIM for deprotonation of [1-13C]-GA (Figure 3B and Table 5). Since (kcat/Km)E is an upper limit, phosphite activation might be as large as the 900-fold activation observed for the wildtype TbbTIM-catalyzed reaction (Table 5).
L232A Mutant
The L232A mutation results in a 17-fold increase in (kcat/Km)E, a 16-fold decrease in Kd for breakdown of the enzyme-phosphite complex, and a 24-fold increase in (kcat/Km)E•HPi/Kd, compared to wildtype TbbTIM. This shows that the mutant enzyme is a substantially more robust catalyst of the reactions of [1-13C]-GA than is wildtype TbbTIM. The larger effect of the mutation on (kcat/Km)E (17-fold increase) compared to (kcat/Km)E•HPi (1.6-fold increase) shows that the mutation results in an 11-fold decrease in the magnitude of enzyme activation by the binding of HPO32- to form E•HPO32- (Scheme 5). We conclude that the catalytic role of L232A is to ensure optimal dianion activation of TIM by a mechanism that causes a corresponding reduction in enzymatic activity towards deprotonation of a substrate that lacks the dianion.31
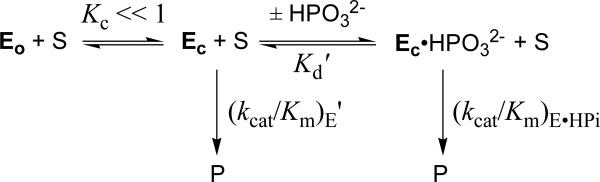
The mechanism for the activation of TIM by dianions
The very different effects of I172A and L232A mutations on the kinetic parameters for reactions of the substrate pieces (Table 5) is an unexpected result that provides a stringent test for models to explain the role of HPO32- in the activation of TIM for catalysis of the reactions of [1-13C]-GA. We have proposed the mechanism in Scheme 6 to rationalize the activation of TIM by HPO32-.5,18,31,38 TIM is shown to exist in two forms: a dominant loop open enzyme EO that is inactive, and a rare active loop-closed enzyme (EC, Kc << 1, Scheme 6) that shows specificity for binding HPO32- or GA† (the transition state for deprotonation of glycolaldehyde). The binding of HPO32- and GA† to the open enzyme EO is proposed to be relatively weak, because a substantial portion of the ligand binding energy is used to drive the change in conformation from EO to EC. Once the energetic price for this conformational change is paid upon binding of the first ligand, the full intrinsic binding energy is observed in binding of the second ligand to form the ternary complex EC•HPO32-•GA†. In the simplest case, the role of phosphite dianion is to hold TIM in the active closed form, without affecting the reactivity of TIM towards catalysis of deprotonation of GA ((kcat/Km)E = (kcat/Km)E•HPi).
| (10) |
| (11) |
Scheme 6.
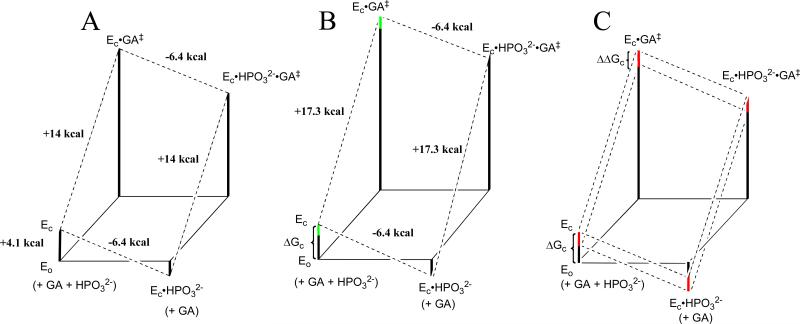
Figure 5A was constructed using the kinetic parameters for wildtype TbbTIM (Table 5). The 6.4 kcal/mol intrinsic phosphite dianion binding energy is the binding energy for association of HPO32- with the transition state complex [E•GA†]. It is calculated as the difference in the barriers to the activated reaction (defined by (kcat/Km)E•HPi/Kd) and to the unactivated reaction (defined by (kcat/Km)E, eq 10). The difference between the intrinsic binding energy (6.4 kcal/mol) and the observed binding energy determined from Kd (2.3 kcal/mol) is the binding energy (4.1 kcal/mol) that is utilized to drive the conversion of EO to EC (ΔGC, eq 11) The barrier to this conformational change is added to the barrier for the reactions of GA catalyzed by the free enzyme, but not to the barrier for the reaction catalyzed by EC•HPO32- (Scheme 6).
