Abstract
Freshwater planaria possess extreme regeneration capabilities mediated by abundant, pluripotent stem cells (neoblasts) in adult animals. Although planaria emerged as an attractive in vivo model system for stem cell biology, gene expression in neoblasts has not been profiled comprehensively and it is unknown how molecular mechanisms for pluripotency in neoblasts relate to those in mammalian embryonic stem cells (ESCs). We purified neoblasts and quantified mRNA and protein expression by sequencing and shotgun proteomics. We identified ∼4000 genes specifically expressed in neoblasts, including all ∼30 known neoblast markers. Genes important for pluripotency in ESCs, including regulators as well as targets of OCT4, were well conserved and upregulated in neoblasts. We found conserved expression of epigenetic regulators and demonstrated their requirement for planarian regeneration by knockdown experiments. Post-transcriptional regulatory genes characteristic for germ cells were also enriched in neoblasts, suggesting the existence of a common ancestral state of germ cells and ESCs. We conclude that molecular determinants of pluripotency are conserved throughout evolution and that planaria are an informative model system for human stem cell biology.
Keywords: epigenetic regulators, neoblasts, planaria, pluripotency, stem cells
Introduction
First described in scientific journals more than 100 years ago (Randolph, 1897), freshwater flatworms are a famous model system for regeneration in animals. Amputated planaria regenerate missing complex body parts within ∼10 days, and even cutting animals into dozens of pieces will result in dozens of regenerated healthy animals. This extreme plasticity of planaria critically depends on stem cells (neoblasts) that are the only proliferating cells in asexual animals and constitute ∼30% of all cells (Baguñà et al, 1989). Neoblasts regenerate all tissues of amputated animals, including the germ line, and continuously replace cells lost to physiological turnover (reviewed in Newmark and Sánchez Alvarado, 2002; Agata, 2003 and Reddien and Sánchez Alvarado, 2004). Recently, transplantation studies have proven the capacity of a single neoblast to regenerate entire stem cell-deficient animals and to transform their host into a genetic clone of the donor (Wagner et al, 2011). Neoblasts display morphological features of undifferentiated cells, that is, large nuclei to cell ratio and extensive regions of open chromatin, and self-renew indefinitely. Moreover, the presence of perinuclear ribonucleoprotein granules, the chromatoid bodies (CBs), is reminiscent of germ cells.
In contrast to a large body of cell biology research describing the function and morphology of neoblasts, much less is known about the molecular mechanisms that control neoblast pluripotency. Recently, sequencing of the ∼800-Mb genome of the planarian Schmidtea mediterranea and several experimental approaches (Umesono et al, 1997; Newmark and Sánchez Alvarado, 2000; Robb et al, 2008; Adamidi et al, 2011), including RNAi loss-of-function studies (Sánchez Alvarado and Newmark, 1999; Reddien et al, 2005a), permitted the identification of a small number of neoblast marker genes. However, gene expression in neoblasts has not been profiled comprehensively, and it is therefore largely unclear which transcripts or proteins are expressed in neoblasts. Previous attempts to analyse gene expression in neoblasts relied on indirect inference of the neoblast transcriptome by comparing gene expression in irradiated animals depleted in neoblasts with wild type (WT) animals using cDNA microarrays (Rossi et al, 2007; Eisenhoffer et al, 2008). These approaches were hampered by technical problems that come with using cDNA microarrays (incomplete gene repertoire, non-specific amplification, cross-hybridization), incomplete gene model annotations, and the problem that irradiation will not just delete stem cells but produce a large number of apoptotic cells and cause indirect effects such as stress responses.
To gain a deeper understanding of neoblast biology, it will be important to know which genes are specifically expressed in these cells. One fundamental question is, if or to what degree molecular mechanisms for controlling pluripotency are conserved between planaria and humans. Answering this question would shed light on the largely unknown evolution of pluripotency across large time scales and may help to identify conserved core regulatory circuits that control pluripotency. Furthermore, such studies may help to characterize the usefulness of planaria as an in vivo model system for human stem cell biology. Perhaps surprisingly, almost nothing is known about how gene expression compares between embryonic stem cells (ESCs) and neoblasts. For instance, the essential mammalian pluripotency transcription factor OCT4 has been described mainly in vertebrates (Hinkley et al, 1992; Morrison and Brickman 2006; Lavial et al, 2007) and it is unknown if it is conserved in planaria.
In this study, we addressed these questions by isolating cell populations enriched in proliferating neoblasts and differentiated cells, respectively, using a published fluorescent activated cell sorting (FACS) protocol (Hayashi et al, 2006). We then profiled mRNA and protein expression in these cells by sequencing (Illumina) and shotgun proteomics based on our previously published comprehensive annotation of the planarian transcriptome comprising ∼20 000 gene loci (Adamidi et al, 2011). We carefully validated our data by in situ hybridization (ISH), qRT–PCR and other methods. Computational cross-species analysis allowed us then to identify homologues of planarian genes in mouse or human. With these expression and homology data in hand, we set out to systematically investigate conservation of gene expression between neoblasts and mammalian ESCs/germ cells. Specifically, we focussed on mammalian (1) epigenetic and chromatin regulatory complexes known to be crucial for stem cell function (2) genes known to regulate key pluripotency transcription factors such as OCT4, SOX2, NANOG, (3) direct regulatory targets of OCT4, SOX2 and NANOG, (4) genes known to be important for pluripotency, and (5) genes characteristic for germ cells, for example, certain RNA-binding proteins (RBPs). For specific epigenetic/chromatin regulators, we then performed knockdown experiments to study their functional importance for regeneration. Overall, we show that genes important for pluripotency in mammals and in particular direct targets as well as upstream regulators of OCT4, SOX2, and NANOG are generally conserved in planaria and enriched in neoblasts. Finally, we present candidates for planarian homologues of OCT4 and SOX2.
Results
FACS sorting allows isolation of neoblast-enriched cell fractions and reproducible transcript quantification by next generation sequencing
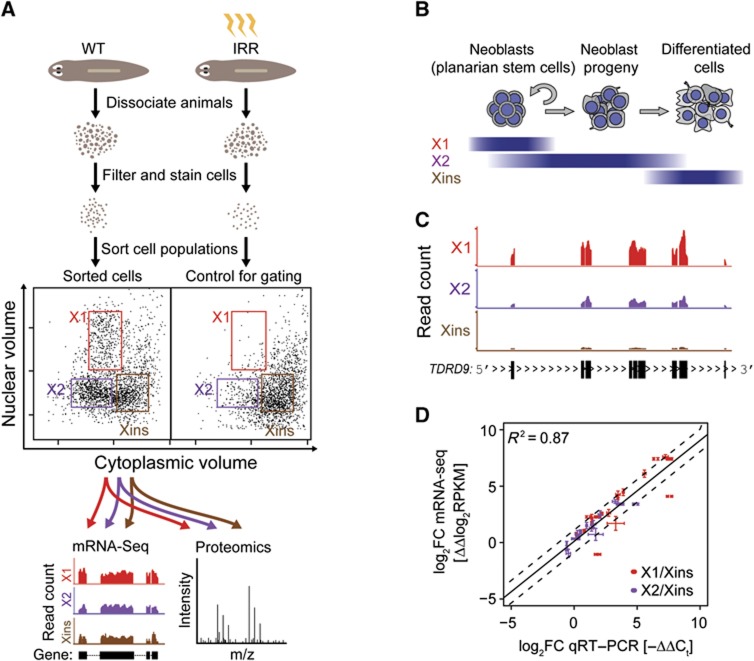
To obtain cell populations enriched and depleted in neoblasts, respectively, we used FACS, following an approach developed by Agata and colleagues (Hayashi et al, 2006; Figure 1A; Materials and methods). Planarian cells selectively eliminated upon X-ray irradiation differ from irradiation-insensitive cells by cytoplasmic volume and DNA content. Based on these properties, dissociated non-irradiated animals were sorted into irradiation-sensitive (X1 and X2) and irradiation-insensitive (Xins) cell fractions. Previous electron microscopic observations indicated that X1 is highly enriched in proliferating neoblasts and contains a small number of differentiating cells, X2 is a mixture of cells that contains fewer neoblasts and both differentiating and differentiated cells, and Xins is primarily composed of differentiated cells (Figure 1B; Higuchi et al, 2007; Eisenhoffer et al, 2008; Hayashi et al, 2010). After validating sample quality by performing qRT–PCR on known neoblast- and tissue-specific markers (Supplementary Figure S1A), samples were subjected to paired-end RNA deep sequencing (mRNA-seq) and mass spectrometry (MS) (see Materials and methods).
Figure 1.
Isolation of planarian cell fractions for transcriptome and proteome analysis. (A) Wild type (WT) and irradiated (IRR) animals were dissociated, filtered and stained for nuclear and cytoplasmic content. Samples were then subjected to FACS. Irradiated animals depleted of neoblasts were used to determine gate settings for the extraction of neoblast-enriched (X1, X2) and depleted (Xins) cell fractions from WT samples. Transcript and protein expression were profiled by mRNA-seq and shotgun proteomics. (B) Schematic representation of the cell composition of FACS fractions. (C) Typical mRNA-seq read coverage profile of a neoblast-specific gene (Smed-TDRD9). Exons are depicted as black boxes at the bottom. (D) Validation of mRNA-seq-based estimates of transcript fold changes by qRT–PCR. mRNA-seq and qRT–PCR derived fold changes for X1 (red symbols) and X2 (purple symbols) versus Xins were compared. qRT–PCR was performed in triplicates. Error bars represent the standard deviation.
mRNA-seq produced ∼70 million 120 bp reads from each fraction, which were mapped to the reference transcriptome assembly (Adamidi et al, 2011; Supplementary data). In total, we confidently measured expression in reads per kilobase of transcript per million mapped reads (RPKM) in at least one of the three cell fractions for 21 865 transcripts corresponding to 20 383 gene loci (Supplementary data; Supplementary Figure S1B). A typical read profile of a neoblast-specific planarian gene (Smed-TDRD9) is shown in Figure 1C. The multi-exonic structure with long introns, reminiscent of higher vertebrates such as humans, is also typical for planarian genes (Abril et al, 2010; Blythe et al, 2010, Adamidi et al, 2011).
To independently validate mRNA-seq data, we performed qRT–PCR assays for a set of 14 genes covering three orders of magnitude of RPKM values (Figure 1D). A comparison of mRNA-seq and qRT–PCR derived log2-fold changes between cell fractions indicated an excellent agreement of the two methods (R2=0.87; Figure 1D). Biological variability was measured by repeating qRT–PCR assays on 17 genes with total RNA extracted from another FACS-sorted sample of independently collected animals. Comparing with mRNA-seq derived log2-fold changes for the original sample (Supplementary Figure S2) yielded a strong correlation (R2=0.80) and thus revealed only a minor impact of biological variability.
