Abstract
Wnt genes and components of Wnt signalling pathways have been implicated in a wide spectrum of important biological phenomena, ranging from early organismal development to cell behaviours to several diseases, especially cancers. Emergence of the field of Wnt signalling can be largely traced back to the discovery of the first mammalian Wnt gene in 1982. In this essay, we mark the thirtieth anniversary of that discovery by describing some of the critical scientific developments that led to the flowering of this field of research.
Keywords: oncogene, signal transduction, tumour formation, Wnts
Introduction: how we learn about what we know
Knowledge differs from the growth of knowledge. To learn the facts about a subject, such as Wnt genes or the Wnt signalling pathway, one can consult an encyclopaedia, a textbook, or a conventional review article. To understand how those facts were unearthed and assembled into coherent concepts, it is necessary to probe the history of a field—to learn about the sequence of events, the logical and illogical connections between those events, and the people who participated in them.
We have approached this essay with more attention to historical development than to a full repertoire of facts. While interesting experimental results about Wnt genes and their effects on cells and organisms continue to appear at an accelerating pace, we believe that there is a great deal to learn about the scientific enterprise more broadly by looking back on the unusual way in which the knowledge about those genes has grown over the past three decades. Some understanding of Wnt signalling is now required of those who aspire to succeed in many prominent fields of biology—including organismal development, cancer research, and stem-cell biology. There is also a sizeable subset of biologists who define themselves primarily as students of Wnt genes, while also aligning themselves with cancer, developmental, or stem-cell biologists or with communities devoted to the fruit fly, worm, amphibians, mouse, or Homo sapiens. This cannot be said of those working on many other genes or signalling pathways, raising the interesting questions of how and why scientists organize themselves in unusual ways and view their subjects through certain kinds of lenses.
We have composed this essay on the occasion of the thirtieth anniversary of the published report that announced our discovery of what proved to be the first mammalian Wnt gene (Nusse and Varmus, 1982). Taking that report as an arbitrary starting point in the history of this field, we have tried to highlight the most significant ways in which the field increased in knowledge, enlarged in scope, and grew in disciples. As we emphasize, some of these advances were logical and straightforward, others were technically difficult and protracted, and yet others were serendipitous and surprising. We also note more briefly how the shape of the field was determined by certain beneficial attitudes and behaviours that may be worthy of emulation.
The ‘pre-history’: mouse models for breast cancer and cancer-causing retroviruses preceded knowledge of Wnt genes
All modern science is built on earlier science. Accordingly, the discovery that launched the intense study of Wnt genes 30 years ago depended on at least two earlier and closely related lines of enquiry: mouse models of cancer and oncogenic retroviruses.
It had been known since the 1930s that certain strains of laboratory mice are highly susceptible to breast cancer, and that the disease is usually transmitted from mothers to offspring mice through the milk (Bittner, 1936; Korteweg, 1936). Later, the tumour-inducing activity was purified from the milk (Lyons and Moore, 1962), and the milk-transmitted factor was shown to be a morphologically atypical retrovirus, called the Mouse Mammary Tumour Virus or MMTV.
Although the study of oncogenic retroviruses can be tracked to the first decade of the 20th century, the basis of their cancer-causing properties came into focus only in the century’s second half. The first great advances came from tissue culture assays for viral growth and cell transforming capacities and from the biochemical and genetic analysis of the RNA genomes of retroviruses isolated from chickens, mice, rats, and other experimental animals. These methods led to the discovery of distinct viral oncogenes, such as Src, Myc, and Ras, and their cellular precursors, called proto-oncogenes, a term denoting any cellular genes that could be converted to active cancer-causing genes (Bishop and Varmus, 1985). Conversion to oncogenicity could occur by the mechanisms that produced highly oncogenic retroviruses or, as was shown in time and described later in this essay, by a variety of other mechanisms, most commonly somatic mutations of several types.
The proto-oncogenes initially discovered by tracing viral oncogenes to their cellular origins were generally not related to each other, but they shared several properties. They had been conserved during evolution and were converted to cancer-causing genes by gain-of-function mutations—as first shown for retroviral oncogenes and later for activated cellular oncogenes found in human cancers. The rapid onset of cancer caused by many retroviruses reflected the ability of active viral oncogenes to transform many infected cells (Bishop and Varmus, 1985).
Before RN came to UCSF in 1980 to work with HV, we had both been interested in breast cancer in the mouse and in the general properties of retroviruses isolated from animals. Such viruses often cause haematological cancers and sarcomas, relatively infrequent types of cancers in human beings; only rarely do they induce epithelial carcinomas, the most common human cancers. So, we were curious about the mechanism by which MMTV might cause carcinoma of the breast. Unlike the most intensively studied cancer-causing retroviruses, however, there was no readily identifiable oncogene in the viral genome. But, like other retroviruses, MMTV was known to insert a DNA copy of its RNA genome into the host cell genome during infection. Moreover, MMTV caused cancer slowly, over the course of several months, unlike oncogene-containing retroviruses, suggesting that viral infection alone, while required for tumour induction, was not sufficient to transform a host cell—a property that had also been recognized in the study of certain leukaemia-inducing retroviruses found in birds and mice (Teich et al, 1982).
It was known, however, that tumours induced by such viruses were composed of cell clones defined by shared proviral integration sites (Cohen et al, 1979; Payne et al, 1981) implying that tumours consisted of the descendants of a single infected cell and were thus the outcome of rare events. The pattern of clonality raised the possibility that one of many infected cells had randomly acquired a provirus that could initiate tumourigenesis. For instance, the insertion of viral DNA might cause a mutation of a gene in the vicinity of the integration site, and the change might confer a growth advantage to that cell. While it was possible that such mutations caused loss-of-function mutations, by disrupting a host cell gene, an alternative and more attractive model was that a host cell gene was transcriptionally activated by the incoming provirus. The latter model was inspired by findings in yeast and prokaryotes, where transposable elements, which are often functionally and structurally similar to retroviral proviruses (Brown and Varmus, 1989), were known to activate residing host cell genes (Errede et al, 1980).
The model of gene activation by proviral insertion gained momentum when Bill Hayward’s group showed that B-cell lymphomas caused by the slowly oncogenic Avian Leukosis Virus (ALV) commonly contained proviruses inserted near the c-myc gene and that c-myc was overexpressed in those tumours (Hayward et al, 1981). Initially, activation of c-myc appeared to be the consequence of a transcriptional promoter present in an ALV provirus inserted upstream of the protein-coding exons of c-myc in the same transcriptional orientation (Hayward et al, 1981), but research in HV’s group showed that proviruses could also enhance c-myc expression from the endogenous c-myc promoter when inserted downstream of the gene or upstream in either orientation (Payne et al, 1982).