Figure 5.
Free energy profiles for turnover of GA by free TIM (EO) and by TIM that is saturated with HPO32- (EC•HPO32-) constructed using the kinetic parameters reported in Table 5. The profiles show the activation free energy changes calculated using the Eyring equation at 298 K for reactions catalyzed by wildtype and mutant forms of TbbTIM. (A) Reactions catalyzed by wildtype TbbTIM. The difference between the total intrinsic phosphite binding energy of -6.4 kcal/mol and ΔG° = -2.3 kcal/mol for binding of HPO32- to the inactive open enzyme EO to give the active closed liganded enzyme EC•HPO32- is attributed to ΔGC = 4.1 kcal/mol for the conformational change that converts EO to EC. (B) Reactions catalyzed by I172A mutant TbbTIM. The overall barrier for conversion of EC•HPO32- to the transition state for reaction of GA is 3.3 kcal/mol higher than for the reaction catalyzed by wildtype TbbTIM. The green bars show the uncertainty in the barrier to the unactivated reaction of GA, that was drawn using the upper limit of (kcat/Km)E < 0.003 M-1 s-1 (Table 5) and a lower limit calculated with the assumption that the total 6.4 kcal/mol intrinsic phosphate dianion binding energy is equal to that for wildtype TbbTIM. (c) Reactions catalyzed by L232A mutant TbbTIM. The red bars show the proposed effect of the L232A mutation on the barrier for the conformational change from EO to EC (ΔΔGC). A comparison of the reaction profiles for wildtype TIM (upper dashed lines) and the L232A mutant (lower dashed lines) shows the effect of this change in ΔGC on the kinetic parameters for the reaction of the substrate pieces.
Figure 5B illustrates the effects of the I172A mutation on the catalyzed reactions of HPO32- and GA. The 170-fold smaller value of (kcat/Km)E•HPi/Kd compared with wildtype TIM corresponds to a 3.0 kcal/mol increase in the barrier for conversion of enzyme and reactants to the transition state complex EC•HPO32-•GA†. The mutation also results in an apparent 0.3 kcal/mol increase in the observed binding energy for HPO32-, so that the overall barrier for conversion of EC•HPO32- to EC•HPO32-•GA† is 3.3 kcal/mol larger than for wildtype TIM. These changes are seen by comparing the right hand sides of Figure 5A and 5B. The left-hand side of Figure 5B was drawn with the assumption that the I172A mutation results in the same decrease in the second-order and third-order rate constants (kcat/Km)E and (kcat/Km)E•HPi/Kd, respectively. However, we have only determined an upper limit for (kcat/Km)E < 0.003 M-1 s-1 (Table 5). The green bar in Figure 5B shows the uncertainty in the barrier to the unactivated reaction. It has been drawn using the upper limit for (kcat/Km)E and a lower limit for this rate constant obtained by making the assumption that the intrinsic HPO32- binding energy cannot be larger than the 6.4 kcal/mol binding energy for wildtype TbbTIM (Figure 5A).
Figure 5C illustrates the effects of the L232A mutation on the catalyzed reactions of the substrate pieces HPO32- and GA.31 The critical observation is the 17-fold increase in (kcat/Km)E for the [supposedly crippled] L232A mutant enzyme-catalyzed reaction of [1-13C]-GA. We propose that the larger value of (kcat/Km)E for the L232A mutant compared to wildtype enzyme reflects the smaller fraction of the ligand binding energy used to drive the change from EO to EC, due to a 1.7 kcal/mol lower barrier to this conformational change. The barrier for conversion of EO to EC (ΔGC = 4.1 kcal/mol) for wildtype TbbTIM is given by the sum of the red and black bars in the lower left hand corner of Figure 5C. This decrease in ΔGC would result in the increase in (kcat/Km)E (17-fold), decrease in Kd (16-fold) and increase in (kcat/Km)E/Kd (24-fold). However, the magnitude of the activation of TIM for deprotonation of GA by the binding of HPO32- is determined by the value of ΔGC, and will therefore decrease.
