Abstract
Eukaryotic RNA polymerase II (Pol II) has evolved an array of heptad repeats with the consensus sequence Tyr1-Ser2-Pro3-Thr4-Ser5-Pro6-Ser7 at the carboxy-terminal domain (CTD) of the large subunit (Rpb1). Differential phosphorylation of Ser2, Ser5, and Ser7 in the 5′ and 3′ regions of genes coordinates the binding of transcription and RNA processing factors to the initiating and elongating polymerase complexes. Here, we report phosphorylation of Thr4 by Polo-like kinase 3 in mammalian cells. ChIPseq analyses indicate an increase of Thr4-P levels in the 3′ region of genes occurring subsequently to an increase of Ser2-P levels. A Thr4/Ala mutant of Pol II displays a lethal phenotype. This mutant reveals a global defect in RNA elongation, while initiation is largely unaffected. Since Thr4 replacement mutants are viable in yeast we conclude that this amino acid has evolved an essential function(s) in the CTD of Pol II for gene transcription in mammalian cells.
Keywords: elongation, phosphorylation, Plk3, Pol II CTD, threonine-4
Introduction
In eukaryotic cells, synthesis of mRNA and small nuclear (sn)RNA is catalysed by the multi-subunit RNA polymerase II (Pol II). The large subunit (Rpb1) of Pol II has evolved a carboxy-terminal domain (CTD) containing tandem repeats with the consensus sequence Tyr1-Ser2-Pro3-Thr4-Ser5-Pro6-Ser7. The number of repeats varies with the complexity of organisms from 26 in yeast to 52 in mammalian cells (Phatnani and Greenleaf, 2006; Corden, 2007; Chapman et al, 2008; Egloff and Murphy, 2008; Buratowski, 2009). CTD is not part of the catalytic domain of Pol II but is required for transcription of chromatin templates and couples transcription with co-transcriptional processes as mRNA capping (Viladevall et al, 2009; Cowling, 2010; Ghosh et al, 2011), splicing (de la Mata and Kornblihtt, 2006; Munoz et al, 2010; David et al, 2011; de Almeida et al, 2011), 3′ processing (Ahn et al, 2004; Egloff et al, 2007; Johnson et al, 2011), and export (Custodio et al, 2007; MacKellar and Greenleaf, 2011). New findings implicate CTD also in other nuclear processes such as chromatin modification (Govind et al, 2010; Spain and Govind, 2011) and transcription-coupled genome stability (Blazek et al, 2011). CTD undergoes continuous structural remodelling during the transcription cycle, which allows the binding and release of a multitude of factors and the coordination of various co-transcriptional processes. The recruitment and release of these factors to CTD is regulated by a complex interplay of kinases and phosphatases that establish and erase specific factor binding platforms in the course of the transcription cycle. In principle, all three amino acids with hydroxy groups (Ser, Thr, Tyr) in the heptad repeat can undergo modification by phosphorylation. The placement and removal of phospho-residues appears to be influenced by the isomeric state of the two proline residues in each heptad repeat that can switch from cis to trans isoforms with the help of peptidyl-propyl isomerases (Singh et al, 2009; Werner-Allen et al, 2011).
Phosphorylation of serine residues is by far the best-studied modification of CTD (for reviews, see Phatnani and Greenleaf, 2006 and Egloff and Murphy, 2008). After binding of Pol II to the promoter Ser5 is phosphorylated by the Cdk7 subunit (Kin28 in S. cerevisiae) of the transcription factor TFIIH in a mediator-dependent manner (Boeing et al, 2010) and subsequently essential for Pol II–Mediator dissociation (Max et al, 2007). Ser5-P is later on required for recruitment of the guanylyltransferase and 5′ capping of the nascent transcript (Cowling, 2010; Ghosh et al, 2011), but also chromatin modifying and splicing factors (for review, see Perales and Bentley, 2009). In yeast, specific inhibition of the analogue-sensitive kinase Kin28 inhibits recruitment of the capping enzyme, but has only little effect on gene transcription (Kanin et al, 2007; Hong et al, 2009). The function of Cdk7 and Kin28 at the promoter is not restricted to Ser5 phosphorylation, since both enzymes were identified also as Ser7-specific kinases (Akhtar et al, 2009; Glover-Cutter et al, 2009; Kim et al, 2009). Ser5-P and Ser7-P marks are placed in CTD already at the promoter, but only Ser7-P marks remain associated with Pol II during the transcription cycle. Ser5-P marks are largely removed when Pol II transcribes sequences downstream of the promoter, while Ser2-P marks are progressively introduced in CTD, if Pol II approaches the 3′ end of genes (Akhtar et al, 2009; Kim et al, 2010; Mayer et al, 2010; Tietjen et al, 2010; Bataille et al, 2012). Cell signalling for the recruitment of Ser2-specific kinase activity is often the critical step in activation of promoter-proximally paused Pol II. At least two kinases can phosphorylate Ser2 in metazoans and yeast. While Cdk9 (kinase subunit of the positive transcription elongation factor P-TEFb) phosphorylates mainly promoter-proximal Pol II, and thereby facilitates the release of Pol II from pause sites, the more recently discovered Cdk12 shows low abundance at the 5′ end of genes, but phosphorylates CTD in the middle and 3′ end of metazoa genes (Bartkowiak et al, 2010; Blazek et al, 2011). For the kinases Bur1 and Ctk1 (Keogh et al, 2003; Jones et al, 2004) similar division of work has been described before for S. cerevisiae (Qiu et al, 2009). The removal of CTD phospho-marks is achieved during different phases of the transcription cycle by phosphatases FCP1, SSU72, and Rtr1 (RPAP2) (Krishnamurthy et al, 2004; Mosley et al, 2009; Werner-Allen et al, 2011; Egloff et al, 2012; Zhang et al, 2012) with specificity to single or several phospho-residues, during mitosis by phosphatase Cdc14 with specificity for Ser2 and Ser5 (Clemente-Blanco et al, 2011), or in a tissue-specific manner by Scp1 in neural cells (Yeo et al, 2005).
Many of the features described above apply to most of Pol II transcribed genes and contribute to the view of a uniform transition of general Pol II complexes with uniform CTD phospho-marks through the transcription cycle in yeast (Mayer et al, 2010; Bataille et al, 2012). First studies in mouse T and embryonic stem cells confirm this general view, but the situation in mammalian cells is also more complex, since genes are much larger, contain many and large introns, and are flanked or interspersed with enhancer and silencer elements (Rahl et al, 2010; Koch and Andrau, 2011; Koch et al, 2011; Brookes et al, 2012).
Some CTD modifications may occur in a gene-specific manner and fulfil gene-specific tasks, but are not required for transcription of all genes. For example, Ser2-P is not essential for transcription, mRNA cleavage, and poly-adenylation (poly-A) of p21(Cip1) in mammalian cells (Gomes et al, 2006), but is required for sexual differentiation in S. pombe (Coudreuse et al, 2010). Ser7-P is not critical for 3′ processing and polyadenylation of protein-encoding genes (Chapman et al, 2007), but for proper 3′ processing of small nuclear (sn)RNAs and the recruitment of the Integrator complex (Egloff et al, 2007, 2010). Methylation of an arginine residue in non-consensus repeats of mammalian CTD regulates the abundance of small nuclear and nucleolar RNAs but not of mRNAs (Sims et al, 2011). Thus, modifications in CTD can serve as general mechanism of gene transcription, but apparently also fulfil gene-specific tasks.
Here, we have studied the modification of CTD Thr4 by phosphorylation in mammalian cells. We show that Thr4 is phosphorylated by Polo-like kinase 3 (Plk3) in vitro and in vivo and that a Thr4/Ala mutant, different to the earlier reported Thr4/Val mutant in chicken DT40 cells (Hsin et al, 2011), has a severe transcription defect characterized by an elongation block proximal to the initiation site.
Results
Residue Thr4 of the CTD consensus repeat is essential for viability of mammalian cells
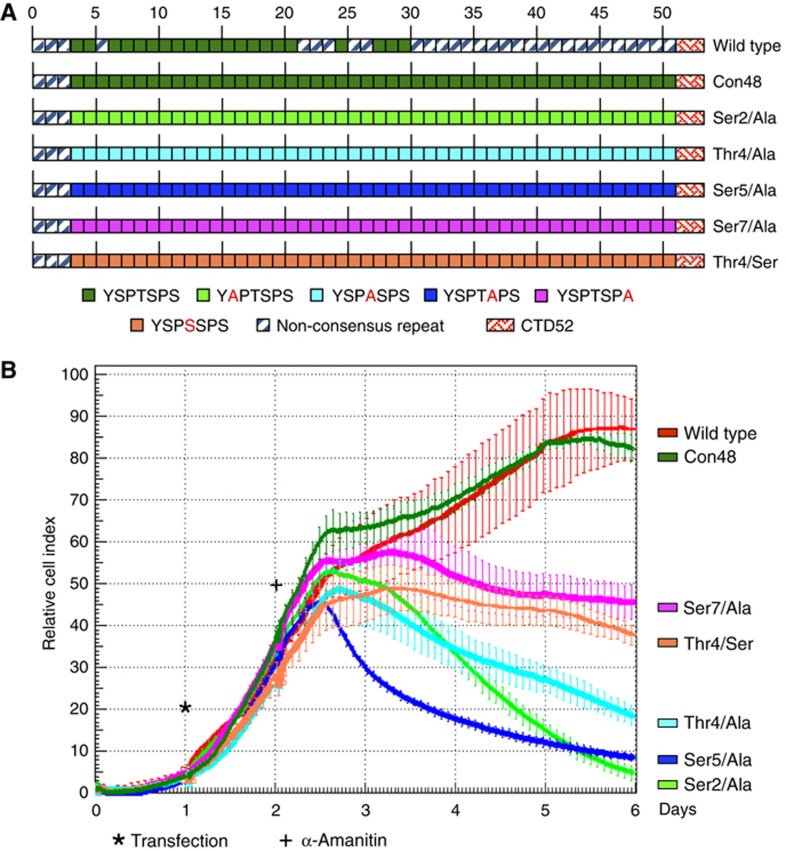
In S. cerevisiae and S. pombe, Thr4 can be replaced by alanine in CTD repeats without affecting cell viability (Stiller et al, 2000; Schwer and Shuman, 2011). To investigate whether Thr4 is essential for the growth of mammalian cells, we transiently transfected HEK293 cells with an expression vector for a mutant of the large subunit (Rpb1) of RNA Pol II resistant to α-amanitin (Gerber et al, 1995; Chapman et al, 2004). This Rpb1 contained either the wild-type sequence of CTD, a CTD with 48 consensus repeats (Con48) (Chapman et al, 2005), or CTDs where positions Ser2, Ser5, Thr4, or Ser7 were replaced by alanine, or Thr4 by Ser (Figure 1A). After transfection and selection with α-amanitin, the viability of cells was measured over a period of 4 days. The Con48 mutant displayed a growth rate similar to recombinant wild-type Pol II. In contrast, mutants comprising replacements of Ser2/Ala, Thr4/Ala, and Ser5/Ala in 48 repeats showed a severe growth defect with a strongly reduced cell count after 4 days. The Thr4/Ser and Ser7/Ala mutants revealed an attenuated phenotype with almost constant cell numbers (Figure 1B). Comparable results were obtained if similar CTD mutants were conditionally expressed in the human B-cell line Raji (Chapman et al, 2007, data not shown). We conclude that Thr4 is an essential residue in mammalian CTD heptad repeat, which cannot be replaced by alanine or serine. The phenotype of the Thr4 mutants could be associated with the failure of proper phosphorylation of CTD at this residue during transcription. Therefore, we next studied if and how Thr4 is phosphorylated in the transcription cycle.