The discoveries concerning c-myc and ALV were of critical importance in cancer research, as it was the first time that a cellular homologue of a viral oncogene was shown to be activated in cancer by mutation. Soon thereafter, many kinds of mutations other than viral insertion mutations—gene amplifications, chromosomal translocations, and point mutations—were shown to activate c-myc and many other progenitors of retroviral oncogenes, in human as well as in animal tumours, without the intervention of retroviruses (Weinberg, 1983). The results with ALV and c-myc also set the stage for looking for a similar mechanism in tumours caused by other retroviruses, such as MMTV, with the prospect of finding proto-oncogenes that are not related to known viral oncogenes.
Molecular cloning of int1, the first novel proto-oncogene identified by proviral tagging
To seek host cell genes that are activated by insertions of MMTV proviruses, we decided to do a systematic screen rather than to look for activation of only those proto-oncogenes known at the time, such as c-myc. We assumed, perhaps naively, that only one or perhaps a few genes in the mouse genome would confer tumourigenic growth when activated by MMTV in mammary cancer. This assumption implied that by comparing MMTV proviruses in multiple different tumours we would find common integration sites, or at least common regions, near those unusual genes. This segment of the genome would represent a so-called ‘common integration site.’ But the commonality would not reflect a predisposition to integrate MMTV at preferred locations in the mouse genome—all data, then and now, suggest that retroviral integration occurs quasi-randomly in host genomes—but instead would result from selection of cells that acquired a growth advantage when a provirus activated a nearby proto-oncogene.
The goal then became to isolate an integrated MMTV provirus plus its adjacent host cell DNA from a tumour by molecular cloning. We reasoned that we should start from a tumour with just one MMTV insertion. Most tumours in the strain we were using (C3H) carried multiple newly acquired proviruses, in addition to a few copies of inherited ‘endogenous’ MMTV DNA that were invariant among the arising tumours because they were products of ancient infections of the mouse germ line (Bentvelzen et al, 1970; Cohen and Varmus, 1979). Since it was difficult to know which of the multiple new proviruses would be functionally important, we looked for tumours carrying only one new provirus; this single insert should, according to the model, be near the relevant gene. Restriction fragments of host DNA derived from the vicinity of the provirus from that tumour would then be used as probes for two purposes: to screen other tumours for integrations in the same domain and to measure transcription of local genes, both in tumours and in normal mammary tissue. Of course, today, with complete sequences of normal mouse genomes and powerful methods for sequencing tumour DNA in hand, precise mapping of insertion sites has become a relatively simple exercise. But at the time, these tools were not available, so a more cumbersome method was required.
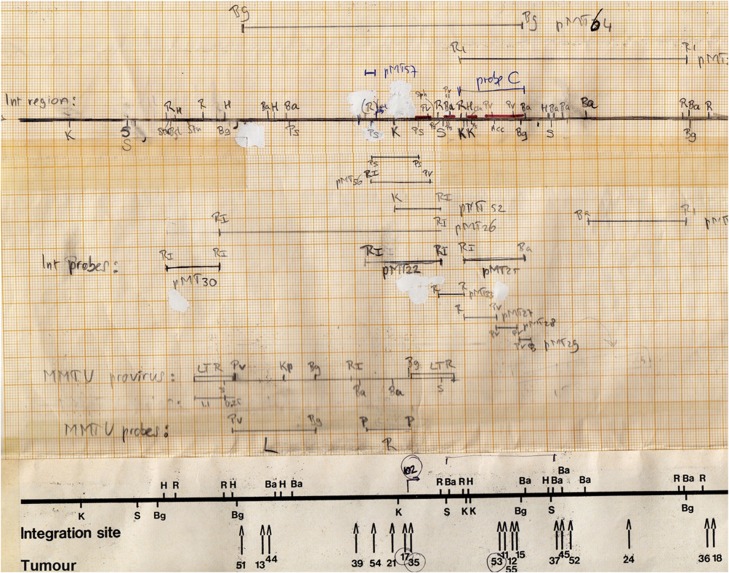
We initiated the screen in October 1980, gathering over 30 MMTV-infected C3H mice with mammary tumours, and we found just one tumour (#18) with a single new provirus. Happily, that single tumour proved to suffice for our purposes. From the genome of this tumour, we cloned a ‘junctional’ fragment, containing a part of an MMTV provirus joined to adjacent chromosomal DNA, then derived smaller fragments containing only host cell DNA. With those fragments as probes, we ‘walked’ along the chromosome in either direction from the original proviral insertion site to obtain more probes for a broader region of the mouse genome (Figure 1). These were then used to examine DNA from other tumours for disruptions caused by MMTV proviruses. Indeed, many tumours in our collection had evidence for MMTV DNA insertions in the region occupied by the single provirus in tumour #18; these were manifested by novel restriction fragments that hybridized with both MMTV and host cell probes. We assembled a map of the relevant region of the mouse genome, with the various proviral insertions in different tumours scattered throughout a region spanning ∼30 KB (Figure 1). It turned out that the original insertion site we cloned from tumour #18 was at one end of the cluster.
Figure 1.
A working map of the mouse int1 locus as drawn by RN and used from 1982 to 1984, with the position of various cloned genomic restriction fragments. Red lines indicate the presence of int1 exons, mapped in 1984. Note the location of Probe C, a genomic fragment hybridizing with int1 mRNA in mouse mammary tumours. At the bottom, the position of MMTV proviruses mapped in different tumours, with the position of the provirus in tumour #18, the starting point of the cloning of the locus, indicated at the right hand end.
But where was the cellular gene that, according to our hypothesis, should have been activated as a result of these integrations? One of the probes (probe C, Figure 1) proved to be particularly helpful. It picked up integration events in many individual tumours and mapped approximately to the middle of the cluster of integrations. More importantly, the same probe also detected a tumour-specific mRNA—that is, a species not found in normal mammary gland tissue—on northern blots. This was the evidence we were looking for—a host gene, a putative proto-oncogene, had been transcriptionally activated by MMTV proviruses in multiple, independent tumours (Nusse and Varmus, 1982). We called the gene int1 (to denote the first common integration site) and promptly submitted a manuscript that was published in the journal Cell in November 1982 (Nusse and Varmus, 1982). The method we used, called proviral tagging, is now widely used to discover proto-oncogenes in cancers induced in mice by retroviruses or transposable elements and in cancers whose growth is accelerated in transgenic animals by these elements (Kool and Berns, 2009; Copeland and Jenkins, 2010).
The paper describing int1 was well received, but its impact at that time was overshadowed by an avalanche of incredibly exciting developments in cancer research. In the same year, 1982, human cancers were found to have mutations in cellular RAS genes, and other human tumours were shown to contain chromosome translocations directly affecting c-myc (Bishop, 1983). Soon thereafter, equally explosive findings were announced, identifying oncogenic proteins as known factors governing growth control: the erbB oncogene was discovered to be derived from the EGF receptor gene, and both encoded protein-tyrosine kinases; and the precursor to the v-sis oncogene was shown to be the gene encoding PDGF, a secreted growth factor (Bishop and Varmus, 1985).