The telling difference between the I172A and L232A mutations is that the former mutation results in similar (100-200)-fold decreases in kcat/Km for the catalyzed reaction of the whole substrate GAP (Table 1) and in (kcat/Km)E•HPi/Kd for the reaction of the substrate pieces (Table 5), while the latter results in a 6-fold decrease in kcat/Km and a 24-fold increase in (kcat/Km)E•HPi/Kd. In other words, mutations at the different sides of the hydrophobic clamp (Figure 2A and 2B) can result in an increase or a decrease in the enzymatic activity towards the catalyzed reactions of substrate pieces. We have not determined the X-ray crystal structures for these mutant enzymes that are required for a full structure-based explanation of the effects of the mutations determined in this work. However, the large body of existing structural data for TIM provides support for the following partial rationalization of these effects.
The ligand-driven conformational change of TIM results in a ca 2 Å shift of the carboxylate side chain of Glu-167 from a “swung-out” position to an active “swung-in” position (Figure 2A) that places the carboxylate anion in a hydrophobic box and aligns the functional group to deprotonate the bound substrate (Figure 2B).21,22 A correlation has been observed between the magnitude of the falloff in the catalytic activity of mutant forms of TIM and the shift in the position of this side chain. For example, the 370-fold and 23-fold decreases, respectively, in kcat/Km for the E165D43,53 and S96P43 mutants of cTIM are associated with 0.7 Å (E165D44) and 0.4 Å (S96P45) shifts in the position of the carboxylate side chain. The P168A mutation of TbbTIM results in a 35-fold decrease in kcat/Km for isomerization of GAP; and, the X-ray crystal structure for the mutant enzyme shows that the side chain for Glu-167 remains in the swung-out position at the enzyme•PGA complex.54 We suggest, similarly, that the loop closure by I172A mutant TIM may not induce the full change in the position of the carboxylate side chain from the inactive “swung-out” to the active “swung-in” position.
Part of the barrier to conversion of EO to EC (ΔGC) is probably desolvation of the solvent exposed active-site of TIM that accompanies ligand binding and the closure of loop 6.5 The I172A mutation creates space at the active site that might be occupied by water, with the possible effect of: (a) Solvating the anionic side chain of Glu-167. This would result in a decrease in side-chain reactivity for deprotonation of carbon, that leads to a decrease in (kcat/Km)E and (kcat/Km)E•HPi/Kd. (b) A decrease in the contribution of the barrier to desolvation of the enzyme active site to ΔGC for the enzyme conformational change. The I172A mutation results in decreases in the values of both (kcat/Km)E•HPi/Kd and (kcat/Km)E (Table 5) that are consistent with a decrease in the reactivity of TIM towards deprotonation of carbon. The value of Kc is defined by ratio of these rate constants (eq 11);31 but, the effect of this mutation on the rate constant ratio is uncertain, because it has only been possible to set limits for (kcat/Km)E.
The 17-fold effect of the L232A mutation on (kcat/Km)E for the reaction of [1-13C]-GA might reflect a decrease in barrier to desolvation of EO during its conversion to EC. This would require that the effect of the mutation be expressed largely on KC (eq 11), and that the effect on side chain basicity/reactivity be relatively small. This is difficult or impossible to reconcile with the proposed function of desolvation of EO, which is to increase the side-chain basicity/reactivity. We suggest that the L232A mutation results in a decrease in ΔGC because the mutation relieves steric interactions between the basic side chain of Glu-167 and the hydrophobic side chain of Leu-232. We advise a healthy skepticism to this proposal, because the factors that control the rate and equilibrium constants for the complex ligand-driven conformational change are poorly understood. This process involves the movement of many atoms, the rotation of both peptide and side chain bonds, and the cleavage and formation of several hydrogen bonds, any of which might be perturbed by the I172A mutation.4,29
Catalysis of isomerization of the whole substrate
The L232A mutation results in a contrasting six-fold decrease in kcat/Km for the TIM-catalyzed isomerization of the whole substrates GAP or DHAP (Table 1) and 17-fold and 24-fold increases, respectively, in (kcat/Km)E and (kcat/Km)E•HPi/Kd for the reactions of GA and of GA + HPO32- (Table 5). This shows that there are substantial differences in the processes that control the kinetic parameters for the reactions of the whole substrate and substrate pieces. We note that the increase in the concentration of EC relative to EO that was proposed to rationalize the effect of the mutation on (kcat/Km)E for the reaction of GA is unlikely to result in an increase in kcat/Km for isomerization of the whole substrate, because this rate constant is at the diffusion-controlled limit.55 A diffusion-limited reaction by the mechanism shown in Scheme 6 would require that conversion of the first-formed EO•GAP complex to the active EC•GAP complex and then to product be faster than dissociation of GAP from EO•GAP.