Figure 1.
Viability of cells expressing CTD mutants. (A) Overview of analysed CTD mutants. (B) HEK293 cells were transfected with an expression vector for Rpb1 carrying a point mutation for α-amanitin resistance (wild type). CTD mutants carry 48 consensus repeats (Con48), or have replaced specific residues in all consensus repeats by alanine (Ser2, Thr4, Ser5, Ser7) or serine (Thr4). Twenty-four hours after transfection, α-amanitin was added (2 μg/ml) and cell growth rates were monitored for 4 days in the xCELLigence system (Roche), error bars show standard deviation of 3 replicates.
Production and characterization of a CTD Thr4-P-specific monoclonal antibody
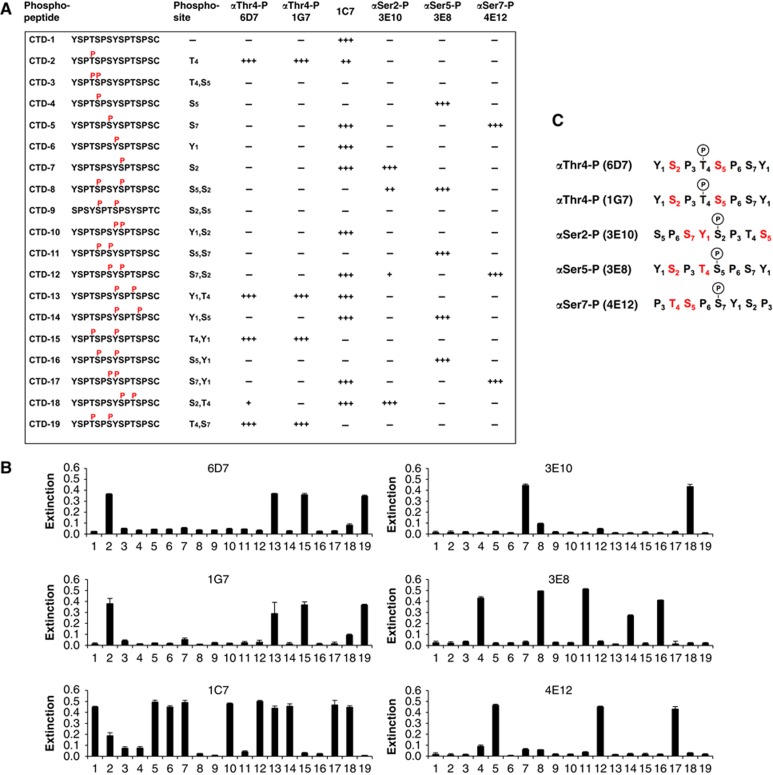
To study Thr4 phosphorylation, two monoclonal antibodies (mAbs) were produced after immunization of rats with the phospho-peptide CTD-2 (Figure 2A) containing a CTD phospho-Thr4 (Thr4-P) epitope. The binding specificity of the two Thr4-P-specific mAbs (6D7 and 1G7), and of the previously generated Ser2-P (3E10), Ser5-P (3E8), and Ser7-P (4E12) mAbs (Chapman et al, 2007) were tested in enzyme-linked immunosorbent assays (ELISAs; Figure 2B). To get further insight how other adjacent modifications can influence or inhibit epitope recognition, a panel of 19 di-heptad peptides (CTD1–19) with various combinations of modifications was included in the test. The analysis gave a comprehensive overview of inhibitory modifications that impair or block binding of specific mAbs to CTD (Figure 2C). For example, the presence of Tyr1-P upstream and downstream of Thr4-P as well as of Ser7-P were not inhibitory for Thr4-P recognition by mAbs 6D7 and 1G7, while phosphorylation of Ser2 or Ser5 next to Thr4-P was inhibitory. A survey of phosphorylated amino-acid residues (in red) with inhibitory activity for binding of CTD-specific mAbs is shown in Figure 2C.
Figure 2.
Thr4-P-specific mAb production. (A) Survey of synthetic phospho-peptides used for characterization of CTD-specific mAbs. Peptide CTD-2 was used for immunization of rats and identification of Thr4-P-specific mAbs (6D7 and 1G7). The binding specificity of Thr4-P-specific mAbs and of previously generated mAbs with specificity for Ser2-P (3E10), Ser5-P (3E8), Ser7-P (4E12), and unmodified CTD (1C7) was determined by ELISA using a panel of 19 CTD peptides with different combinations of phosphorylated amino acids. Phosphorylation of amino acids adjacent to the phospho site used for immunization either inhibited (−), did not inhibit (+++), or inhibited binding of mAbs to various degree (++, +). (B) Quantitative ELISA data for CTD peptides 1–19. Reactivity below 0.05 indicates background. (C) Overview of conditions of phospho-CTD recognition by mAbs. Red amino acids indicate full or partial inhibition of mAb binding. Phosphorylated amino acids in black colour (Tyr1, Ser, Thr) did not inhibit mAb binding. Error bars show standard deviation of three experiments.
RNA Pol II CTD is phosphorylated at residue Thr4 in vivo
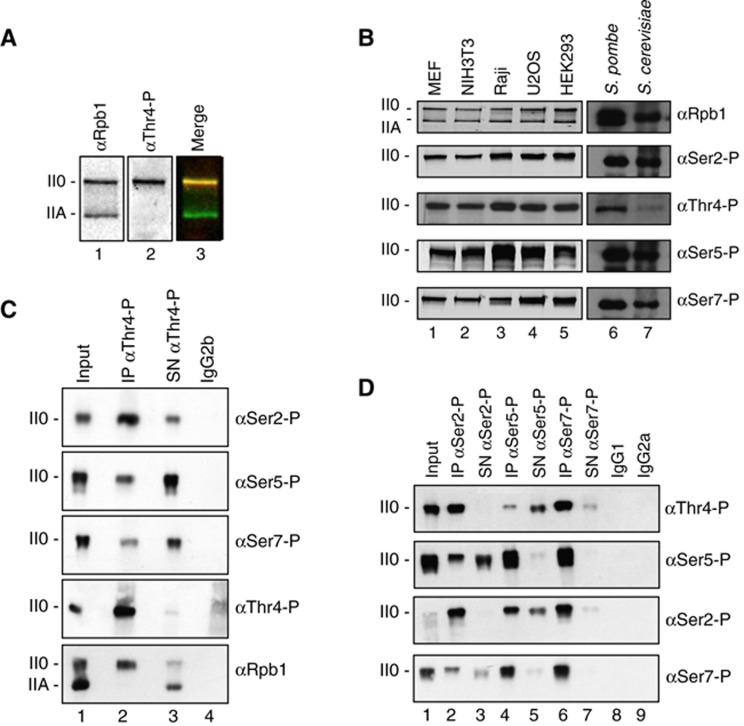
Modification of Thr4 by phosphorylation was detectable in HeLa cells (Figure 3A), in other cell lines of human and mouse origin, as well as in S. pombe and S. cerevisiae (Figure 3B). The occurrence of Thr4-P apparently was restricted to the hyper-phosphorylated II0 form of Pol II. Divergent from Ser2-P and Ser5-P, phosphorylation at residue Thr4 requires a minimal length of the CTD and is not or only hardly observed for CTD truncation mutants with only 8, 16, 20, or 24 consensus repeats (Supplementary Figure S1). The similar length restriction has been observed for phosphorylation of Ser7 in CTD truncation mutants before (Chapman et al, 2007).
Figure 3.
RNA polymerase II CTD is phosphorylated at residue Thr4. (A) Western blot analysis of HeLa cell extracts with Rpb1-specific mAb (POL 3/3) and the Thr4-P-specific mAb (6D7). II0 and IIA designate the hyperphosphorylated and hypophosphorylated forms of the large subunit Rpb1 of Pol II. (B) Extracts of mouse (MEF, NIH3T3) and human cell lines (Raji, U2OS, HEK293), or S. pombe and S. cerevisiae were analysed with mAbs specific for the various phospho-residues in CTD described in Figure 2A. (C, D) Immunoprecipitation (IP) experiments with phospho-specific CTD mAbs for Thr4-P, Ser2-P, Ser5-P, and Ser7-P in extracts of HeLa cells. Analysis of western blots with indicated mAbs. SN, supernatant; IgG1, IgG2a, and IgG2b are isotype controls.