The difficulties of defining the mechanism of action of the int1 gene and its encoded protein
In the midst of all of the excitement about retroviral oncogenes and their progenitors, int1 attracted relatively little attention beyond the confines of our laboratories, despite the many open questions that seemed important and interesting to us. Initially, in the absence of a nucleotide sequence, we had no clue how the gene would function. Work done in both our laboratories (HV at UCSF; and RN after his return to the Netherlands Cancer Institute in Amsterdam in 1982) elucidated the structure and sequence of the int1 gene (van Ooyen and Nusse, 1984) and its cDNA (Fung et al, 1985), revealing no homology with any other gene or protein known at the time. We did notice, however, that the predicted protein sequence started with a signal sequence, indicating that the int1 protein would be secreted. This opened up the exciting possibility that this protein might be an extracellular growth factor. But direct proof of this prediction turned out to be very hard to obtain. For many years, no one was able to produce or isolate significant quantities of the int1 protein, a problem that was not solved until 2003 (see below). To make matters worse, generating useful antibodies to int1 was an equally frustrating enterprise; in fact, detecting int1 protein in cells or tissues remains an elusive goal even today. These problems, in particular the lack of active protein for experimental use, precluded conventional signalling assays in cell culture. As a result, indirect assays, such as those dependent on gene transfer, had to be used to study signalling events in cells expressing int1. In particular, the identification of specific int1 cell surface receptors by binding assays was not possible until much later.
On the other hand, we did establish that expression of the int1 gene could affect cell behaviour in a fashion that resembled conventional transformation and provided a biological assay: various mammary epithelial cell lines could be morphologically altered by overexpression of int1, albeit in a subtle way and rarely leading to formation of cells capable of growing into a tumour (Brown et al, 1986; Rijsewijk et al, 1987b). More dramatically, Ann Tsukamoto in HV’s laboratory was able to recapitulate the oncogenic effect of int1 in mice without resorting to virus infection: mice expressing an int1 transgene under the influence of an MMTV transcriptional regulator developed cancer in the mammary gland within about 6 months of age (Tsukamoto et al, 1988). This established that int1 is a bona fide proto-oncogene. These mice—and several others expressing int1 under the control of inducible promoters—have since become widely used mouse models for studying breast carcinogenesis and for finding genes that can cooperate with the int1 transgene during oncogenesis (MacArthur et al, 1995).
Developmental genetics helped to reveal the function of int1 when the gene was discovered to be the homologue of the Drosophila segment polarity gene, Wingless
How could the function of int1 protein and the components of its signalling pathway be deciphered without a direct, convenient biochemical or cell-based assay? Fortunately, int1 did have one advantage: a high degree of conservation across species. The human int1 protein sequence turned out to be almost completely (99%) identical to that of the mouse homologue (Van Ooyen et al, 1985). Moreover, int1-related sequences appeared to be present in the DNA of Drosophila melanogaster as judged from molecular hybridization (Nusse et al, 1984).
At the time, the new molecular methods that were responsible for unveiling the genes central to cancer research had also re-energized efforts to understand embryogenesis, using the rich treasury of Drosophila developmental mutants. During the 1970s, genetic screens in Drosophila had unveiled a set of genes that were essential for the development of the embryo. Nüsslein-Volhard and Wieschaus (1980) showed specific patterning defects, ranging from abnormal segment numbers to polarity changes, in mutants for many of these genes. They coined the term ‘segment polarity genes’ for one class of mutants that shared a similar patterning phenotype during embryogenesis. One of the genes in this group was called Wingless; others, Armadillo and Arrow. The Wingless gene had actually been identified earlier as a weak mutant allele leading to loss of wing tissue, hence the name Wingless (Sharma and Chopra, 1976). Subsequent to the genetic screens, many of the segmentation genes were molecularly cloned, generating a treasure trove of reagents to study developmental mechanisms. For example, the expression patterns of these genes often produced stripes corresponding to body segments. By examining the expression pattern of one gene in the background of mutations in other genes, hierarchies of genetic interactions were uncovered, providing unparalleled insights into how embryos develop (Ingham, 1988).
RN and his colleagues cloned the Drosophila int1 homologue and used polytene chromosome mapping (the genomics technology at the time) to locate the gene. It turned out to map close to Wingless, one of the segment polarity genes; a striped expression pattern observed with a Drosophila int1 probe also suggested a role in segmentation (Rijsewijk et al, 1987a). Around the same time, Baker (1987) had cloned the Wingless gene by a P-element transposon tagging, a method akin to the proviral tagging methods we had used for int1. The gene he cloned had restriction maps matching our Drosophila int1 clone. The genes were identical; the int1 homologue in Drosophila was Wingless, one of the first examples of a gene involved in development and also activated in cancer (Rijsewijk et al, 1987a). This was an exciting discovery in its own right. In addition, the membership of Wingless in the segment polarity group promised to open doors to discovering the mechanisms of action of int1/Wingless, since it seemed likely that other genes in the group would interact with int1/Wingless genetically and biochemically. As we now understand, the core of its signalling pathway is indeed based on genetic relationships between segment polarity genes, with a key role for Armadillo (see below) (Figure 2).
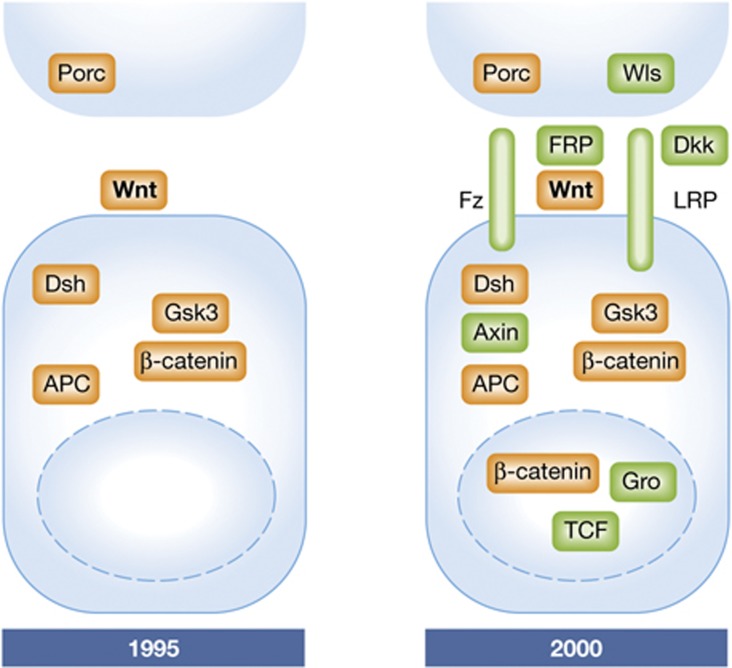
Figure 2.
Wnt signalling components as known in 1995 and 2000.
