Abstract
New genes originate frequently across diverse taxa. Given that genetic networks are typically comprised of robust, co-evolved interactions, the emergence of new genes raises an intriguing question: how do new genes interact with pre-existing genes? Here, we show that a recently originated gene rapidly evolved new gene networks and impacted sex-biased gene expression in Drosophila. This 4–6 million-year-old factor, named Zeus for its role in male fecundity, originated through retroposition of a highly conserved housekeeping gene, Caf40. Zeus acquired male reproductive organ expression patterns and phenotypes. Comparative expression profiling of mutants and closely related species revealed that Zeus has recruited a new set of downstream genes, and shaped the evolution of gene expression in germline. Comparative ChIP-chip revealed that the genomic binding profile of Zeus diverged rapidly from Caf40. These data demonstrate, for the first time, how a new gene quickly evolved novel networks governing essential biological processes at the genomic level.
Keywords: gene regulation, network evolution, new gene origination
Introduction
As major contributors to adaptation, genes with novel biological functions often emerge during evolution (Long et al, 2003; Kaessmann et al, 2009), acting as new catalytic enzymes (Burki and Kaessmann, 2004; Zhang et al, 2004; Matsuno et al, 2009), new cellular components (Marques et al, 2008; Rosso et al, 2008) or acquiring antiviral immunity functions (Stremlau et al, 2004). These genes frequently recruited regulatory elements from different sources and acquired expression in various tissues (Kaessmann, 2010), especially in male reproductive organs (Vibranovski et al, 2009a; Kaessmann, 2010). Recently, the evolutionary roles of several new genes have been characterized. They can contribute to substrate specificity alteration (Zhang et al, 2004), diet shift (Zhang, 2006), resolving intralocus sexual antagonistic conflict (Gallach et al, 2010), or modulating male-courtship behaviour (Dai et al, 2008). Furthermore, although genetic networks are typically viewed to be robust (Kitano, 2004; Wagner, 2005), evidence shows that some small and local structures in certain pathways are evolutionarily mutable (Matsuno et al, 2009; Ding et al, 2010).
However, it is unclear if gene networks can undergo extensive changes on short evolutionary time scales, or just evolve slowly under constraint by the robustness. It is tempting to ask if new genes could integrate into complex biological networks and bring evolutionary innovation to the global network structure. Specifically, how and why new genes interact with pre-existing genes are poorly understood. Here, we describe how a young gene rapidly evolved extensive new gene–gene interactions in the genome and reshaped global sex-biased gene expression. The Zeus (CG9573, Drcd-1-r) gene was initially identified as a young retrogene that is linked to a male-fertility locus (Bai et al, 2007; Quezada-Diaz et al, 2010). We functionally and phenotypically characterized Zeus as well as Caf40, the protein encoded by its parental locus (Caf40, CG14213, or Drcd-1), and examined the role of Zeus in the evolution of the male reproductive network. The interactions of Zeus with the genome provided an opportunity to understand the evolutionary targets of a new gene that has integrated into pre-existing global gene networks.
Results
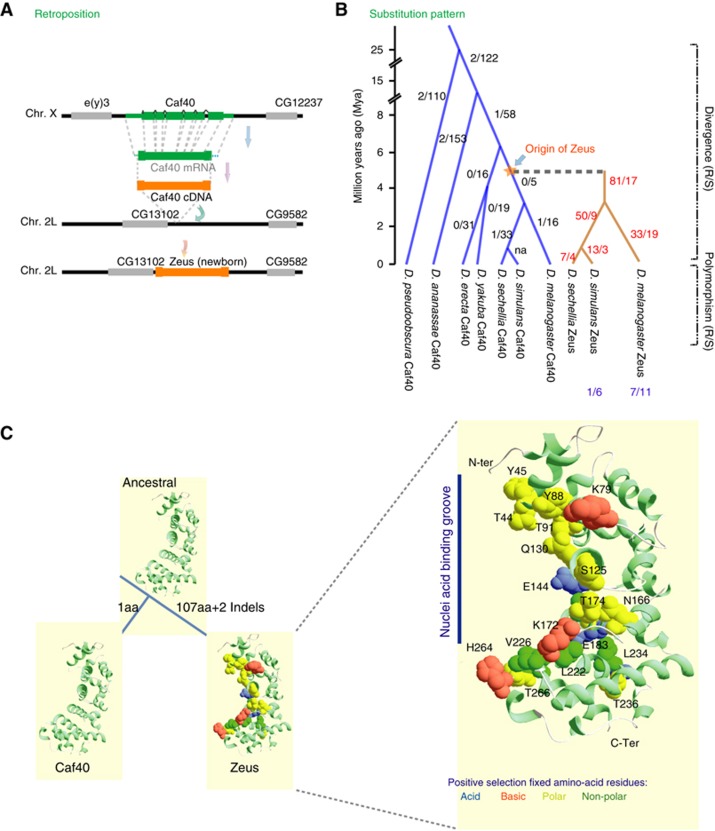
Origin and rapid evolution of a nascent retrogene, Zeus
Zeus originated 4–6 million years (Myr) ago, from a parental gene Caf40 (CG14213) by an out-of-X chromosome retroposition event in Drosophila (Bai et al, 2007; Figure 1A) The parental gene Caf40 is ancient, shared by a broad range of taxa including yeasts, worms, flies, and mammals (Chen et al, 2001; Garces et al, 2007). While Caf40 is highly conserved in all eukaryotes, the new duplicate, Zeus, evolved rapidly (Quezada-Diaz et al, 2010). We mapped the nucleotide substitutions of Zeus and Caf40 along all major lineages of the D. melanogaster group, and detected an initial burst of 81 replacement substitutions in 1–2 Myr (Figure 1B; Supplementary Figure S1). We sequenced 13 African and cosmopolitan D. melanogaster lines and observed a significant excess of between-species non-synonymous substitutions compared with within-species polymorphisms (Supplementary Table S1; McDonald–Kreitman (MK) test, P=0.00007). These data suggest that strong positive selection has dominated the evolution of Zeus.
Figure 1.
Origin and adaptive evolution of Zeus. Schematic representation of the origin and subsequent evolution of Zeus. (A) Retroposition event that led to the origin and gene structure evolution of Zeus. Green boxes represent exons of the parental gene Caf40; orange boxes represent exons of the new gene Zeus; grey boxes represent adjacent genes; boxes were not drawn to scale. (B) Sequence evolution of Zeus and Caf40 in the Drosophila phylogeny; replacement (R)/silent (S) substitutions are shown near major internal branches; R/S polymorphism data are shown at external branches where applicable. (C) Substitutions in binding domain and binding evolution: the left panel shows the homology models of the 3D structures of Zeus, Caf40 and the inferred ancestral protein; the numbers on tree branches indicate amino-acid substitutions in those particular lineages, ‘2 indels’ stands for a 9 amino-acid (aa) deletion at the N-terminus and a 6-aa insertion at the C-terminus; the right panel shows the zoom-in view for the amino-acid (aa) substitutions in the nucleic acid binding groove of the Zeus protein; aa substitutions fixed by positive Darwinian selection are represented as spheres and colour coded according to charge/polarity properties.