Analysis of global gene expression profiles of FACS-sorted cells recovers neoblast-enriched transcript and protein expression of known markers
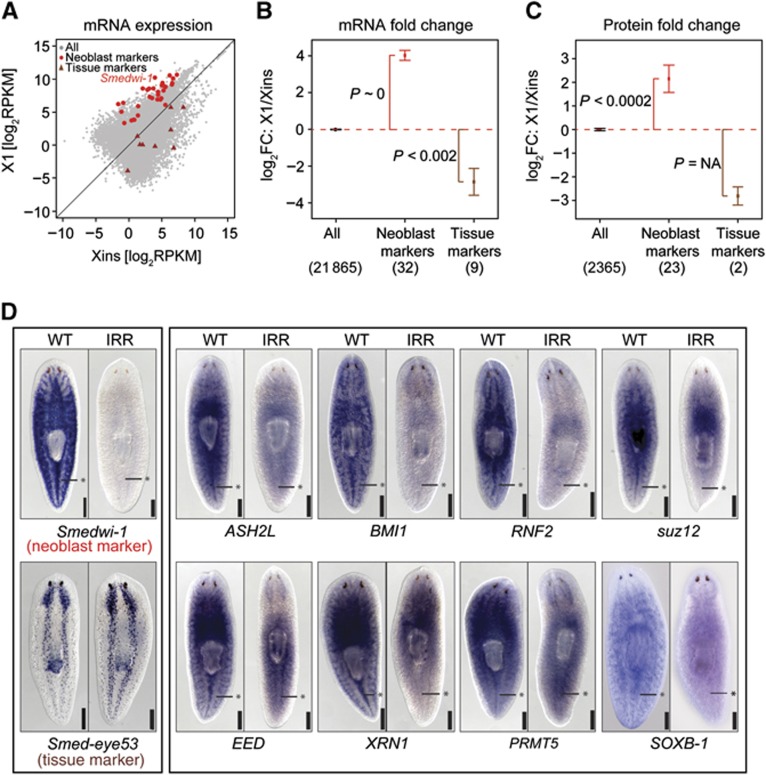
Global differences of the transcriptome between different FACS-sorted fractions were quantified by the correlation of transcript expression (Supplementary Figure S3). Consistent with the expected cellular composition, the highest correlation was observed between the neoblast-enriched fractions X1 and X2 (Spearman’s correlation coefficient ρ=0.85). In comparison, the correlation between X2 and Xins was reduced (ρ=0.71), but still substantially higher than the correlation between X1 and Xins (ρ=0.48), reflecting the strong dissimilarity in global gene expression profiles between neoblasts and differentiated cells (Figure 2A). To confirm that the observed dissimilarities were indeed due to differential enrichment of neoblasts in X1, X2, and Xins, we screened our data for 32 known bona fide neoblast marker genes (Supplementary Table S1). Consistently, all markers were found to be overexpressed in X1 versus Xins (hypergeometric P-value P∼0; Figure 2A and B); except for Smed-VLGA1, all of them were at least four-fold upregulated. In contrast, tissue-specific markers were either downregulated in X1 versus Xins (P<2·10−3; Figure 2A and B; Supplementary Table S1) or did not exhibit significant changes (i.e., Smed-FOXG). In summary, the composition of the transcriptome is highly dissimilar between X1, X2, and Xins and these differences reflect the enrichment of neoblasts in X1 and X2, and of differentiated cells in Xins.
Figure 2.
Validation of transcript and protein quantification and identification of neoblast-specific genes. (A) Comparison of transcript expression in X1 and Xins. Neoblast markers (red dots) and tissue markers (brown triangles) are highlighted. (B, C) Average fold changes (FC) of transcripts (B) and proteins (C) for neoblast and tissue markers. Error bars represent the standard error of the mean. The number of genes is shown in parentheses. Enrichment of upregulated or downregulated genes (P) was assessed by Fisher’s exact test. (D) Validation of neoblast enrichment by ISH. Asterisks mark the midline posterior to the pharynx where neoblasts are concentrated. ISH for the known neoblast marker Smedwi-1 and the nervous system marker Smed-eye53 were used as controls.
To investigate the neoblast proteome, we measured protein abundance by MS runs on all three cell fractions (Adamidi et al, 2011; Supplementary data). We identified ∼2400 proteins in each fraction and 2730 proteins in total. A comparison of protein fold changes between all pairs of cell fractions revealed, in contrast to our observation for transcript fold changes, a similar correlation in each case (ρ∼0.75). This observation potentially reflects a bias for highly expressed proteins that might be more homogeneously expressed across all fractions. Indeed, if the 50% most highly expressed proteins are discarded the correlation generally decreases, and more pronounced differences between the fractions emerge (X1 versus Xins: ρ=0.39; X1 versus X2: ρ=0.5; X2 versus Xins: ρ=0.46; correlations are significantly different: P<0.001). Furthermore, 18 out of 20 neoblast markers with measured protein fold changes (90%) were upregulated on the protein level in X1 versus Xins (P<2·10−4), while two identified tissue-specific markers were downregulated (Figure 2C). Thus, the cellular composition of X1, X2, and Xins is also reflected on the protein level.
Identification of novel neoblast marker genes with functionally characterized homologues
We searched for novel genes with neoblast-enriched expression and homology to genes with known functions in other organisms. Using stringent BLAST protein alignments (alignment E-value <10−10), we were able to identify candidate homologues in at least one out of six well-studied animals (C. elegans, D. melanogaster, D. rerio, H. sapiens, M. musculus, and X. tropicalis) for 12 309 planarian transcripts (56%). For 11 584 transcripts (52%), we identified homologues in human or mouse. To validate the neoblast-enriched expression of the newly identified X1-associated transcripts with homology to well-characterized metazoan genes, we performed ISH for eight candidates conservatively selected by medium expression strengths in X1 (between 7 and 90 RPKM; Figure 2D). In all cases, ISH confirmed enhanced expression in neoblasts. As observed earlier for several neoblast markers, we also detected expression in the brain (e.g., Guo et al, 2006; Solana et al, 2009; Scimone et al, 2010). We conclude that genes with upregulated transcripts in X1 display indeed enriched expression in neoblasts.
Epigenetic regulators associated with stem cell plasticity are required for neoblast function
Among the many transcripts that are upregulated in X1 we identified a large number of homologues of mammalian epigenetic factors and chromatin proteins (Supplementary Table S2). We first validated neoblast-enriched expression by ISH for planarian homologues of chromatin-modifying factors such as the myeloid/lymphoid or mixed lineage leukaemia (MLL) complex methyltransferase SET1/ASH2L, the polycomb complex components BMI1, RNF2, SUZ12, and EED (Figure 2D), the histone lysine methyltransferase SETD8 as well as SSRP1, a member of the high-mobility group (HMG) protein (see below).
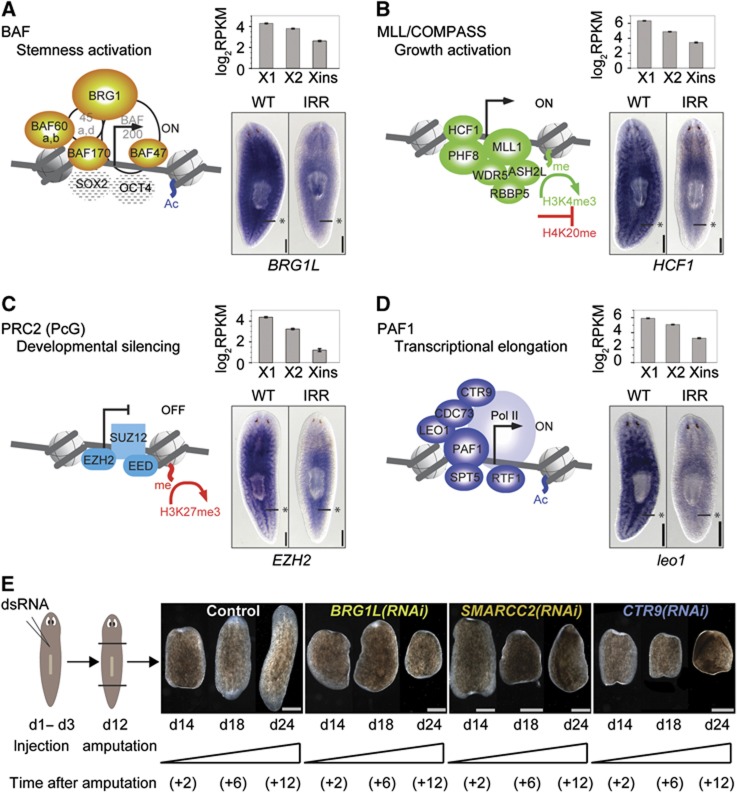
To more systematically analyse the conservation and expression of mammalian epigenetic factors and chromatin regulators in S. mediterranea, we compiled from the literature transcriptional regulation and chromatin-associated complexes with crucial function in mammalian stem cell regulation (Figure 3A–D). These four complexes include (1) Brahma-associated factors (BAFs), (2) MLL/COMPASS genes, (3) the Polycomb Group (PcG), and (4) RNA polymerase II-associated factors (PAFs).
Figure 3.
Homologues of chromatin-modifying complexes regulating mammalian ESCs are overexpressed in neoblasts and functionally important. (A–D) Core components of four chromatin remodelling complexes with known functions in mammalian ESCs. Solid symbols represent genes for which we could identify homologues in planaria. Components with unclear homology are shaded. (A) BAF, (B) MLL/COMPASS, (C) PRC2 (PcG), and (D) PAF1. For a single component of each complex, expression in the three FACS fractions is indicated and ISH shows enhanced neoblast expression. Asterisks mark the midline posterior to the pharynx where neoblasts are concentrated. (E) RNAi knockdown of Smed-BRG1L, Smed-SMARCC2 (BAF170 homologue), and Smed-CTR9 completely blocked regeneration of animals amputated at day 12 (d12). Trunk region of control and RNAi worms is shown at day 2, 6, and 12 after amputation. The phenotype was penetrant in each case (observed for >5 animals). Scale bars are 0.5 mm in (A–D) and 1 mm in (E).