It had long been argued by some that cancer can be considered akin to a developmental abnormality, a disease caused by cells that have escaped from the normal developmental constraints on proliferation and differentiation (Boveri, 2008). Implicated in cancer as well as in embryogenesis, int1 became a poster child for these connections. Interest in the roles of int1 in development was further strengthened by reports by McMahon and Moon (1989) showing that int1 was implicated in embryonic axis formation in another animal, Xenopus laevis. Xenopus had been used classically to study morphological changes in development; more recently, injection methods had been used to perturb development by introduction of wild-type and mutant genes and proteins into early stage Xenopus embryos. McMahon and Moon (1989) found that ectopic expression of int1 duplicated the dorsal axis, suggesting a role for int1 as an organizer and an embryonic ‘inducer’, a signal between germ layers that would lead to pattern formation. Because of the speed and clarity of the embryological assays, Xenopus embryology became a mainstay of research on int1 and other developmental signals.
In other organisms, int1 expression patterns in embryos and other tissues suggested a diversity of functions in the development of tissues. For instance, int1 was initially found to be expressed mainly near the midbrain and cerebellum at the mid-stage of mouse development and in a specific stage of male germ cell maturation (Shackleford and Varmus, 1987; Wilkinson et al, 1987).
Int1 was one of the first genes to be knocked out by homologous gene targeting in mice, producing a developmental phenotype
During the late 1980s, Mario Capecchi and Oliver Smithies developed the groundbreaking technique for generating gene-specific mouse mutants by homologous recombination. After learning about the highly restricted pattern of expression of int1, Capecchi’s group chose the gene as one of its initial targets for application of this amazing technology. After breeding to homozygosity, disruption of int1 produced a dramatically diminished cerebellum, accompanied by severe ataxia (Thomas and Capecchi, 1990). In McMahon’s laboratory, int1 null mutants caused an embryonic lethal phenotype in the mid-brain also affecting the development of the cerebellum (McMahon and Bradley, 1990). Soon thereafter, it was also shown that a classical mouse mutation, Swaying (Lane, 1967), was an allele of the int1 gene; when int1 was cloned from Swaying mice, it proved to have a frameshift mutation (Thomas et al, 1991).
These several discoveries about int1 mutant organisms in flies and mice were very striking. In a review article published in 1992 (Nusse and Varmus, 1992), we wrote: ‘With the benefit of hindsight, we now recognize that phenomena studied for several decades are the consequences of Wnt gene mutations. Viral insertion mutations regularly promote mammary tumours in laboratory mice (Bittner, 1936; Korteweg, 1936), a spontaneous frameshift mutation of mice (swaying) impairs cerebellar structure and function (Lane, 1967; Thomas et al, 1991) and wingless mutations in Drosophila can transform a wing to a notum or disrupt segment polarity (Sharma and Chopra, 1976; Nüsslein-Volhard and Wieschaus, 1980)’.
Renaming int1 as Wnt1 and recognition of a Wnt gene family
Around 1990, it became clear that the int nomenclature had become inadequate and confusing. For example, additional screens for MMTV proviral insertion sites in tumours had yielded other activated genes, called int2, int3, and int4 (Dickson et al, 1984; Gallahan and Callahan, 1987; Roelink et al, 1990). But by sequence comparisons, these genes were not usually related to int1. One MMTV target gene, initially called int4, did prove to be related to int1 (Roelink et al, 1990). But the frequently activated int2 gene, first identified by Clive Dickson and Gordon Peters (Dickson et al, 1984), turned out to be a member of the FGF family (Dickson and Peters, 1987). Interestingly, FGF genes were also implicated in normal development at about this time, sometimes in coordination with int1-related genes, as in mesoderm formation (Kimelman et al, 1992). Moreover, int1 and int2 are sometimes co-activated in MMTV-induced breast tumours (Peters et al, 1986). int3 was shown to be related to Notch, another important developmental regulator (Gallahan and Callahan, 1997).
At the same time that the int gene nomenclature was becoming unworkable, various experiments, including PCR-based homology screens, had revealed a large family of genes related to int1 (Gavin et al, 1990). It would have been confusing to christen all these genes with the term ‘int’, whether or not they had been activated by proviral integration. To avoid further confusion, all those working on int1 and its relatives, including Wingless, consented to a new hybrid name ‘Wnt’ (for Wingless-related integration site) to denote genes belonging to the int1/Wingless family, with int1, now called Wnt1, as the founding member (Nusse et al, 1991). (In accord with other recognized relationships, int2 is now called FGF3, the int3 gene is Notch4, and int4 is Wnt3A.)
The Wnt family as a vantage point to study gene evolution and development in metazoans
With the complete sequences of the genomes of many multicellular animals in hand, we now realize that vertebrates contain a family of 19 Wnt-related genes; pairs of these genes can often be placed in subfamilies that are highly similar to each other, perhaps reflecting gene duplications relatively recently in evolution (Gavin et al, 1990). Each of these genes seems likely to have a specific role in development or other processes; they are generally expressed in different cells and at different times in maturation (Gavin et al, 1990). Many of the Wnt genes have now been deliberately mutated in the mouse, almost always leading to striking phenotypes, including limb polarity defects and sexual dimorphic abnormalities (Parr and McMahon, 1995, 1998).
Even representatives of the earliest branches of the animal kingdom, such as Hydra and Nematostella, have the same number of Wnt subfamilies as vertebrates (Kusserow et al, 2005). Readily recognized orthologues of specific Wnt genes—for example, Wnt1—have been found throughout the entire animal kingdom, often expressed in tantalizing patterns (Kusserow et al, 2005). As a result, evolutionary biologists speculate that the early amplification and diversification of the Wnt family were at the roots of the increased complexity of animal body plans (Sidow, 1992; Holstein, 2012). It appears from such findings that Wnt genes were probably present in genomes prior to the split of the animal kingdom into protostomes and deuterostomes, are therefore at least 600 M years old, and may have a universal role in setting up the primary axis of animals (Petersen and Reddien, 2009; Niehrs, 2010; Holstein, 2012). However, it is also clear that single-cell organisms do not contain Wnt genes, nor do plants.
In the midst of all of these genes and families, it remains striking that Wnt1 is one of the key Wnt family members and may have been the primordial one. Wnt1 is the true orthologue of Wingless, a gene in Drosophila with numerous functions in later development as well as early embryogenesis. While there are six other Wnts in Drosophila, the others each play a minor role compared with Wingless. Moreover, there are very useful temperature-sensitive alleles of Wingless to study its numerous functions. As a result of these attributes, Wingless has been a rich source for understanding developmental processes. In other organisms that have multiple Wnts, Planaria in particular, the true Wnt1 orthologue also has a special place because of the requirement for it in regeneration (Gurley et al, 2010). While we do not understand why Wnt1 is the most frequently activated gene in MMTV-induced breast cancer, it should be noted that we now know that Wnt1 is closely linked to another Wnt gene that is often insertionally activated, Wnt10B (Lee et al, 1995). Thus, some MMTV inserts may have activated both genes, providing a greater growth advantage.
