Abstract
Iron deficiency (ID) remains a public health concern affecting ~25% of the world’s population. Metabolic consequences of ID include elevated plasma glucose concentrations consistent with increased reliance on glucose as a metabolic substrate, though the mechanisms controlling these responses remain unclear. To further characterize the metabolic response to ID, weanling male Sprague–Dawley rats were fed either a control (C; 40 mg Fe/kg diet) or iron-deficient (ID; 3 mg Fe/kg diet) diet or were pair-fed (PF) the C diet to the level of intake of the ID group for 21 days. In addition to reductions in hemoglobin, hematocrit, and plasma iron, the ID group also exhibited higher percent body fat and plasma triglycerides compared to the PF group. Steady-state levels of both plasma glucose and insulin increased 40 and 45%, respectively, in the ID group compared to the PF group. Plasma cortisol levels were decreased 67% in the ID group compared to the PF diet group. The systematic evaluation of the expression of genes involved in insulin signaling, glucose metabolism, and fatty acid metabolism in the liver and skeletal muscle revealed significant alterations in the expression of 48 and 52 genes in these tissues, respectively. A significant concurrent increase in lipogenic gene expression and decrease in gene expression related to β-oxidation in both the liver and skeletal muscle, in combination with differential tissue expression of genes involved in glucose metabolism, provides novel insight into the adaptive metabolic response in rodent models of severe iron deficiency anemia.
Keywords: Anemia, Lipotoxicity, Insulin, PCR array, Skeletal muscle, Liver
Introduction
Iron is an essential nutrient as it is vital for many life-preserving processes including oxygen transport and DNA synthesis, as well as cellular proliferation and energy metabolism. Iron deficiency (ID) remains a major public health concern affecting some 1.6 billion people worldwide and significantly contributing to the global burden of disease (Worldwide prevalence of anaemia 1993–2005: WHO global database on anaemia 2008). Symptoms of ID include weakness, fatigue, listlessness, and a compromised immune response. In laboratory animal models of ID, another observed but less understood physiological response to an inadequate iron status is an elevation in plasma glucose levels (Borel et al. 1991; Davies et al. 1984). ID hyperglycemic animals also exhibit elevated insulin levels, increased insulin sensitivity, and altered glucose utilization in peripheral tissues, particularly skeletal muscle (Borel et al. 1993; Brooks et al. 1987; Farrell et al. 1988).
In addition to altered glucose utilization, significant changes in lipid homeostasis have been reported, though mechanistic studies investigating the utilization, uptake, and storage of lipids in ID animals have offered mixed results (Amine et al. 1976; Jain et al. 1982; Sherman et al. 1978). For instance, although hypertriglyceridemia is commonly reported in ID animals, tracer studies examining the extent to which these changes in plasma lipids are a result of iron-dependent effects on endogenous triacylglycerol synthesis remain unclear (Amine et al. 1976; Amine and Hegsted 1971; Jain et al. 1982; Sherman et al. 1978). Despite variable changes in hepatic triacylglycerol content in response to ID, an increased abundance of lipid droplets has been observed in the skeletal muscles of ID rats (Ross and Eisenstein 2002; Sherman et al. 1978; Stangl and Kirchgessner 1998). The disparity of these reports may be due to strain differences and/or the degrees of diet-induced ID that appear to have significant effects on the metabolic response to an impaired iron status (Borel et al. 1991; Stangl and Kirchgessner 1998; Yamagishi et al. 2000). Therefore, even though lipemia and hyperglycemia have been described in ID animals, relatively few studies have focused on characterizing alterations in gene expression in the liver and skeletal muscle that might contribute to the metabolic responses observed in ID (Amine et al. 1976; Borel et al. 1991; Farrell et al. 1988; Linderman et al. 1994; Sherman et al. 1978; Collins 2006; Kamei et al. 2010; Tosco et al. 2010).
Recently, investigations into the coordination of the metabolic adaptation to ID in Saccharomyces cerevisiae have produced intriguing results demonstrating that targeted mRNA degradation via the coordination of two mRNA binding proteins drives the metabolic adaption to iron deprivation (Puig et al. 2005, 2008). Although the types of studies conducted in S. cerevisiae will likely prove more difficult using animal models, they do suggest that at least part of the metabolic adaption to ID occurs at the level of mRNA expression and stability. Therefore, in an effort to begin to further characterize the metabolic response to ID, we examined the expression of genes involved in glucose and fatty acid metabolism in both the liver and skeletal muscle. We hypothesized that dietary iron restriction would significantly alter metabolic gene expression and would correspond to metabolic adaptations observed in response to ID such as hyperglycemia and hypertriglyceridemia. Our results suggest that lipogenic gene expression is significantly up-regulated and that gene expression related to β-oxidation is significantly down-regulated in response to ID. The findings presented herein provide insight into the potential mechanisms contributing to the alterations in fuel utilization and energy metabolism associated with ID.
Study design and methods
Twenty-four 21-day-old weanling male Sprague–Dawley (Harlan, IN, USA) rats were housed individually in stainless-steel, wire-bottomed cages at the Oklahoma State University Laboratory Animal Research facility in a temperature- and humidity-controlled environment and maintained on a 12-h light:dark cycle with ad libitum access to deionized water. Rats in each group were allowed access to the control diet for 3 days prior to starting dietary treatments. After the acclimation period, rats were randomly assigned to one of three diet groups (n = 8/group) for 21 days: control (C; 40 mg Fe/kg diet), iron-deficient (ID; <3 mg Fe/kg diet), or pair-fed (PF; fed the control diet at the level of intake of the ID group). The powdered diets were purchased from Harlan Teklad (Madison, WI, USA; C-TD.89300 and ID-TD.80396) (Report of the American Institute of Nutrition ad hoc Committee on Standards for Nutritional Studies 1977). Individual body weights and food intake were measured daily. After the 21-day experimental period, rats were anesthetized with a mixture of ketamine/xylazine (75 mg ketamine and 7.5 mg xylazine/kg body weight). Body composition was determined by dual-energy X-ray absorptiometry (DXA, Hologic QDR Series 4500). Rats were then killed by exsanguination between 8:00 and 10:00 a.m., and tissues were excised, snap frozen in liquid nitrogen, and stored at −80°C until further analysis.
Assessment of iron status
Whole blood was collected from the abdominal aorta into EDTA-coated tubes and assayed for hemoglobin and hematocrit concentrations using an electronic hematology analyzer (Max M Model; Coulter Corporation). To obtain plasma, whole blood was collected into EDTA-coated tubes, mixed on a rotator at 25°C for 20 min, centrifuged at 800×g for 20 min at 4°C, and then stored at −80°C until further analysis. Plasma iron was determined using an ELAN 9000 ICP-Mass Spectrometer (PerkinElmer, IL, USA). Hemolyzed samples were excluded from the analysis.
Metabolic indices
Plasma glucose and triglycerides were measured by previously described enzymatic methods using ACE glucose and triglyceride reagents, respectively, (Alfa Wassermann, NJ, USA) on an ACE Clinical Analyzer (In Vitro Diagnostic Products for Human Use, Proposed Establishment of Glucose 1974; Bucolo and David 1973). Plasma insulin (Crystal Chem Inc., IL, USA) and cortisol (R&D Systems, MN, USA) were measured by ELISA according to the manufacturer’s instructions.