In mammals, a distinct form of the BAF complex, esBAF, is known to be associated with the pluripotency regulators SOX2, OCT4, and NANOG at the promoter of most of their target genes (Ho et al, 2009). Components of the MLL complex are required for self-renewal (Ang et al, 2011) and proper cell-fate specification upon differentiation (Jiang et al, 2011) of ESCs. The two main PcG repressive complexes, termed as PRC1 and PRC2, are critical for the appropriate silencing of tissue-specific genes in ESCs (reviewed in Schwartz and Pirrotta, 2007). Moreover, a recent study demonstrated that BAF and PcG complexes can act synergistically to enforce the pluripotent state (Ho et al, 2011). Finally, the PAF1 complex is required to maintain the chromatin structure of key pluripotency genes in ESCs (Ding et al, 2009). We next studied conservation and expression of these complexes in S. mediterranea. For mammalian genes that have been shown to be critical for the activity of these four complexes in stem cells, we were able to identify homologues in planaria (Figure 3A–D, solid symbols). Strikingly, all of them were strongly upregulated in X1 and X2 (Supplementary Table S2). Even more, homologues that belong to the same functional complex had comparable fold changes between X1 and Xins as well as between X2 and Xins (Supplementary Table S2). Furthermore, we could validate neoblast-enriched expression for components of each complex via ISH (Figure 3A–D).
To analyse the function of these complexes in planaria, we performed RNAi-based knockdown experiments and monitored regeneration and tissue turnover kinetics for planarian homologues of the BAF complex components BRG1L and SMARCC2/BAF170 and the PAF1 complex component CTR9 (Figure 3E). Twelve days after the first dsRNA injection, worms were either amputated to monitor regeneration or left intact to observe effects on tissue turnover. Drastic regeneration defects were already observed 6 days after amputation. Knockdowns of the BRG1 homologue Smed-BRG1L resulted in regeneration defects characteristic for neoblast dysfunction (Figure 3E; note that a different BRG1 homologue, Smed-BRG1, which functions in germ cell development, was described previously; Wang et al, 2010). A similar regeneration phenotype was observed upon knockdown of Smed-SMARCC2, a homologue of the BAF complex component SMARCC2/BAF170 (Figure 3E). Knockdown of a gene homologous to the PAF1 complex component CTR9, Smed-CTR9, also resulted in a strong regeneration phenotype indicative of neoblast alteration (Figure 3E). When repeating knockdown experiments without amputation in order to measure effects on homeostatic tissue turnover, animals failed to replace old tissue, leading to ventral curling, head regression and eventually lysis and death (Supplementary Figure S4A). These are typical signs of neoblast dysfunction, which can be due to a failure in maintenance/proliferation and/or differentiation (Reddien et al, 2005a).
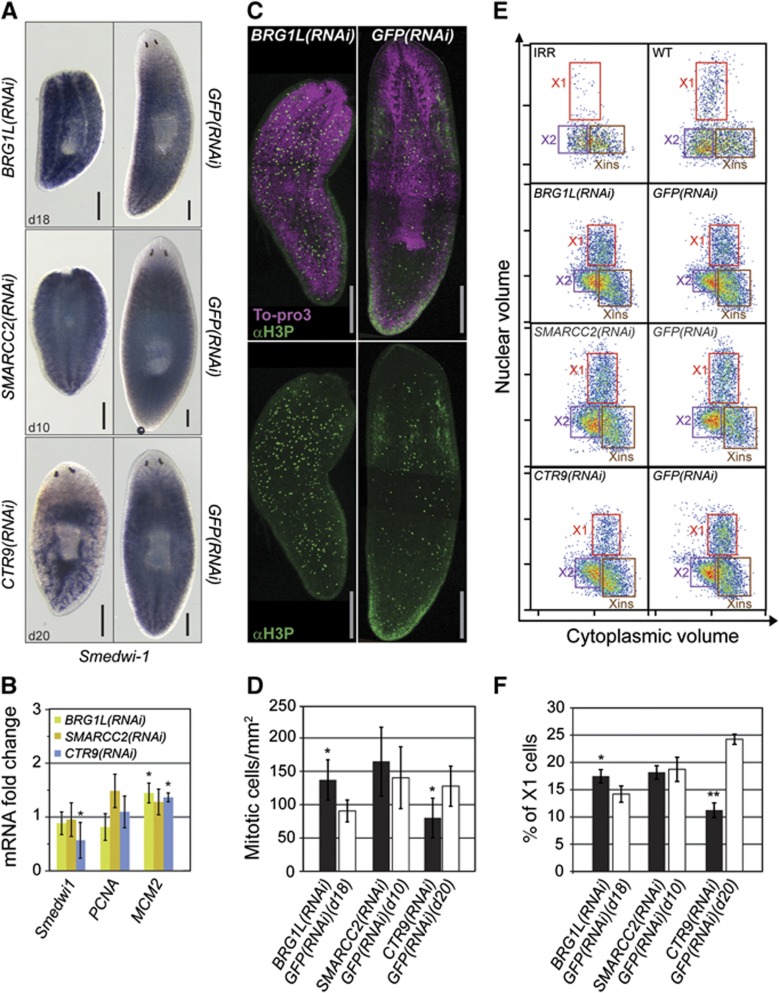
To understand whether the observed defects were more trivially explained by a lack of neoblast proliferation/maintenance, we performed additional experiments using three different approaches. First, we measured expression of neoblast marker genes (Smedwi-1, Smed-PCNA, and Smed-MCM2) in whole animals by ISH and RT–qPCR. BRG1L(RNAi) and SMARCC2(RNAi) animals displayed levels of Smedwi-1 expression similar to control animals (Figure 4A and B). In contrast, CTR9(RNAi) animals showed reduced expression of Smedwi-1 (Figure 4A and B). However, expression of Smed-PCNA and Smed-MCM2 did not decrease in any of the knockdown experiments (Figure 4B). Second, we tested mitotic activity by labelling mitotic cells with an anti-phosphohistone H3(Ser10) antibody (αH3P) and counted labelled cells for at least eight animals per group (Figure 4C). BRG1L(RNAi) and SMARCC2(RNAi) animals showed slightly increased mitotic activity, whereas the number of mitotic cells was reduced in CTR9(RNAi) animals (Figure 4D; Supplementary Figure S4B). Third, flow-cytometric detection of proliferation indicated that X1 cell numbers were stable in BRG1L(RNAi) and SMARCC2(RNAi) compared with control animals, whereas a significant reduction was observed in CTR9(RNAi) animals 20 days after RNAi (Figure 4E and F). These experiments demonstrate that neoblasts were still present and proliferating after RNAi against Smed-BRG1L and Smed-SMARCC2 at times when RNAi animals failed to replace the old tissue. Hence, these genes appear to be not essential for proliferation and maintenance of neoblasts, but required during differentiation. Although CTR9(RNAi) animals showed a decreased number of proliferating neoblasts, these cells still displayed mitotic activity. This suggests that a loss of neoblasts is not the only reason for failed regeneration and tissue turnover in CTR9(RNAi) animals and that Smed-CTR9 might also be required for neoblast differentiation. Furthermore, at an early RNAi time point (10 days after injection) we did not observe any reduction in proliferating neoblast number, suggesting that neoblast depletion occurs in late phases of the CTR9(RNAi) phenotype (Supplementary Figure S5). Taken together, these data provide evidence that the planarian homologues of chromatin-associated factors involved in regulation of pluripotency in mammalian ESCs are upregulated in neoblasts and implicated in the maintenance and differentiation of these cells.
Figure 4.
Pluripotency-associated chromatin modification factors are important for neoblast differentiation and maintenance. (A) Neoblast presence was detected by Smedwi-1 ISH in RNAi animals (BRG1L(RNAi), SMARCC2(RNAi), and CTR9(RNAi)) fixed at the indicated number of days after the first RNAi injection. GFP(RNAi) animals are shown as controls. (B) Change in relative mRNA expression of neoblast markers in RNAi versus control animals. qRT–PCR was performed in triplicates and error bars represent the standard deviation. (C, D) RNAi animals were assayed for mitoses by labelling with αH3P and counting the labelled cells. (C) Representative images of αH3P staining with and without nuclear labelling. (D) Average number of mitotic cells per surface area in RNAi animals (>8 animals per RNAi experiment). Error bars represent the standard deviation. (E, F) RNAi animals were assessed for the presence of proliferative neoblasts by flow cytometry. (E) Representative images of flow-cytometry profiles of cells dissociated from RNAi and control animals. The profile from animals 1 day after irradiation (IRR) is shown as a measure of neoblast loss. (F) Percentage of cells in the X1 fraction, averaged across three biological replicates. Error bars represent the standard deviation. Scale bars are 0.5 mm. *P<0.05, **P<0.001 (t-test).
Chromatin-associated factors are globally upregulated in neoblasts
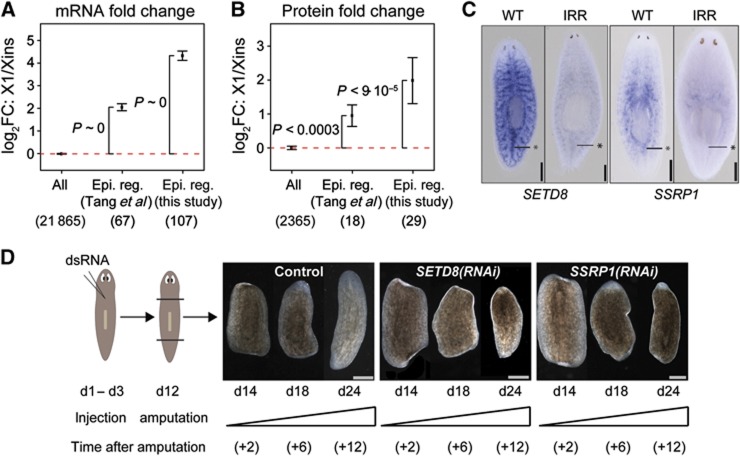
To investigate the expression of chromatin-associated factors on a systems level, we extracted 114 epigenetic regulators from the literature that are known to be robustly expressed in mouse ESCs (Tang et al, 2010). We recovered homologues for 67 of these genes and found their transcripts to be on average four-fold upregulated in X1 versus Xins (P<2.2·10−16; Figure 5A). Protein expression was also enriched in neoblasts (P<3·10−4), although upregulation was less pronounced (∼2-fold; Figure 5B). Next, we performed a literature search to extract other members of human and mouse chromatin remodelling complexes and associated factors and identified their planarian homologues. These 107 putative chromatin regulators (Supplementary Table S2) overlapped with the set of mouse epigenetic regulators extracted from Tang et al (2010) by only 30%. Nevertheless, expression of these chromatin regulators was even more strongly enhanced in X1 versus Xins than for the set extracted from Tang et al (2010) with an average seven-fold upregulation of transcripts (P<2.2·10−16) and proteins (P<9·10−5) (Figure 5A and B).
Figure 5.