Unexpected findings reveal the importance of the Wnt pathway in human cancers
From the time that Wnt1 was discovered as an initiating gene in mouse mammary carcinogenesis, it remained of great interest and importance to establish whether Wnt genes were involved in any human cancer. Early tests for aberrant expression of Wnt genes in human breast cancer gave ambiguous results, at best, and no Wnt genes appeared to be mutated in any kinds of human tumours by DNA rearrangements or (as more recently documented by next generation sequencing of whole exomes or whole genomes) by point mutations ( http://www.sanger.ac.uk/genetics/CGP/cosmic).
But work from a different angle changed our perceptions about the role of Wnt genes in human cancer, demonstrating that downstream components of the Wnt signalling pathway, rather than the Wnt genes themselves at the upstream end of the pathway, were commonly altered in several types of human cancer. The first news came from the study of colon cancer, and it was dramatic.
Around 1990, significant advances had been made in positional cloning of inherited human disease genes, including genes predisposing to several types of cancer. Among the hereditary forms of human cancer, adenomatous polyposis coli (APC), a trait associated with multiple polyps in the colon, often leads to colon cancer at a relatively early age. The corresponding mutations—often non-sense or frameshift mutations that produce truncated proteins—were found in an enormous gene called APC, which was cloned from human chromosome 5 in 1991 (Groden et al, 1991; Kinzler et al, 1991). Identifying the human APC gene led to the cloning of a mouse homologue, subsequently shown by Bill Dove’s group to be mutated in a mouse strain called Min (Multiple intestinal neoplasia) (Su et al, 1993). Just as in human families, the cancer trait in the Min mouse is produced by an APC truncating mutation, inherited in an autosomal dominant manner. But despite the new genetic insights into intestinal cancer, the function of the large APC protein posed a biochemical mystery.
Soon thereafter, the groups of Paul Polakis, Bert Vogelstein, and Ken Kinzler established that APC interacted in cells with a protein called β-catenin (Rubinfeld et al, 1993; Su et al, 1993). At the time, β-catenin had just been characterized by Masatoshi Takeichi and Rolf Kemler as a protein binding to the cytoplasmic domain of the adhesion molecule E-cadherin (Ozawa et al, 1989; Takeichi, 1990). Intriguingly, Pierre McCrea and Barry Gumbiner had found that β-catenin gene was a vertebrate homologue of the segment polarity gene, Armadillo (McCrea et al, 1991), while Peifer and Wieschaus (1990) had established similarity between Plakoglobin (a β-catenin-related adhesion complex member) and Armadillo. Together, these findings suggested that APC and β-catenin/Armadillo were involved in regulating adhesion between vertebrate cells. Given the role of Armadillo in segment polarity, a function shared with Wingless, a model emerged in which Wnt/Wingless signalling controlled cell adhesion in development (Peifer and Wieschaus, 1990; Peifer et al, 1993), and the same adhesion-based mechanism could control growth of cells in tissues and cause cancer when misregulated.
While this was tantalizing, there were also reports that β-catenin/Armadillo was present in the nucleus, as well as at the cell membrane (Funayama et al, 1995). Other publications mentioned that injection of antibodies to β-catenin/Armadillo could induce dorsal axis duplication in Xenopus (McCrea et al, 1993), possibly by stabilizing the β-catenin/Armadillo protein; yet others claimed that depletion of maternal β-catenin/Armadillo could eliminate the dorsal axis (Heasman et al, 1994). These observations would ultimately all make sense: β-catenin is a key participant of Wnt signalling, but the molecular mechanisms remained unexplained until a few years later.
In deciphering the cascade of events between the Wnt signal and the role of β-catenin/Armadillo, the genetic interactions between a protein kinase called Glycogen Synthase Kinase 3 (GSK3) and other Wnt components proved to be of critical importance. Norbert Perrimon and colleagues showed that GSK3 (the Drosophila homologue was called zeste-white 3, also known as shaggy) was a negative regulator of the pathway; at the genetic level, Wnt/Wingless acted as a GSK3 inhibitor (Siegfried et al, 1992). Until then, GSK3 was known for its role in glucose metabolism (Dent et al, 1989), so its newly discovered role in developmental signalling was certainly surprising.
A Wnt signalling cascade from the cell surface to the nucleus, an unusual pathway experimentally assembled from several different models
By 1995, the combined results from fly and mouse genetics, Xenopus embryology, and fly and mammalian cell culture experiments had generated an outline of a Wnt signalling pathway (Figure 2). It became clear that Wnt signalling was unusual compared with the other pathways known at the time: those consisted mostly of successions of protein phosphorylations, with protein associations based on recognition of phosphorylated domains. In Wnt signalling, the most upstream known component was a cytoplasmic protein of uncertain biochemical function, Dishevelled, which then was proposed to inhibit the abundant GSK3 protein kinase (Peifer et al, 1991; Siegfried et al, 1992, 1994; Noordermeer et al, 1994). GSK3 was known to be a negative regulator of β-catenin/Armadillo and was found in a complex with β-catenin/Armadillo, together with the APC protein (Rubinfeld et al, 1996). The role of the GSK3 kinase activity as a suppressor of Wnt action was confirmed by injecting dominant-negative (kinase-dead) mutants of GSK3 into early Xenopus embryos; this maneuver produced a phenocopy of the effect of Wnt1, duplication of the dorsal axis, that had been reported previously by Moon and MacMahon (Dominguez et al, 1995; He et al, 1995; Pierce and Kimelman, 1996).
A critical next step was to determine how phosphorylation by GSK3 governed β-catenin. This seemed likely to occur by control of the level of β-catenin protein. In cells activated by Wnt, levels of β-catenin are commonly increased (Riggleman et al, 1990; Peifer et al, 1994; Van Leeuwen et al, 1994) by stabilizing the β-catenin protein, not by an increase in its synthesis. Several highly conserved Ser/Thr phosphorylation sites near the amino terminus of β-catenin were proposed as possible targets for phosphorylation by GSK3 (Peifer et al, 1994). As shown by Rolf Kemler’s and Randall Moon’s groups, phosphorylated β-catenin is targeted for degradation by the ubiquitination/proteasome pathway (Yost et al, 1996; Aberle et al, 1997; Orford et al, 1997), with a critical role for F-box proteins (Jiang and Struhl, 1998). Eliminating one or more of the N-terminal phosphorylation sites stabilizes β-catenin, producing abundant protein highly active in Xenopus axis formation assays (Yost et al, 1996). As a result, there are striking parallels between the Wnt, Hedgehog, and NF-kB signalling pathways; in all three cases, regulated signalling depends on degradation of a key pathway component by the ubiquitination/proteasome pathway after phosphorylation (Jiang and Struhl, 1998; Maniatis, 1999).