Gene expression pattern of Zeus
We then examined the expression pattern of Zeus using enhancer trap line, in which the Gal4 or LacZ element were inserted at the promoter region of 5′-UTR of the gene (Materials and methods). We found that Zeus is mainly expressed in the male reproductive system, including the testes and accessory glands (Figure 2A; Supplementary Figure S2). High levels of Zeus expression were detected during the entire process of spermatogenesis from mitotic spermatogonia, primary spermatocytes, differentiating spermatids, to mature sperm (Figure 2A; Supplementary Figure S2). We confirmed the expression patterns with RT–PCR (Supplementary Figure S2). This expression pattern is consistent with public microarray data (Chintapalli et al, 2007; Supplementary Figure S2) and high-throughput in-situ data from the literature (Tomancak et al, 2002). The male gonad expression of Zeus implies a specific role in male reproduction, and we went on to further test this hypothesis.
Figure 2.
Characterization of expression and phenotype of Zeus. (A) Expression pattern of Zeus in adult testis, blue arrows point at primary spermatocytes (left), early spermatids (middle) and mature sperms (right). (B) Testis phenotype of Zeus: top panel, wild-type testis overall morphology (left panel, phase contrast) and nuclei (right panel, DAPI staining); bottom panel, constitutive (Act5C-Gal4 driven) Zeus-RNAi testis overall morphology showing ectopic misgrowth at the tip (left panel, phase contrast) and irregular sperm bundles (right panel, DAPI staining). Red arrowheads point to representative abnormalities. (C) Sperm development phenotype shows misorganized sperm bundles (yellow arrows) in a Zeus-RNAi testis. (D) RT–PCR showing in-vivo transcription levels of Zeus, Caf40, CG13102 and gapdh1 in constitutive Zeus-RNAi and control animals. (E) Fecundity phenotype of constitutive Zeus-RNAi animals showing increased male sterility (left), reduced male fertility (middle), and normal female fertility (right); genotypes are shown below each data column; error bars on the column represent standard errors of the mean (s.e.m.); brackets denote statistical comparisons (ANOVA, *P<0.01). (F) Fecundity phenotype of tissue-specific Zeus-RNAi (left), RNAi-rescue (middle) and Zeus EMS mutants (right): nanos-Gal4-Zeus-RNAi; Acp26Aa(X)-Gal4-Zeus-RNAi; nos-Zeus-RNAi; Caf40-Overexpress, nanos-Gal4-Zeus-RNAi with Caf40 overexpression; Q94*, P50L, L182F, and Q110* are mutants with respective mutations in the Zeus protein sequences.
Phenotypic characterization of Zeus in male reproduction
To investigate the phenotype of Zeus, we used RNA interference (RNAi) to knockdown its expression (Dietzl et al, 2007). Constitutive RNAi against Zeus caused a 70% fertility reduction in male flies compared with wild-type and related controls (ANOVA, P<0.001 in each comparison; Figure 2E), with no detectable effect in females (Figure 2E). Germline-specific (nos-Gal4) Zeus RNAi also produced strong and significant male fertility defect, while accessory gland-specific (Acp26Aa-Gal4) RNAi did not (Figure 2F), suggesting that the primary fecundity function of Zeus lies in the testes. We carried out RT–PCR and verified that in Zeus-RNAi animals, the mRNA levels of Zeus were significantly reduced, while neither the parental gene Caf40 nor its overlapping gene (CG13102) was affected (Figure 2D). Moreover, the Zeus reproductive phenotype was recapitulated by presumptive Zeus null mutants, including two premature stop codons (Q94*, Q110*) and two missense mutations (P50L, L182F) that changed the polarity of amino acids (Figure 2F; Supplementary Figure S2). These point mutation alleles failed to complement each other, or previously isolated P-element insertion at the 5′-UTR of Zeus, which also caused male sterility (Castrion, 1993; Bai et al, 2007) as we confirmed independently (Supplementary Figure S2). Furthermore, testes from Zeus knockdown male flies showed specific defects in structure, including the disorganized cysts, tumour formation, and/or misoriented sperm bundles (Figure 2B and C). Both Zeus knockdown and mutant males can produce motile sperm, but they either fail to fertilize wild-type female eggs, or the fertilized eggs fail to develop (Supplementary Figure S2). These data suggest that Zeus plays an important role in male reproduction, probably functioning in the late stages of spermatogenesis or fertilization. Thus, despite of its recent origin, Zeus is essential for male fitness.
Expression pattern and phenotype of the parental gene, Caf40
As a comparison, we also investigated the function and phenotype of the parental gene Caf40, which is highly conserved, and thus possibly performs the ancestral function. The expression patterns and phenotypes of Caf40 are dramatically different from Zeus. First, Caf40 is expressed throughout the life cycle in most animal tissues, particularly at high levels in the musculature, digestive system, appendages, and central and peripheral nervous systems but only weakly in the reproductive tract (Figure 3A). Second, constitutive knockdown of Caf40 led to lethality at the onset of pupation (Figure 3B; Supplementary Table S2), and tissue-specific knockdown in the adult eye lead to missing sensory bristles and aberrant ommatidia development (Supplementary Figure S3). Third, germline knockdown of Caf40 did not produce significant male fertility defect (Supplementary Figure S3). Finally, ectopic expression of Caf40 failed to rescue Zeus male reproduction defects (Figure 2F). These data suggest that, instead of being a simple and redundant copy of Caf40, the new gene Zeus has evolved the capability to influence a distinct phenotype and established a unique role in the organism.
Figure 3.
Characterization of expression and phenotype of the parental gene Caf40. (A) Caf40 expression during several major stages of the life cycle, including early larvae (a1), late larvae (a2), pupae (a3) and adult (a4), and in several adult tissues nervous system (a5, MB, mushroom body; OL, optic lobe), digestive system (a6), musculature (a7), and weakly in reproductive organs (a8); (a4) note the Caf40::GFP fly showing strong constitutive GFP signal (red arrowhead) and the control fly showing no fluorescence (blue arrow). (B) Prepupae lethal phenotype of Caf40: top panel, normal pupae development of wild-type Can-S (top left) and non-induced control animals (in the same cross as Caf40-RNAi, top right); black arrow heads point to developing head/eye structures; bottom panel, disrupted pupae development of Caf40-RNAi animals under a constitutive driver (Act5C-Gal4), black arrow heads point to necrotic larval head structures (without formation of adult-like head/eye structures); blue arrows point to the empty pupal cuticle of control flies, which fully developed and hatched 7 days after pupation in the same cross; d AP, days after pupation.
Perturbation of global gene expression pattern in Zeus animals
The new phenotype of the young gene Zeus suggested that the genetic network has changed since its origin, because obviously the ancestral organisms were able to reproduce without the gene. This allowed us to test the concept of network evolution driven by new gene origination. To understand how the global gene expression network was affected when Zeus was perturbed, we carried out expression profiling of the transcriptome of Zeus-RNAi testes, where Zeus is primarily expressed (Materials and methods). In the set of genes differentially expressed between Zeus knockdown and controls (Supplementary Table S5), we found that female-biased genes were highly enriched among the RNAi upregulated genes, whereas male-biased genes were highly enriched in the RNAi downregulated genes (Supplementary Figure S4; Supplementary Table S6, χ2 tests, P=0 for both female- and male-biased gene tests). Moreover, the upregulated, female-biased genes were enriched on the X chromosome, whereas the downregulated, male-biased genes were overrepresented on autosomes (Supplementary Table S6, χ2 test, P=0.0001), consistent with previously reported chromosomal distribution patterns of sex-biased genes (Vibranovski et al, 2009b). These data suggest that Zeus tends to repress female-biased genes and activate male-biased genes in male reproductive organs.