Homologues of mammalian epigenetic regulators are globally enriched in neoblasts. (A, B) Average transcript (A) and protein (B) fold changes between X1 and Xins for homologues of mouse epigenetic regulators (Tang et al, 2010) and homologues of manually compiled mammalian chromatin-associated factors (Supplementary Table S2). The number of mammalian genes with planarian homologues and measured fold changes is shown in parentheses. Error bars represent the standard error of the mean. Enrichment of upregulated genes (P) was assessed by Fisher’s exact test. (C) ISH in wild type (WT) versus irradiated (IRR) animals reveals neoblast expression of genes homologous to SETD8 and SSRP1. Asterisks mark midline posterior to the pharynx where neoblasts are concentrated. (D) RNAi knockdown of Smed-SETD8 and Smed-SSRP1 blocked regeneration of animals amputated at day 12 (d12). Trunk region of control and RNAi worms is shown at day 2, 6, and 12 after amputation. The phenotype was penetrant in each case (observed for >5 animals). Scale bars are 0.5 mm in (C) and 1 mm (D).
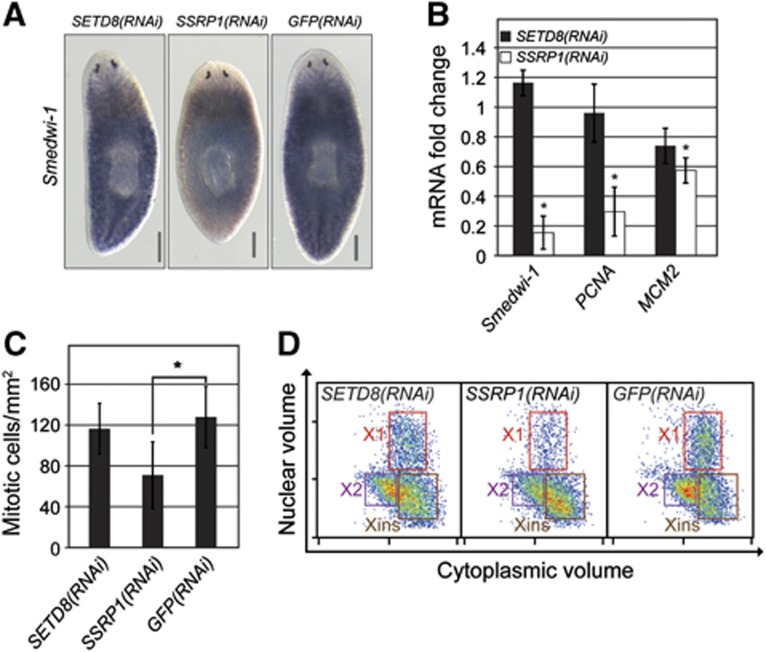
We next focussed on the histone lysine methyltransferase SETD8 and the HMG protein SSRP1. SETD8 monomethylates lysine 20 of histone 4 (Fang et al, 2002) and lack of SETD8 in mammalian cells results in substantial DNA damage, improper chromatin condensation, inability to progress to the G2 phase of the cell cycle, and embryonic lethality (reviewed in Beck et al, 2012). SSRP1 is associated with a widely conserved chromatin-associated complex named as FACT (facilitates chromatin transcription). FACT has been implicated in numerous processes such as DNA replication, DNA repair, basal and regulated transcription (Okuhara et al, 1999; Orphanides et al, 1999; Keller et al, 2001) and is essential for viability of mouse embryos (Cao et al, 2003). We could validate enhanced neoblast expression of SETD8 and SSRP1 homologues by ISH (Figure 5C). Since SETD8 and SSRP1 are chromatin regulators with a crucial role in the cell cycle, we investigated the function of their planarian homologues in regeneration and tissue turnover by RNAi experiments. Knockdown of both genes led to a failure in regeneration (Figure 5D) and tissue turnover (Supplementary Figure S4). To again investigate if these phenotypes were due to a failure to differentiate or simply due to neoblast depletion, we analysed the presence and proliferation of neoblasts. Reduced expression of neoblast markers Smedwi-1, Smed-PCNA and Smed-MCM2 indicated that SSRP1(RNAi) animals were significantly depleted in neoblasts (Figure 6A and B). Concordantly, αH3P staining and flow-cytometry analysis revealed a depletion of mitotic cells (Figure 6C and D). Hence, SSRP1 appears to have a non-redundant function in neoblast proliferation and/or maintenance. In contrast, SETD8 depletion did not cause a detectable decrease in neoblast number as indicated by stable expression of neoblast markers (Figure 6A and B) and no depletion of mitotic cells (Figure 6C and D). Taken together, our data suggest that SETD8 is not required for neoblast proliferation and/or maintenance, but rather implicated in control of differentiation during homeostasis and regeneration.
Figure 6.
SETD8 and SSRP1 are required for neoblast differentiation and/or maintenance. (A) Neoblast presence was detected by Smedwi-1 ISH on SETD8(RNAi) and SSRP1(RNAi) animals fixed 23 days after first RNAi injection. GFP(RNAi) animals are shown as control. (B) Change in the relative mRNA expression of neoblast markers in RNAi versus control animals. qRT–PCR was performed in triplicates and error bars represent the standard deviation. (C) Average number of mitotic cells per surface area in RNAi animals (>8 animals per RNAi experiment) labelled with αH3P. Error bars represent the standard deviation. (D) RNAi animals were assessed for the presence of proliferating neoblasts by flow cytometry. Representative images of flow-cytometry profiles of cells dissociated from RNAi and control animals are shown. Scale bars are 0.5 mm. *P<0.05 (t-test).
Altogether, our data demonstrate a global enrichment of chromatin regulators in neoblasts, consistent with the hyperdynamic state of chromatin in many types of stem cells (reviewed in Meshorer and Misteli, 2006), and indicate a functional role of these genes in maintenance of neoblasts and/or differentiation.
RBPs with predicted functions in maintaining genome integrity and totipotency in metazoan germ line are highly enriched in neoblasts
Most of the previously identified neoblast marker genes encode for RBPs that belong to gene families homologous to germ granule components, such as piwi, bruno, DDX6, vasa, and pumilio (Supplementary Table S1) and have been linked to the totipotent nature of germ cells in metazoans (reviewed in Kimmins and Sassone-Corsi, 2005 and Seydoux and Braun, 2006). Thus, it has been proposed that planarian neoblasts maintain pluripotency by employing mechanisms of post-transcriptional regulation displayed in animal germ cells and early development (Juliano et al, 2010; Rouhana et al, 2010; Shibata et al, 2010).
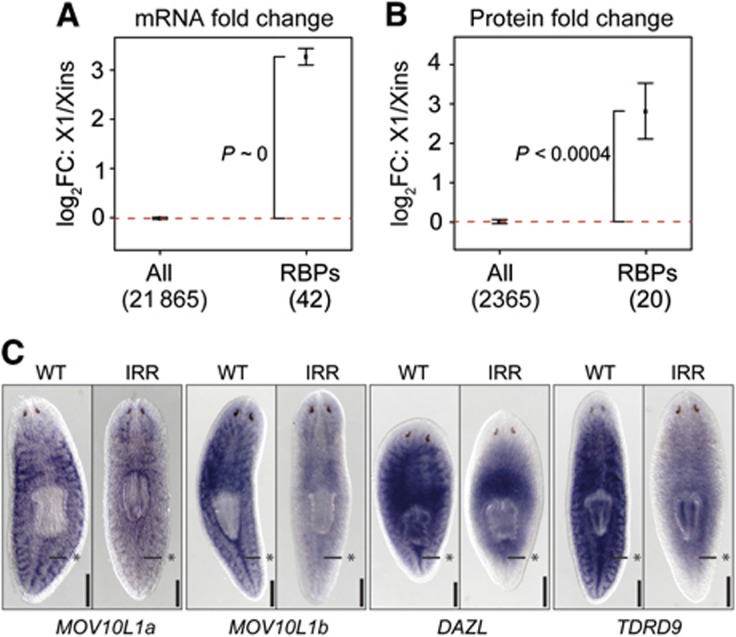
To systematically check the expression of germ line-associated RNA regulators in neoblasts, we searched for homologues of metazoan RBPs that either localize to or interact with germ granules and more generally associate with RNA localization and processing. Transcripts of 40 identified homologues were highly upregulated in neoblasts, on average >8-fold (P<2.2·10−16; Figure 7A; Supplementary Table S3). For 20 homologues, we could also measure protein fold changes between X1 and Xins and observed on average a six-fold upregulation (P<4·10−4; Figure 7B). Among newly identified RBPs, we report two MOV10 homologues (Smed-MOV10L1a and b), one Deleted-in-AZoospermia (DAZ)-like gene (Smed-DAZL), five Tudor-domain containing genes (Smed-TDRD1L1-3, Smed-TDRKH, and Smed-TDRD9) and several DExD-like RNA helicases. MOV10 is a putative RNA helicase homologue of Drosophila Armitage and has been shown to participate in post-transcriptional regulation (Cook et al, 2004; Tomari et al, 2004) and in PcG-mediated gene silencing (El Messaoudi-Aubert et al, 2010). Smed-DAZL encodes a homologue of the DAZ RNA binding family of proteins, which are essential for germ line development in several species and function in translation regulation (Xu et al, 2001; Collier et al, 2005). DAZL is also expressed in undifferentiated human ESC lines (Clark et al, 2004; Moore et al, 2004). Smed-TDRD9 is a homologue of mouse TDRD9 and Drosophila Spindle-E, a known germ granule component that contains a Tudor and a DEAD box helicase domain and cooperates with the piwi-associated RNA (piRNA) pathway (Aravin et al, 2009; Shoji et al, 2009) in silencing of transposable elements (reviewed in Siomi et al, 2011).
Figure 7.
Homologues of metazoan germ granule-associated genes are upregulated in neoblasts. (A, B) Average transcript (A) and protein (B) fold changes between X1 and Xins for homologues of metazoan germ granule-associated RNA-binding proteins (RBPs). The number of metazoan genes with planarian homologues and measured fold changes is shown in parentheses. Error bars represent the standard error of the mean. Enrichment of upregulated genes (P) was assessed by Fisher’s exact test. (C) ISH in wild type (WT) versus irradiated (IRR) animals reveals neoblast expression of four genes homologous to granule components (MOV10L1a, MOV10L1b, TDRD9, and DAZL). Asterisks mark midline posterior to the pharynx where neoblasts are concentrated. Scale bars are 0.5 mm in (C).
We validated neoblast-enriched expression by ISH for four RBPs. As seen for the CB markers Spoltud-1 (Solana et al, 2009) and DjCBC1 (Yoshida-Kashikawa et al, 2007), the RBPs tested showed irradiation-sensitive neoblast-enriched expression as well as irradiation-insensitive brain-specific expression (Figure 7C). Brain-specific expression is likely due to the presence of neuronal granules in the planarian brain (Oosaki and Ishii, 1965), which are similar in composition to CBs (reviewed in Anderson and Kedersha, 2009).