The observed decrease in kcat/Km for isomerization of GAP catalyzed by the L232A mutant may be due to an increase in the chemical barrier to isomerization that we cannot easily rationalize. Alternatively, it might reflect an increase in the barrier to the formation and breakdown of a reactive encounter complex between TIM and substrate, since a substantial variation in these barriers has been observed for other enzyme-catalyzed reactions. For example, the value of kcat/Km for yeast orotidine 5'-monophosphate decarboxylase-catalyzed decarboxylation of 5-fluoroorotidine 5'-monophosphate decreases with increasing solvent viscosity in the manner predicted for an encounter-limited reaction.56 The value of kcat/Km = 2.6 × 107 M-1 s-1 for the catalyzed reaction in water is smaller than kcat/Km = 3.8 × 107 for isomerization of the carbonyl form of GAP catalyzed by L232A TbbTIM (Table 1). The Y208F mutation of cTIM results in the loss of an intraloop hydrogen bond to the backbone amide of Ala-176. The mutation leads to a 2000-fold decrease in kcat/Km for isomerization of GAP, to a value that is well below the diffusion-controlled limit,57 but which decreases with increasing solvent viscosity.58 It was proposed that loop closure to trap the initial substrate complex is rate-determining for the Y208F mutant TIM-catalyzed reaction. We suggest, by analogy, that the decrease in kcat/Km observed for the L232A mutant TIM-catalyzed reactions of GAP and DHAP reflects an increase in the kinetic barrier to conversion of the complex between open enzyme and ligand to the active closed form.
Acknowledgement
We acknowledge the National Institutes of Health Grant GM39754 for generous support of this work. We thank Professor Gerald Koudelka for advice on the preparation of the I172A, I172V and L232A mutants of Tbb TIM.
REFERENCES
- 1.Knowles JR, Albery WJ. Acc. Chem. Res. 1977;10:105–111. [Google Scholar]
- 2.Knowles JR. Nature. 1991;350:121–124. doi: 10.1038/350121a0. [DOI] [PubMed] [Google Scholar]
- 3.Knowles JR. Philos. Trans. R. Soc. London, Ser. B. 1991;332:115–121. doi: 10.1098/rstb.1991.0039. [DOI] [PubMed] [Google Scholar]
- 4.Wierenga RK. Cell. Mol. Life Sci. 2010;67:3961–3982. doi: 10.1007/s00018-010-0473-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Richard JP. Biochemistry. 2012;51:2652–2661. doi: 10.1021/bi300195b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rieder SV, Rose IA. J. Biol. Chem. 1959;234:1007–1010. [PubMed] [Google Scholar]
- 7.Orosz F, Olah J, Ovadi J. Biochim. Biophys. Acta, Mol. Basis Dis. 2009;1792:1168–1174. doi: 10.1016/j.bbadis.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 8.Orosz F, Olah J, Ovadi J. IUBMB Life. 2006;58:703–715. doi: 10.1080/15216540601115960. [DOI] [PubMed] [Google Scholar]
- 9.Richard JP. J. Am. Chem. Soc. 1984;106:4926–4936. [Google Scholar]
- 10.Waley SG, Miller JC, Rose IA, O'Connell EL. Nature. 1970;227:181. doi: 10.1038/227181a0. [DOI] [PubMed] [Google Scholar]