Pathway-focused PCR array and qPCR
Changes in gene expression were analyzed by pathway-focused insulin signaling, glucose metabolism, and fatty acid metabolism PCR arrays for rat (SABiosciences, MD). Briefly, total RNA was isolated from a portion of the liver or gastrocnemius muscle using STAT-60 (Tel-test, Inc., TX). The concentration of RNA was determined using a Nanodrop spectrophotometer (Thermo Fisher Scientific, DE, USA), and integrity of the RNA was determined by examining 18S and 28S rRNA by agarose gel electrophoresis. The RNA was then treated with DNase I (Roche, IN, USA) and reverse-transcribed using SuperScript II (Invitrogen, CA, USA) in a final volume of 120 μL. The cDNA was used as a template for qPCR according to the array instructions using SYBR green chemistry on an ABI 7900HT system (Applied Biosystems, CA, USA). Array data were analyzed using SABiosciences RT2 Profiler PCR Data Analysis software at http://pcrdataanalysis.sabiosciences.com/pcr/arrayanalysis.php and were considered significant at ≥1.5-fold change and P < 0.05. This level of significance was set based on previous work examining the validity and sensitivity of PCR arrays (Gaj et al. 2008; Jae-Eun Pie et al. 2010; Swali et al. 2011). Findings from these studies indicate that transcriptional responses to dietary intervention(s) tend to be modest yet biologically meaningful, even when fold changes are <2.0 and/or when P values >0.05 (Gaj et al. 2008; Jae-Eun Pie et al. 2010; Swali et al. 2011). Relative quantitation for each gene was determined by normalizing to 5 housekeeping genes (RPLP1, HPRT1, RPL13A, LDHA, and ACTB) comparing the ID and PF groups using the 2−ΔΔCt method (User Bulletin no. 2, Applied Biosystems). For gene expression analysis by qPCR, cDNA was prepared as described and analyzed using the 2−ΔΔCt method with Cyclophilin B (Cyclo) as the invariant control.
Statistical analysis
Comparisons among the three treatment groups were made using one-way ANOVA followed by least significant difference as the post hoc test using SPSS software version 17.0 (IBM-SPSS, IL, USA). Student’s t tests were employed to determine significance between the ID and PF groups for PCR arrays and qPCR analyses. All tests were done at the 95% confidence interval and presented as means ± SEM.
Results
Hemoglobin, hematocrit, and plasma iron were significantly reduced by 50, 55, and 74%, respectively, in the ID group compared to rats in either the PF and C groups (Table 1) (P < 0.05). Others have previously described that rats fed an iron-restricted diet have decreased food intake compared to rats fed a control diet, likely as a result of a diminished appetite in response to iron deficiency (Amine et al. 1970; Beard et al. 1995; Chen et al. 1997). In the present study, the ID group consumed less food than the C group, and rats in the PF and ID groups weighed ~10% less than the rats in the C group (Table 2) (P < 0.05). Based on the differences in food intake and body weight, the PF group was selected as the more appropriate control to ensure that observed biological changes were the result of iron deficiency and not simply due to decreased nutrient intake. Interestingly, despite no differences in lean body mass or total body mass between the ID and PF groups, rats in the ID group exhibited higher percent body fat and liver weights compared to the PF group (Table 2) (P < 0.05).
Table 1.
Hematologic indices of iron status
| Diet | Hemoglobin (g/L) | Hematocrit (%) | Plasma iron (μmol/L) |
|---|---|---|---|
| Control | 135.0 ± 3.0a | 37.0 ± 0.9a | 57.7 ± 5.7a |
| Pair-fed | 133.0 ± 2.0a | 38.1 ± 0.5a | 49.9 ± 6.5a |
| Iron-deficient | 66.0 ± 2.0b | 16.8 ± 0.7b | 12.7 ± 3.0b |
Values are means ± SEM, n = 8/group
abMeans in the same column not sharing common superscripts are significantly different among treatment groups, P < 0.05
Table 2.
Anthropometric data
| Diet | Body wt. (g) | Liver wt. (g/100 g body wt.) | Lean mass (g) | Body fat (%) |
|---|---|---|---|---|
| Control | 196.3 ± 4.2a | 5.02 ± 0.1a | 182.7 ± 3.3a | 8.4 ± 0.4a |
| Pair-fed | 177.5 ± 2.2b | 4.05 ± 0.1b | 168.3 ± 7.1b | 7.0 ± 0.4b |
| Iron-deficient | 180.6 ± 6.9b | 4.68 ± 0.1c | 167.4 ± 1.7b | 8.5 ± 0.6a |
Values are means ± SEM, n = 8/group
abcMeans in the same column not sharing common superscripts are significantly different among treatment groups, P < 0.05
Steady-state levels of both plasma glucose and plasma insulin were increased 1.4- and 1.6-fold, respectively, in the ID group compared to the PF group (Table 3) (P < 0.05). To determine whether a stress-mediated increase in cortisol levels in response to iron deficiency was partially responsible for contributing to hyperglycemia, plasma cortisol levels were assessed (Campos et al. 1998; Weinberg et al. 1980). Plasma cortisol levels in the ID group were reduced by 67% in comparison to the PF group (P < 0.05) (Table 3). Plasma triacylglycerols were 40% higher in the ID group compared to the PF group (P < 0.05) (Table 3).
Table 3.