To investigate the evolution of gene regulation after retroposition, we knocked down Zeus and Caf40 in parallel with a germline-specific driver and profiled the global gene expression in testes samples (Materials and methods). Compared with controls, Zeus-RNAi reduced 80% of Zeus mRNA, but did not affect Caf40 expression level; and vice versa for Caf40-RNAi. These data sets enabled us to identify distinct downstream genes for each factor. We identified a large set of genes that were significantly differentially expressed (multiple testing corrected P-value of <0.01) between Zeus-RNAi and control (Figure 4A; Supplementary Tables S3 and S4). The expression levels of these genes altered at least two-fold (absolute log-2 ratio>1). We also identified a set of downstream genes for Caf40. We found that 49.4% (444/899) of Zeus’ downstream genes are distinct from Caf40’s (Figure 4A), indicating that half of the Zeus downstream genes are not regulated by Caf40. For the genes co-regulated by both factors, the magnitudes, or even the directions of downstream gene expression changes may differ (i.e., upregulation in Zeus and downregulation in Caf40; Figure 4A). These data suggest that Zeus evolved to regulate a largely distinct gene set, implying its new role related to gene regulation in the male germline.
Figure 4.
Evolution of global gene regulation of Zeus in male germline. (A) Heatmap of male-germline RNA-seq expression profiling showing evolution of globally target genes and altered gene expression between Zeus and Caf40; top panel, expression profile of Zeus-specific target genes; middle panel, Zeus/Caf40 shared target genes; bottom panel, Caf40-specific target genes; left columns, nanosGal4»Zeus-RNAi; middle column, control; right column, nanosGal4»Caf40-RNAi; colour key and histogram with respect to the expression distribution were shown at bottom-right corner; microarray data (not shown) recapitulate consistent patterns; (B) heatmap of male-germline RNA-seq expression profiling of closely related species showing the influence of Zeus on its target genes after gene origination and speciation; top panel, expression profile of Zeus-suppressed orthologous target genes; bottom panel, expression profile of Zeus-activated orthologous target genes; left columns, nanosGal4»Zeus-RNAi; middle column, D. melanogaster wild type; right column, D. simulans wild type; rightmost column, D. yakuba wild type; colour key and histogram with respect to the expression distribution were shown at bottom-right corner; microarray data (not shown) recapitulate consistent patterns. (heatmaps represent difference between samples, gene number not drawn to scale)
Genome-wide binding profile of Zeus
To understand how Zeus integrated into the biological network, we then investigated how Zeus physically interacts with existing genes in the genome. We first examined where the Zeus protein is localized in the cell. We generated transgenic fly lines expressing GFP-tagged Zeus recombinant protein and found that the majority of Zeus protein is localized in the nucleus (Figure 5A). Being descended from Caf40, the Zeus protein has an Rcd1-like domain (Rcd, Retinoid acid-induced Cell Differentiation). Rcd1 homologues function as transcriptional regulators of cellular differentiation (Liu et al, 1998; Okazaki et al, 1998; Hiroi et al, 2002; Garces et al, 2007). Previous structural analysis revealed that an Rcd1 domain has six Armadillo-like repeats, forming a nucleic acid binding groove (Garces et al, 2007). Gel-shift assays demonstrated that Rcd1 domain physically binds to DNA (Chen et al, 2001; Garces et al, 2007). The putative nucleic acid binding groove of Zeus has experienced excessive amino-acid substitutions fixed by positive selection (Figure 1C; Supplementary Figure S1). Most (91%) of these novel amino acids were either charged or polar residues on the surface of the groove, forming a distinct surface conformation of Zeus (Figure 1C). We then set out to determine where the Zeus protein associates with chromosomes in the cell, and, ultimately how these adaptive amino-acid changes might have led to evolution in binding. We generated transgenic fly lines that expressed 3 × FLAG-tagged Zeus protein in vivo, under the control of an upstream activating sequence (UAS) promoter. We expressed the recombinant proteins either constitutively (Act5C-Gal4 driver) or in a tissue-specific manner (germline driver or accessory gland driver), and confirmed the detection of the recombinant protein (Figure 5B).
Figure 5.
Chromosomal binding profiles of Zeus and Caf40. (A) Confocal images of Zeus–GFP (green, GFP) localization with respect to the nucleus (blue, DAPI), scale bars are 10 μm. (B) purification and detection of FLAG-tagged Zeus protein in transgenic Drosophila male adults. (C) Overview of genome-wide binding profiles of Zeus (red) and Caf40 (blue) on all four chromosomes in D. melanogaster; black scale bar represents 5 Mb of chromosome. (D) Representative conserved (left), Caf40-specific (middle) and Zeus-specific (right) peaks/binding sites; track colour coding: (from top to bottom) Zeus-IP track (red), Zeus binding profile; Caf40-IP track (blue), Caf40 binding profile coordinate (black), base positions in the chromosome; Ref-seq(+/−) of the annotated D. melanogaster genome (green). (E) Venn diagrams showing the number and overlap of binding target genes of Zeus and Caf40.
We performed ChIP-chip (chromatin immunoprecipitation followed by microarray hybridization) to identify the global binding profile of Zeus, using an anti-FLAG antibody. This avoids cross-detection of endogenous Caf40 binding due to protein sequence similarity. We identified 363 peaks (putative binding sites/regions) for Zeus at a cutoff P-value of 1 × 10−3 (Figure 5C–E; Supplementary Figure S5; Supplementary Tables S7 and S8). Almost all of them (362/363, 99.7%) can be found within or near annotated genes (Figure 5C–E; Supplementary Figure S5). With this criterion, we identified 322 genes with Zeus binding sites as Zeus-binding genes (Figure 5E). Unexpectedly, although these binding sites/genes were distributed across all major chromosomal arms, they were highly overrepresented on the X chromosome (Supplementary Table S9) (120/363, 77% excess over random expectation, χ2 test, P=4.0 × 10−11). Because the X chromosome is enriched with female-biased genes (Sturgill et al, 2007; Vibranovski et al, 2009b), this binding pattern is in concordance with the sex-biased regulation of Zeus downstream genes.
Evolution of Zeus downstream targets
To study the evolution of Zeus downstream targets, we carried out transgenic experiments and ChIP-chip for Caf40 in parallel. By comparing the genome-wide binding profiles of Zeus and Caf40, we found many clear examples of conservation and divergence (Figure 5C and D; Supplementary Figure S5; Supplementary Tables S7 and S8). We found that Zeus’ putative targets, the genes bound by Zeus protein as revealed from ChIP, are largely different from those of Caf40’s, diverging by 60%. These data revealed that, after origination, Zeus quickly recruited many pre-existing genes as its targets in the genome and established novel functional gene–gene interactions.