Neoblast CBs, like their germ cell or neuron counterparts, are known to interact with nuclear pores and mitochondria, as well as other RNP complexes involved in mRNA transport, decay, and translation. Consistent with the high expression of RBPs in neoblasts, we also found nuclear pore and mitochondrial transport components and many RBPs involved in RNA splicing, post-transcriptional or translational regulation, nucleocytoplasmic transport and mitochondrial export to be highly upregulated in neoblasts. In summary, our genome-wide profiling data concur and broaden previous findings demonstrating the enrichment of post-transcriptional regulators in neoblasts and their contribution to the maintenance of stem cell identity (Rouhana et al, 2010).
Clustering of transcript expression profiles identifies a large number of putative neoblast-enriched genes and differentiation markers
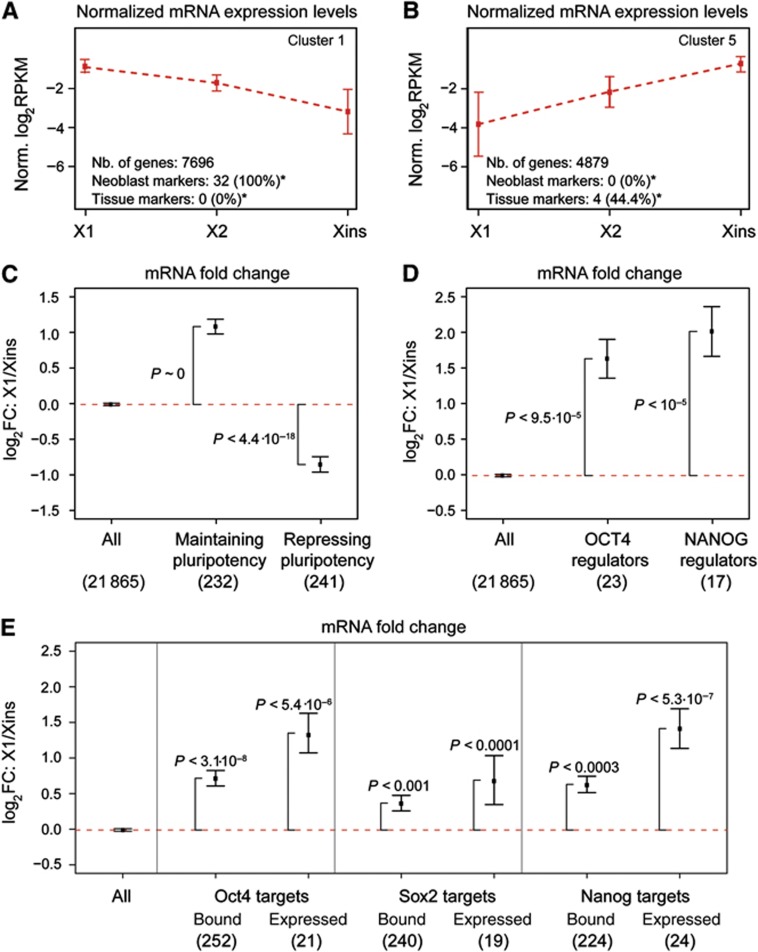
To identify groups of genes specifically expressed in neoblasts and differentiated cells, respectively, we performed hierarchical clustering of all genes based on the correlation of their transcript expression profiles (Supplementary data). We identified six distinct clusters (clusters 1–6), each containing >1000 genes (Figure 8A and B; Supplementary Figure S6; Supplementary Table S4). Two clusters comprise genes that are upregulated strongly (cluster 1; Figure 8A) or more mildly (cluster 2) in neoblast-enriched fractions. Two other clusters contain genes that are most highly expressed in differentiated cells (clusters 5 and 6; Figure 8B) and the remaining clusters (clusters 3 and 4) have similar expression levels in X1 and Xins. To infer biological function of the clusters, we performed gene ontology (GO) analysis for the human homologues of each cluster (Supplementary data). Overrepresented GO terms (multiple testing corrected P<0.05; Supplementary Table S5) revealed a major activity of cluster 1 in (1) DNA replication and cell-cycle regulation, (2) transcriptional regulation and chromatin organization, and (3) RNP-mediated post-transcriptional regulation. These functional categories are consistent with the morphological features that have classically defined embryonic-type stem cells in general (high mitotic activity and a dynamic chromatin state) and neoblasts in particular (presence of CBs reminiscent of germ cells). In conjunction with the observed expression profile, the functional classification suggests a pronounced overrepresentation of neoblast-specific genes in cluster 1. Consistently, all known neoblast markers but none of the tissue-specific markers fall into this cluster (P<2.2·10−16 and P<2·10−2, respectively). To obtain a more stringent set of genes with enhanced transcript expression in neoblasts, we selected genes from cluster 1 with at least two-fold upregulation in X1 versus Xins and enhanced expression in X1 versus X2 after discarding genes with low confidence fold change quantification (confidence level≤0.8, see Supplementary data). This final set of 4032 genes contained 28 out of 32 (88%) of the known neoblast markers and thus represents a bona fide set of candidate genes for studying neoblast biology (Supplementary Table S6). In contrast, cluster 5 and cluster 6, which comprise transcripts that are upregulated in Xins, contain a significantly enhanced number of genes involved in cell-cell signalling, neuronal differentiation, and morphogenesis of neuronal tissue (Supplementary Table S5), reflecting the presence of a population of differentiated cells in the Xins population. Indeed, seven out of nine tissue-specific markers fall into these clusters. Hence, the two clusters comprise 7800 candidate genes that in the future could be screened for repressors of pluripotency and differentiation markers.
Figure 8.
Homologues of mammalian pluripotency genes are enriched in the neoblast transcriptome. (A, B) Two of the six transcript expression clusters. Cluster 1 (A) contains genes with enhanced neoblast expression and cluster 5 (B) contains genes upregulated in differentiated cells. Average expression (broken red line) and standard deviation (error bars) are shown. (C–E) Average transcript fold changes between X1 and Xins for homologues of (C) mouse genes required for maintenance and repression of pluripotency, (D) human regulators of OCT4 and NANOG expression in ESCs and (E) Oct4, Nanog, and Sox2 targets in mouse ESCs. In (E), data are shown for all targets (Bound), and for the subset of targets upregulated in pluripotent versus differentiated cells (Expressed). In (C–E), error bars represent the standard error of the mean. Enrichment of upregulated or downregulated genes (P) was assessed by Fisher’s exact test. The number of mammalian genes with planarian homologues is shown in parentheses. *P<0.05 (Fisher’s exact test).
Finally, the only cluster with an expression peak in X2, cluster 3, comprises 9 out of 10 previously known early and late cell division progeny markers that were shown to exhibit enriched expression in X2 (Eisenhoffer et al, 2008). A single progeny marker was most highly expressed in Xins and fell into cluster 5. Therefore, our expression analysis of FACS-sorted cell fractions discriminates efficiently genes expressed in neoblasts from those expressed in their progeny and cluster 3 could be mined for more progeny markers.
Comparison of neoblast and ESC transcriptomes reveals deep conservation of pluripotency-associated gene expression
Having identified a global set of genes with neoblast-enriched transcript expression, we next compared their homologues with known pluripotency-associated genes in mouse ESCs. We extracted genes from the literature (Tang et al, 2010) that were shown to be at least four-fold upregulated on the transcript level in ESCs and the inner cell mass (ICM) in comparison with cells that were obtained from differentiated ICM outgrowths (Supplementary Table S7A). The distinction between ESCs and ICM is critical, since ESCs, which are isolated from the ICM, are highly proliferative and are grown in culture, while the ICM has only limited self-renewal capacity (reviewed in Niwa, 2007). Genes upregulated in ICM and ESCs versus differentiated ICM outgrowths are thus expected to be preferentially involved in maintaining pluripotency instead of exclusively regulating self-renewal. We also identified the homologues of genes expressed upon differentiation (Supplementary Table S7B), that is, genes associated with repression of pluripotency in mouse ESCs (Tang et al, 2010). Strikingly, in comparison with all planarian genes, homologues of mouse genes associated with the maintenance of pluripotency are on average about two-fold upregulated (P∼0), whereas homologues of pluripotency repressors exhibit on average almost two-fold downregulation in X1 versus Xins (P<4.4·10−18; Figure 8C). This observation provides further evidence for deep conservation of gene expression between mammalian ESCs and neoblasts.
Homologues of genes affecting OCT4 and NANOG expression in human ESCs are overexpressed in neoblasts
We performed a similar analysis for homologues of genes that were identified by RNAi-based assays as positive direct or indirect regulators of OCT4 and NANOG expression in human ESCs (Supplementary Table S7C and D; Chia et al, 2010). The POU domain transcription factor OCT4 is crucial for the induction of pluripotency in mammalian ESCs and promoters of OCT4-regulated genes are frequently co-occupied by the homeodomain transcription factor NANOG, a key factor for maintaining pluripotency (reviewed in Niwa, 2007). Genes required for expression of OCT4 and NANOG with identified homologues in planaria include components of the basal transcriptional machinery (e.g., TAF7, EIF2S2, EIF2B4, and EDF1), several nucleoporins (e.g., TPR and NUP107), post-transcriptional regulators (e.g., PCF11 and NCBP1) and among those, in particular, splicing factors (SF3A1 and SF3A3). Note that planarian homologues of NANOG and OCT4 regulators strongly overlap (due to the way these genes were identified by Chia et al): all 17 homologues of NANOG regulators are contained within the set of 23 homologues of OCT4 regulators. In comparison with all planarian genes, the average upregulation in X1 versus Xins was three-fold for homologues of OCT4 regulators (22 of 23 genes upregulated; P<9.5·10−5) and four-fold for homologues of genes that co-regulate NANOG expression (16 of 17 genes upregulated; P<10−5; Figure 8D). Hence, expression of genes that positively regulate OCT4 and NANOG expression in human ESCs is enhanced in neoblasts.