Plasma levels of glucose, insulin, cortisol and triglycerides
| Diet | Glucose (mmol/L) | Insulin (pmol/L) | Cortisol (nmol/L) | Triglycerides (mmol/L) |
|---|---|---|---|---|
| Control | 16.2 ± 0.8a | 100.8 ± 15.5ab | 193.9 ± 32.9ab | 1.1 ± 0.1a |
| Pair-fed | 12.1 ± 0.9b | 93.6 ± 11.7b | 261.4 ± 31.3b | 0.5 ± 0.0b |
| Iron-deficient | 17.5 ± 1.0a | 147.8 ± 13.7a | 113.9 ± 32.3a | 0.8 ± 0.1c |
Values are means ± SEM, n = 8/group
abcMeans in the same column not sharing common superscripts are significantly different among treatment groups, P < 0.05
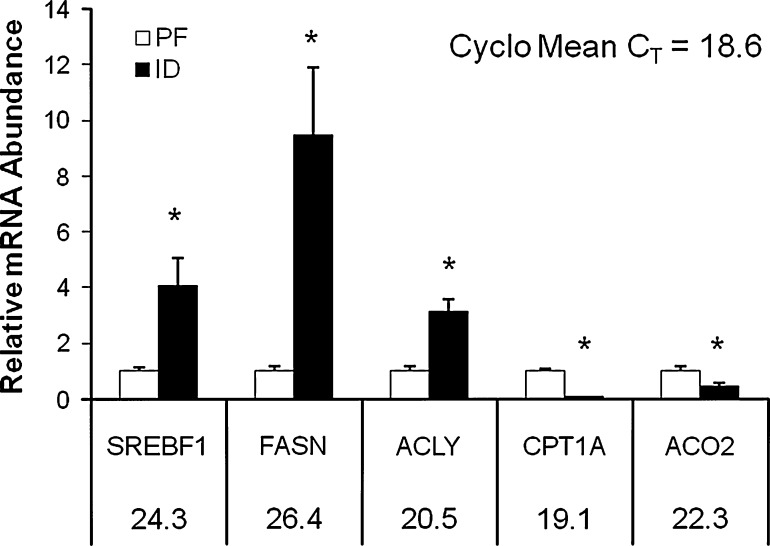
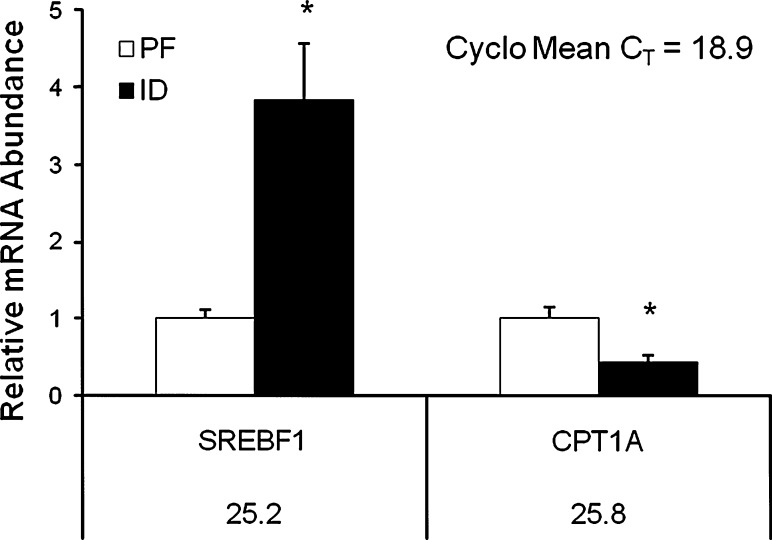
Gene expression analyses from the insulin signaling array showed a significant increase in the expression of insulin-1 (INS1) and insulin-2 (INS2) mRNA in the livers of the ID rats (P < 0.05) (Table 4). Compared to the PF group, the ID group also exhibited an increase in the hepatic expression of lipogenic insulin target genes such as sterol regulatory element binding transcription factor (SREBF1), fatty acid synthase (FASN), and acetyl-CoA carboxylase alpha (ACACA) (P < 0.05) (Table 4). Interestingly, SREBF1 expression was also significantly increased in the skeletal muscle of the ID animals (P < 0.05) (Table 6). Changes in SREBF1 and FASN mRNA expression were validated by qPCR. In response to ID, hepatic FASN mRNA expression was increased ninefold, and SREBF1 expression was increased fourfold in both liver and muscle (P < 0.05) (Figs. 1, 2).
Table 4.
Genes that increased in livers of iron-deficient rats compared to pair-fed rats
| Gene name | Gene symbol | Fold regulation | P value |
|---|---|---|---|
| Insulin signaling array | |||
| Insulin1 | Ins1 | +11.8 | 0.026 |
| Fatty acid synthase | Fasn | +9.2 | 0.052 |
| Insulin2 | Ins2 | +8.4 | 0.006 |
| Glucose-6-phosphatase, catalytic subunit | G6pc | +4.0 | 0.008 |
| Sterol regulatory element binding transcription factor 1 | Srebf1 | +2.9 | 0.082 |
| Low density lipoprotein receptor | Ldlr | +2.9 | 0.012 |
| Nitric oxide synthase 2, inducible | Nos2 | +2.1 | 0.037 |
| Acetyl-Coenzyme A carboxylase alpha | Acaca | +2.1 | 0.034 |
| Neuropeptide Y | Npy | +1.8 | 0.047 |
| V-akt murine thymoma viral oncogene homolog 2 | Akt2 | +1.5 | 0.017 |
| Glycogen Synthase kinase 3 beta | Gsk3b | +1.5 | 0.016 |
| Bcl2-like 1 | Bcl2l1 | +1.5 | <0.001 |
| Glucose metabolism array | |||
| Amylo-1,6-glucosidase, 4-alpha-glucanotransferase | Agl | +5.9 | 0.035 |
| Phosphorylase, glycogen, liver | Pygl | +5.0 | 0.001 |
| Pyruvate dehydrogenase kinase, isozyme 1 | Pdk1 | +4.8 | 0.053 |
| ATP citrate lyase | Acly | +4.7 | 0.005 |
| Phosphoglycerate kinase 1 | Pgk1 | +4.0 | <0.001 |
| Glyceraldehyde-3-phosphate dehydrogenase | Gapdh | +3.3 | 0.038 |
| Pyruvate kinase, liver and RBC | Pklr | +3.1 | 0.005 |
| Glucose-6-phosphate dehydrogenase | G6pd | +2.5 | 0.014 |
| Transkeletose | Tkt | +2.4 | <0.001 |
| Ribokinase | Rbks | +2.2 | 0.050 |
| Pyruvate dehydrogenase kinase, isozyme 4 | Pdk4 | +2.1 | 0.008 |
| Pyruvate dehydrogenase (lipoamide) beta | Pdhb | +1.9 | 0.035 |
| Phosphofructokinase, liver | Pfkl | +1.8 | 0.004 |
| Dihydrolipoamide dehydrogenase | Dld | +1.8 | 0.039 |
| Transaldolase 1 | Taldo1 | +1.6 | 0.027 |
Fold regulation indicates relative fold change in mRNA abundance in the ID group compared to the PF group
Table 6.
Genes that increased in gastrocnemius muscles of iron-deficient rats compared to pair-fed rats
| Gene name | Gene symbol | Fold regulation | P value |
|---|---|---|---|
| Insulin signaling array | |||
| Sterol regulatory element binding transcription factor 1 | Srebf1 | +2.7 | 0.055 |
| Harvey rat sarcoma virus oncogene, subgroup R | Rras2 | +1.5 | 0.033 |
| Phosphofructokinase, liver | Pfkl | +1.7 | 0.036 |
| Fatty acid metabolism array | |||
| Fatty acid binding protein 5, epidermal | Fabp5 | +5.5 | <0.001 |
| Acyl-CoA synthetase long-chain family member 3 | Acsl3 | +4.5 | <0.001 |
| Protein kinase, AMP-activated, gamma 2 non-catalytic subunit | Prkag2 | +3.6 | <0.001 |
| Acetyl-Coenzyme A acetyltransferase 2 | Acat2 | +3.1 | 0.002 |
| Acyl-CoA synthetase medium-chain family member 3 | Acsm3 | +1.9 | 0.009 |
| Protein kinase, cAMP-dependent, catalytic, alpha | Prkaca | +1.9 | <0.001 |
| Acyl-CoA synthetase long-chain family member 5 | Acsl5 | +1.7 | 0.003 |
| Protein kinase, AMP-activated, beta 1 non-catalytic subunit | Prkab1 | +1.7 | 0.042 |
Fold regulation indicates relative fold change in mRNA abundance in the ID group compared to the PF group
Fig. 1.
Validation of select hepatic gene expression changes by qRT-PCR. mRNA levels were normalized to Cyclophilin B (Cyclo) mRNA as the invariant control. Numbers beneath gene names indicate CT value obtained for the PF group. Asterisk indicates statistical significance between the PF and ID groups (P < 0.05). Error bars show SEM
Fig. 2.