By intersecting the ChIP-chip and expression profiling data sets, we sought to discover putative Zeus direct targets, which were physically associated with Zeus protein and differentially expressed upon Zeus knockdown (Supplementary Tables S10 and S11). Interestingly, we found that Zeus direct targets were predominantly upregulated (51 out of 63 genes) in Zeus-RNAi samples (Fisher’s exact test, P=0.0003; Supplementary Figure S6), suggesting that the direct action of Zeus tends to repress. We analysed Zeus and its downstream genes during spermatogenesis. Zeus expression decreases from the meiotic stage to the post-meiotic stage. Zeus downstream genes were enriched in the increasing expression category at the stage transition compared with randomly simulated genes (37.9% versus 28.7%, χ2 test, P=0.0003), consistent with putatative, repressive action.
Role of Zeus in the evolution of male germline gene expression
Structural modelling with evolutionary inference revealed that the putative nucleic acid binding groove of Zeus experienced excessive amino-acid substitutions that have been fixed by positive selection. Most (91%) of these novel amino acids are either charged or polar residues on the surface of the groove, forming a distinct surface conformation of Zeus (Figure 1C). In contrast, the Caf40 binding domain did not change in 25 Myr, and the whole protein has remained evolutionarily stagnant. Caf40 downstream genes are also much more conserved than Zeus downstream genes or randomly simulated genes (Supplementary Figure S5). These data imply that Zeus gained a large set of distinct gene–gene interactions in the last several million years. We also analysed the Zeus and Caf40 target gene sets with the stage-specific spermatogenesis expression data (Vibranovski et al, 2009a). During spermatogenesis, the relative expression level of Zeus compared with Caf40 (Zeus/Caf40) decreases during meiosis/post-meiosis stage (Supplementary Table S12). Accordingly, Zeus’ target genes’/downstream genes’ expression tend to increase compared with Caf40’s (Supplementary Table S12), in concordance with the fact that Zeus primarily acts as a repressor.
We then sought to detect evolutionary signatures of Zeus in the evolution of gene regulation across multiple Drosophila species. Using RNA-seq, we performed expression profiling with testes samples from four representative species with or without Zeus, which are, D. melanogaster, D. simulans, and D. yakuba. We analysed the orthologues of Zeus downstream genes in these species. Among genes activated by Zeus in D. melanogaster, most (73.6%, or 167/227) are expressed at significantly lower levels in D. yakuba (Figure 4B; Supplementary Figure S4; Supplementary Table S13). On the other hand, among genes repressed by Zeus in D. melanogaster, the majority (58.4%, or 115/197) has higher expression in D. yakuba (Figure 4B; Supplementary Figure S4; Supplementary Table S13). These results revealed that the evolution of the gene expression of Zeus targets is correlated with the presence/absence of Zeus in closely related species (Fisher’s exact test, P=4.0 × 10–12). These results were confirmed by expression profiling with custom-designed microarrays for these species. These data suggested that, although the divergence between these species is substantial, effects of Zeus origination on the evolution of gene expression were retained.
Discussion
New genes frequently arose and quickly established functional essentiality (Chen et al, 2010; Ding et al, 2010), where removal or perturbation of these previously non-existing genes resulted in severe phenotypes in extant organisms, suggesting that new genes have integrated into functional gene networks. Many new genes encode putative regulatory proteins including transcription factors, DNA/RNA-binding proteins and molecular chaperones (Chen et al, 2010, 2012), implying their potential to interact with other genes in the genome, and possible mechanisms by which they might gain essential functions via regulatory integration. Our integrative analyses revealed that, shortly after the birth of a new gene, Zeus, a novel reproductive subnetwork was assembled in the gene network.
The targets of Zeus could have been recently gained during adaptive evolution, or simply inherited from Caf40. The former is more likely because Caf40 is highly conserved and might represent the ancestral protein function, though the alternative remains a possibility. The binding domain of the parental gene had no change of amino-acid residue, and the non-binding domain of Caf40 changed only 0.66% of replacement sites during the last 25 Myr. In contrast, the new gene Zeus changed 114 replacement sites with 2 in-frame indels, diverged 35% from the parental gene in 5 Myr, equivalent to the divergence of Caf40 orthologues between Drosophila and human. This is a reminiscent of the fact that Caf40 downstream genes are much more conserved than the genome average, while Zeus downstream genes are not (Supplementary Figure S5D), suggesting a more conserved role for Caf40 in gene regulation during the evolutionary period we examined. It is intriguing that such extensive new binding sites have evolved in synchrony with a new DNA binding domain in Zeus. The molecular mechanism at work here awaits future study. It has been shown that transcription circuit evolution can also be driven by evolving protein–protein interactions (Tuch et al, 2008), implying that the origin of new binding sites of Zeus is possibly a combination of co-evolving, protein–nucleic acid and protein–protein interactions.
Although the stepwise processes of the Zeus network evolution in ancestral species has yet to be elucidated, our data provided the first example of a newborn gene quickly integrated into the gene network governing essential biological processes by regulating hundreds of existing genes, and rapidly building new pathways that shaped organismal phenotypes.
Materials and methods
Molecular evolutionary analyses for Zeus
New genes were previously computationally identified (Bai et al, 2007; Chen et al, 2010; Zhang et al, 2010). Orthologous sequences from D. melanogaster, D. simulans, and D. yakuba were retrieved from Flybase. Genome and protein sequences were retrieved from Flybase (fly genome assembly 2006, Dm3). Primary polymorphism data were collected from the Drosophila Population Genomics Project (DPGP, http://www.dpgp.org/) (Begun and Lindfors, 2005). Coding sequences of each gene from the 37 D. melanogaster lines and that from the D. yakuba or D. simulans orthologues were aligned (ClustalW) to estimate the non-synonymous divergence, synonymous divergence, and polymorphic frequency spectra using the Polymorphorama program (Haddrill et al, 2008). Estimation of the prevalence of natural selection, or the proportion of non-synonymous substitutions driven by positive selection (α values), was carried out with the polymorphism data using the DoFE package (Eyre-Walker and Keightley, 2009b). The proportion of non-synonymous substitutions driven by positive selection (α values) was calculated with the DoFE package (Bierne and Eyre-Walker, 2004; Eyre-Walker and Keightley, 2009a). In breif, DoFE implemented the framework of MK test which compared within-population polymorphism (P) at two categories of sites and the corresponding levels of between-species divergence. Usually synonymous (s) and non-synonymous (n) sites are used. Assuming that synonymous sites are neutral while non-synonymous sites could be neutral, highly deleterious and highly beneficial, the ratio Dn/Ds should be equal to the ratio Pn/Ps (Bierne and Eyre-Walker, 2004). Thus, α is equal to 1−DsPn/(DnPs) (Bierne and Eyre-Walker, 2004). As for the P-values, it is the possibility that α is significantly different from 0. In other words, it reflects whether neutrality should be rejected.