Characterization of a Thr4-P-positive sub-population of Pol II0
The panel of mAbs with specificity for different CTD phosphorylation marks allowed addressing the question, whether specific marks occur in Pol II0 preferentially in certain combinations. For this analysis, we first optimized the protocol for immunoprecipitation (IP) of Pol II (see Materials and methods). Under optimized conditions, all modification-specific antibodies precipitated Pol II quantitatively without leaving significant amounts of Pol II0 with the respective modification in the supernatant (Figure 3C and D). The supernatant was further analysed for Pol II0 residues with other modifications. If Pol II0 was precipitated with the Thr4-P-specific mAb (Figure 3C), then a significant amount of Pol II0 remained in the supernatant. The leftover in the supernatant was Ser5-P and Ser7-P positive, but showed only little reactivity for Ser2-P. Inverse IP experiments with Ser2-P-, Ser5-P-, and Ser7-P-specific mAbs confirmed these results (Figure 3D). The Ser2-P-specific mAb co-precipitated Pol II with the Thr4-P mark quantitatively, while large amounts of Thr4-P-positive Pol II0 remained in the supernatant after precipitation with the Ser5-P-specific mAb. These experiments demonstrate the existence of three major populations of Pol II0 regarding their CTD marks. A population associated preferentially with (i) Ser5-P marks, (ii) Ser5-P and Ser2-P marks, and (iii) Ser2-P and Thr4-P marks. All three populations apparently occur together with the Ser7-P mark albeit small fractions of Ser2-P and Thr4-P-positive Pol II0 is left in the supernatant of Ser7-P-specific precipitates (Figure 3D, lane 7). The presence of differently modified populations of Pol II was also supported by immunofluorescence microscopy analysis of nuclei (Supplementary Figure S2). To summarize, the Thr4-P mark is detectable only in a subpopulation of polymerases and is strictly associated with the occurrence of the Ser2-P mark while the association with the Ser5-P mark is not stringent. The conclusion is subject to the caution that masked CTD epitopes are not recognized by antibodies.
CTD Thr4 is target of Plk3 kinase in vitro and in vivo
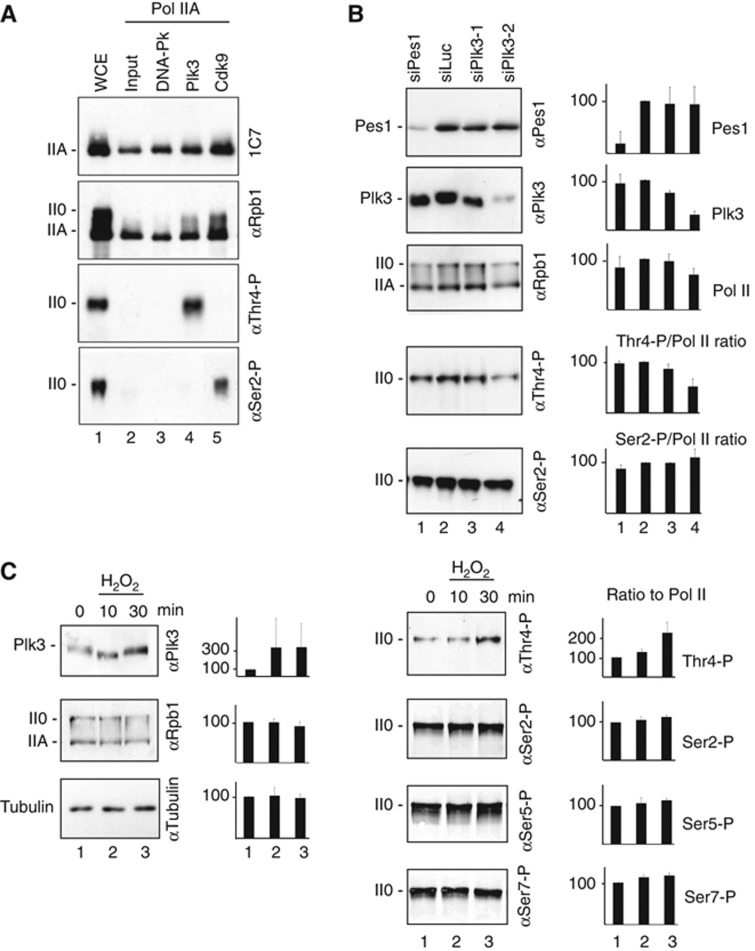
In order to identify Thr4-specific kinases, we screened a library of 81 recombinant serine/threonine kinases (Supplementary Figure S3) for specificity of Thr4 phosphorylation of a CTD peptide in ELISA experiments. This pre-screen yielded a few kinases, however, only Plk3 strongly phosphorylated purified Pol IIA and caused a shift to the II0 form (Figure 4A). The reactivity of Plk1, a related kinase, towards Thr4 was ∼10-fold lower (data not shown), while other kinases did not phosphorylate Thr4 of purified Pol IIA. Phosphorylation of Ser2 by Cdk9 served as a positive control in our experiments. Cdk9 has previously been claimed as a Thr4-specific kinase, because Thr4 phosphorylation is sensitive to the kinase inhibitor flavopiridol (Hsin et al, 2011). We could confirm this sensitivity for Cdk9 but not for Plk3 in vitro (Supplementary Figure S4). Cdk9 failed to phosphorylate Thr4 in vitro (Figure 4A, lane 5). Since Cdk9 can phosphorylate Ser2 and Ser5 residues, the recognition of Thr4-P by mAbs 6D7 and 1G7 could be inhibited by these adjacent modifications. Therefore, the contribution of Cdk9 to Thr4 phosphorylation remains elusive. Short interfering RNA (siRNA) knockdown experiments were performed in HeLa cells to test if Plk3 is required for phosphorylation of Thr4 in vivo. The knockdown reduced Plk3 protein levels to <40% of control cells and the ratio Thr4-P/Pol II by ∼50%, while the ratio Ser2-P/Pol II slightly increased (Figure 4B). Thus, knockdown of Plk3 specifically reduces Thr4-P levels in cells. The remaining Thr4-P signal may be due to the incomplete knockdown of Plk3 and/or other not yet identified kinases, which can contribute to Thr4 phosphorylation in vivo.
Figure 4.
Identification of Plk3 as a CTD Thr4-specific kinase. (A) Immunoprecipitated Pol IIA from HeLa cell extracts was used as substrate for DNA-Pk, Plk3, and Cdk9, and subsequently analysed by western blot with mAbs specific for CTD modifications Thr4-P and Ser2-P, or Rpb1 (POL 3/3 and 1C7, latter recognizes non-modified CTD). (B) Knockdown of Plk3 by siRNA reduces Plk3 protein levels in HeLa cells and concomitantly the levels of Thr4-P in Pol II CTD. (C) Treatment of HeLa cells with 0.2 mM H2O2 induces increased Plk3 protein levels after 30 min and concomitantly a significant increase in Thr4-P levels, while Ser2-P, Ser5-P, and Ser7-P levels remained unaffected. Experiments were performed with at least three replicates; error bars show standard deviation.
Plk3 can be activated by hypoxia/reoxygenation stress and may fulfil a specialized role in transcriptional gene regulation upon stress (Wang et al, 2008; Yang et al, 2008). Therefore, we tested whether oxidative stress can specifically modulate Thr4-P levels. Indeed, treatment of HeLa cells with H2O2 for 30 min upregulated Plk3 protein and Thr4-P levels in CTD concomitantly, but did not affect the phosphorylation levels of other residues in CTD (Figure 4C). We conclude that Plk3 phosphorylates Thr4 residues of CTD in vivo and can contribute to Thr4 phosphorylation in a stress-dependent manner.
Thr4-phosphorylated RNA Pol II is enriched in the 3′ region of genes
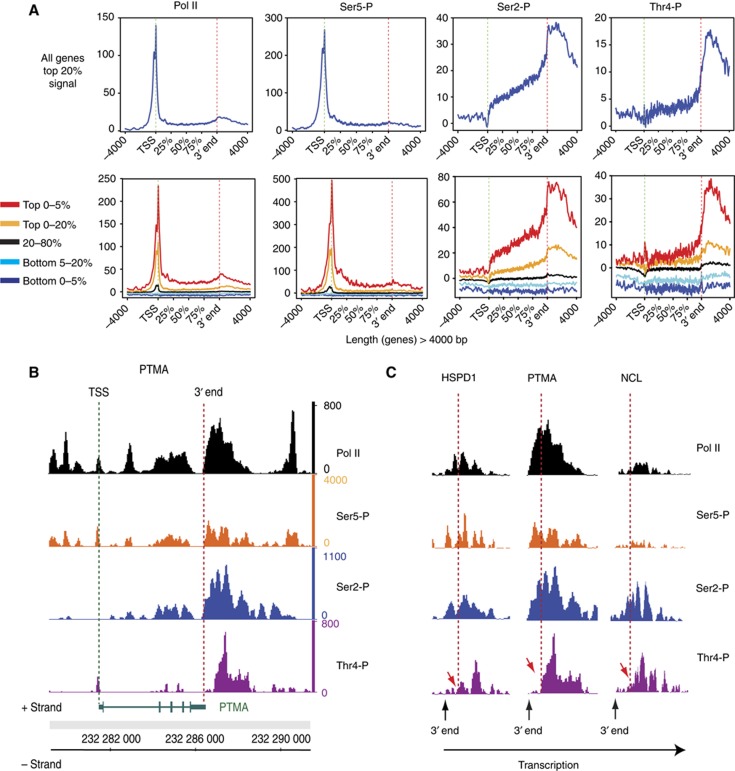
Chromatin immunoprecipitation (ChIP) experiments were performed in the human B-cell line Raji to identify the step(s) of the transcription cycle involving Thr4 phosphorylation. The occupancy profiles of total Pol II and of the three CTD marks Ser2-P, Ser5-P, and Thr4-P were examined across the genome of Raji cells by deep sequencing. The genome-wide occupancy analysis revealed high levels of Pol II and Ser5-P at transcription start sites (TSSs), while signals for Ser2-P were essentially absent at TSS (Figure 5A). The signal for Ser2-P steadily increased within the gene body and reached a maximum at the poly-A site (3′ end), and dropped slowly thereafter. Signals for Thr4-P were low but detectable for few genes at the TSS (see top 5% of the signal; Figure 5A, lower panel), slightly rose in the body of genes, but strongly increased at the 3′ end of genes and reached a maximum between 500 and 2000 nt downstream of the poly-A site. Inspection of individual genes (HSPD1, PTMA, and NCL) revealed significant differences in the local occurrence of the Ser2-P and Thr4-P at the 3′ end of genes. While Ser2-P signals increased concomitantly with Pol II levels at the 3′ end of genes, the increase of Thr4-P levels was apparently delayed and occurred with a local distance of several 100 nt later (Figure 5B and C). This suggests that phosphorylation of CTD Ser2 probably precedes the phosphorylation of Thr4 at the 3′ end of genes. For more gene ChIP profiles, see Supplementary Figure S5. As outlined above, the Thr4-P-specific mAb cannot discriminate between de-novo phosphorylation of a Thr4 residue and the unmasking of a pre-existing Thr4-P mark, thus the strong increase of the Thr4-P mark in the 3′ region of genes may derive from either or both options. We conclude that Thr4-P-associated structural changes occur in the CTD downstream of the poly-A site.