Validation of select skeletal muscle gene expression changes by qRT-PCR. mRNA levels were normalized to Cyclophilin B (Cyclo) mRNA as the invariant control. Numbers beneath gene names indicate CT value obtained for the PF group. Asterisk indicates statistical significance between the PF and ID groups (P < 0.05). Error bars show SEM
Results from the pathway array focused on genes related to glucose metabolism revealed a number of genes whose expression was differentially altered between liver and skeletal muscle in response to dietary ID. For example, the expression of pentose phosphate pathway enzymes such as glucose-6-phosphate dehydrogenase (G6PD), transaldolase 1 (TALDO1), and transketolase (TKT) were increased in the livers of the ID group, while hexose-6-phosphate dehydrogenase (H6PD) and Taldo1 expression were decreased in the muscles of the ID group compared to the PF group (P < 0.05) (Tables 4, 7). Similarly, the expression of glycolytic genes including glyceraldehyde-3-phosphate dehydrogenase (GAPDH), phosphoglycerate kinase 1 (PGK1), and pyruvate kinase, (PKLR) were increased in the livers of ID rats, but the expression of the glycolytic genes Enolase-2 and -3 (ENO) were decreased in ID skeletal muscles (P < 0.05) (Tables 4, 7). Additionally, the expression of pyruvate dehydrogenase kinase 4 (PDK4) mRNA was significantly increased in the livers in the ID group, whereas it was significantly decreased in ID skeletal muscle compared to the PF group (P < 0.05) (Tables 4, 7).
Table 7.
Genes that decreased in gastrocnemius muscles of iron-deficient rats compared to pair-fed rats
| Gene name | Gene symbol | Fold regulation | P value |
|---|---|---|---|
| Insulin signaling array | |||
| Fructose-1,6- biphosphatase 1 | Fbp1 | −2.1 | 0.013 |
| Eukaryotic translation initiation factor 4E binding protein 1 | Eif4ebp1 | −1.9 | 0.006 |
| Bcl2-like 1 | Bcl2l1 | −1.9 | 0.000 |
| Insulin Receptor | Insr | −1.8 | 0.018 |
| V-akt murine thymoma viral oncogene homolog 1 | Akt1 | −1.5 | 0.049 |
| Glucose metabolism array | |||
| Pyruvate dehydrogenase kinase, isozyme 4 | Pdk4 | −4.4 | 0.021 |
| Hexokinase 3, white cell | Hk3 | −3.1 | 0.012 |
| Frucotse = 6,6-bisphosphatase 2 | Fbp2 | −2.9 | 0.010 |
| Enolase 3, beta, muscle | Eno3 | −2.3 | 0.008 |
| Hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) | H6pd | −2.2 | 0.036 |
| Glucose 6 phosphatase, catalytic, 3 | G6pc3 | −2.2 | 0.024 |
| Enolase 2, gamma, neuronal | Eno2 | −2.2 | 0.0499 |
| Phosphoglycerate mutase 2 (muscle) | Pgam2 | −2.1 | 0.019 |
| Galactose mutarotase (aldose 1-epimerase) | Galm | −2.1 | 0.002 |
| Phosphoenolpyruvate carboxykinase 2 | Pck2 | −2.1 | 0.012 |
| Glycogen synthase 1, muscle | Gys1 | −2.2 | 0.051 |
| Succinate-CoA ligase, GDP-forming, beta subunit | Suclg2 | −2.0 | 0.017 |
| Oxoglutarate dehydrogenase-like | Ogdhl | −1.9 | 0.037 |
| Transaldolase 1 | Taldo1 | −1.9 | 0.025 |
| Glycogen synthase kinase 3 alpha | Gsk3a | −1.7 | 0.031 |
| Glycogen synthase kinase 3 beta | Gsk3b | −1.7 | 0.029 |
| Isocitrate dehydrogenase 2 (NADP +), mitochondrial | Idh2 | −1.6 | 0.014 |
| Aconitase 2, mitochondrial | Aco2 | −1.6 | 0.012 |
| Isocitrate dehydrogenase 3 (NAD +) beta | Idh3b | −1.5 | 0.046 |
| Fatty acid metabolism array | |||
| Carnitine palmitoyltransferase 1a, liver | Cpt1a | −8.1 | 0.000 |
| Carnitine palmitoyltransferase 2 | Cpt2 | −4.4 | 0.000 |
| 3-hydroxybutyrate dehydrogenase, type 2 | Bdh2 | −2.9 | 0.000 |
| 3-hydroxy-3-methylglutaryl-Coenzyme A synthase 2 (mitochondrial) | Hmgcs2 | −2.8 | 0.000 |
| Solute carrier family 27 (fatty acid transporter), member 2 | Slc27a2 | −2.5 | 0.000 |
| Acyl-CoA thioesterase 12 | Acot12 | −2.3 | 0.005 |
| 2,4-dienoyl CoA reductase 1, mitochondrial | Decr1 | −2.2 | 0.000 |
| Acyl-CoA thioesterase 2 | Acot2 | −2.2 | 0.003 |
| Acyl-Coenzyme A dehydrogenase family, member 11 | Acad11 | −2.2 | 0.003 |
| Acetyl-Coenzyme A acyltransferase 2 | Acaa2 | −1.9 | 0.002 |
| Hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), alpha subunit | Hadha | −1.9 | 0.002 |
| Carnitine acetyltransferase | Crat | −1.7 | 0.009 |
| Glycerol-3-phosphate dehydrogenase 2, mitochondrial | Gpd2 | −1.7 | 0.006 |
| Acyl-Coenzyme A oxidase 3, pristanoyl | Acox3 | −1.6 | 0.015 |
| Acyl-Coenzyme A dehydrogenase family, member 10 | Acad10 | −1.5 | 0.008 |
| Solute carrier family 27 (fatty acid transporter), member 4 | Slc27a4 | −1.5 | 0.002 |
Fold regulation indicates relative fold change in mRNA abundance in the ID group compared to the PF group
Analysis of data from the fat metabolism array showed genes involved in β-oxidation, including acyl-CoA thioesterase 3 (ACOT3), carnitine palmitoyltransferase 1a (CPT1A), and carnitine palmitoyltransferase 1b (CPT1B), were significantly decreased in both the liver and skeletal muscle of the ID group (P < 0.05) (Tables 5, 7). The array findings for CPT1A were further examined by qPCR. CPT1A expression in the liver and skeletal muscles of the ID group was 90 and 54% lower, respectively, than that of the PF group (P < 0.05) (Figs. 1, 2). Concurrently, genes involved in the regulation of fatty acid synthesis such as acyl-CoA synthetase long-chain family member 5 (ACSL5) and acyl-CoA synthetase medium-chain family member 3 (ASCM3) were increased in the skeletal muscle in response to ID (P < 0.05) (Table 6). Compared to the PF group, the expression of the fatty acid transporter (FATP) genes solute carrier (SLC) family 27 member 1 (SLC27A1, FATP1) and SLC27A3 (FATP3) in the liver and SLC271A2 (FATP2) and SLC27A4 (FATP4) in the muscle were also significantly decreased in the ID group (P < 0.05) (Tables 5, 7).
Table 5.