Orthologous sequences of Zeus (CG9573) and Caf40 (CG14213) from D. melanogaster, D. simulans, D. sechellia, D. yakuba, D. erecta, D. annanassae, and D. pseudoobscura were retrieved from Flybase. Zeus polymorphism data were collected from 13 D. melanogaster strains (Emerson et al, 2008) by PCR sequencing and from six publicly available in-bred D. simulans strains (DPGP; http://www.dpgp.org/). Alignments were performed in ClustalX 1.83 (Higgins et al, 1996). Non-synonymous (R) and synonymous (S) substitutions were mapped to the phylogeny using PAML (Yang, 1997). The probabilities of positive selection in substitutions were calculated with a Bayes Empirical Bayes (BEB) estimation implemented in PAML. Branch-site new model A (model=2 NSsites=2) was used to estimate whether Zeus is subject to positive selection upon its origination (Yang, 2007). The null model is still model A but the ratio Dn/Ds (or ω) of the branch of the interest is fixed as 1 (fix_omega=1; omega=1). The tree topology is specified as: (zeus_mel, (zeus_sim,zeus_sec))#1,(caf40_mel,caf40_sec)),(caf40_ere,caf40_yak)),caf40_ana), (caf40_pse,caf40_per)),caf40_wil),(caf40_vir,caf40_gri)); the estimated ω is as high as 6.1 and the corresponding P of likelihood ratio test is 0.004, suggesting that positive selection acts on Zeus copy upon its origination. Homology-based structural modelling was run in Swiss-Model (Arnold et al, 2006). Amino-acid substitutions that were fixed by positive selection (with BEB probability of >0.95) were mapped to the nucleic acid binding groove of the Zeus homology-based 3D structures with SPDBV (Guex and Peitsch, 1997). MK test was conducted in DNASP4.0 (Rozas et al, 2003).
Genomic PCR and Southern blot
Genomic DNA was extracted using standard molecular biology protocols. PCRs were carried out with region-specific primers. For Southern blots, first, the Zeus CDS was cloned into the pBSK(−) backbone; second, probes were synthesized by in-vitro transcription incorporating DIG, hybridized to genomic DNAs, and then visualized by anti-DIG-AP staining.
Enhancer trap lines and expression patterns
RNA extraction, standard and semi-quantitative RT–PCR were performed using RNeasy mini-Kit (Qiagen), SuperscriptIII RT (Invitrogen) with manufacturer’s protocols. The same amount of cDNA was used for each sample and PCR was carried out with Platinum Taq Polymerase (Invitrogen). Cycle series from 18 to 32 were performed for each PCR and non-saturation cycle was used for quantification by agarose gel electrophoresis and ImageJ. Primer sequences for Zeus and Caf40 are
- CG9573F55
5′-CACCATGAGTGCGGAACCAAGTCCG-3′,
- CG9573F50
5′-CACCATGAGTGCGGAACCAAGTC-3′,
- CG9573R55
5′-CTAGGAGGAGCCCATTGGGT-3′,
- CG9573R50
5′-CTAGGAGGAGCCCATTG-3′,
- CG14213F55
5′-CACCATGAGTGCTCAACCGAGTCCG-3′,
- CG14213F50
5′-CACCATGAGTGCTCAACCGAGTC-3′,
- CG14213R55
5′-CTAGGAGCCCAGTGGCGAC-3′,
- CG14214R50
5′-CTAGGAGCCCAGTGGC-3′.
Enhancer trap lines for Zeus (P ms(2)29F07717) and Caf40 (P{GawB}NP1003/FM7c) were ordered from the Bloomington stock center and GETDB (Hayashi et al, 2002). The enhancer trap line P{PZ}ms(2)29F07717 is annotated as being associated with Zeus and CG13102. The P element-carrying lacZ construct was inserted in the 5′-UTR of Zeus. NP1003 is annotated as associated with Caf40, e(y)3, and CG12237. Its P{GawB} is on the sense strand, ∼5 kilobases (kb) upstream of Caf40, within an intron of e(y)3 but on the opposite strand as e(y)3 and ∼8 kb from and on the opposite strand as CG12237. The lacZ-based enhancer trap line was directly dissected and stained with a b-galactosidase staining kit (Invitrogen). The Gal4-based NP enhancer trap lines were crossed to reporter lines yw;UAS-mCD8-GFP/CyO and/or yw,UAS-2xEYFP. F1s with both enhancer trap Gal4 and reporter alleles were collected and dissected to observe fluorescence signals. Expression patterns for both genes were consistent with the FlyAtlas microarray data (Chintapalli et al, 2007) and BDGP high-throughput embryo in-situ data (Tomancak et al, 2007).
Zeus RNAi and phenotype
UAS-IR RNAi lines were obtained from the VDRC and TRiP RNAi libraries (Dietzl et al, 2007; Ni et al, 2009). UAS-IR lines were crossed to constitutive driver line yw;Actin5C::Gal4/CyO,y+, germline-specific driver nanos-Gal4 or accessory gland driver Acp26Aa-Gal4 to produce RNAi progeny. Rescue of Zeus by Caf40 was done by crossing nanos-Gal4»Zeus-RNAi flies to UAS-Caf40-CDS flies. Flies were aged at 25 or 29°C. Testes and ovaries were dissected, stained, and examined under a fluorescence microscope. For the fertility test, all flies were raised in a 25°C incubator with a 13 h:11 h light:dark cycle and assay with mating and offspring counting. For example, the RNAi line (GD49820) was used for Zeus RNAi knockdown in this study. In the male fertility test, virgin Zeus-RNAi males (UAS::IR-Zeus/Y;Actin5C::Gal4/+ and UAS::IR-Zeus/Y;;TubP::Gal4/+) and control males (UAS::IR-Zeus, yw;Actin5C::Gal4/CyO,y+) and wild-type D. melanogaster (Oregon-R)) were collected and aged 4–7 days before mating. Single male flies of a particular genotype were crossed to five of 4- to 7-day-old wild-type D. melanogaster virgin females (Oregon-R). The flies in these crosses were allowed to mate and lay eggs for 9 days and were then cleared on day 10. The numbers of offspring eclosed from day 12 to day 24 were counted. The female fertility test was carried out similarly, with a single virgin female of the desired genotype crossed to five wild-type males. Ten to forty replicates were carried out for each genotype. Statistically significant differences between genotypes were calculated by ANOVA.
Zeus mutant screen and phenotypic analysis
The S. Hawley lines were screened using FlyTILL and mutations in the Zeus locus were identified by genomic PCR and sanger sequencing using multiple primer pairs. The P-element insertion mutant P{PZ}ms(2)29F07717 was obtained from Bloomington stock center and the insertion position was verified by genomic PCR using multiple primers. Presumptive null mutants were subjected to fertility tests as described ahead. Complementation was carried out by crossing mutants to generate trans-heterozygotes.
Caf40 RNAi and phenotype
The males from the Caf40 RNAi line (KK101462) were crossed to the constitutive driver lines yw;Actin5C::Gal4/CyO,y+ and yw;;TubP::Gal4/Tm3,Sb to produce RNAi progeny. In constitutive RNAi crosses, RNAi progeny (UAS-IR/Gal4) and non-RNAi control progeny (UAS-IR/Balancer) were identified by phenotypic markers on balancers in hatched adults. The number of progeny in each genotype was counted. If no RNAi progeny hatched while control progeny hatched normally, the RNAi treatment was considered as lethal. Multiple crosses were performed. The developmental stage(s) when lethality occurred were examined and imaged under a stereoscope. The Caf40 RNAi line was also crossed to a tissue-specific driver line w;GMR::Gal4/CyO-GFP. RNAi progeny was collected, and adult cuticles were mounted, coated with Pt/Pd alloy and imaged with a scanning electron microscope.