Figure 5.
Genome-wide Pol II and CTD phosphorylation profiles. (A) Diagram summarizing the average transcription unit with representative clusters for Pol II, Ser5-P, Ser2-P, and Thr4-P occupancy profiles across the genome using average transcription unit analysis (see Materials and methods). The top panel represents the profiles for the top 20% of the signal for each Pol II population and the bottom panel various level of binding depending on the signal range. The profiles are built for genes >4 kb in length, which represent the large majority of the protein-coding genes. (B) Protymosin-alpha (PTMA) gene is an example with high Thr4-P levels in the 3′ region of the gene. (C) HSPD1, PTMA, and NCL are examples for Thr4 phosphorylation subsequent to Ser2 phosphorylation at the 3′ end of genes. Normalized ChIPseq signals for each experiment are shown. Genes on positive (+) and negative (−) strand are indicated below the ChIPseq lane. Dashed lines indicate the position in the 3′ region of genes with increase of Thr4-P (red arrows).
CTD Thr4/Ala mutants display an elongation defect
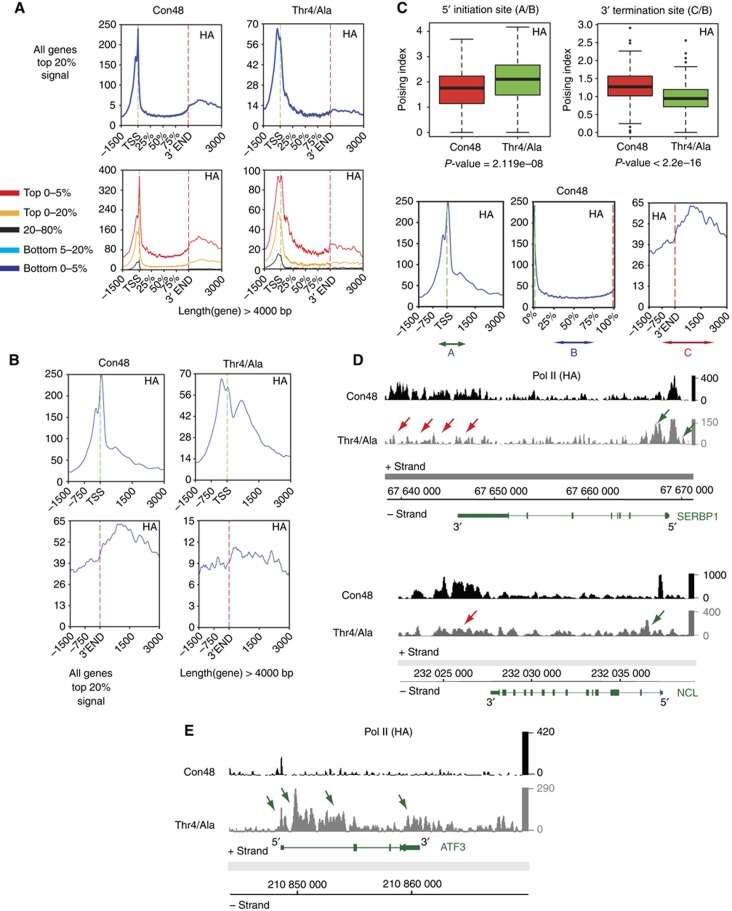
To study the role of specific amino-acid residues in the heptad structure of CTD, we have established a system that allows the comparison of CTDs with different repeat composition in stably transfected Raji cells. Recombinant, HA-tagged polymerases are engineered with a point mutation conferring the resistance to α-amanitin, allowing the endogenous polymerase to be inhibited and degraded (Meininghaus and Eick, 1999) after addition of α-amanitin, but without affecting recombinant polymerase activity (Meininghaus et al, 2000). The system is useful to study the activity of all kinds of CTD mutants. However, the analysis of the phenotype of CTD mutants is limited to a short time window of few days, if the mutant cannot sustain expression of the recombinant polymerase. In this case, the mRNA for the recombinant Pol II has prior to be synthesized by the endogenous Pol II in the absence of α-amanitin (Supplementary Figure S6). The lethal phenotype of some CTD mutants, however, need not imply that these mutants have a general defect in gene transcription. It could well be that only specific classes of genes are affected, or that maturation of mRNA or other small RNAs is impaired. To get deeper insight in the potential transcriptional defect of the Thr4/Ala mutant, we compared the genome-wide occupancy profile of this mutant with the viable all consensus CTD mutant Con48 in ChIP experiments (for mutants, see Figure 1). The analysis of both mutants was performed with an HA-tag-specific mAb to ensure that possible residual amounts of endogenous Pol II do not affect the results. The gene-specific profiles of both mutants showed striking differences. While both Con48 and Thr4/Ala mutants showed a strong peak at the TSS, the peak in the Thr4/Ala mutant was less sharp and characterized by a broad shoulder downstream of the TSS (Figure 6A). A high-resolution profile of the TSS region (Figure 6B, upper panel) revealed an additional broad peak for mutant Thr4/Ala ∼500 bp downstream of TSS that was largely absent in Con48. The second large difference was seen in the 3′ region of genes. While the level of Pol II in the Con48 mutant significantly increased at the 3′ end as observed for wt Pol II (Figure 5A), this increase was almost absent in the Thr4/Ala mutant (Figure 6A and B, lower panel). Genome-wide quantification of Pol II densities by boxplots (Figure 6C) confirmed higher amounts of Pol II at the TSS and lower amounts at the 3′ end of genes in the Thr4/Ala mutant. SERBP1 (Serpine1 mRNA binding protein 1) and NCL (nucleolin) are shown as representative genes with an altered Pol II occupancy profile in mutant Thr4/Ala (Figure 6D). The additional peak of Pol II downstream of TSS and the lack of Pol II in the 3′ region of the genes are indicated by green and red arrows, respectively. For more profiles, see Supplementary Figure S7. Previously, a 3′ processing defect of histone mRNAs has been described in chicken DT40 expressing a CTD Thr4/Val mutant, while histone gene transcription apparently was not affected in nuclear run-on experiments (Hsin et al, 2011). Consistent with the genome-wide elongation defect of mutant Thr4/Ala in Raji cells in mRNA encoding genes, this defect was also observed in several histone genes (Supplementary Figure S8). Finally, we found few genes that become activated in the Thr4/Ala mutant. High amounts of Pol II are detectable on the ATF3 (activation transcription factor 3) gene in the Thr4/Ala but not in the Con48 mutant (Figure 6E). This implies that CTD residue Thr4 may also be involved in the negative regulation of gene transcription by preventing initiation. Expression levels of Pol II in Thr4/Ala mutants are reduced compared with Con48 mutants (Supplementary Figure S7); therefore, the impact of this mutation on global initiation remains however uncertain. In conclusion, the Thr4/Ala mutant displays a complex transcriptional phenotype and affects gene activity probably at different levels. The most prominent phenotype constitutes a block to elongation downstream of TSS.
Figure 6.
Genome-wide profiles of CTD mutants Con48 and Thr4/Ala. (A) Average transcription unit occupancy profiles with representative clusters for HA-tagged CTD mutants. Genes with length >4 kb are represented with different selections of signal intensities (different percentages are represented by different colours). (B) Distribution of Pol II upstream and downstream of TSS and 3′ end of genes in Thr4/Ala mutant cell line for the top 20% of binding signal of genes >4 kb. (C) Boxplots of the poising of Pol II at 5′ initiation site and 3′ termination site. In the mutant Thr4/Ala, the poising index is increasing at 5′ whereas it is decreasing at 3′. The profile at bottom shows the areas used for calculating the poising: A=−500/+1000 at TSS, B=50% to end of gene body, C=0–3000 after 3′. Poising at termination site (TS): A/B, poising at 3′: C/B. Genes with Top 5% of binding signal were considered. The significance of the difference of the median was assessed using a non-parametric Mann–Whitney–Wilcoxon test and returned significant P-values of 2.119e−08 and <2.2e−16 for the TSS and 3′ end, respectively. (D) Example of genes reflecting an altered Pol II distribution (SERBP1 and NCL) in the Thr4/Ala mutant. Red arrows indicate reduced Pol II signal at the 3′ end and green arrows increased Pol II at TSSs. More individual examples are shown in Supplementary Figure S7. For all examples shown here and further in the manuscript, DNA is oriented with the chromosomal coordinates direction so that the plus strand shows genes in the 5′–3′ and the minus strand in the 3′–5′ orientation. (E) Example for the minority of genes, ATF3, reflecting an increased Pol II density on the gene body and after the 3′ end, when Thr4 is mutated to alanine.