Genes that decreased in livers of iron-deficient rats compared to pair-fed rats
| Gene name | Gene symbol | Fold regulation | P value |
|---|---|---|---|
| Glucose metabolism array | |||
| Aconitase 2, mitochondrial | Aco2 | −2.3 | <0.001 |
| Fatty Acid Metabolism Array | |||
| 3-hydroxy-3-methylglutaryl-Coenzyme A synthase 2 (mitochondrial) | Hmgcs2 | −3.0 | 0.016 |
| Enoyl-Coenzyme A, hydratase/3-hydroxyacyl-Coenzyme A dehydrogenase | Ehhadh | −2.6 | 0.003 |
| Acyl-CoA-thioesterase 3 | Acot3 | −2.5 | 0.016 |
| Rattus norvegicus acyl-Coenzyme A dehydrogenase family, member 10 | Acad10 | −2.1 | 0.004 |
| 2,4-dienoyl CoA reductase 2, peroxisomal | Decr2 | −2.0 | 0.007 |
| Protein kinase, AMP-activated, gamma 3 non-catalytic subunit | Prkag3 | −1.8 | <0.001 |
| Acetyl-Coenzyme A acyltransferase 1 | Acaa1 | −1.8 | 0.019 |
| Enoyl-Coenzyme A hydratase, short chain 1, mitochondrial | Echs1 | −1.8 | 0.012 |
| Acyl-CoA synthetase long-chain family member 4 | Acsl4 | −1.7 | <0.001 |
| Lipoprotein lipase | Lpl | −1.7 | 0.019 |
| Acyl-Coenzyme A dehydrogenase family, member 9 | Acad9 | −1.7 | 0.017 |
| Carnitine palmitoyltransferase 1a, liver | Cpt1a | −1.6 | 0.032 |
| Solute carrier family 27 (fatty acid transporter), member 1 | Slc27a1 | −1.6 | 0.005 |
| 3-hydroxymethyl-3-methylglutaryl-Coenzyme A lyase | Hmgcl | −1.5 | 0.002 |
| Acyl-CoA-thioesterase 9 | Acot9 | −1.5 | 0.025 |
| Solute carrier family 27 (fatty acid transporter), member 3 | Slc27a3 | −1.5 | 0.051 |
| Carnitine palmitoyltransferase 2 | Cpt2 | −1.5 | 0.023 |
| Glutaryl-Coenzyme A dehydrogenase | Gcdh | −1.5 | 0.049 |
| Lipase, hormone sensitive | Lipe | −1.5 | 0.019 |
Fold regulation indicates relative fold change in mRNA abundance in the ID group compared to the PF group
Table 8 highlights key findings regarding changes in metabolic gene expression in response to dietary iron deficiency including significant decreases in β-oxidative gene expression, such as CPT1A, in both the liver and muscle. A concurrent increase in lipogenic gene expression was also observed in both of these tissues as hepatic FASN expression was increased, and SREBF1 expression was increased in both the liver and muscle of ID animals. Expression of the TCA cycle gene ACO2 was also significantly increased, while expression of the glycolytic gene PFKL was significantly decreased in both the liver and skeletal muscle of the ID group. Interestingly, expression of the insulin responsive gene PDK4 was significantly increased in the liver, but decreased in the skeletal muscle in response to dietary ID.
Table 8.
Key tissue changes in gene expression in response to a dietary iron deficiency potentially contributing to a pro-lipogenic phenotype
| Gene | Liver | Skeletal muscle | Function |
|---|---|---|---|
| SREBF1 | ↑ | ↑ | Lipogenesis |
| FASN | ↑ | NC | β-oxidation |
| CPT1A | ↓ | ↓ | β-oxidation |
| CPT2 | ↓ | ↓ | β-oxidation |
| ACAD10 | ↓ | ↓ | β-oxidation |
| HMGCS2 | ↓ | ↓ | Ketogenesis |
| ACO2 | ↓ | ↓ | TCA Cycle |
| PFKL | ↑ | ↑ | Glycolysis |
| PDK4 | ↑ | ↓ | Gluconeogenesis |
Discussion
The precise mechanisms responsible for the relative hyperglycemia associated with ID anemia remain unclear (Brooks et al. 1987; Davies et al. 1984; Farrell et al. 1988; Yamagishi and Komabayashi 2003). Increased plasma glucose levels may be attributed to a decreased capacity for aerobic metabolism resulting from severely repressed hemoglobin levels characteristic of anemia, though the extent to which this metabolic phenotype is dependent upon iron alone is unclear. Even a modest reduction in hemoglobin induced by dietary copper deficiency impairs glucose tolerance, though this response is largely dependent on the primary source of dietary carbohydrate (Fields et al. 1983, 1984; Hassel et al. 1983). Because the animals in the present study were not fasted prior to sample and tissue collection, an impaired glucose tolerance, as opposed to an increase in fasting glucose levels, cannot be ruled out, though evidence from previous studies suggests that ID primarily alters fasting glucose levels (Borel et al. 1991; Brooks et al. 1987; Farrell et al. 1988). In fact, blood glucose levels are inversely related to hemoglobin levels, with more severe anemia associated with a more pronounced elevation in fasting blood glucose (Borel et al. 1991). With only a moderately depressed hemoglobin concentration, there is a concomitant decrease in the oxidative capacity of peripheral tissues and an increased reliance on the anaerobic metabolism as evidenced by the increased blood lactate levels observed in ID animals (Brooks et al. 1987; Farrell et al. 1988; Linderman et al. 1994). Thus, elevated blood glucose levels provide a means of adapting to decreased oxidative capacity by ensuring that ample substrate is available for energy production (Brooks et al. 1987; Farrell et al. 1988; Linderman et al. 1994).
In addition to elevated plasma glucose, ID animals also exhibit a relative elevation in plasma insulin. Interestingly, extrapancreatic insulin gene expression has previously been reported in the livers of hyperglycemic mice and in iron-deficient rats (Chen et al. 2010; Kojima et al. 2004; Kamei et al. 2010). The increased hepatic expression of INS1 and INS2 mRNA may contribute to the increased steady-state level of insulin observed in the present study and those reported by others in similar experimental models (Beard 2001; Borel et al. 1993; Chen et al. 2010; Farrell et al. 1988; Kojima et al. 2004). The increased insulin could serve to facilitate the entry of glucose into insulin-dependent tissues (e.g., skeletal muscle). Indeed, insulin sensitivity is enhanced in ID, particularly when hemoglobin falls to 60 g/L or below, allowing for both increased glucose clearance and disposal rates (Borel et al. 1993; Farrell et al. 1988; Linderman et al. 1994). Moderate elevations in both glucose and insulin levels observed in ID presents an interesting question: Why do blood glucose levels remain elevated if circulating levels of plasma insulin are increased and peripheral insulin sensitivity is enhanced?
In addition to stimulating insulin-dependent glucose uptake, insulin also coordinates glucose homeostasis through the regulation of gluconeogenic gene expression via the insulin-induced phosphorylation of the transcription factor forkhead box protein O1 (FOXO1) (Nakae et al. 2002). Phosphorylation of FOXO1 prevents its nuclear translocation resulting in decreased expression of target genes such as PCK1, G6PASE, and PDK4 (Granner et al. 1983; Nakae et al. 2002; Gross et al. 2008). In the fed state when insulin levels are elevated, insulin also activates hepatic lipogenesis through the increased expression of genes such as ACACA and FASN via the transcription factor SREBF1 (Horton et al. 2002; Shimomura et al. 1999b). In contrast to the fed state, chronic hyperinsulinemia may alter normal insulin signaling. Obese diabetic (ob/ob) mice with chronic hyperinsulinemia exhibit “mixed insulin resistance”, where a combination of hepatic insulin resistance (e.g., impaired repression of gluconeogenesis) and sensitivity (e.g., enhanced lipogenesis) exists (Kerouz et al. 1997; Li et al. 2010; Shimomura et al. 2000).