Expression profiling
Testes from nanos-Zeus RNAi, nanos-Caf40 RNAi, Act5C-Zeus RNAi, and wild-type D. melanogaster (Oregon-R), D. simulans (HD01) and D. yakuba (Tai6) were dissected from 1- to 7-day-old males. Biological quadruplicate RNA samples were purified using Trizol/choloform extraction followed by RNeasy-mini kit purification (Qiagen). RNA concentrations were measured with a NanoDrop, and RNA integrity was verified using a Bioanalyzer RNA chip (Agilent). For RNA-seq, library preparation was performed according to Illumina’s standard protocols, and sequencing were performed on Illumina HiSeq. For expression arrays, RNA samples were then reverse transcribed into cDNAs, which were then used to generate Cy3-/Cy5-cRNAs using the two-colour Quick Amp labeling Kit (Agilent). These labelled cRNA samples were hybridized to Drosophila Gene Expression Microarrays (Agilent) using a Tecan HS 4800 Pro Hybridization Station at the High-throughput Genome Analysis Core (HGAC) in the Argonne National Laboratory.
RNA-seq expression profiling data analysis
For RNA-seq, reads were mapped to reference transcriptomes (Flybase) of the respective species using the Bowtie program (Langmead et al, 2009), with unique mapping allowing a maximum of two mismatches per read. After unique mapping, we obtained transcriptome profiles for these samples with ∼30-fold coverage. We calculated the relative expression level for each gene in the genome, using the reads per million total reads (RPM) and reads per kilobase transcript per million total reads (RPKM) statistics as described previously (Mortazavi et al, 2008). We calculated Spearman correlation and ensured high reproducibility between biological replicates (R>0.99). Orthologous relationships across species were obtained from Flybase annotations. Differential expression was called using Likelihood-ratio tests were performed with our own scripts for R (http://www.r-project.org/). The P-value and the log ratio of each gene (comparison) were calculated according to this principle. The false discovery rate (FDR) method was used for multiple testing corrections. As stated in the text, differentially expression was called for a gene if q-value <0.01 and absolute fold change>2.
Microarray expression profiling data analysis
For expression arrays, probe intensity data were extracted using Feature Extraction software (Agilent). All arrays passed all quality controls (QC), with a high quality of data readout and high linear correlation of spike-ins (R>0.98). Extra QC and analyses were performed using our own scripts together with Bioconductor packages limma and marray. Briefly, background correction was performed using the backgroundCorrect function with the ‘normexp’ method and offset; Loess normalization was then performed to correct for different dye effects; intraarray normalization was carried out using the ‘Aquantile’ option. All ‘flag’ probes by Agilent QC were filtered out as ‘NA’. Next, we calculated the Spearman correlations of within-array duplicate probes (R>0.99) and the pairwise correlations between quadruplicates (R>0.97). We then pooled multiple probes for each gene, calculated the mean M value (log-2 scale probe intensity ratios) and performed a two-tail Wilcoxon rank sum test between control and RNAi samples. Differentially expressed genes were indentified using the cutoff P-value of <0.05 after multiple test correction (FDR method). For RNA-seq, we used bowtie to map the reads to the annotated Drosophila genomes (genome sequence plus the exon–exon conjunction sequence according to transcript annotation as described in Mortazavi et al (2008) using Bowtie (Langmead et al, 2009), allowing a maximum of two mismatches. We calculated the relative expression level for each gene in the genome, using the number of RPKM as described previously (Mortazavi et al, 2008). The information of orthologous genes between species were retrieved from Flybase. Likelihood-ratio tests were performed in a generalized linear model framework as described in Marioni et al (2008) with our own R script as described previously (Chen et al, 2012).
Generation of GFP-tagged protein expression lines and determination of cellular localization
The Zeus ORF was amplified with primers 5′-ATGAGTGCGGAACCAAGTCCG-3′ and 5′-CTAGGAGGAGCCCATTGGGT-3′, cloned into a pTGW vector (Murphy; http://www.ciwemb.edu/labs/murphy/Gateway%20vectors.html) and then injected into w1118 to generate transgenic fly lines (UASt::GFP-Zeus). Males from these lines were crossed to virgin females from the constitutive Gal4 driver line yw;Actin5C::Gal4/CyO,y+. F1 males (4–7 days old), which contained the UASt::GFP-Zeus and Actin5C::Gal4 driver and thus constitutively expressed the GFP-tagged Zeus protein, were collected. Their male reproductive organs were dissected and co-stained with DAPI. Visualization of GFP-tagged Zeus protein and the nucleus was performed with an SP5 confocal microscope (Leica).
Generation of Zeus FLAG-tagged protein expression lines
The coding region of Zeus was cloned into a pTFMW backbone and microinjected into w1118 embryos to establish FLAG-tag fusion protein expression lines under the UASt promoter (genotype UASt::FLAG-Zeus) These males were crossed to virgin females with the genotype yw;Actin5C::Gal4/CyO,y+ to generate constitutive FLAG–Zeus expression flies (Actin5C::Gal4»UASt::FLAG–Zeus). The same cloning, transgenic, and crossing schemes were carried out in parallel for the parental gene Caf40 (the primer pairs for Caf40 were 5′-ATGAGTGCTCAACCGAGTCCG-3′ and 5′-CTAGGAGCCCAGTGGCGAC-3′).
Zeus recombinant protein expression and purification
Adult flies (4–7 days old) from constitutive FLAG–Zeus expression lines were collected. Proteins were extracted from these adults with RIPA buffer (Boston BioProducts). FLAG–Zeus recombinant protein was successfully purified from the crude extract by immunoprecipitation (IP) with a FLAG-IP kit (Sigma) and then visualized by western blot using an anti-FLAG antibody (Sigma) and Commassie-Blue stain, following the standard molecular biology protocols. All protein extraction and purification was performed at 4°C.
Chromatin immunoprecipitation followed by array hybridization (ChIP-chip) of Zeus and Caf40
Chromatin was collected from 4- to 7-day-old whole adult flies. IP was performed in triplicate using a rabbit anti-FLAG antibody (Sigma). The reference sample, in our case input DNA (DNA purified directly from chromatin without antibody incubation), and IP DNA samples were then amplified using Klenow (Invitrogen) to incorporate dUTP. Fragmentation and labelling were then performed using an Affymetrix labeling kit. Hybridization was carried out using Affymetrix Drosophila Tilling Arrays (1.0). Data were collected at the Functional Genomic Facility (FGF) at the University of Chicago. All procedures were performed according to standard fly ChIP-chip protocol (Roy et al, 2010; Negre et al, 2011).
ChIP-chip data analysis
ChIP-chip data were analysed in TAS (Affymetrix) using ‘Two Sample Comparison Analysis’ of IP triplicates versus input triplicates, with the following parameters: Probe Analysis (Bandwidth=500 bp, Test type=One-sided upper, Intensities=PM only) and Interval Analysis (Threshold=1e−005, Threshold option=Less than threshold, Maximum Gap=80 bp, Minimum run=40 bp). The major analysis of binding sites (peaks) identification was done with a P-value cutoff of 0.001, and all the patterns were robust with other P-value cutoffs (0.01 or 0.0001).
For peak-calling FDR estimation, three samples from the six input samples were randomly chosen as ‘pseudo-IP’ with the remaining three as ‘input’ to identify pseudo binding sites (pseudo peaks) using the same parameters.