Discussion
Our knowledge about the presence and gene-specific changes of CTD modifications originate mainly from the application of mAbs in genome-wide ChIP experiments. However, constraints for epitope binding of the antibodies are often not well characterized. Usually antibodies are selected for their ability to recognize a specific CTD modification in peptides, but recognition of the same epitope in Pol II prepared from cell extracts can be impaired or inhibited by other modifications occurring next to the epitope in vivo. In this study, we have tested the conditions of epitope recognition for Ser2-P, Ser5-P, Ser7-P as well as for the new Thr4-P mark in CTD. As expected, all CTD-specific mAbs underlie specific restrictions in recognition of their respective epitope. For example, occurrence of Thr4-P in the same repeat together with Ser2-P or Ser5-P inhibits recognition by the Thr4-P-specific mAb, whereas phosphorylation of Tyr1 and Ser7 residues next to Thr4-P was not inhibitory. The restriction of epitope recognition by mAbs has an eminent impact on the interpretation of data generated by ChIP (discussed below), or IP and immunoblot experiments. For the recognition of CTD by an mAb, a single epitope in CTD may be sufficient that fulfils the criteria described above. Hence, the signal strength in immunoblots depends solely on the number of accessible CTD marks and not on the number of marks physically present in CTD. Likewise, absence of a CTD mark can indicate either its physical absence or its masking by other modifications. Currently, we have to deal with these limitations but hopefully, mass spectroscopic analyses of CTD modifications will help to elucidate this complex issue in the future.
Thr4 is the fourth identified amino acid in the CTD heptad repeat, which undergoes modification by phosphorylation. Apparently, a significant portion of the Pol II0 fraction is positive for this mark. Ser2-P-specific antibodies precipitate the fraction of Thr4-P-positive Pol II0 entirely, demonstrating that all Thr4-P-positive Pol II0 is also positive for Ser2-P. Inversely, Thr4-P-specific antibodies precipitated the fraction of Ser2-P marks only incompletely, leaving a substantial fraction of Ser2-P-positive Pol II0 in the supernatant. This indicates that only a subfraction of Ser2-P-positive Pol II0 is target for Thr4 phosphorylation. The Thr4-P-specific mAb also failed to precipitate the fractions of Ser5-P- and Ser7-P-positive Pol II0 quantitatively, confirming previous reports that Ser5-P/Ser-7 and Ser2-P marks are associated with different populations of Pol II0 in cells. Differences in Pol II0 populations were also detected after immunostaining of HeLa cells with antibodies for CTD modifications. The Ser5-P- and Ser7-P-specific mAbs stained accentuated nuclear spots, which were much less apparent after staining with Ser2-P- and Thr4-P-specific antibodies. Taken together, Thr4-P is a new phosphorylation mark in CTD that occurs tightly associated with Ser2-P marks in CTD. In addition, the antibodies described in this study enable us to purify and define different fractions of Pol II0 biochemically.
The Thr4-P mark is strictly associated with the transcriptionally engaged Pol II0 form. To analyse whether the occurrence of this mark is associated with a specific group of genes, or specific positions within transcription units we performed ChIP experiments and compared the occurrence of Thr4-P marks with Ser5-P and Ser2-P marks in CTD. The Ser5-P marks are highly abundant at TSSs and strongly decline in the body of genes. In contrast, Ser2-P marks are absent at TSS, slowly increase in the body of genes and peak close to the poly-A site of genes. This distribution pattern of Ser5-P and Ser2-P marks has been described for genes in low (Kim et al, 2010; Mayer et al, 2010; Tietjen et al, 2010; Bataille et al, 2012) and higher eukaryotes (Rahl et al, 2010; Koch and Andrau, 2011; Koch et al, 2011; Brookes et al, 2012). Likewise, Thr4-P marks show a very striking distribution pattern on genes. They are low or absent at the TSS, remain low in the gene body, but strongly increase downstream of the poly-A site. Interestingly, the profile of Thr4-P increase does not follow exactly the increase of Ser2-P marks, and generally reaches its maximum ∼300 bp downstream of the Ser2-P peaks. This suggests that the increase of Ser2-P marks in CTD might be a prerequisite for the subsequent phosphorylation of Thr4-P. This assumption is consistent with the results of our co-IP experiments for Ser2-P and Thr4-P, and also with the failure of Thr4 phosphorylation of the Ser2/Ala mutant (Supplementary Figure S9).
What could be the function of Thr4-P at the 3′ end of genes? Screens for CTD binding factors have been performed with CTD peptides phosphorylated at Ser2, Ser5, and Ser7 residues, but not with Thr4 phosphorylated peptides. The interaction domains of Pcf11 (Meinhart and Cramer, 2004), Pin1 (Verdecia et al, 2000), Cgt1 (Fabrega et al, 2003), Set2 (Vojnic et al, 2006), and Spt6 (Sun et al, 2010) with Ser5-P and/or Ser2-P CTD peptides have been characterized, while the structure of the Ser7-P binding domain of the Integrator subunit (Egloff et al, 2010) is still unknown. The function of Thr4-P could be two-fold: (i) Thr4-P could act as an inhibitory mark for CTD binding of factors that require the unphosphorylated state of Thr4 or (ii) Thr4-P establishes a new binding platform for new factors.
The Thr4-P mark becomes detectable very late in the transcription cycle still after the appearance of the Ser2-P mark. As discussed above, we cannot discriminate whether Thr4 becomes newly phosphorylated in the 3′ region of genes or whether an unmasking of a pre-existing Thr4-P mark occurs at this position. Regardless of this uncertainty, however, the increased reactivity of the Thr4-P-specific mAb in the 3′ region of genes clearly indicates an alteration in the CTD structure that is associated with Thr4-P. If and how this Thr4-P-associated structural change can be linked to termination or 3′ processing events remains to be elucidated.
Raji or HEK293 cells expressing the Thr4/Ala mutant of Pol II are not viable. A lethal phenotype was also reported for chicken DT40 cells expressing a Thr4/Val mutant of Pol II (Hsin et al, 2011). Analysis of the transcriptome of mutant DT40 cells revealed a specific defect in processing of non-polyadenylated histone mRNAs, while abundance and processing of other mRNAs was not affected. We studied the genome-wide distribution of Pol II in the Thr4/Ala mutant in ChIP experiments. Since cells lose their viability after the switch from the endogenous to the recombinant polymerase, the time window for a genome-wide analysis was narrow and restricted to the first 2 days. Nonetheless, the window was large enough to allow ChIP experiments. IPs were performed with the HA-tag-specific antibody to ensure that only the distribution of the mutant polymerase was analysed. The analysis showed striking differences between the mutant Thr4/Ala and control polymerase Con48. While the levels for both polymerases were high at the TSS, levels for Thr4/Ala markedly declined in the body and 3′ region of genes indicating an elongation defect in the mutant. This defect was in accordance with a local accumulation of polymerases immediately downstream of the initiation site in mutant cells that was not seen in wild-type cells. A similar elongation defect was also observed for various histone genes, indicating no specific defect at these loci as compared with mRNA encoding genes (Supplementary Figure S8) as previously reported for the Thr4/Val mutant in DT40 chicken cells. Whether the difference is based on the nature of the mutants (Thr4/Ala versus Thr4/Val), the different cellular background, or other reasons remains to be elucidated.
While Pol II profiling at most genes is impaired, we observed few genes that become apparently activated in the Thr4/Ala cells and display high levels of Pol II in the body of the gene. This suggests that Thr4 may also be involved in regulation of transcription initiation. The vast majority of genes, however, display an elongation defect. Since Thr4-P signals are not detectable at TSSs or directly downstream thereof in wild-type cells, it remains questionable whether a lack of Thr4 phosphorylation is really causative for the elongation in the Thr4/Ala mutant. It appears also probable that the unmodified Thr4 residue or other modification(s) of Thr4 could be critical for initiation and the early phase of elongation.
We identified Plk3 as a Thr4-specific kinase. None of the cyclin-dependent kinases, including Cdk9 (Cdk12 was not tested), with specificity towards CTD residues Ser2, Ser5, and Ser7 revealed specificity towards Thr4 in vitro. In concordance with the in-vitro assays, Thr4-P levels responded to upregulation and downregulation of Plk3 levels by H2O2 and siRNA in vivo, while phosphorylation levels of other CTD residues did not change significantly. While the activity of other Plk family members (Plk1) is strictly associated with specific phases of mitosis, Plk3 is present in cells probably throughout the entire cell cycle (Bahassi el et al, 2002). Therefore, Plk3 may regulate and coordinate gene activity by Thr4 phosphorylation under physiological conditions but also play a role for adaption of gene activity after stress signals.
We also observed that Thr4-P levels are sensitive to the Cdk9 inhibitor flavopiridol. However, as shown in this work Cdk9 could properly phosphorylate Ser2 in vitro but failed to phosphorylate Thr4. Therefore, the previously reported inhibitory effect of flavopiridol on Thr4 phosphorylation (Hsin et al, 2011) could be indirect and caused by inhibition of Ser2 phosphorylation, if Ser2-P is a prerequisite for priming Thr4 phosphorylation. For reasons discussed above, however, we cannot rule out that phosphorylation of Thr4 by Cdk9 is not detectable by our antibodies.
How is Plk3 recruited to CTD? All Polo-like kinases contain a Polo-box next to the kinase domain that recognizes specific protein motifs. The binding motif for Plk1 is characterized by an Ser, Thr-P, Pro motif (Elia et al, 2003a, 2003b) but binding to an Ser, Ser-P, Pro motif has also been reported (Lowery et al, 2005). The latter motif is present several times in the sequence of mammalian CTD. It will be interesting to study if and how specific CTD hepad repeats are involved in the recruitment of Plk3 to CTD and whether phosphorylation of specific CTD residues is a prerequisite for Plk3 recruitment and Thr4 phosphorylation.
In conclusion, Plk3 is a member of a new class of CTD-specific kinases, which is involved in Thr4 phosphorylation of CTD under physiological but also under stress condition. Plk3 has previously been described as tumour suppressor gene. CTD-Thr4 could be a critical target for the tumour suppressor activity of Plk3, since it has a global impact on gene activity in mammalian cells.
Materials and methods
Cell lines
Raji cells were grown in RPMI-1640 medium supplemented with 10% fetal calf serum (FCS), 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM L-glutamine (Gibco/Invitrogen) at 37°C and 5% CO2. The stable-transfected Raji cell lines were produced and maintained by selection with G418 (Invitrogen) (Chapman et al, 2005). Cell lines were induced to express recombinant Rbp1 through removal of tetracycline. NIH 3T3, mouse embryonic fibroblast (MEF), U2OS, HeLa, and HEK293 cells were incubated in Dulbecco's modified Eagle's medium supplemented with 10% FCS, 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM L-glutamine (DMEM, Gibco) at 37°C and 8% CO2.