This model of mixed insulin resistance likely results from a bifurcation of the insulin signaling pathway at mammalian target of rapamycin complex 1 (mTORC1) (Li et al. 2010). In this model, mTORC1 continues to activate the SREBF1-dependent increase in lipogenesis, while FOXO1 target genes are inadequately repressed due to impaired insulin signaling that results in decreased phosphorylation of FOXO1 (Li et al. 2010). Although hepatic mTORC1 and FOXO1 phosphorylation were not assessed in this study, alterations in hepatic gene expression are consistent with impaired insulin signaling in response to severe ID. The increased hepatic expression of SREBF1, ACACA, and FASN mRNA observed in the current study suggests that normal insulin signaling through mTorc1 was retained. Despite increased lipogenic gene expression and increased plasma triacylglycerols, hepatic expression of the FOXO1 target gene PDK4 was increased and may indicate an impairment in insulin signaling (i.e., insulin resistance) in this part of the signaling pathway. Potential differences in insulin signaling in response to ID may provide an explanation for how blood glucose levels remain elevated even when both serum insulin levels and peripheral insulin sensitivity are increased (Beard 2001; Borel et al. 1993; Farrell et al. 1988). Iron-dependent factors or mechanisms contributing to altered insulin signaling remain to be characterized.
Ectopic lipid deposition in liver and skeletal muscle as a result of inappropriately elevated lipogenesis in ID may contribute to a condition of relative lipotoxicity and subsequent insulin resistance. The increased expression of SREBF1 mRNA is of particular interest in light of its role in stimulating lipogenic gene expression thereby promoting lipid accumulation in both the liver and skeletal muscle (Ducluzeau et al. 2001; Guillet-Deniau et al. 2002; Horton et al. 2003; Shimomura et al. 1999a). An imbalance of lipogenic gene expression coupled with a decrease in the expression of genes involved in β-oxidation of fatty acids in the liver, and the skeletal muscle may enhance lipid accumulation in these tissues.
If a relative lipotoxicity contributes to insulin resistance in response to ID, then one prediction of this model is that abnormal fat accumulation and increased plasma fatty acids and triacylglycerols might be observed. Indeed, in animal models of severe ID, lipids accumulate in both liver and skeletal muscle (Johnson et al. 1990; Sherman et al. 1978). Elevated plasma triacyglycerols are also observed in a severely ID animals (Amine et al. 1976; Sherman et al. 1978; Yamagishi et al. 2000). Although neither hepatic nor skeletal muscle lipid accumulation was assessed in the present study, overall body composition was examined. Despite a similarity in body mass between the ID and PF groups, total body fat was higher in the ID group compared to the PF group. Because the diets between the ID and PF groups were isocaloric and differed only in iron content, increased lipogenesis and improper storage of excess fat may be a metabolic consequence of ID.
It is difficult to predict whether this proposed ID-induced mixed insulin resistance would result in further negative metabolic consequences similar to those observed in metabolic syndrome and type 2 diabetes as other more pressing problems begin to arise after an extended time on a severely iron-restricted diet. In fact, examining the metabolic response to ID is complicated by the observation that mild to moderate levels of ID are not associated with the pronounced alterations in glucose homeostasis observed in models where hemoglobin levels fall to ≤60 g/L (Borel et al. 1993, 1991; Linderman et al. 1994). The degree of anemia in most clinical cases of ID anemia is not as extreme as those typically induced in animal models (McLean et al. 2009). Nonetheless, ID remains the single most common micronutrient deficiency affecting not only under-nourished populations, but over-nourished populations as well (Lecube et al. 2006; Pinhas-Hamiel et al. 2003; Yanoff et al. 2007). As overweight and obesity already place individuals at risk for metabolic disease, future studies may be required to investigate the potential of ID to exacerbate conditions where normal glucose and fat metabolism are already disrupted. In fact, recent studies have found that ID may be more prevalent in overweight children and adolescents, as well as obese postmenopausal women, populations that are already at increased risk for developing metabolic disease (Lecube et al. 2006; Nead et al. 2004; Pinhas-Hamiel et al. 2003).
In the present study, we offer insight to the potential factors involved in the hyperglycemia and hyperinsulinemia observed in rodent models of ID anemia. As ID animals shift their reliance from fat to glucose as the preferred metabolic substrate for peripheral tissues, this increased reliance on glucose may contribute to elevated blood glucose levels that subsequently promote enhanced insulin secretion. In severe ID anemia, plasma glucose levels remain elevated despite both increased peripheral insulin sensitivity and glucose clearance rates. Iron-dependent alterations in glucose and lipid metabolism observed by us and others are consistent with a bifurcated insulin signaling pathway in the liver resulting in the continuous activation of lipogenesis, and thereby contributing to ectopic lipid accumulation and potential lipotoxicity. While the lipogenic pathway remains insulin-sensitive, the hepatic FOXO1 branch of the pathway becomes more insulin-resistant, resulting in a derepression of PDK4 gene expression subsequently increasing the availability of gluconeogenic substrate. Indeed, enhanced gluconeogenesis has been observed as a result of dietary iron deficiency (Borel et al. 1993; Linderman et al. 1994). Although much remains to be known about molecular mechanisms underlying the metabolic response to iron deficiency, the results presented herein provide evidence consistent with altered hepatic insulin signaling promoting enhanced lipogenesis and impaired lipid oxidation. Future studies further interrogating changes in the hepatic insulin signaling pathway in response to iron deficiency will enhance our understanding of metabolic adaptations that occur as a result inadequate iron intake.
Acknowledgments
This research was supported by National Institutes of Health 1R15DK088721-01 (Clarke SL) and National Research Initiative Grant no. 2008-35200-04445 from the United States Department of Agriculture National Institute of Food and Agriculture Bioactive Food Components for Optimal Health Program (Clarke SL).
Conflict of interest
None.