- 11.Miller JC, Waley SG. Biochem. J. 1971;123:163–170. doi: 10.1042/bj1230163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nickbarg EB, Davenport RC, Petsko GA, Knowles JR. Biochemistry. 1988;27:5948–5960. doi: 10.1021/bi00416a019. [DOI] [PubMed] [Google Scholar]
- 13.Komives EA, Chang LC, Lolis E, Tilton RF, Petsko GA, Knowles JR. Biochemistry. 1991;30:3011–3019. doi: 10.1021/bi00226a005. [DOI] [PubMed] [Google Scholar]
- 14.Joseph-McCarthy D, Lolis E, Komives EA, Petsko GA. Biochemistry. 1994;33:2815–2823. doi: 10.1021/bi00176a010. [DOI] [PubMed] [Google Scholar]
- 15.Lodi PJ, Chang LC, Knowles JR, Komives EA. Biochemistry. 1994;33:2809–2814. doi: 10.1021/bi00176a009. [DOI] [PubMed] [Google Scholar]
- 16.Go MK, Amyes TL, Richard JP. J. Am. Chem. Soc. 2010;132:13525–13532. doi: 10.1021/ja106104h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Go MK, Koudelka A, Amyes TL, Richard JP. Biochemistry. 2010;49:5377–5389. doi: 10.1021/bi100538b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malabanan MM, Amyes TL, Richard JP. Cur. Opin. Struct. Biol. 2010;20:702–710. doi: 10.1016/j.sbi.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amyes TL, O'Donoghue AC, Richard JP. J. Am. Chem. Soc. 2001;123:11325–11326. doi: 10.1021/ja016754a. [DOI] [PubMed] [Google Scholar]
- 20.Amyes TL, Richard JP. Biochemistry. 2007;46:5841–5854. doi: 10.1021/bi700409b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jogl G, Rozovsky S, McDermott AE, Tong L. Proc. Natl. Acad. Sci. 2003;100:50–55. doi: 10.1073/pnas.0233793100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kursula I, Wierenga RK. J. Biol. Chem. 2003;278:9544–9551. doi: 10.1074/jbc.M211389200. [DOI] [PubMed] [Google Scholar]
- 23.Alahuhta M, Wierenga RK. Proteins Struct., Funct., Bioinf. 2010;78:1878–1888. doi: 10.1002/prot.22701. [DOI] [PubMed] [Google Scholar]
- 24.Nobel MEM, Zeelen JP, Wierenga RK. Proteins Struct., Funct., Genet. 1993;16:311–326. doi: 10.1002/prot.340160402. [DOI] [PubMed] [Google Scholar]
- 25.Alber T, Banner DW, Bloomer AC, Petsko GA, Phillips D, Rivers PS, Wilson IA. Philos. Trans. R. Soc. London, Ser. B. 1981;293:159–171. doi: 10.1098/rstb.1981.0069. [DOI] [PubMed] [Google Scholar]
- 26.Lolis E, Petsko GA. Biochemistry. 1990;29:6619–6625. doi: 10.1021/bi00480a010. [DOI] [PubMed] [Google Scholar]
- 27.Davenport RC, Bash PA, Seaton BA, Karplus M, Petsko GA, Ringe D. Biochemistry. 1991;30:5821–5826. doi: 10.1021/bi00238a002. [DOI] [PubMed] [Google Scholar]
- 28.Pompliano DL, Peyman A, Knowles JR. Biochemistry. 1990;29:3186–3194. doi: 10.1021/bi00465a005. [DOI] [PubMed] [Google Scholar]
- 29.Kursula I, Salin M, Sun J, Norledge BV, Haapalainen AM, Sampson NS, Wierenga RK. Prot. Eng., Des. & Sel. 2004;17:375–382. doi: 10.1093/protein/gzh048. [DOI] [PubMed] [Google Scholar]
- 30.Wierenga RK, Noble MEM, Vriend G, Nauche S, Hol WGJ. J. Mol. Biol. 1991;220:995–1015. doi: 10.1016/0022-2836(91)90368-g. [DOI] [PubMed] [Google Scholar]
- 31.Malabanan MM, Amyes TL, Richard JP. J. Am. Chem. Soc. 2011;133:16428–16431. doi: 10.1021/ja208019p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bergemeyer HU, Haid E, Nelboeck-Hochstetter M. 1972. Office, U. P., Ed. US.