Antibodies
mAbs against Rpb1 (POL 3/3), haemagglutinin (HA) tag (3F10, Roche) and the different CTD phosphorylations (3E10, 3E8, 4E12) were used as described previously (Chapman et al, 2007). Affinity purified antibodies against Plk3 (D14F12), α-tubulin (T6557) and (HA) tag (ab9110) were obtained from Cell Signaling, Sigma, and Abcam, respectively. The rat mAbs 6D7 (IgG2b) and 1G7 (IgG2a) against CTD-Thr4-P were generated as described previously (Chapman et al, 2007). For immunization, we used CTD-specific phosphopeptides (YSPTPSPSYSPTSPSC) coupled to ovalbumin (Peptide Specialty Laboratories GmbH, Heidelberg, Germany). The rat mAb 1C7 (IgG2a) recognizes the unphosphorylated CTD peptides (Figure 2). The Pes1 (pescadillo1)-specific antibody (8E9) is described elsewhere (Holzel et al, 2005). Generation of the CTD K7-me-specific antibody will be described elsewhere.
Cloning of CTD mutants
Synthetic CTD constructs were created essentially as described (Chapman et al, 2005), and transferred to a tetracycline-regulated expression vector. In brief, pUC19-CTD modified to contain an Nhe I site in repeat 3 was digested with Nhe I and Sty I, to which synthetic linkers with the sequence for three heptads with, for example, Ser7/Ala mutations were unidirectionally introduced. Isoschizomers of Nhe I (Avr II and Spe I) included in the linkers permit sequential cloning of multimers from (Nhe I–Cla I) back in to the same vector (Spe I–Cla I), leading to a doubling of repeats at each step. Joining of Spe I to Nhe I codes for Thr-Ser, allowing seamless joining and expansion of repeat multimers.
Cell viability assay
For cell viability of Rpb1 mutants, we analysed transiently transfected HEK293 cells expressing the different constructs. Cell proliferation was continuously monitored throughout the experiment with the xCELLigence System (Roche). In all, 3000 HEK293 cells were plated in a final volume of 150 μl DMEM (10% FCS) per well on an E-plate 96. After 24 h, cells were transfected with 0.1 μg plasmid DNA and 0.4 μl FuGENE HD Transfection Reagent (Roche). After a further 24 h, cells were treated with α-amanitin 2 μg/ml (Sigma) to inhibit endogenous Pol II and the cell index (CI) was continuously monitored for 4 days.
Immunofluorescence microscopy
HeLa cells were grown on a coverslip for 24 h with DMEM/10% FCS complete medium. Cells were fixed with 2% paraformaldehyde at room temperature for 5 min and permeabilized with 0.15% Triton X-100 for 15 min at room temperature. All samples were blocked with 1% BSA for 30 min and incubated with the appropriate primary antibody overnight at 4°C. Cells were washed with PBS, 0.15% Triton X-100 for 10 min at room temperature, blocked with 1% BSA for 7 min and incubated with Cy3-conjugated goat anti-rat immunoglobulin (Dianova) and Alexa Fluor 488 goat anti-mouse immunoglobulin (Molecular Probes) in the dark for 45 min. Cells were washed again with PBS containing 0.15% Triton X-100, stained with DAPI (4′, 6-diamidino-2-phenylindole) (Roth), and mounted on slides using fluorescent mounting medium (Dako). Confocal microscopy was performed on a Leica LSCM SP2 fluorescence microscope. Images were taken with objective HCX PL APO × 63 1.4 and processed using Image J 1.37V software.
Immunoprecipitation
1 × 106 HeLa cells were lysed in 200 μl IP buffer (50 mM Tris–HCl, pH 8.0, 150 mM NaCl, 1% NP-40 (Roche), 1 × PhosSTOP (Roche), 1 × protease inhibitor cocktail (Roche) for 20 min on ice. All samples were sonicated on ice using a BRANSON Sonifier 250 (15 s on, 15 s off, 50% duty) and centrifuged at 14 500 r.p.m. for 15 min at 4°C. The supernatant was incubated with antibody-coupled protein G/A-sepharose (1:1) beads (2.5 μg of antibodies for 4 h at 4°C, followed by two washes with 1 ml IP buffer) rotating overnight. Beads were washed several times with 1 ml IP buffer and proteins were boiled off Sepharose beads in laemmli buffer containing 8 M urea for SDS–PAGE.
In-vitro kinase assay
For ELISA, the CTD peptide (YSPTSPS YSPTSPS YSPTSPS YSPTSPSC; Peptide Specialty Laboratories GmbH) was coupled on 96-well maleimide plates for 60 min at 37°C in carbonate buffer at pH 9.5. After washing, the kinase assay was performed using recombinant kinase (100 ng) in 25 μl kinase buffer (20 mM Tris–HCl (pH 7.4), 20 mM NaCl, 10 mM MgCl2, 1 μM DTT and 2 μM ATP) at 28°C for 60 min followed by washing and blocking with PBS/milk (1%) for 30 min. Primary antibodies were added and incubated for 30 min. After an additional washing and blocking step, biotin-coupled secondary antibodies were added for 30 min. Following another washing and blocking step, peroxidase attached to avidin was added to the wells. After washing five times with PBS, 50 μl of substrate buffer (o-phenylenediamine and H2O2; pH 5.0) were added and after the colour change, samples OD were measured at 405 nm in the ELISA reader.
For the kinase assay with the endogenous Pol II as substrate, we immuno-purified Pol II from whole cell extracts with an antibody recognizing unphosphorylated CTD (1C7). In all, 10 μl of the substrate coupled Sepharose G beads were incubated with 40 μl kinase buffer B (50 mM Hepes (pH 7.9), 100 mM KCl, 10 mM MgCl2, 200 μM EGTA, 100 μM EDTA, 1 mM DTT, 200 μM ATP, 1 μg BSA, and 200 ng of the recombinant kinase) at 30°C for 30 min. Specific kinase activity specified by the supplier in pmol/μg × min: Plk3 (131), Cdk9/CycT (26), DNA-Pk (50). Laemmli buffer was added (six-fold) and samples were incubated for 5 min at 95°C followed by western blot analysis.
RNA interference
Synthetic siRNA oligonucleotides (Eurofins/MWG) were double transfected into cells with Oligofectamine (Invitrogen). HeLa cells were seeded at a density of 100 000 cells in 6-well plates the day before transfection. In all, 100 nM siRNA oligonucleotide diluted in 300 μl Opti-MEM (Invitrogen) was incubated with 3 μl Oligofectamine (Invitrogen) for 25 min and then mixed with 600 μl Opti-MEM. Transfection mixture was added to cells for 6 h. The following siRNA sequences were used:
Luciferase: 5′-CGUACGCGGAAUACUUCGATT-3′;
Pes1: 5′-AGGUCUUCCUGUCCAUCAATT-3';
human Plk3-1: 5′-CGGCCUCAUGCGCACAUCCTT-3′;
human Plk3-2: 5′-CUGCGUGACAGUCCCAGACTT-3′.
Western blots
Samples of protein were harvested following treatment using 2 × laemmli buffer. Protein equivalent to 200 000 cells was loaded in 20 μl laemmli, per lane, and subjected to SDS–PAGE on a 6.5% gel before transfer onto nitrocellulose (GE Healthcare). Membranes were either stained with affinity purified, IR-labelled secondary antibodies against rat (680 nm; Alexa, Invitrogen) and mouse (800 nm; Rockford, Biomol), and revealed using the Odyssey (Licor), or stained with HRP-conjugated secondary antibodies against rat (Sigma), mouse (Promega) or rabbit (Promega), and revealed by enhanced chemiluminescence.
Chromatin preparation and ChIPSeq
CTD mutants carry 48 consensus repeats (Con48), or have replaced Thr4 residues in all consensus repeats by alanine (Thr4/Ala) were induced for 24 h and then treated with α-amanitin (2 μg/ml) for 48 h. Wild-type or stable-transfected Raji cells were crosslinked at room temperature with the addition of 1/10th volume of crosslinking solution (11% formaldehyde, 100 mM NaCl, 1 mM EDTA pH 8.0, 0.5 mM EGTA pH 8.0, 50 mM Hepes pH 7.8) in growth medium for 10 min. The reaction was quenched with the addition of 250 mM glycine for 5 min. Cells were washed twice with cold DPBS and 5 × 107 cells/aliquot were pelleted.
Final concentrations of 1 × phosphatase inhibitors, 1 × EDTA-free protease inhibitors (Roche), 0.2 mM PMSF and 1 μg/ml pepstatin were added to all LB buffers. Centrifugations and lysis steps were carried out at 4°C. Each cell aliquot was lysed in 2.5 ml LB1 (50 mM Hepes pH 7.5, 140 mM NaCl, 1 mM EDTA pH 8.0, 10% glycerol, 1% NP-40, 0.5% Triton X-100) for 20 min on a rotating wheel. Chromatin was collected by centrifugation for 5 min at 1350 g and washed in 2.5 ml LB2 (200 mM NaCl, 1 mM EDTA pH 8.0, 0.5 mM EGTA pH 8.0, 10 mM Tris pH 8.0) for 20 min. Chromatin was collected, resuspended in 1.5 ml of LB3 (1 mM EDTA pH 8.0, 0.5 mM EGTA pH 8, 10 mM Tris pH 8.0, 100 mM NaCl, 0.1% Na-Deoxycholate, 0.5% N-lauroylsarcosine) and sonicated using a Misonix 4000 (Misonix Inc.) sonicator for 14 cycles (30 s on, 30 s off, amplitude 40). After the addition of Triton X-100 to a final concentration of 1%, particulates were collected through centrifugation at 20 000 g for 20 min a 50-μl aliquot was taken to serve as input control. The input was combined with an equal volume of 2 × elution buffer (100 mM Tris pH 8.0, 20 mM EDTA pH 8.0, 2% SDS) and incubated overnight at 65°C for 13–15 h. SDS was diluted by the addition of an equal volume of TE (10 mM Tris pH 8.0, 1 mM EDTA pH 8.0), followed by 2 h of RNase A (0.2 μg/ml) and 2 h of Proteinase K (0.2 μg/ml) digestions at 37 and 55°C, respectively. DNA was purified by two subsequent phenol:chloroform:isoamylalcohol (25:24:1, pH 8) extractions, followed by a PCR purification column (Qiagen). DNA was eluted using 50 μl H2O and the concentration was measured on a Nanodrop 1000 (Thermo Scientific). To verify DNA fragmentation, 500 ng was run on a 2% agarose gel. ChIPseq and data processing are described in detail in Supplementary data.