References
- Amine EK, Hegsted DM. Iron deficiency lipemia in the rat and chick. J Nutr. 1971;101(11):1575–1582. doi: 10.1093/jn/101.11.1575. [DOI] [PubMed] [Google Scholar]
- Amine EK, Corey J, Hegsted DM, Hayes KC. Comparative hematology during deficiencies of iron and vitamin A in the rat. J Nutr. 1970;100(9):1033–1040. doi: 10.1093/jn/100.9.1033. [DOI] [PubMed] [Google Scholar]
- Amine E, Desilets EJ, Hegsted DM. Effects of dietary fats on lipogenesis in iron deficiency anemic chicks and rats. J Nutr. 1976;106:405–411. [Google Scholar]
- Beard JL. Iron biology in immune function, muscle metabolism and neuronal functioning. J Nutr. 2001;131(2S-2):568S–579S. doi: 10.1093/jn/131.2.568S. [DOI] [PubMed] [Google Scholar]
- Beard JL, Zhan CS, Brigham DE. Growth in iron-deficient rats. Proc Soc Exp Biol Med Soc (New York, NY) 1995;209(1):65–72. doi: 10.3181/00379727-209-43879. [DOI] [PubMed] [Google Scholar]
- Borel MJ, Smith SH, Brigham DE, Beard JL. The impact of varying degrees of iron nutriture on several functional consequences of iron deficiency in rats. J Nutr. 1991;121(5):729–736. doi: 10.1093/jn/121.5.729. [DOI] [PubMed] [Google Scholar]
- Borel MJ, Beard JL, Farrell PA. Hepatic glucose production and insulin sensitivity and responsiveness in iron-deficient anemic rats. Am J Physiol. 1993;264(3 Pt 1):E380–E390. doi: 10.1152/ajpendo.1993.264.3.E380. [DOI] [PubMed] [Google Scholar]
- Brooks GA, Henderson SA, Dallman PR. Increased glucose dependence in resting, iron-deficient rats. Am J Physiol. 1987;253(4 Pt 1):E461–E466. doi: 10.1152/ajpendo.1987.253.4.E461. [DOI] [PubMed] [Google Scholar]
- Bucolo G, David H. Quantitative determination of serum triglycerides by the use of enzymes. Clin Chem. 1973;19(5):476–482. [PubMed] [Google Scholar]
- Campos MS, Barrionuevo M, Alferez MJ, Gomez-Ayala AE, Rodriguez-Matas MC, Lopez Aliaga I, Lisbona F. Interactions among iron, calcium, phosphorus and magnesium in the nutritionally iron-deficient rat. Exp Physiol. 1998;83(6):771–781. doi: 10.1113/expphysiol.1998.sp004158. [DOI] [PubMed] [Google Scholar]
- Chen OS, Schalinske KL, Eisenstein RS. Dietary iron intake modulates the activity of iron regulatory proteins and the abundance of ferritin and mitochondrial aconitase in rat liver. J Nutr. 1997;127(2):238–248. doi: 10.1093/jn/127.2.238. [DOI] [PubMed] [Google Scholar]
- Chen X, Larson CS, West J, Zhang X, Kaufman DB. In vivo detection of extrapancreatic insulin gene expression in diabetic mice by bioluminescence imaging. PloS one. 2010;5(2):e9397. doi: 10.1371/journal.pone.0009397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins JF. Gene chip analyses reveal differential genetic responses to iron deficiency in rat duodenum and jejunum. Biol Res. 2006;39(1):25–37. [PMC free article] [PubMed] [Google Scholar]
- Davies KJ, Donovan CM, Refino CJ, Brooks GA, Packer L, Dallman PR. Distinguishing effects of anemia and muscle iron deficiency on exercise bioenergetics in the rat. Am J Physiol. 1984;246(6 Pt 1):E535–E543. doi: 10.1152/ajpendo.1984.246.6.E535. [DOI] [PubMed] [Google Scholar]
- Ducluzeau PH, Perretti N, Laville M, Andreelli F, Vega N, Riou JP, Vidal H. Regulation by insulin of gene expression in human skeletal muscle and adipose tissue. Evidence for specific defects in type 2 diabetes. Diabetes. 2001;50(5):1134–1142. doi: 10.2337/diabetes.50.5.1134. [DOI] [PubMed] [Google Scholar]
- Farrell PA, Beard JL, Druckenmiller M. Increased insulin sensitivity in iron-deficient rats. J Nutr. 1988;118(9):1104–1109. doi: 10.1093/jn/118.9.1104. [DOI] [PubMed] [Google Scholar]
- Fields M, Ferretti RJ, Smith JC, Jr, Reiser S. Effect of copper deficiency on metabolism and mortality in rats fed sucrose or starch diets. J Nutr. 1983;113(7):1335–1345. doi: 10.1093/jn/113.7.1335. [DOI] [PubMed] [Google Scholar]
- Fields M, Ferretti RJ, Smith JC, Jr, Reiser S. Impairment of glucose tolerance in copper-deficient rats: dependency on the type of dietary carbohydrate. J Nutr. 1984;114(2):393–397. doi: 10.1093/jn/114.2.393. [DOI] [PubMed] [Google Scholar]
- Gaj S, Eijssen L, Mensink RP, Evelo CT. Validating nutrient-related gene expression changes from microarrays using RT(2) PCR-arrays. Genes Nutr. 2008;3(3–4):153–157. doi: 10.1007/s12263-008-0094-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granner D, Andreone T, Sasaki K, Beale E. Inhibition of transcription of the phosphoenolpyruvate carboxykinase gene by insulin. Nature. 1983;305(5934):549–551. doi: 10.1038/305549a0. [DOI] [PubMed] [Google Scholar]
- Gross DN, Heuvel AP, Birnbaum MJ. The role of FoxO in the regulation of metabolism. Oncogene. 2008;27(16):2320–2336. doi: 10.1038/onc.2008.25. [DOI] [PubMed] [Google Scholar]
- Guillet-Deniau I, Mieulet V, Le Lay S, Achouri Y, Carre D, Girard J, Foufelle F, Ferre P. Sterol regulatory element binding protein-1c expression and action in rat muscles: insulin-like effects on the control of glycolytic and lipogenic enzymes and UCP3 gene expression. Diabetes. 2002;51(6):1722–1728. doi: 10.2337/diabetes.51.6.1722. [DOI] [PubMed] [Google Scholar]
- Hassel CA, Marchello JA, Lei KY. Impaired glucose tolerance in copper-deficient rats. J Nutr. 1983;113(5):1081–1083. doi: 10.1093/jn/113.5.1081. [DOI] [PubMed] [Google Scholar]
- Horton JD, Goldstein JL, Brown MS. SREBPs: transcriptional mediators of lipid homeostasis. Cold Spring Harb Symp Quant Biol. 2002;67:491–498. doi: 10.1101/sqb.2002.67.491. [DOI] [PubMed] [Google Scholar]
- Horton JD, Shah NA, Warrington JA, Anderson NN, Park SW, Brown MS, Goldstein JL. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc Natl Acad Sci USA. 2003;100(21):12027–12032. doi: 10.1073/pnas.1534923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- In Vitro Diagnostic Products for Human Use, Proposed Establishment of Glucose (1974). US Dept. of Health, Education and Welfare, FDA
- Jae-Eun Pie Y-RK, Kim In-Kyoung, Seo Sang-Hui, Lee Seung Ho, Lee Hee Ra, Yoo Young, Chung Ji Tae, Youn Jong Pil, MoonJu Oh, Hwang Seung Yong, Kim Meyoung-Kon. Correlation between nutrition intake and gene expression profiles in children with asthma. Mol Cell Toxicol. 2010;6:313–319. doi: 10.1007/s13273-010-0042-1. [DOI] [Google Scholar]
- Jain SK, Yip R, Pramanik AK, Dallman PR, Shohet SB. Reduced plasma cholesterol esterifying activity in iron-deficient rats: its possible role in the lipemia of iron deficiency. J Nutr. 1982;112(6):1230–1232. doi: 10.1093/jn/112.6.1230. [DOI] [PubMed] [Google Scholar]