The study of the molecular pathology of colon cancers then offered a remarkable example of the predictive power of knowledge about signalling. As mentioned earlier, inherited mutations in the APC gene were known to cause the familial disease APC, and somatic mutations in APC were found in most (ca. 85%) but not in all sporadic colorectal cancers. Why not all? Could mutations affecting other components of the Wnt signalling pathway substitute for APC mutations? Or did other mutant signalling pathways drive those tumours? Paul Polakis, Bert Vogelstein, Hans Clevers and their colleagues looked specifically for altered β-catenin genes in APC wild-type tumours, on the supposition that β-catenin protein could be stabilized by mutations affecting the N-terminus as well as by loss of APC. Indeed they found that about 5–10% of sporadic colon cancers had mutations, often short deletions, that removed or changed the phosphorylation sites that target β-catenin for degradation (Korinek et al, 1997; Morin et al, 1997; Rubinfeld et al, 1997). Subsequently, mutations have also been found in another component of the degradation complex, Axin, in colorectal and other types of cancers (Satoh et al, 2000).
This other component of the β-catenin/Armadillo/GSK3/APC complex has an interesting history. Cloned by Frank Costantini as a mouse developmental mutant called fused (Zeng et al, 1997), the Axin gene encodes a protein that shares homology with Dishevelled, suggesting possible participation in the Wnt pathway. This turned out to be the case. Axin is now known to participate in the β-catenin destruction complex, together with APC and GSK3 (Behrens et al, 1998; Ikeda et al, 1998). Using a novel cell-free system for studying β-catenin degradation, Marc Kirschner and colleagues showed that Axin is the rate-determining component of the complex, even though it was the most recently identified (Lee et al, 2003). Axin has a similar role in intact mammalian cells (Li et al, 2012).
After these several elements were implicated in Wnt signalling, two apparent and important gaps in the pathway remained, one at each end of the pathway (Figure 2). On the upstream end, there were no proteins known to recognize extracellular Wnt proteins and transmit a signal to the cell’s interior (Wnt receptors). On the downstream end, the anticipated effects on gene expression through transcriptional control could not be explained because β-catenin does not have the expected physical attributes of a transcription factor, and no established transcription factor was known to partner with it. Then, in a single year, 1996, these gaps were closed, generating excitement in the growing Wnt field (Figure 2).
Wnt receptor proteins were known in other contexts before their roles in Wnt signalling were uncovered
After many trial-and-error searches, one class of the elusive Wnt receptors was identified: the Frizzled transmembrane proteins. Originally found in mutant screens by Calvin Bridges (Bridges and Brehme, 1944), Frizzled had been identified in Drosophila as a gene required for planar polarity (Gubb and Garcia-Bellido, 1982; Vinson and Adler, 1987), the orientation of cells in tissues. Paul Adler and colleagues showed Frizzled to encode a seven-pass transmembrane protein (Vinson et al, 1989). Genetically, at least, Frizzled interacted with Dishevelled, which was shown by Perrimon and Mahowald (1987) to be involved in Wingless signalling as well. While this suggested that Frizzled could mediate Wingless signalling, the absence of an embryonic segment polarity phenotype in Frizzled mutants indicated otherwise.
Here serendipity stepped in. Jeremy Nathans and his colleagues found a Frizzled homologue among components of a human retinal cDNA library that had been made to pursue their interests in the molecular biology of vision. When the cDNA was used to seek homologues in a library of Drosophila DNA, a second Drosophila Frizzled gene (Dfz2) was cloned, and the Dfz2 gene displayed a striped pattern of gene expression in the embryo, implying that it might be directly involved in segment polarity. A collaboration between the Nathans and RN laboratories revealed that the Wingless protein, which RN’s laboratory had solubilized at the time, could bind to Dfz2 and, more weakly, to Frizzled itself (Bhanot et al, 1996). Moreover, in cultured Drosophila cells that did not express Frizzled genes, transfection of an expression vector containing Frizzled genes conferred active signalling, as demonstrated by an increase in Armadillo (β-catenin) levels (Bhanot et al, 1996). Genetic and other interactions between Frizzleds and Wnts were also reported by the groups of Randall Moon and Robert Horvitz (Sawa et al, 1996; Yang-Snyder et al, 1996).
Just like Wnts, Frizzleds form a large gene family in all branches of metazoan animals. Genetically, Frizzled genes are often redundant and display phenotypes only when mutated in combination with other family members (Ye et al, 2011). In Drosophila, this was shown for Frizzled and Dfz2 using dsRNA interference technology (Kennerdell and Carthew, 1998). (Interestingly, this occurred in the same year that this revolutionizing method to inhibit gene expression was first reported by Andy Fire and Craig Mello; Fire et al, 1998). By using loss-of-function mutations in Frizzled and Dfz2, Eric Wieschaus, Gary Struhl, Ken Cadigan, and Krishna Bhat uncovered a segment polarity phenotype indistinguishable from phenotypes characteristic of the other genes in that class—but only as double mutants, explaining why these receptor genes were not in the original Nüsslein-Volhard/Wieschaus collection (Bhat, 1998; Bhanot et al, 1999; Chen and Struhl, 1999; Muller et al, 1999). Despite the evidence for redundancy, Frizzled proteins have different affinities for different Wnts (Rulifson et al, 2000), indicating a high degree of specificity in their interactions. However, persistent experimental problems with the biochemistry of Wnt proteins have hampered systematic surveys of the interactions.
To complement the Wnt receptor story, the Drosophila gene Arrow, one of the last segment polarity genes to be identified, was cloned by Stephen DiNardo and colleagues a few years later. Arrow proved to be a member of the Low density lipoprotein receptor-Related Protein (LRP) family of receptors (Wehrli et al, 2000). Based on additional genetic data from Bill Skarnes, who made LRP mouse mutants, and biochemical experiments from Xi He’s laboratory, a model emerged in which Arrow/LRP is a co-receptor for Wnts, physically adjacent to Frizzleds in the cell membrane (Pinson et al, 2000; Tamai et al, 2000). When signalling to downstream components, however, Arrow/LRP may be the key player. Its cytoplasmic tail is phosphorylated as a consequence of Wnt binding and interacts directly with GSK3 and Axin (Mao et al, 2001b; Tamai et al, 2004; Davidson et al, 2005; Zeng et al, 2005) and Frizzled’s intracellular role in signalling may be limited to binding Dishevelled (Macdonald et al, 2009). Arrow/LRP is also the target of several Wnt antagonists including the protein Dickkopf, isolated by Christof Niehrs (Glinka et al, 1998; Bafico et al, 2001; Mao et al, 2001a; Semenov et al, 2001). The Dickkopf-Wnt antagonism is conserved across many animal phyla (Guder et al, 2006), illustrating the ancient nature of Wnt signalling in animal development and evolution.
Eddy De Robertis, Jeremy Nathans, Jeff Rubin, and their co-workers have uncovered several other Wnt antagonists, in addition to Dickkopf. These are secreted molecules usually consisting of Wnt receptor domains that bind to Wnt itself. Some of these molecules have names such as FRP or FRZB, reflecting their similarity to the Frizzled receptor (Finch et al, 1997; Leyns et al, 1997; Rattner et al, 1997). Others, such as the WIF protein (Hsieh et al, 1999), are unrelated to Frizzled. These proteins are likely involved in fine-tuning the concentration of active Wnt outside cells.