Accession codes
All Chipseq data are deposited at Gene Expression Omnibus: GSE37519.
Supplementary Material
Acknowledgments
This work is supported by Deutsche Forschungsgemeinschaft Grants SFB-Transregio5 and SFB684 to DE. Work in the PF laboratory is supported by institutional grants from Institut National de la Santé et de la Recherche Médicale and the Centre National de la Recherche Scientifique (CNRS), and by specific grants from the Fondation Princesse Grace de Monaco, the Agence Nationale de la Recherche (ANR), the Institut National du Cancer (INCa) and the Commission of the European Communities. FK was supported by grants from ‘Chromatin Plasticity’ Marie Curie Research Training Network (MRTN-CT-2006-0357733) Association pour la Recherche sur le Cancer and Agence Nationale de la recherche (ANR ‘ChromaTin’), RF by Genopole and CNRS, and PC by grants from INCa and Fondation pour la Recherche Médicale. The work was also supported by a Regulome grant from the ANR. The work was also granted for sequencing costs by an ESGI consortium grant of the EU to MG and IG. We thank J Manley for sharing data before publication.
Author contributions: CH, MH, J-CA, and DE conceived the project and designed experiments. CH, MH, FK, and RDC conducted and analysed the wet-lab experiments. FK, IG, MG, J-CA, and PF conceived the framework for the ChIP-seq studies. CH, FK, J-CA, and DE designed the ChIP-seq experiments. MG and IG conducted all ChIP-seq experiments. ND and RF carried out the bioinformatics of processing of ChIP-seq data. AF and EK produced monoclonal antibodies, MH, CH, and AF characterized specificity of antibodies. DE, CH, and J-CA wrote the manuscript. All authors reviewed the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ahn SH, Kim M, Buratowski S (2004) Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol Cell 13: 67–76 [DOI] [PubMed] [Google Scholar]
- Akhtar MS, Heidemann M, Tietjen JR, Zhang DW, Chapman RD, Eick D, Ansari AZ (2009) TFIIH kinase places bivalent marks on the carboxy-terminal domain of RNA polymerase II. Mol Cell 34: 387–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahassi el M, Conn CW, Myer DL, Hennigan RF, McGowan CH, Sanchez Y, Stambrook PJ (2002) Mammalian Polo-like kinase 3 (Plk3) is a multifunctional protein involved in stress response pathways. Oncogene 21: 6633–6640 [DOI] [PubMed] [Google Scholar]
- Bartkowiak B, Liu P, Phatnani HP, Fuda NJ, Cooper JJ, Price DH, Adelman K, Lis JT, Greenleaf AL (2010) CDK12 is a transcription elongation-associated CTD kinase, the metazoan ortholog of yeast Ctk1. Genes Dev 24: 2303–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bataille AR, Jeronimo C, Jacques PE, Laramee L, Fortin ME, Forest A, Bergeron M, Hanes SD, Robert F (2012) A universal RNA polymerase II CTD cycle is orchestrated by complex interplays between kinase, phosphatase, and isomerase enzymes along genes. Mol Cell 45: 158–170 [DOI] [PubMed] [Google Scholar]
- Blazek D, Kohoutek J, Bartholomeeusen K, Johansen E, Hulinkova P, Luo Z, Cimermancic P, Ule J, Peterlin BM (2011) The cyclin K/Cdk12 complex maintains genomic stability via regulation of expression of DNA damage response genes. Genes Dev 25: 2158–2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeing S, Rigault C, Heidemann M, Eick D, Meisterernst M (2010) RNA polymerase II C-terminal heptarepeat domain Ser-7 phosphorylation is established in a mediator-dependent fashion. J Biol Chem 285: 188–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookes E, de Santiago I, Hebenstreit D, Morris KJ, Carroll T, Xie SQ, Stock JK, Heidemann M, Eick D, Nozaki N, Kimura H, Ragoussis J, Teichmann SA, Pombo A (2012) Polycomb associates genome-wide with a specific RNA polymerase II variant, and regulates metabolic genes in ESCs. Cell Stem Cell 10: 157–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S (2009) Progression through the RNA polymerase II CTD cycle. Mol Cell 36: 541–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman RD, Conrad M, Eick D (2005) Role of the mammalian RNA polymerase II C-terminal domain (CTD) nonconsensus repeats in CTD stability and cell proliferation. Mol Cell Biol 25: 7665–7674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman RD, Heidemann M, Albert TK, Mailhammer R, Flatley A, Meisterernst M, Kremmer E, Eick D (2007) Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science 318: 1780–1782 [DOI] [PubMed] [Google Scholar]
- Chapman RD, Heidemann M, Hintermair C, Eick D (2008) Molecular evolution of the RNA polymerase II CTD. Trends Genet 24: 289–296 [DOI] [PubMed] [Google Scholar]
- Chapman RD, Palancade B, Lang A, Bensaude O, Eick D (2004) The last CTD repeat of the mammalian RNA polymerase II large subunit is important for its stability. Nucleic Acids Res 32: 35–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemente-Blanco A, Sen N, Mayan-Santos M, Sacristan MP, Graham B, Jarmuz A, Giess A, Webb E, Game L, Eick D, Bueno A, Merkenschlager M, Aragon L (2011) Cdc14 phosphatase promotes segregation of telomeres through repression of RNA polymerase II transcription. Nat Cell Biol 13: 1450–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corden JL (2007) Transcription. Seven ups the code. Science 318: 1735–1736 [DOI] [PubMed] [Google Scholar]
- Coudreuse D, van Bakel H, Dewez M, Soutourina J, Parnell T, Vandenhaute J, Cairns B, Werner M, Hermand D (2010) A gene-specific requirement of RNA polymerase II CTD phosphorylation for sexual differentiation in S. pombe. Curr Biol 20: 1053–1064 [DOI] [PubMed] [Google Scholar]
- Cowling VH (2010) Regulation of mRNA cap methylation. Biochem J 425: 295–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Custodio N, Vivo M, Antoniou M, Carmo-Fonseca M (2007) Splicing- and cleavage-independent requirement of RNA polymerase II CTD for mRNA release from the transcription site. J Cell Biol 179: 199–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David CJ, Boyne AR, Millhouse SR, Manley JL (2011) The RNA polymerase II C-terminal domain promotes splicing activation through recruitment of a U2AF65-Prp19 complex. Genes Dev 25: 972–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida SF, Grosso AR, Koch F, Fenouil R, Carvalho S, Andrade J, Levezinho H, Gut M, Eick D, Gut I, Andrau JC, Ferrier P, Carmo-Fonseca M (2011) Splicing enhances recruitment of methyltransferase HYPB/Setd2 and methylation of histone H3 Lys36. Nat Struct Mol Biol 18: 977–983 [DOI] [PubMed] [Google Scholar]
- de la Mata M, Kornblihtt AR (2006) RNA polymerase II C-terminal domain mediates regulation of alternative splicing by SRp20. Nat Struct Mol Biol 13: 973–980 [DOI] [PubMed] [Google Scholar]
- Egloff S, Murphy S (2008) Cracking the RNA polymerase II CTD code. Trends Genet 24: 280–288 [DOI] [PubMed] [Google Scholar]
- Egloff S, O'Reilly D, Chapman RD, Taylor A, Tanzhaus K, Pitts L, Eick D, Murphy S (2007) Serine-7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science 318: 1777–1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff S, Szczepaniak SA, Dienstbier M, Taylor A, Knight S, Murphy S (2010) The integrator complex recognizes a new double mark on the RNA polymerase II carboxyl-terminal domain. J Biol Chem 285: 20564–20569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff S, Zaborowska J, Laitem C, Kiss T, Murphy S (2012) Ser7 phosphorylation of the CTD recruits the RPAP2 Ser5 phosphatase to snRNA genes. Mol Cell 45: 111–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia AE, Cantley LC, Yaffe MB (2003a) Proteomic screen finds pSer/pThr-binding domain localizing Plk1 to mitotic substrates. Science 299: 1228–1231 [DOI] [PubMed] [Google Scholar]
- Elia AE, Rellos P, Haire LF, Chao JW, Ivins FJ, Hoepker K, Mohammad D, Cantley LC, Smerdon SJ, Yaffe MB (2003b) The molecular basis for phosphodependent substrate targeting and regulation of Plks by the Polo-box domain. Cell 115: 83–95 [DOI] [PubMed] [Google Scholar]
- Fabrega C, Shen V, Shuman S, Lima CD (2003) Structure of an mRNA capping enzyme bound to the phosphorylated carboxy-terminal domain of RNA polymerase II. Mol Cell 11: 1549–1561 [DOI] [PubMed] [Google Scholar]
- Gerber HP, Hagmann M, Seipel K, Georgiev O, West MA, Litingtung Y, Schaffner W, Corden JL (1995) RNA polymerase II C-terminal domain required for enhancer-driven transcription. Nature 374: 660–662 [DOI] [PubMed] [Google Scholar]