- Johnson JA, Willis WT, Dallman PR, Brooks GA. Muscle mitochondrial ultrastructure in exercise-trained iron-deficient rats. J Appl Physiol. 1990;68(1):113–118. doi: 10.1152/jappl.1990.68.1.113. [DOI] [PubMed] [Google Scholar]
- Kamei A, Watanabe Y, Ishijima T, Uehara M, Arai S, Kato H, Nakai Y, Abe K. Dietary iron-deficient anemia induces a variety of metabolic changes and even apoptosis in rat liver: a DNA microarray study. Physiol Genom. 2010;42(2):149–156. doi: 10.1152/physiolgenomics.00150.2009. [DOI] [PubMed] [Google Scholar]
- Kerouz NJ, Horsch D, Pons S, Kahn CR. Differential regulation of insulin receptor substrates-1 and -2 (IRS-1 and IRS-2) and phosphatidylinositol 3-kinase isoforms in liver and muscle of the obese diabetic (ob/ob) mouse. J Clin Invest. 1997;100(12):3164–3172. doi: 10.1172/JCI119872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima H, Fujimiya M, Matsumura K, Nakahara T, Hara M, Chan L. Extrapancreatic insulin-producing cells in multiple organs in diabetes. Proc Natl Acad Sci USA. 2004;101(8):2458–2463. doi: 10.1073/pnas.0308690100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecube A, Carrera A, Losada E, Hernandez C, Simo R, Mesa J (2006) Iron deficiency in obese postmenopausal women. Obesity (Silver Spring, MD) 14(10):1724–1730 [DOI] [PubMed]
- Li S, Brown MS, Goldstein JL. Bifurcation of insulin signaling pathway in rat liver: mTORC1 required for stimulation of lipogenesis, but not inhibition of gluconeogenesis. Proc Natl Acad Sci USA. 2010;107(8):3441–3446. doi: 10.1073/pnas.0914798107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linderman JK, Brooks GA, Rodriguez RE, Dallman PR. Maintenance of euglycemia is impaired in gluconeogenesis-inhibited iron-deficient rats at rest and during exercise. J Nutr. 1994;124(11):2131–2138. doi: 10.1093/jn/124.11.2131. [DOI] [PubMed] [Google Scholar]
- McLean E, Cogswell M, Egli I, Wojdyla D, Benoist B. Worldwide prevalence of anaemia, WHO Vitamin and Mineral Nutrition Information System, 1993–2005. Pub Health Nutr. 2009;12(4):444–454. doi: 10.1017/S1368980008002401. [DOI] [PubMed] [Google Scholar]
- Nakae J, Biggs WH, 3rd, Kitamura T, Cavenee WK, Wright CV, Arden KC, Accili D. Regulation of insulin action and pancreatic beta-cell function by mutated alleles of the gene encoding forkhead transcription factor Foxo1. Nat Genet. 2002;32(2):245–253. doi: 10.1038/ng890. [DOI] [PubMed] [Google Scholar]
- Nead KG, Halterman JS, Kaczorowski JM, Auinger P, Weitzman M. Overweight children and adolescents: a risk group for iron deficiency. Pediatrics. 2004;114(1):104–108. doi: 10.1542/peds.114.1.104. [DOI] [PubMed] [Google Scholar]
- Pinhas-Hamiel O, Newfield RS, Koren I, Agmon A, Lilos P, Phillip M. Greater prevalence of iron deficiency in overweight and obese children and adolescents. Int J Obes Relat Metab Disord. 2003;27(3):416–418. doi: 10.1038/sj.ijo.0802224. [DOI] [PubMed] [Google Scholar]
- Puig S, Askeland E, Thiele DJ. Coordinated remodeling of cellular metabolism during iron deficiency through targeted mRNA degradation. Cell. 2005;120(1):99–110. doi: 10.1016/j.cell.2004.11.032. [DOI] [PubMed] [Google Scholar]
- Puig S, Vergara SV, Thiele DJ. Cooperation of two mRNA-binding proteins drives metabolic adaptation to iron deficiency. Cell Metab. 2008;7(6):555–564. doi: 10.1016/j.cmet.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Report of the American Institute of Nutrition ad hoc Committee on Standards for Nutritional Studies (1977). J Nutr 107 (7):1340–1348 [DOI] [PubMed]
- Ross KL, Eisenstein RS. Iron deficiency decreases mitochondrial aconitase abundance and citrate concentration without affecting tricarboxylic acid cycle capacity in rat liver. J Nutr. 2002;132(4):643–651. doi: 10.1093/jn/132.4.643. [DOI] [PubMed] [Google Scholar]
- Sherman AR, Guthrie HA, Wolinsky I, Zulak IM. Iron deficiency hyperlipidemia in 18-day-old rat pups: effects of milk lipids, lipoprotein lipase, and triglyceride synthesis. J Nutr. 1978;108(1):152–162. doi: 10.1093/jn/108.1.152. [DOI] [PubMed] [Google Scholar]
- Shimomura I, Bashmakov Y, Horton JD. Increased levels of nuclear SREBP-1c associated with fatty livers in two mouse models of diabetes mellitus. J Biol Chem. 1999;274(42):30028–30032. doi: 10.1074/jbc.274.42.30028. [DOI] [PubMed] [Google Scholar]
- Shimomura I, Bashmakov Y, Ikemoto S, Horton JD, Brown MS, Goldstein JL. Insulin selectively increases SREBP-1c mRNA in the livers of rats with streptozotocin-induced diabetes. Proc Natl Acad Sci USA. 1999;96(24):13656–13661. doi: 10.1073/pnas.96.24.13656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura I, Matsuda M, Hammer RE, Bashmakov Y, Brown MS, Goldstein JL. Decreased IRS-2 and increased SREBP-1c lead to mixed insulin resistance and sensitivity in livers of lipodystrophic and ob/ob mice. Mol cell. 2000;6(1):77–86. [PubMed] [Google Scholar]
- Stangl GI, Kirchgessner M. Different degrees of moderate iron deficiency modulate lipid metabolism of rats. Lipids. 1998;33(9):889–895. doi: 10.1007/s11745-998-0285-8. [DOI] [PubMed] [Google Scholar]
- Swali A, McMullen S, Hayes H, Gambling L, McArdle HJ, Langley-Evans SC (2011) Cell cycle regulation and cytoskeletal remodelling are critical processes in the nutritional programming of embryonic development. PloS One 6 (8):e23189 [DOI] [PMC free article] [PubMed]
- Tosco A, Fontanella B, Danise R, Cicatiello L, Grober OM, Ravo M, Weisz A, Marzullo L (2010) Molecular bases of copper and iron deficiency-associated dyslipidemia: a microarray analysis of the rat intestinal transcriptome. Genes Nutr 5(1):1–8 [DOI] [PMC free article] [PubMed]
- Weinberg J, Dallman PR, Levine S. Iron deficiency during early development in the rat: behavioral and physiological consequences. Pharmacol Biochem Behav. 1980;12(4):493–502. doi: 10.1016/0091-3057(80)90179-3. [DOI] [PubMed] [Google Scholar]
- Worldwide prevalence of anaemia 1993–2005 : WHO global database on anaemia (2008). WHO
- Yamagishi H, Komabayashi T. Alteration of glucose metabolism and increased fructosamine in iron-deficiency anemic rats. Nutr Res. 2003;23(11):1547–1553. doi: 10.1016/S0271-5317(03)00159-3. [DOI] [Google Scholar]
- Yamagishi H, Okazaki H, Shimizu M, Izawa T, Komabayashi T. Relationships among serum triacylglycerol, fat pad weight, and lipolysis in iron-deficient rats. J Nutr Biochem. 2000;11(9):455–460. doi: 10.1016/S0955-2863(00)00113-3. [DOI] [PubMed] [Google Scholar]
- Yanoff LB, Menzie CM, Denkinger B, Sebring NG, McHugh T, Remaley AT, Yanovski JA (2007) Inflammation and iron deficiency in the hypoferremia of obesity. Int J Obes (2005) 31(9):1412–1419 [DOI] [PMC free article] [PubMed]