Homologues of direct targets of Oct4 and Nanog are overexpressed in neoblasts
Finally, we analysed transcript expression for planarian homologues of genes that are targeted by Oct4, Nanog, and Sox2 in mouse ESCs (Chen et al, 2008). Chen et al defined an association score between transcription factor binding sites inferred by chromatin immunoprecipitation followed by sequencing (ChIP-seq), and putative neighbouring target genes. For each transcription factor, we extracted the top 10% of all genes ranked by their association score and identified their planarian homologues (Supplementary Table S7E–G). Although the three factors are known to co-target many genes (Boyer et al, 2005), a substantial fraction of targets (∼70%) is specifically targeted by only one factor (Supplementary Figure S9A). Strikingly, out of 252 homologues of Oct4 targets (224 and 240 homologues of Nanog and Sox2 targets, respectively), a major subset of 202 genes (80%, P<3.1·10−8) had increased expression in X1 compared with Xins (177 (79%, P<3·10−4) and 176 (73%, P<10−3) genes for Nanog and Sox2, respectively). We then retrieved more confident predictions for activated targets of each factor by only considering genes found to be upregulated in ICM and ESCs by Tang et al (2010) (Supplementary Figure S9B). In case of Oct4 and Nanog, upregulation in X1 was much more pronounced for homologues of these activated targets (Figure 8E). In case of Sox2, upregulation was also enhanced but the effect was relatively weak. In mammals, Oct4, Nanog, and Sox2 co-regulate a substantial number of targets (Boyer et al, 2005; Loh et al, 2006; Chen et al, 2008). Hence, we tested if homologues of genes co-targeted by different factors were even more upregulated in neoblasts. While this was observed for shared targets of Oct4 and Nanog, co-targets of Oct4 and Sox2 did not display increased upregulation (Supplementary Figure S9C).
In summary, our transcriptome analysis suggests deep conservation of both upstream and downstream components of the networks controlled by key pluripotency factors SOX2, OCT4, and NANOG. This finding is perhaps surprising, since homologues of these factors have not been described in planaria and NANOG is most likely not conserved beyond vertebrate organisms (Theunissen et al, 2011). Consistently, we could not identify a planarian homologue of NANOG in our transcriptome assembly (Supplementary data). However, we were able to identify candidate genes with putative homology to SOX2 and OCT4, respectively (see Supplementary data and Discussion).
Discussion
In this study, we used a FACS-based approach combined with quantitative RNA sequencing and shotgun proteomics to for the first time directly profile and quantify gene expression in planarian neoblasts. Using these data, we identified and validated a large number of genes with enhanced neoblast expression. By screening their homology annotations, we observed various functional categories reminiscent of stem and germ cell biology. Most prominent was a strong enrichment of epigenetic regulators.
Epigenetic regulators in neoblasts
Among the multitude of stem cells investigated in different systems, the hyperdynamic chromatin state currently emerges as one of the few unifying molecular features (reviewed in Meshorer and Misteli, 2006 and Gaspar-Maia et al, 2011). Consistently, the disruption of chromatin remodelling functions often leads to the inability of ESCs to differentiate appropriately and eventually results in embryonic lethal phenotypes (reviewed in Ho and Crabtree, 2010 and Young, 2011). The expression of a few planarian chromatin remodelling or epigenetic factors was previously shown to be enhanced in neoblasts (Reddien et al, 2005a; Bonuccelli et al, 2010; Scimone et al, 2010). We confirmed these data but identified many more (∼100) epigenetic regulators enriched in neoblasts (Supplementary Table S2).
Specifically, we found that neoblasts overexpress epigenetic complexes known to be required for maintaining pluripotency in ESCs (esBAF, MLL1, PRC2 and PAF1 complex) and we were able to demonstrate functional requirement during regeneration and tissue turnover for homologues of the esBAF and PAF1 complex components. Importantly, we could show that neoblast maintenance and proliferation is unaffected in BRG1L(RNAi) and SMARCC2(RNAi) animals and only moderately affected in CTR9(RNAi) animals. Hence, these complexes appear to be implicated in the control of differentiation. We made a similar observation for Smed-SETD8. However, while this study was under review, it was shown that in growing neoblast colonies after survival from sublethal irradiation, SETD8 is required for persistence of cell division (Wagner et al, 2012). Thus, under the extreme condition of neoblast loss, Smed-SETD8 might function in modified ways. Notably, Wagner and colleagues observed a regeneration phenotype of PRC2 complex components, consistent with our finding that this complex is enriched in neoblasts. They also validated enhanced expression in neoblasts for 28 genes, comprising homologues of chromatin modifiers and RBPs, but also other classes. Remarkably, in our data all of these genes were highly upregulated in X1 (Supplementary Figure S10).
Post-transcriptional gene regulation in neoblasts
Another class of genes with increased expression in neoblasts was functionally associated with post-transcriptional regulation in germ granules. Our observations add to accumulating evidence for conservation of post-transcriptional regulation by RBPs between CBs in planaria and germ granules in other metazoan model organisms (Rouhana et al, 2010; Shibata et al, 2010). Thus, planarian neoblasts may provide interesting insights into a possible common evolutionary origin of germ cells and pluripotent stem cells.
We were impressed by the striking conservation of stem-cell expression of RBPs and epigenetic regulators, indicating that these modes of gene regulatory control of pluripotency are extremely old and perhaps even better conserved than transcriptional regulatory relationships. However, the targets of RBPs and epigenetic regulators of course may have extensively changed during evolution. We further note that microRNAs which are specifically expressed in neoblasts (Friedländer et al, 2009) do not appear to be specifically conserved and expressed in mammalian embryonic stem cells.
Conservation of pluripotency regulatory networks and identification of candidates for planarian OCT4 and SOX2
We also analysed the evolutionary conservation and neoblast expression of mammalian genes associated with maintenance and repression of pluripotency. While homologues of mammalian genes associated with maintenance of pluripotency were upregulated, homologues of pluripotency repressors were downregulated in neoblasts. Consistently, we observed conserved transcript expression for homologues of genes that regulate expression of OCT4 and NANOG in human ESCs and for homologues of Oct4, Sox2, and Nanog targets in mouse. Expression of Oct4 is essential for inducing pluripotency (Takahashi and Yamanaka, 2006), while Nanog is dispensable for induction but required for maintenance of pluripotency (Chambers et al, 2003; Mitsui et al, 2003).
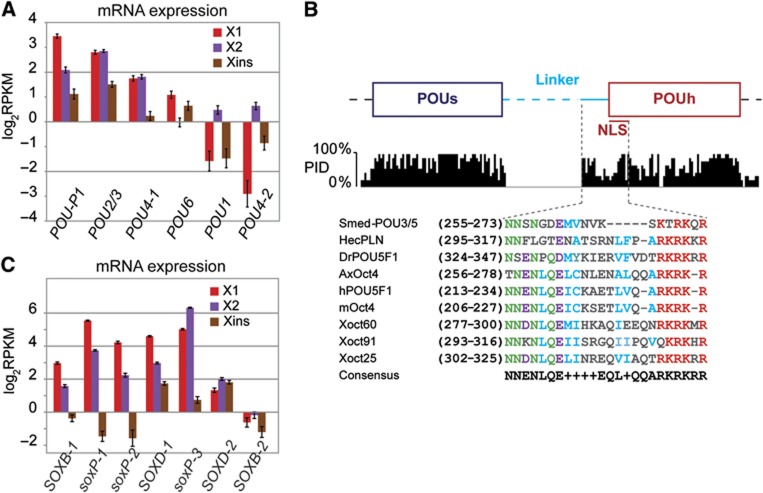
We applied state-of-the-art methods to screen our transcriptome assembly for planarian homologues of OCT4, NANOG, and SOX2 (Supplementary Table S8). While we were unable to identify a NANOG homologue, our search for OCT4 homologues revealed several POU domain-containing genes (Supplementary data; Supplementary Figure S7). One of those, Smed-POU-P1, showed an expression profile similar to known neoblast markers (Figure 9A). It contains a conserved POU specific domain (POUs) and a conserved POU homeodomain (POUh). Although the linker region that connects the two subdomains is unusually long, it is also partially conserved (Figure 9B).
Figure 9.
Identification of POU and SOX homologues in planaria. (A) Comparison of transcript expression of selected Smed-POU genes (see Supplementary data). Smed-POU-P1 shows the characteristic profile of neoblast markers. (B) Conserved section of the Smed-POU-P1 alignment to vertebrate homologues of Oct4 (see Supplementary Figure S7C) spanning the POU domain. The Percentage Identity (PID) for each aligned position is shown below the structural scheme. Conserved residues within the linker region and nuclear localization signal (NLS) are shown in magnified section, protein sequences are coloured according to conserved residues. (C) Comparison of mRNA expression of selected Smed-SOX genes (see Supplementary data).
We identified several planarian genes with homology to SRY-related HMG-box domains (Figure 9C; Supplementary data; Supplementary Figure S8). Three of these genes (Smed-soxP-1, -2, and -3) have recently been described (Wagner et al, 2012). SOX2 belongs to the class B family of SOX proteins and two of our candidates, Smed-SOXB-1 and Smed-SOXB-2, yielded reciprocal best BLAST matches to mammalian members of this class. Smed-SOXB-1 displayed an expression profile similar to known neoblast markers, which we validated by ISH (Figure 2D).
These putative POU- and SOXB-like transcription factors require further functional characterization to assess their role in neoblast biology and planarian regeneration. The apparent lack of a Nanog homologue contrasts with extensive conservation of gene expression observed for its targets and indicates that in planaria, and likely also ancestrally, its role might be played by a different transcription factor. It should be noted that even highly conserved regulatory networks can sometimes differ in key upstream transcription factors. For example, a key transcription factor that drives early embryonic development in Drosophila is Bicoid, while this factor is substituted by Orthodenticle and Hunchback in the beetle Tribolium (Schröder; see Supplementary data for another example). Gene networks of pluripotency control could have undergone similar changes during evolution, which, for instance, could have led to the inclusion of Nanog or other pluripotency regulators in the vertebrate lineage without changing the expression of downstream genes. Alternatively, it is possible that upregulation of Nanog target homologues is due to co-targeting by Oct4 and Sox2. However, after removing shared targets from the set of analysed genes, a significant upregulation was still observed (Supplementary Figure S9C), suggesting that homologues of Nanog targets may represent a set of pluripotency-associated genes in planaria.
Deep conservation of stem cell expression of pluripotency-associated genes
In summary, the data presented here provide evidence for global conservation of expression of pluripotency-associated genes. The underlying gene regulatory network, while presumably having been rewired during evolution, still shows remarkable conservation as a whole. In the future, it will be important to perform experiments which can further elucidate regulatory interactions. Unfortunately, the use of planaria as a model organism for stem cell biology is currently still affected by certain experimental limitations: it is still unknown whether neoblasts represent a heterogeneous mixture of cells and which cells within this population are pluripotent. Moreover, it is currently impossible to culture these cells and perform controlled perturbation experiments. Finally, trans-genetics have not yet been established in planaria. It is conceivable that some of these limitations will be overcome in the future. The use of biochemical methods such as iPAR-CLIP (Jungkamp et al, 2011) could help to identify, in vivo, direct targets of conserved post-transcriptional regulators of pluripotency to study conservation and evolution of these ‘wirings’. Similarly, if future work confirms essential function of the putative Oct4 and Sox2 homologues identified in our study, it will be very interesting to identify their targets via ChIP-seq. These experiments will potentially reveal to what extent regulatory interactions are conserved and identify core interactions under strong evolutionary constraint as well as evolutionary innovations. It will be helpful to extend these analyses to other species to be able to ultimately identify key regulatory circuits that control various aspects of pluripotency throughout animal life. In this regard, an evolutionary perspective on pluripotency control, such as outlined here, will help to dissect and better understand gene regulatory networks in mammalian ESCs.