- Ghosh A, Shuman S, Lima CD (2011) Structural insights to how mammalian capping enzyme reads the CTD code. Mol Cell 43: 299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover-Cutter K, Larochelle S, Erickson B, Zhang C, Shokat K, Fisher RP, Bentley DL (2009) TFIIH-associated Cdk7 kinase functions in phosphorylation of C-terminal domain Ser7 residues, promoter-proximal pausing, and termination by RNA polymerase II. Mol Cell Biol 29: 5455–5464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes NP, Bjerke G, Llorente B, Szostek SA, Emerson BM, Espinosa JM (2006) Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes Dev 20: 601–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govind CK, Qiu H, Ginsburg DS, Ruan C, Hofmeyer K, Hu C, Swaminathan V, Workman JL, Li B, Hinnebusch AG (2010) Phosphorylated Pol II CTD recruits multiple HDACs, including Rpd3C(S), for methylation-dependent deacetylation of ORF nucleosomes. Mol Cell 39: 234–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzel M, Rohrmoser M, Schlee M, Grimm T, Harasim T, Malamoussi A, Gruber-Eber A, Kremmer E, Hiddemann W, Bornkamm GW, Eick D (2005) Mammalian WDR12 is a novel member of the Pes1-Bop1 complex and is required for ribosome biogenesis and cell proliferation. J Cell Biol 170: 367–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SW, Hong SM, Yoo JW, Lee YC, Kim S, Lis JT, Lee DK (2009) Phosphorylation of the RNA polymerase II C-terminal domain by TFIIH kinase is not essential for transcription of Saccharomyces cerevisiae genome. Proc Natl Acad Sci USA 106: 14276–14280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsin JP, Sheth A, Manley JL (2011) RNAP II CTD phosphorylated on threonine-4 is required for histone mRNA 3′ end processing. Science 334: 683–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SA, Kim H, Erickson B, Bentley DL (2011) The export factor Yra1 modulates mRNA 3' end processing. Nat Struct Mol Biol 18: 1164–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JC, Phatnani HP, Haystead TA, MacDonald JA, Alam SM, Greenleaf AL (2004) C-terminal repeat domain kinase I phosphorylates Ser2 and Ser5 of RNA polymerase II C-terminal domain repeats. J Biol Chem 279: 24957–24964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanin EI, Kipp RT, Kung C, Slattery M, Viale A, Hahn S, Shokat KM, Ansari AZ (2007) Chemical inhibition of the TFIIH-associated kinase Cdk7/Kin28 does not impair global mRNA synthesis. Proc Natl Acad Sci USA 104: 5812–5817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keogh MC, Podolny V, Buratowski S (2003) Bur1 kinase is required for efficient transcription elongation by RNA polymerase II. Mol Cell Biol 23: 7005–7018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Erickson B, Luo W, Seward D, Graber JH, Pollock DD, Megee PC, Bentley DL (2010) Gene-specific RNA polymerase II phosphorylation and the CTD code. Nat Struct Mol Biol 17: 1279–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Suh H, Cho EJ, Buratowski S (2009) Phosphorylation of the yeast Rpb1 C-terminal domain at serines 2, 5, and 7. J Biol Chem 284: 26421–26426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch F, Andrau JC (2011) Initiating RNA Polymerase II and TIPs as hallmarks of enhancer activity and tissue-specificity. Transcription 2: 263–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch F, Fenouil R, Gut M, Cauchy P, Albert TK, Zacarias-Cabeza J, Spicuglia S, de la Chapelle AL, Heidemann M, Hintermair C, Eick D, Gut I, Ferrier P, Andrau JC (2011) Transcription initiation platforms and GTF recruitment at tissue-specific enhancers and promoters. Nat Struct Mol Biol 18: 956–963 [DOI] [PubMed] [Google Scholar]
- Krishnamurthy S, He X, Reyes-Reyes M, Moore C, Hampsey M (2004) Ssu72 Is an RNA polymerase II CTD phosphatase. Mol Cell 14: 387–394 [DOI] [PubMed] [Google Scholar]
- Lowery DM, Lim D, Yaffe MB (2005) Structure and function of Polo-like kinases. Oncogene 24: 248–259 [DOI] [PubMed] [Google Scholar]
- MacKellar AL, Greenleaf AL (2011) Cotranscriptional association of mRNA export factor Yra1 with C-terminal domain of RNA polymerase II. J Biol Chem 286: 36385–36395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Max T, Sogaard M, Svejstrup JQ (2007) Hyperphosphorylation of the C-terminal repeat domain of RNA polymerase II facilitates dissociation of its complex with mediator. J Biol Chem 282: 14113–14120 [DOI] [PubMed] [Google Scholar]
- Mayer A, Lidschreiber M, Siebert M, Leike K, Soding J, Cramer P (2010) Uniform transitions of the general RNA polymerase II transcription complex. Nat Struct Mol Biol 17: 1272–1278 [DOI] [PubMed] [Google Scholar]
- Meinhart A, Cramer P (2004) Recognition of RNA polymerase II carboxy-terminal domain by 3'-RNA-processing factors. Nature 430: 223–226 [DOI] [PubMed] [Google Scholar]
- Meininghaus M, Chapman RD, Horndasch M, Eick D (2000) Conditional expression of RNA polymerase II in mammalian cells. Deletion of the carboxyl-terminal domain of the large subunit affects early steps in transcription. J Biol Chem 275: 24375–24382 [DOI] [PubMed] [Google Scholar]
- Meininghaus M, Eick D (1999) Requirement of the carboxy-terminal domain of RNA polymerase II for the transcriptional activation of chromosomal c-fos and hsp70A genes. FEBS Lett 446: 173–176 [DOI] [PubMed] [Google Scholar]
- Mosley AL, Pattenden SG, Carey M, Venkatesh S, Gilmore JM, Florens L, Workman JL, Washburn MP (2009) Rtr1 is a CTD phosphatase that regulates RNA polymerase II during the transition from serine 5 to serine 2 phosphorylation. Mol Cell 34: 168–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz MJ, de la Mata M, Kornblihtt AR (2010) The carboxy terminal domain of RNA polymerase II and alternative splicing. Trends Biochem Sci 35: 497–504 [DOI] [PubMed] [Google Scholar]
- Perales R, Bentley D (2009) ‘Cotranscriptionality’: the transcription elongation complex as a nexus for nuclear transactions. Mol Cell 36: 178–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phatnani HP, Greenleaf AL (2006) Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev 20: 2922–2936 [DOI] [PubMed] [Google Scholar]
- Qiu H, Hu C, Hinnebusch AG (2009) Phosphorylation of the Pol II CTD by KIN28 enhances BUR1/BUR2 recruitment and Ser2 CTD phosphorylation near promoters. Mol Cell 33: 752–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA (2010) c-Myc regulates transcriptional pause release. Cell 141: 432–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B, Shuman S (2011) Deciphering the RNA polymerase II CTD code in fission yeast. Mol Cell 43: 311–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ 3rd, Rojas LA, Beck D, Bonasio R, Schuller R, Drury WJ 3rd, Eick D, Reinberg D (2011) The C-terminal domain of RNA polymerase II is modified by site-specific methylation. Science 332: 99–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N, Ma Z, Gemmill T, Wu X, Defiglio H, Rossettini A, Rabeler C, Beane O, Morse RH, Palumbo MJ, Hanes SD (2009) The Ess1 prolyl isomerase is required for transcription termination of small noncoding RNAs via the Nrd1 pathway. Mol Cell 36: 255–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spain MM, Govind CK (2011) A role for phosphorylated Pol II CTD in modulating transcription coupled histone dynamics. Transcription 2: 78–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiller JW, McConaughy BL, Hall BD (2000) Evolutionary complementation for polymerase II CTD function. Yeast 16: 57–64 [DOI] [PubMed] [Google Scholar]
- Sun M, Lariviere L, Dengl S, Mayer A, Cramer P (2010) A tandem SH2 domain in transcription elongation factor Spt6 binds the phosphorylated RNA polymerase II C-terminal repeat domain (CTD). J Biol Chem 285: 41597–41603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tietjen JR, Zhang DW, Rodriguez-Molina JB, White BE, Akhtar MS, Heidemann M, Li X, Chapman RD, Shokat K, Keles S, Eick D, Ansari AZ (2010) Chemical-genomic dissection of the CTD code. Nat Struct Mol Biol 17: 1154–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdecia MA, Huang H, Dutil E, Kaiser DA, Hunter T, Noel JP (2000) Structure of the human anti-apoptotic protein survivin reveals a dimeric arrangement. Nat Struct Biol 7: 602–608 [DOI] [PubMed] [Google Scholar]
- Viladevall L St, Amour CV, Rosebrock A, Schneider S, Zhang C, Allen JJ, Shokat KM, Schwer B, Leatherwood JK, Fisher RP (2009) TFIIH and P-TEFb coordinate transcription with capping enzyme recruitment at specific genes in fission yeast. Mol Cell 33: 738–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojnic E, Simon B, Strahl BD, Sattler M, Cramer P (2006) Structure and carboxyl-terminal domain (CTD) binding of the Set2 SRI domain that couples histone H3 Lys36 methylation to transcription. J Biol Chem 281: 13–15 [DOI] [PubMed] [Google Scholar]
- Wang L, Gao J, Dai W, Lu L (2008) Activation of Polo-like kinase 3 by hypoxic stresses. J Biol Chem 283: 25928–25935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner-Allen JW, Lee CJ, Liu P, Nicely NI, Wang S, Greenleaf AL, Zhou P (2011) cis-Proline-mediated Ser(P)5 dephosphorylation by the RNA polymerase II C-terminal domain phosphatase Ssu72. J Biol Chem 286: 5717–5726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Bai J, Shen R, Brown SA, Komissarova E, Huang Y, Jiang N, Alberts GF, Costa M, Lu L, Winkles JA, Dai W (2008) Polo-like kinase 3 functions as a tumor suppressor and is a negative regulator of hypoxia-inducible factor-1 alpha under hypoxic conditions. Cancer Res 68: 4077–4085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo M, Lee SK, Lee B, Ruiz EC, Pfaff SL, Gill GN (2005) Small CTD phosphatases function in silencing neuronal gene expression. Science 307: 596–600 [DOI] [PubMed] [Google Scholar]
- Zhang DW, Mosley AL, Ramisetty SR, Rodriguez-Molina JB, Washburn MP, Ansari AZ (2012) Ssu72 phosphatase dependent erasure of phospho-Ser7 marks on the RNA polymerase II C-terminal domain is essential for viability and transcription termination. J Biol Chem 287: 8541–8551 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.