- 33.O'Donoghue AC, Amyes TL, Richard JP. Biochemistry. 2005;44:2610–2621. doi: 10.1021/bi047954c. [DOI] [PubMed] [Google Scholar]
- 34.Borchert TV, Pratt K, Zeelen JP, Callens M, Noble MEM, Opperdoes FR, Michels PAM, Wierenga RK. Eur. J. Biochem. 1993;211:703–710. doi: 10.1111/j.1432-1033.1993.tb17599.x. [DOI] [PubMed] [Google Scholar]
- 35.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. Proteomics Protocol Handbook. 2005:571–607. [Google Scholar]
- 36.Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Glasoe PK, Long FA. J. Phys. Chem. 1960;64:188–190. [Google Scholar]
- 38.Go MK, Amyes TL, Richard JP. Biochemistry. 2009;48:5769–5778. doi: 10.1021/bi900636c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malabanan MM, Go MK, Amyes TL, Richard JP. Biochemistry. 2011;50:5767–5779. doi: 10.1021/bi2005416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Plaut B, Knowles JR. Biochem. J. 1972;129:311–320. doi: 10.1042/bj1290311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lambeir AM, Opperdoes FR, Wierenga RK. Eur. J. Biochem. 1987;168:69–74. doi: 10.1111/j.1432-1033.1987.tb13388.x. [DOI] [PubMed] [Google Scholar]
- 42.Trentham DR, McMurray CH, Pogson CI. Biochem. J. 1969;114:19–24. doi: 10.1042/bj1140019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blacklow SC, Knowles JR. Biochemistry. 1990;29:4099–4108. doi: 10.1021/bi00469a012. [DOI] [PubMed] [Google Scholar]
- 44.Joseph-McCarthy D, Rost LE, Komives EA, Petsko GA. Biochemistry. 1994;33:2824–2829. doi: 10.1021/bi00176a011. [DOI] [PubMed] [Google Scholar]
- 45.Zhang Z, Komives EA, Sugio S, Blacklow SC, Narayana N, Xuong NH, Stock AM, Petsko GA, Ringe D. Biochemistry. 1999;38:4389–4397. doi: 10.1021/bi9826759. [DOI] [PubMed] [Google Scholar]
- 46.Eigen M. Angew. Chem., Int. Ed. Engl. 1964;3:1–72. [Google Scholar]
- 47.Tu C, Paranawithana SR, Jewell DA, Tanhauser SM, LoGrasso PV, Wynns GC, Laipis PJ, Silverman DN. Biochemistry. 1990;29:6400–6405. doi: 10.1021/bi00479a009. [DOI] [PubMed] [Google Scholar]
- 48.O'Donoghue AC, Amyes TL, Richard JP. Org. Biomol. Chem. 2008;6:391–396. doi: 10.1039/b714304d. [DOI] [PubMed] [Google Scholar]
- 49.Zhang Z, Sugio S, Komives EA, Liu KD, Knowles JR, Petsko GA, Ringe D. Biochemistry. 1994;33:2830–2837. doi: 10.1021/bi00176a012. [DOI] [PubMed] [Google Scholar]
- 50.Rose IA, Fung WJ, Warms JVB. Biochemistry. 1990;29:4312–4317. doi: 10.1021/bi00470a008. [DOI] [PubMed] [Google Scholar]
- 51.Go MK, Malabanan MM, Amyes TL, Richard JP. Biochemistry. 2010;49:7704–7708. doi: 10.1021/bi101118g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arya R, Lalloz MRA, Bellingham AJ, Layton DM. Hum. Mutat. 1997;10:290–294. doi: 10.1002/(SICI)1098-1004(1997)10:4<290::AID-HUMU4>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 53.Straus D, Raines R, Kawashima E, Knowles JR, Gilbert W. Proc. Natl. Acad. Sci. 1985;82:2272–2276. doi: 10.1073/pnas.82.8.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Casteleijn MG, Alahuhta M, Groebel K, El-Sayed I, Augustyns K, Lambeir A-M, Neubauer P, Wierenga RK. Biochemistry. 2006;45:15483–15494. doi: 10.1021/bi061683j. [DOI] [PubMed] [Google Scholar]
- 55.Blacklow SC, Raines RT, Lim WA, Zamore PD, Knowles JR. Biochemistry. 1988;27:1158–1165. doi: 10.1021/bi00404a013. [DOI] [PubMed] [Google Scholar]
- 56.Wood BM, Chan KK, Amyes TL, Richard JP, Gerlt JA. Biochemistry. 2009;48:5510–5517. doi: 10.1021/bi9006226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sampson NS, Knowles JR. Biochemistry. 1992;31:8482–8487. doi: 10.1021/bi00151a014. [DOI] [PubMed] [Google Scholar]
- 58.Sampson NS, Knowles JR. Biochemistry. 1992;31:8488–8494. doi: 10.1021/bi00151a015. [DOI] [PubMed] [Google Scholar]