Materials and methods
Planarian culture
Planaria from clonal asexual strain of CIW4 were maintained at 20°C in autoclaved planarian water (Cebrià and Newmark, 2005) and were starved for 1 week before experiments. For irradiation, 60 Gy of gamma rays were used.
Cell sorting and flow-cytometric analyses
Sample preparation for FACS was adapted from Hayashi et al (2006) with modifications (Supplementary data). Cells were sorted using FACS Vantage SE equipped with UV laser (Becton Dickinson) directly into TRIzol LS (Invitrogen) for RNA and/or protein isolation. Flow-cytometric analyses were performed with BD LSRII (BD Biosciences).
Quantitative RT–PCR
Total RNA was isolated using TRIzol LS or TRIzol reagent according to the manufacturer's protocol. cDNA was synthesized from total RNA of sorted cells and WT and irradiated intact worms. ura4 (CUFF.232566.1) served as an internal control (normalization) (Reddien et al, 2005b; Friedländer et al, 2009). PCRs were performed using the SYBR-Green RT–PCR kit (Applied Biosystems).
mRNA sequencing
In all cases, 0.5 μg of total RNA was purified, polyA selection was performed, mRNA was fragmented and adapter ligation and amplification was performed according to the manufacturer’s protocol for Solexa/Illumina sequencing. mRNA sequencing was performed on the Genome Analyzer (GA II) following the manufacturer's instructions using the Paired-End Sample Preparation Kit (Illumina).
Shotgun proteomics
The details of the extraction and LC-MS/MS protocol can be found in Supplementary data.
Whole-mount ISH, immunostaining
ISH was carried out as described previously (Umesono et al, 1997) with modifications (Felix and Aboobaker, 2010). The detailed protocol for ISH and immunostaining can be found in Supplementary data.
RNAi
dsRNA was in vitro transcribed with Ambion Maxiscript by using PCR-amplified templates with flanking T7 promoters. Planaria were injected with 3 × 30 nl of 1 μg/μl dsRNA for 3 consecutive days and either cut 12 days after first injection for regeneration or left uncut for homeostasis assays. Water or GFP dsRNA was used as negative control. See Supplementary data.
Supplementary Material
Acknowledgments
We acknowledge the technical assistance of Salah Ayoub and thank all other members of the Rajewsky laboratory for discussions. We also want to thank Tetsutaro Hayashi for his helpful comments on FACS experiments. DG received funding from the European Community's Seventh Framework Programme (FP7/2007–2013) under grant agreement No HEALTH-F4-2010-241504 (EURATRANS). PO received funding from the European Initial Training Network (EU-ITN): EVONET. We are indebted to Christine Kocks for helpful discussions and comments on the manuscript.
Author contributions: PO conducted most of the experiments under the supervision of CA. DG contributed the entire computational analysis. JS provided crucial help with RNAi and histone labelling experiments. JS and AR helped with ISH. CA, DG, PO, JS, UZ, and NR analysed the data. GM provided mass-spectrometry measurements, supervised by SK. YW prepared the sequencing library, supervised by WC. HR provided extensive help with FACS. NR conceived and supervised the project. DG and NR wrote the paper with help from CA, PO, JS, and UZ.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abril JF, Cebrià F, Rodríguez-Esteban G, Horn T, Fraguas S, Calvo B, Bartscherer K, Saló E (2010) Smed454 dataset: unravelling the transcriptome of Schmidtea mediterranea. BMC Genomics 11: 731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamidi C, Wang Y, Gruen D, Mastrobuoni G, You X, Tolle D, Dodt M, Mackowiak SD, Gogol-Doering A, Oenal P, Rybak A, Ross E, Sánchez Alvarado A, Kempa S, Dieterich C, Rajewsky N, Chen W (2011) De novo assembly and validation of planaria transcriptome by massive parallel sequencing and shotgun proteomics. Genome Res 21: 1193–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agata K (2003) Regeneration and gene regulation in planarians. Curr Opin Genet Dev 13: 492–496 [DOI] [PubMed] [Google Scholar]
- Anderson P, Kedersha N (2009) RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol 10: 430–436 [DOI] [PubMed] [Google Scholar]
- Ang YS, Tsai SY, Lee DF, Monk J, Su J, Ratnakumar K, Ding J, Ge Y, Darr H, Chang B, Wang J, Rendl M, Bernstein E, Schaniel C, Lemischka IR (2011) Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell 145: 183–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, van der Heijden GW, Castañeda J, Vagin VV, Hannon GJ, Bortvin A (2009) Cytoplasmic compartmentalization of the fetal piRNA pathway in mice. PLoS Genet 5: e1000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baguñà J, Saló E, Auladell C (1989) Regeneration and pattern formation in planarians. III. That neoblasts are totipotent stem cells and the source of blastema cells. Development 107: 77–86 [Google Scholar]
- Beck DB, Oda H, Shen SS, Reinberg D (2012) PR-Set7 and H4K20me1: at the crossroads of genome integrity, cell cycle, chromosome condensation, and transcription. Genes Dev 26: 325–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blythe MJ, Kao D, Malla S, Rowsell J, Wilson R, Evans D, Jowett J, Hall A, Lemay V, Lam S, Aboobaker AA (2010) A dual platform approach to transcript discovery for the planarian Schmidtea mediterranea to establish RNAseq for stem cell and regeneration biology. PLoS One 5: e15617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonuccelli L, Rossi L, Lena A, Scarcelli V, Rainaldi G, Evangelista M, Iacopetti P, Gremigni V, Salvetti A (2010) An RbAp48-like gene regulates adult stem cells in planarians. J Cell Sci 123: 690–698 [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch R, Young RA (2005) Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 22: 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S, Bendall H, Hicks GG, Nashabi A, Sakano H, Shinkai Y, Gariglio M, Oltz EM, Ruley HE (2003) The high-mobility-group box protein SSRP1/T160 is essential for cell viability in day 3.5 mouse embryos. Mol Cell Biol 23: 5301–5307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebrià F, Newmark PA (2005) Planarian homologs of netrin and netrin receptor are required for proper regeneration of the central nervous system and the maintenance of nervous system architecture. Development 132: 3691–3703 [DOI] [PubMed] [Google Scholar]
- Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A (2003) Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 113: 643–655 [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, Loh YH, Yeo HC, Yeo ZX, Narang V, Govindarajan KR, Leong B, Shahab A, Ruan Y, Bourque G, Sung WK et al. (2008) Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133: 1106–1117 [DOI] [PubMed] [Google Scholar]
- Chia NY, Chan YS, Feng B, Lu X, Orlov YL, Moreau D, Kumar P, Yang L, Jiang J, Lau MS, Huss M, Soh BS, Kraus P, Li P, Lufkin T, Lim B, Clarke ND, Bard F, Ng HH (2010) A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature 468: 316–320 [DOI] [PubMed] [Google Scholar]
- Clark AT, Bodnar MS, Fox M, Rodriquez RT, Abeyta MJ, Firpo MT, Pera RA (2004) Spontaneous differentiation of germ cells from human embryonic stem cells in vitro. Hum Mol Genet 13: 727–739 [DOI] [PubMed] [Google Scholar]
- Collier B, Gorgoni B, Loveridge C, Cooke HJ, Gray NK (2005) The DAZL family proteins are PABP-binding proteins that regulate translation in germ cells. EMBO J 24: 2656–2666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook HA, Koppetsch BS, Wu J, Theurkauf WE (2004) The Drosophila SDE3 homolog armitage is required for oskar mRNA silencing and embryonic axis specification. Cell 116: 817–829 [DOI] [PubMed] [Google Scholar]
- Ding L, Paszkowski-Rogacz M, Nitzsche A, Slabicki MM, Heninger AK, de Vries I, Kittler R, Junqueira M, Shevchenko A, Schulz H, Hubner N, Doss MX, Sachinidis A, Hescheler J, Iacone R, Anastassiadis K, Stewart AF, Pisabarro MT, Caldarelli A, Poser I et al. (2009) A genome-scale RNAi screen for Oct4 modulators defines a role of the Paf1 complex for embryonic stem cell identity. Cell Stem Cell 4: 403–415 [DOI] [PubMed] [Google Scholar]
- Eisenhoffer GT, Kang H, Sánchez Alvarado A (2008) Molecular analysis of stem cells and their descendants during cell turnover and regeneration in the planarian Schmidtea mediterranea. Cell Stem Cell 3: 327–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Messaoudi-Aubert S, Nicholls J, Maertens GN, Brookes S, Bernstein E, Peters G (2010) Role for the MOV10 RNA helicase in polycomb-mediated repression of the INK4a tumor suppressor. Nat Struct Mol Biol 17: 862–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J, Feng Q, Ketel CS, Wang H, Cao R, Xia L, Erdjument-Bromage H, Tempst P, Simon JA, Zhang Y (2002) Purification and functional characterization of SET8, a nucleosomal histone H4-lysine 20-specific methyltransferase. Curr Biol 12: 1086–1099 [DOI] [PubMed] [Google Scholar]
- Felix DA, Aboobaker AA (2010) The TALE class homeobox gene Smed-prep defines the anterior compartment for head regeneration. PLoS Genet 6: e1000915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedländer MR, Adamidi C, Han T, Lebedeva S, Isenbarger TA, Hirst M, Marra M, Nusbaum C, Lee WL, Jenkin JC, Sánchez Alvarado A, Kim JK, Rajewsky N (2009) High-resolution profiling and discovery of planarian small RNAs. Proc Natl Acad Sci USA 106: 11546–11551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar-Maia A, Alajem A, Meshorer E, Ramalho-Santos M (2011) Open chromatin in pluripotency and reprogramming. Nat Rev Mol Cell Biol 12: 36–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo T, Peters AH, Newmark PA (2006) A Bruno-like gene is required for stem cell maintenance in planarians. Dev Cell 11: 159–169 [DOI] [PubMed] [Google Scholar]
- Hayashi T, Asami M, Higuchi S, Shibata N, Agata K (2006) Isolation of planarian X-ray-sensitive stem cells by fluorescence-activated cell sorting. Dev Growth Differ 48: 371–380 [DOI] [PubMed] [Google Scholar]
- Hayashi T, Shibata N, Okumura R, Kudome T, Nishimura O, Tarui H, Agata K (2010) Single-cell gene profiling of planarian stem cells using fluorescent activated cell sorting and its "index sorting" function for stem cell research. Dev Growth Differ 52: 131–144 [DOI] [PubMed] [Google Scholar]