The tandem arrays of Frizzled and LRP are not the only Wnt receptors, as there are various members of the trans-membrane tyrosine kinase family that serve to receive Wnts; these include the ROR (Oishi et al, 2003; Mikels and Nusse, 2006) and Derailed/RYK (Yoshikawa et al, 2003) proteins. Interestingly, these two classes of molecules have different Wnt binding modules: the RORs contain a CRD domain similar to the Frizzled CRD, while Derailed/RYK is related to the WIF protein mentioned above (Patthy, 2000). Wnt interactions with these receptors often lead to effects in cells that are unrelated to β-catenin, possibly mediating ‘non-canonical Wnt signalling’ (van Amerongen et al, 2008).
Another unexpected but previously well-known protein, TCF/LEF1, explains the role of β-catenin in the Wnt signalling pathway
The TCF/Lef1 protein proved to be the long-sought Wnt transcription factor in the nucleus Discovery of the critical interaction between this protein and β-catenin highlights one of the themes of this essay: historically, the map of Wnt signalling was assembled by merging evidence from several different cell types and organisms. TCF/Lef1, an HMG box-containing transcription factor, was first implicated in immune T-cell gene expression (Travis et al, 1991; van de Wetering et al, 1991; Waterman et al, 1991) without any evident link to Wnt signalling. Working separately on C. elegans, Jim Priess and colleagues identified an HMG-box family member, POP1, involved in mesoderm specification in the worm embryo (Lin et al, 1995), initially also without connections to the Wnt pathway.
Soon thereafter, a surprising discovery was reported: TCF/Lef1 could interact with β-catenin, considered at that time to be an adhesion molecule. Hans Clevers extended his earlier work on TCF/Lef1 to make this finding (Molenaar et al, 1996), while Walter Birchmeier (Behrens et al, 1996) and Rolf Kemler (Huber et al, 1996) started from β-catenin to establish binding to TCF/Lef1. Using Drosophila, Mariann Bienz and Rudi Grosschedl (Riese et al, 1997) found that Wingless signalling was mediated by TCF/Lef1 while Konrad Basler used mutagenesis screens to find a gene called Pangolin (Brunner et al, 1997), the single TCF/Lef1 homologue in Drosophila. Around the same time, continued investigations into C. elegans embryogenesis by Jim Priess, Craig Mello, and Bruce Bowerman unveiled that the set of MOM genes implicated in lineage choices were members of the Wnt pathway, including Wnt itself (MOM2), Porcupine (MOM1), and Frizzled (MOM5). All of these MOMs converged on POP1 as a transcription factor and WRM1 as a β-catenin-related gene (Rocheleau et al, 1997, 1999; Thorpe et al, 1997).
In many contexts, TCF/Lef1 can switch between two states. When bound to Groucho, it acts as a repressor of target genes; but when Groucho is displaced by β-catenin, the same target genes are transcriptionally activated (Cavallo et al, 1998; Daniels and Weis, 2005). Crystallographic studies by Bill Weis and Wenqing Xu revealed the molecular details of the binding between TCF, β-catenin, and other proteins. The structure of β-catenin contains a groove made by the ‘Armadillo’ repeats in the protein, explaining how β-catenin can interact with several different partners, including TCF/Lef1, E-cadherin, and APC (Huber et al, 1997; Graham et al, 2000).
These discoveries about the interaction between β-catenin and TCF/Lef1 were in more than one respect very significant, as they not only closed the gaps in the Wnt pathway but also provided unparalleled tools for experiments. TCF/Lef1 recognizes a well-defined DNA binding site. By multimerizing this sequence, Hans Clevers and colleagues generated very convenient luciferase-based Wnt reporters, called Top-FLASH and now widely used in the Wnt field to measure signalling (Korinek et al, 1997). There are now numerous genes known to have TCF/Lef1 binding sites in their promoters and hence likely to be transcriptional targets for the Wnt signalling pathway in at least some cell types; among these genes are several implicated in cancer, such as c-myc (He et al, 1998).
At last, the purification of active Wnt protein
As we have recounted, by the end of the 20th century we had a blueprint of the Wnt signalling pathway and a readout for the pathway, both of which were missing in the two previous decades in which Wnt genes were intensively studied (Figure 2). Still lacking, however, was the purification of any active Wnt protein, a problem that we and many other researchers had been working on since the initial cloning of Wnt1. Why were Wnt proteins so much more refractory to biochemical purification than many other secretory proteins? Were they modified in a fashion that rendered them insoluble or highly adherent?
It was known from Norbert Perrimon’s work and Drosophila genetics that export likely required a specialized protein encoded by Porcupine. Evidence that Porcupine encoded a putative acyltransferase (Kadowaki et al, 1996; Hofmann, 2000) suggested that detergents might be needed to keep lipid-modified Wnt proteins soluble during extraction from cells. With the help of assays that judged Wnt activity in extracts based on increased β-catenin levels in cells, Karl Willert in RN’s laboratory finally managed to break through the purification barriers (Willert et al, 2003). (Coincidentally, Willert had obtained his PhD working with HV at UCSF.) Wnts were found to be indeed covalently attached to lipids, explaining to some extent their resistance to biochemical manipulation (Willert et al, 2003). It is now recognized that secretion of Wnt proteins is a complex process, involving a dedicated enzyme (Porcupine; Kadowaki et al, 1996) and secretory proteins that are specific for Wnt signals. Among these is the multiple-pass transmembrane protein Wntless/Evi, identified by the groups of Konrad Basler and Michael Boutros, once again using the Drosophila genetic resources that have over the years been so instrumental in understanding the details of Wnt signalling (Banziger et al, 2006; Bartscherer et al, 2006; Port and Basler, 2010).
A growing and very active field of Wnt signalling
The availability of Wnt proteins and more quantitative reporters (e.g., TopFlash) as reliable end points for signalling have simplified the study of Wnt signalling in cell culture, attracting many new investigators. The generation of Wnt reporter mice, initially Top-gal animals from Elaine Fuchs (DasGupta and Fuchs, 1999) and later animals with transgenic markers driven by Axin 2 promoters (Lustig et al, 2002; van Amerongen et al, 2012), provided yet more experimental opportunities. One can now trace Wnt-responding cells in any tissue of the mouse, examine the origin of these cells, and follow their fate in normal settings or after injury (Barker et al, 2007, 2010; van Amerongen et al, 2012). These new experimental tools have led to a rapidly growing list of Wnt signalling components, built on the core pathway.
Wnt signalling is clearly complicated and unusual when compared with other growth factor cascades. At various nodes in the Wnt pathway, there are links to cyto-architectural proteins, such as those involved in adhesion and cell polarity (Nelson and Nusse, 2004). The Wnt pathway is clearly important for cell fate changes and the control of gene expression, but Wnt signalling can also influence how cells are shaped and polarized and how they divide (Veeman et al, 2003). Hitoshi Sawa, Bruce Bowerman, and Craig Mello have provided conclusive evidence for a major role for Wnt signalling in asymmetric cell division in C. elegans and the annelid worm Platynereis dumerilii (Rocheleau et al, 1999; Schneider and Bowerman, 2007; Sugioka et al, 2011). Given the multiple roles of the Wnt pathway in development, these cell biological phenotypes are perhaps not surprising and have opened fertile ground for further research.