The binding site distribution and overlap was analysed with CisGenome (Ji et al, 2008). Because >97% of the binding sites overlapped with annotated genes, these genes were defined as Zeus/Caf40 binding genes. Binding gene identification and overlap were calculated by our own scripts. Peak centre coordinates were mapped to the to D. melanogaster genome release 5 using LiftOver (Kuhn et al, 2009). Enriched motifs of Zeus/Caf40 were identified using MEME (Bailey, 2002) from ±250 bp sequences flanking the peak centre.
Sex-biased gene expression analysis
Male/female-biased genes were defined as genes that are expressed significantly higher in male/female whole body or reproductive organs (testes/ovaries). In our analyses, we use (1) data sets of male/female-biased genes from SEBIDA (Gnad and Parsch, 2006) and/or (2) ovary and testis data from FlyAtlas (Chintapalli et al, 2007), where genes with high expression in testis (FDR<0.05) were defined as male-biased genes and vice versa (Vibranovski et al, 2009a; Zhang et al, 2010). Results from the analysis with either of these two data sets showed identical patterns. Spermatogenesis stage-specific gene expression analyses of Zeus/Caf40 targets were carried out using the SpermPress database (Vibranovski et al, 2009a) and our own scripts. Briefly, using Baysian inference of expression profiling data, genes were classified into downregulated (Under), stable (Equal) or upregulated (Over) during stage transitions (Mitosis—Meiosis—Post-meiosis); statistical patterns were calculated by comparing the categories of different gene groups.
Accession codes
The microarray and high-throughput sequencing data have been deposited to GEO (GSE36920; GSE36573; GSE36574; GSE36764).
Supplementary Material
Acknowledgments
We thank Terence Murphy and DGRC for providing the Drosophila Gateway vectors, Perrimon laboratory for the Valium vectors and nanos-Gal4 line, Dr Claudia and Dr Chapman for Acp26Aa-Gal4 line, Masatoshi Tomaru and GETDB for Zeus enhancer trap lines, the VDRC and TRiP for providing the Zeus RNAi lines, FlyTILL and Dr S Hawley for mutant library screen, and the fly community for providing many related lines. We thank Siming Shou, Jaejung Kim, Tifani Eshoo, and Marc Domanus for technical help with genomics. We thank Zhongzhou Zheng, Wenjun Xiong, Yang Liu, Bejamin Ross, and Amanda Neisch for technical assistance in molecular biology. We also thank Marty Kreitman, Ilya Ruvinsky, Richard Hudson, Wei Du, Tim Karr, Harmit Malik, Kevin Thornton and all members of Long and White laboratories for stimulating discussions about genetic and evolutionary analysis and reading the manuscript. This work was supported by US National Science Foundation awards (NSFCAREER MCB0238168 and MCB1051826), US National Institutes of Health R01 grants (R01GM065429-01A1 and 1R01GM078070-01A1) and a Packard Fellowship for Science and Engineering to ML, an NSF Doctoral Dissertation Improvement Grant (DEB-1110607) to SC, an NIH Genetics and Regulation Training Grant T32 GM007197 and a Department of Education Evolutionary Genomics GAANN fellowship to BHK, an NIH grant 1P50GM081892 and Chicago Biomedical Consortium Searle Funds at the Chicago Community Trust to KPW.
Author contributions: ML and KPW supervised the work; SC, ML, XN, and KPW designed the experiments; SC, XN, BHK, and MDV performed the experiments. XN, SC, YEZ, MDV, and BHK analysed the data; SC, ML, XN, BHK, and KPW wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22: 195–201 [DOI] [PubMed] [Google Scholar]
- Bai Y, Casola C, Feschotte C, Betran E (2007) Comparative genomics reveals a constant rate of origination and convergent acquisition of functional retrogenes in Drosophila. Genome Biol 8: R11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL (2002) Discovering novel sequence motifs with MEME. Curr Protoc Bioinformatics Chapter 2: Unit 2 4 [DOI] [PubMed] [Google Scholar]
- Begun DJ, Lindfors HA (2005) Rapid Evolution of genomic Acp complement in the melanogaster subgroup of Drosophila. Mol Biol Evol 22: 2010–2021 [DOI] [PubMed] [Google Scholar]
- Bierne N, Eyre-Walker A (2004) The genomic rate of adaptive amino acid substitution in Drosophila. Mol Biol Evol 21: 1350–1360 [DOI] [PubMed] [Google Scholar]
- Burki F, Kaessmann H (2004) Birth and adaptive evolution of a hominoid gene that supports high neurotransmitter flux. Nat Genet 36: 1061–1063 [DOI] [PubMed] [Google Scholar]
- Castrillon DH, Gönczy P, Alexander S, Rawson R, Eberhart CG, Viswanathan S, DiNardo S, Wasserman SA (1993) Toward a molecular genetic analysis of spermatogenesis in Drosophila melanogaster: characterization of male-sterile mutants generated by single P element mutagenesis. Genetics 135: 489–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Rappsilber J, Chiang YC, Russell P, Mann M, Denis CL (2001) Purification and characterization of the 1.0 MDa CCR4-NOT complex identifies two novel components of the complex. J Mol Biol 314: 683–694 [DOI] [PubMed] [Google Scholar]
- Chen S, Spletter M, Ni X, White KP, Luo L, Long M (2012) Frequent recent origination of brain genes shaped the evolution of foraging behavior in Drosophila. Cell Rep 1: 118–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Zhang YE, Long M (2010) New genes in Drosophila quickly become essential. Science 330: 1682–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintapalli VR, Wang J, Dow JA (2007) Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet 39: 715–720 [DOI] [PubMed] [Google Scholar]
- Dai H, Chen Y, Chen S, Mao Q, Kennedy D, Landback P, Eyre-Walker A, Du W, Long M (2008) The evolution of courtship behaviors through the origination of a new gene in Drosophila. Proc Natl Acad Sci USA 105: 7478–7483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin C (1859) On the Origin of Species by Means of Natural Selection London: J. Murray, [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, Couto A, Marra V, Keleman K, Dickson BJ (2007) A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448: 151–156 [DOI] [PubMed] [Google Scholar]
- Ding Y, Zhao L, Yang S, Jiang Y, Chen Y, Zhao R, Zhang Y, Zhang G, Dong Y, Yu H, Zhou Q, Wang W (2010) A young Drosophila duplicate gene plays essential roles in spermatogenesis by regulating several Y-linked male fertility genes. PLoS Genet 6: e1001255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson JJ, Cardoso-Moreira M, Borevitz JO, Long M (2008) Natural selection shapes genome-wide patterns of copy-number polymorphism in Drosophila melanogaster. Science 320: 1629–1631 [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A, Keightley PD (2009a) Estimating the rate of adaptive molecular evolution in the presence of slightly deleterious mutations and population size change. Mol Biol Evol 26: 2097–2108 [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A, Keightley PD (2009b) Estimating the rate of adaptive molecular evolution in the presence of slightly deleterious mutations and population size change. Mol Biol Evol 26: 2097–2108 [DOI] [PubMed] [Google Scholar]
- Gallach M, Chandrasekaran C, Betran E (2010) Analyses of nuclearly encoded mitochondrial genes suggest gene duplication as a mechanism for resolving intralocus sexually antagonistic conflict in Drosophila. Genome Biol Evol 2: 835–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garces RG, Gillon W, Pai EF (2007) Atomic model of human Rcd-1 reveals an armadillo-like-repeat protein with in vitro nucleic acid binding properties. Protein Sci 16: 176–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnad F, Parsch J (2006) Sebida: a database for the functional and evolutionary analysis of genes with sex-biased expression. Bioinformatics 22: 2577–2579 [DOI] [PubMed] [Google Scholar]
- Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18: 2714–2723 [DOI] [PubMed] [Google Scholar]
- Haddrill PR, Bachtrog D, Andolfatto P (2008) Positive and negative selection on noncoding DNA in Drosophila simulans. Mol Biol Evol 25: 1825–1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S, Ito K, Sado Y, Taniguchi M, Akimoto A, Takeuchi H, Aigaki T, Matsuzaki F, Nakagoshi H, Tanimura T, Ueda R, Uemura T, Yoshihara M, Goto S (2002) GETDB, a database compiling expression patterns and molecular locations of a collection of Gal4 enhancer traps. Genesis 34: 58–61 [DOI] [PubMed] [Google Scholar]
- Higgins DG, Thompson JD, Gibson TJ (1996) Using CLUSTAL for multiple sequence alignments. Methods Enzymol 266: 383–402 [DOI] [PubMed] [Google Scholar]
- Hiroi N, Ito T, Yamamoto H, Ochiya T, Jinno S, Okayama H (2002) Mammalian Rcd1 is a novel transcriptional cofactor that mediates retinoic acid-induced cell differentiation. EMBO J 21: 5235–5244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Jiang H, Ma W, Johnson DS, Myers RM, Wong WH (2008) An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat Biotechnol 26: 1293–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaessmann H (2010) Origins, evolution, and phenotypic impact of new genes. Genome Res 20: 1313–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaessmann H, Vinckenbosch N, Long M (2009) RNA-based gene duplication: mechanistic and evolutionary insights. Nat Rev Genet 10: 19–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano H (2004) Biological robustness. Nat Rev Genet 5: 826–837 [DOI] [PubMed] [Google Scholar]
- Kuhn RM, Karolchik D, Zweig AS, Wang T, Smith KE, Rosenbloom KR, Rhead B, Raney BJ, Pohl A, Pheasant M, Meyer L, Hsu F, Hinrichs AS, Harte RA, Giardine B, Fujita P, Diekhans M, Dreszer T, Clawson H, Barber GP et al. (2009) The UCSC Genome Browser Database: update 2009. Nucleic Acids Res 37: Database issueD755–D761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HY, Badarinarayana V, Audino DC, Rappsilber J, Mann M, Denis CL (1998) The NOT proteins are part of the CCR4 transcriptional complex and affect gene expression both positively and negatively. EMBO J 17: 1096–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M, Betran E, Thornton K, Wang W (2003) The origin of new genes: glimpses from the young and old. Nat Rev Genet 4: 865–875 [DOI] [PubMed] [Google Scholar]
- Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y (2008) RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res 18: 1509–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques AC, Vinckenbosch N, Brawand D, Kaessmann H (2008) Functional diversification of duplicate genes through subcellular adaptation of encoded proteins. Genome Biol 9: R54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuno M, Compagnon V, Schoch GA, Schmitt M, Debayle D, Bassard JE, Pollet B, Hehn A, Heintz D, Ullmann P, Lapierre C, Bernier F, Ehlting J, Werck-Reichhart D (2009) Evolution of a novel phenolic pathway for pollen development. Science 325: 1688–1692 [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Negre N, Brown CD, Ma L, Bristow CA, Miller SW, Wagner U, Kheradpour P, Eaton ML, Loriaux P, Sealfon R, Li Z, Ishii H, Spokony RF, Chen J, Hwang L, Cheng C, Auburn RP, Davis MB, Domanus M, Shah PK et al. (2011) A cis-regulatory map of the Drosophila genome. Nature 471: 527–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni JQ, Liu LP, Binari R, Hardy R, Shim HS, Cavallaro A, Booker M, Pfeiffer BD, Markstein M, Wang H, Villalta C, Laverty TR, Perkins LA, Perrimon N (2009) A Drosophila resource of transgenic RNAi lines for neurogenetics. Genetics 182: 1089–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki N, Okazaki K, Watanabe Y, Kato-Hayashi M, Yamamoto M, Okayama H (1998) Novel factor highly conserved among eukaryotes controls sexual development in fission yeast. Mol Cell Biol 18: 887–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quezada-Diaz JE, Muliyil T, Rio J, Betran E (2010) Drcd-1 related: a positively selected spermatogenesis retrogene in Drosophila. Genetica 138: 925–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso L, Marques AC, Weier M, Lambert N, Lambot MA, Vanderhaeghen P, Kaessmann H (2008) Birth and rapid subcellular adaptation of a hominoid-specific CDC14 protein. PLoS Biol 6: e140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Ernst J, Kharchenko PV, Kheradpour P, Negre N, Eaton ML, Landolin JM, Bristow CA, Ma L, Lin MF, Washietl S, Arshinoff BI, Ay F, Meyer PE, Robine N, Washington NL, Di Stefano L, Berezikov E, Brown CD, Candeias R et al. (2010) Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330: 1787–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R (2003) DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19: 2496–2497 [DOI] [PubMed] [Google Scholar]
- Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J (2004) The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature 427: 848–853 [DOI] [PubMed] [Google Scholar]
- Sturgill D, Zhang Y, Parisi M, Oliver B (2007) Demasculinization of X chromosomes in the Drosophila genus. Nature 450: 238–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomancak P, Beaton A, Weiszmann R, Kwan E, Shu S, Lewis SE, Richards S, Ashburner M, Hartenstein V, Celniker SE, Rubin GM (2002) Systematic determination of patterns of gene expression during Drosophila embryogenesis. Genome Biol 3: RESEARCH0088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomancak P, Berman BP, Beaton A, Weiszmann R, Kwan E, Hartenstein V, Celniker SE, Rubin GM (2007) Global analysis of patterns of gene expression during Drosophila embryogenesis. Genome Biol 8: R145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuch BB, Li H, Johnson AD (2008) Evolution of eukaryotic transcription circuits. Science 319: 1797–1799 [DOI] [PubMed] [Google Scholar]
- Vibranovski MD, Lopes HF, Karr TL, Long M (2009a) Stage-specific expression profiling of Drosophila spermatogenesis suggests that meiotic sex chromosome inactivation drives genomic relocation of testis-expressed genes. PLoS Genet 5: e1000731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vibranovski MD, Zhang Y, Long M (2009b) General gene movement off the X chromosome in the Drosophila genus. Genome Res 19: 897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A (2005) Robustness and Evolvability in Living Systems Princeton, NJ: Princeton University Press, [Google Scholar]
- Yang Z (1997) PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci 13: 555–556 [DOI] [PubMed] [Google Scholar]
- Yang Z (2007) PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24: 1586–1591 [DOI] [PubMed] [Google Scholar]
- Zhang J (2006) Parallel adaptive origins of digestive RNases in Asian and African leaf monkeys. Nat Genet 38: 819–823 [DOI] [PubMed] [Google Scholar]
- Zhang J, Dean AM, Brunet F, Long M (2004) Evolving protein functional diversity in new genes of Drosophila. Proc Natl Acad Sci USA 101: 16246–16250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YE, Vibranovski MD, Krinsky BH, Long M (2010) Age-dependent chromosomal distribution of male-biased genes in Drosophila. Genome Res 20: 1526–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.