- Higuchi S, Hayashi T, Hori I, Shibata N, Sakamoto H, Agata K (2007) Characterization and categorization of fluorescence activated cell sorted planarian stem cells by ultrastructural analysis. Dev Growth Differ 49: 571–581 [DOI] [PubMed] [Google Scholar]
- Hinkley CS, Martin JF, Leibham D, Perry M (1992) Sequential expression of multiple POU proteins during amphibian early development. Mol Cell Biol 12: 638–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Crabtree GR (2010) Chromatin remodelling during development. Nature 463: 474–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Miller EL, Ronan JL, Ho WQ, Jothi R, Crabtree GR (2011) esBAF facilitates pluripotency by conditioning the genome for LIF/STAT3 signalling and by regulating polycomb function. Nat Cell Biol 13: 903–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Ronan JL, Wu J, Staahl BT, Chen L, Kuo A, Lessard J, Nesvizhskii AI, Ranish J, Crabtree GR (2009) An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc Natl Acad Sci USA 106: 5181–5186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Shukla A, Wang X, Chen WY, Bernstein BE, Roeder RG (2011) Role for Dpy-30 in ES cell-fate specification by regulation of H3K4 methylation within bivalent domains. Cell 144: 513–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliano CE, Swartz SZ, Wessel GM (2010) A conserved germline multipotency program. Development 137: 4113–4126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungkamp AC, Stoeckius M, Mecenas D, Grün D, Mastrobuoni G, Kempa S, Rajewsky N (2011) In vivo and transcriptome-wide identification of RNA binding protein target sites. Mol Cell 44: 828–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller DM, Zeng X, Wang Y, Zhang QH, Kapoor M, Shu H, Goodman R, Lozano G, Zhao Y, Lu H (2001) A DNA damage-induced p53 serine 392 kinase complex contains CK2, hSpt16, and SSRP1. Mol Cell 7: 283–292 [DOI] [PubMed] [Google Scholar]
- Kimmins S, Sassone-Corsi P (2005) Chromatin remodelling and epigenetic features of germ cells. Nature 434: 583–589 [DOI] [PubMed] [Google Scholar]
- Lavial F, Acloque H, Bertocchini F, Macleod DJ, Boast S, Bachelard E, Montillet G, Thenot S, Sang HM, Stern CD, Samarut J, Pain B (2007) The Oct4 homologue PouV and Nanog regulate pluripotency in chicken embryonic stem cells. Development 134: 3549–3563 [DOI] [PubMed] [Google Scholar]
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, Wong KY, Sung KW, Lee CW, Zhao XD, Chiu KP, Lipovich L, Kuznetsov VA, Robson P, Stanton LW, Wei CL et al. (2006) The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet 38: 431–440 [DOI] [PubMed] [Google Scholar]
- Meshorer E, Misteli T (2006) Chromatin in pluripotent embryonic stem cells and differentiation. Nat Rev Mol Cell Biol 7: 540–546 [DOI] [PubMed] [Google Scholar]
- Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S (2003) The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113: 631–642 [DOI] [PubMed] [Google Scholar]
- Moore FL, Jaruzelska J, Dorfman DM, Reijo-Pera RA (2004) Identification of a novel gene, DZIP (DAZ-interacting protein), that encodes a protein that interacts with DAZ (deleted in azoospermia) and is expressed in embryonic stem cells and germ cells. Genomics 83: 834–843 [DOI] [PubMed] [Google Scholar]
- Morrison GM, Brickman JM (2006) Conserved roles for Oct4 homologues in maintaining multipotency during early vertebrate development. Development 133: 2011–2022 [DOI] [PubMed] [Google Scholar]
- Newmark PA, Sánchez Alvarado A (2000) Bromodeoxyuridine specifically labels the regenerative stem cells of planarians. Dev Biol 220: 142–153 [DOI] [PubMed] [Google Scholar]
- Newmark PA, Sánchez Alvarado A (2002) Not your father's planarian: a classic model enters the era of functional genomics. Nat Rev Genet 3: 210–219 [DOI] [PubMed] [Google Scholar]
- Niwa H (2007) How is pluripotency determined and maintained? Development 134: 635–646 [DOI] [PubMed] [Google Scholar]
- Okuhara K, Ohta K, Seo H, Shioda M, Yamada T, Tanaka Y, Dohmae N, Seyama Y, Shibata T, Murofushi H (1999) A DNA unwinding factor involved in DNA replication in cell-free extracts of Xenopus eggs. Curr Biol 9: 341–350 [DOI] [PubMed] [Google Scholar]
- Oosaki T, Ishii S (1965) Observations on the ultrastructure of nerve cells in the brain of the planarian, Dugesia gonocephala. Z Zellforsch Mikrosk Anat 66: 782–793 [DOI] [PubMed] [Google Scholar]
- Orphanides G, Wu WH, Lane WS, Hampsey M, Reinberg D (1999) The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature 400: 284–288 [DOI] [PubMed] [Google Scholar]
- Randolph H (1897) Observations and experiments on regeneration in planarians. Arch Entw Mech Org 5: 352–372 [Google Scholar]
- Reddien PW, Bermange AL, Murfitt KJ, Jennings JR, Sánchez Alvarado A (2005a) Identification of genes needed for regeneration, stem cell function, and tissue homeostasis by systematic gene perturbation in planaria. Dev Cell 8: 635–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien PW, Oviedo NJ, Jennings JR, Jenkin JC, Sánchez Alvarado A (2005b) SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science 310: 1327–1330 [DOI] [PubMed] [Google Scholar]
- Reddien PW, Sánchez Alvarado A (2004) Fundamentals of planarian regeneration. Annu Rev Cell Dev Biol 20: 725–757 [DOI] [PubMed] [Google Scholar]
- Robb SM, Ross E, Sánchez Alvarado A (2008) SmedGD: the Schmidtea mediterranea genome database. Nucleic Acids Res 36: D599–D606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi L, Salvetti A, Marincola FM, Lena A, Deri P, Mannini L, Batistoni R, Wang E, Gremigni V (2007) Deciphering the molecular machinery of stem cells: a look at the neoblast gene expression profile. Genome Biol 8: R62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhana L, Shibata N, Nishimura O, Agata K (2010) Different requirements for conserved post-transcriptional regulators in planarian regeneration and stem cell maintenance. Dev Biol 341: 429–443 [DOI] [PubMed] [Google Scholar]
- Sánchez Alvarado A, Newmark PA (1999) Double-stranded RNA specifically disrupts gene expression during planarian regeneration. Proc Natl Acad Sci USA 96: 5049–5054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz YB, Pirrotta V (2007) Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet 8: 9–22 [DOI] [PubMed] [Google Scholar]
- Scimone ML, Meisel J, Reddien PW (2010) The Mi-2-like Smed-CHD4 gene is required for stem cell differentiation in the planarian Schmidtea mediterranea. Development 137: 1231–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydoux G, Braun RE (2006) Pathway to totipotency: lessons from germ cells. Cell 127: 891–904 [DOI] [PubMed] [Google Scholar]
- Shibata N, Rouhana L, Agata K (2010) Cellular and molecular dissection of pluripotent adult somatic stem cells in planarians. Dev Growth Differ 52: 27–41 [DOI] [PubMed] [Google Scholar]
- Shoji M, Tanaka T, Hosokawa M, Reuter M, Stark A, Kato Y, Kondoh G, Okawa K, Chujo T, Suzuki T, Hata K, Martin SL, Noce T, Kuramochi-Miyagawa S, Nakano T, Sasaki H, Pillai RS, Nakatsuji N, Chuma S (2009) The TDRD9-MIWI2 complex is essential for piRNA-mediated retrotransposon silencing in the mouse male germline. Dev Cell 17: 775–787 [DOI] [PubMed] [Google Scholar]
- Schröder R. (2003) The genes orthodenticle and hunchback substitute for bicoid in the beetle Tribolium. Nature 422: 621–625 [DOI] [PubMed] [Google Scholar]
- Siomi MC, Sato K, Pezic D, Aravin AA (2011) PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol 12: 246–258 [DOI] [PubMed] [Google Scholar]
- Solana J, Lasko P, Romero R (2009) Spoltud-1 is a chromatoid body component required for planarian long-term stem cell self-renewal. Dev Biol 328: 410–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676 [DOI] [PubMed] [Google Scholar]
- Tang F, Barbacioru C, Bao S, Lee C, Nordman E, Wang X, Lao K, Surani MA (2010) Tracing the derivation of embryonic stem cells from the inner cell mass by single-cell RNA-Seq analysis. Cell Stem Cell 6: 468–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theunissen TW, Costa Y, Radzisheuskaya A, van Oosten AL, Lavial F, Pain B, Castro LF, Silva JC (2011) Reprogramming capacity of Nanog is functionally conserved in vertebrates and resides in a unique homeodomain. Development 138: 4853–4865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomari Y, Du T, Haley B, Schwarz DS, Bennett R, Cook HA, Koppetsch BS, Theurkauf WE, Zamore PD (2004) RISC assembly defects in the Drosophila RNAi mutant armitage. Cell 116: 831–841 [DOI] [PubMed] [Google Scholar]
- Umesono Y, Watanabe K, Agata K (1997) A planarian orthopedia homolog is specifically expressed in the branch region of both the mature and regenerating brain. Dev Growth Differ 39: 723–727 [DOI] [PubMed] [Google Scholar]
- Wagner DE, Ho JJ, Reddien PW (2012) Genetic regulators of a pluripotent adult stem cell system in planarians identified by RNAi and clonal analysis. Cell Stem Cell 10: 299–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner DE, Wang IE, Reddien PW (2011) Clonogenic neoblasts are pluripotent adult stem cells that underlie planarian regeneration. Science 332: 811–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Stary JM, Wilhelm JE, Newmark PA. (2010) A functional genomic screen in planarians identifies novel regulators of germ cell development. Genes Dev 24: 2081–2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu EY, Moore FL, Pera RA (2001) A gene family required for human germ cell development evolved from an ancient meiotic gene conserved in metazoans. Proc Natl Acad Sci USA 98: 7414–7419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida-Kashikawa M, Shibata N, Takechi K, Agata K (2007) DjCBC-1, a conserved DEAD box RNA helicase of the RCK/p54/Me31B family, is a component of RNA-protein complexes in planarian stem cells and neurons. Dev Dyn 236: 3436–3450 [DOI] [PubMed] [Google Scholar]
- Young RA (2011) Control of the embryonic stem cell state. Cell 144: 940–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.