Sociology of the field of Wnt signalling: annual meetings, sharing information, and a dedicated website
A history of Wnt signalling would be incomplete without a few comments on the sociology of the field, which, we believe, has several unusual aspects. Those features have contributed to one of the overarching characteristics of this field: the propensity of investigators working on Wnt genes and Wnt signalling to identify themselves, at least in part and often primarily, as students of Wnts, regardless of whether they are cancer biologists, developmental biologists, or biochemists. First and foremost among the unifying activities are the annual (or sometimes bi-annual) Wnt meetings. These are organized by working scientists in the field, not by institutions or meetings specialists, in a very informal, low-cost, but effective way. No one, except a keynote speaker, receives an invitation accompanied by a promise of reimbursements; all others are expected to get there, find food and lodging, and arrange to cover expenses. Nevertheless, the meetings are well attended by many principal investigators, not by just trainees. The meetings started in on a small scale in 1990 as regular gatherings of our two nearby laboratories, after RN had moved to Stanford from Amsterdam. We then asked members of other laboratories to attend as well; Figure 3 shows most of the attendees at the 1991 meeting at UCSF. After HV moved to the NIH in 1993, we continued the gatherings; attendance soon grew to ∼300 people, as our own trainees started their own laboratories to work on Wnt genes, and others joined the field. Many of the discoveries we have presented in this essay were first made public during Wnt meetings, including the identification of Frizzleds as Wnt receptors and TCFs/Lefs as transcription factors in Wnt signalling, both at the 1996 meeting at Stanford (Figure 4); and the Arrow/LRP findings were first reported at another Wnt meeting, at Stanford in 1999. The meetings have covered an increasingly wide range of subjects and biological systems related to Wnt signalling, and their popularity attests to the loyalty of Wnt researchers to the subject matter in its many manifestations.
Figure 3.
Participants of the 1991 Wnt meeting at UCSF. From left: RN, Andrew McMahon, Arend Sidow, Vladimir Pecenka, John Mason, Lee Fradkin, HV, Henk Roelink, Jasprina Noordermeer, Supriya Shivakumar, Frank van Leeuwen, Cindy Harryman, Jean-Paul Vincent, Jackie Papkoff, two unidentified people, Tony Brown, a third unidentified person, Helen Kwan. Top row, from left: Karl Willert, Neil Parkin, and Jan Kitajewski.
Figure 4.
Celebration of discovery of Frizzleds as Wnt receptors at the 1996 Wnt meeting at Stanford. From left: Jeremy Nathans, Matthew Scott, RN, and HV.
Wnt meetings have also helped to establish a culture of sharing information and reagents. An example of the congenial relationship was the unanimous and friction-free acceptance of the new nomenclature (from int to Wnt) when it was felt that this would benefit the field and the general comprehension of its work. In parallel to the informal Wnt meetings in the Unites States, there have been numerous other Wnt conferences throughout the world, perhaps a bit more official but still in the same spirit of open exchange and camaraderie.
In 1996, around the time that the Internet became easier to navigate with browsers such as Netscape, RN started and still maintains a website, the Wnt homepage ( http://wnt.stanford.edu). The various pages list genes and signalling diagrams, in many cases linked to other genome databases. But the site is also used to announce meetings and to supply information on Wnt technology and reagents, and has become a popular resource for the world-wide Wnt community and for others seeking information about Wnt genes, as appreciation of their importance has expanded.
Contemporary Wnt signalling systems, including stem cells, and an outlook
It is now clear that Wnt signalling is widely implicated in diverse biological processes. For instance, the large majority of developmental decisions that cells make during embryogenesis and thereafter appears to be coordinated, in large part, by Wnts. Scientists are beginning to understand how organs grow and regenerate after injury, and it is clear that Wnt signalling has major functions in these processes as well. A particularly prominent example of the centrality of Wnt signalling is the recently recognized role of Wnts in maintaining stem cells. The choices that stem cells make to self-renew or to differentiate are very much dependent on extrinsic signalling factors coming from a niche. Wnt signals are widely active as niche factors, as illustrated by the identification of the LGR5 receptor as a Wnt target gene in many different kinds of adult stem cells (Barker et al, 2007) and the requirement for TCF4 to maintain stem cells in the intestine (Korinek et al, 1998).
The discoveries related to stem cells have also illuminated the connections between Wnt and cancer: in a simple but likely correct view, stem cells are normally dependent on external Wnts for self-renewal, but when a negative Wnt regulator such as APC is mutated in stem cells, cells with markers of early lineage development proliferate in an uncontrolled manner, producing cancers of the colon and other organs. In the mammary gland, where we first identified Wnt1 as an oncogene, stem cells are also Wnt dependent (Shackleton et al, 2006; Zeng and Nusse, 2010; van Amerongen et al, 2012), and Wnt1-induced tumours bear hallmarks of normal mammary stem cells (Li et al, 2003).
Wnt signalling mutations have also been implicated in a growing list of degenerative diseases. Important among these are bone density abnormalities with dysfunctional LRP receptors (Gong et al, 2001) and retinal degeneration with Frizzled mutations (Robitaille et al, 2002). Some metabolic disorders, including diabetes mellitus, have been associated with alterations in Wnt pathway genes (Grant et al, 2006).
It has been gratifying to witness growth of the Wnt field, from the finding of a single cancer gene in a mouse model to a rich system branching out to influence so many aspects of metazoan biology and human disease. While outsiders may be intimidated by the current size of the field and the biochemical complexities of Wnt signalling, we suggest that there are still many fundamental aspects of Wnt-related biology to be discovered, understood, and exploited. Increasingly, structures of Wnt signalling components are being elucidated, often in complexes with their partners (Dann et al, 2001; Schwarz-Romond et al, 2007; Ahn et al, 2011; Chen et al, 2011; Cheng et al, 2011); but important aspects of the pathway’s molecular machinery and biochemical regulators remain incompletely defined.
Progress in the Wnt field is much more rapid today than it was in the early history of this field, thanks to more sophisticated tools: Wnt-specific assays and materials and more general methods in structural biology, genetics, and cell biology. For instance, recognition of the role of Wnt signalling in stem-cell regulation has already led to the use of Wnt proteins or Wnt agonists to expand stem cells in culture (Sato et al, 2010; Zeng and Nusse, 2010; ten Berge et al, 2011). On the other hand, despite the evidence for widespread involvement of Wnt signaling in human carcinogenesis, the kinds of targeted cancer therapies that are now being developed against components of several other signaling pathways have not yet been produced to interfere with the Wnt pathway. Among the most significant challenges in future research in the Wnt field is the identification of effective and specific Wnt pathway inhibitors for use in cancer and other diseases. We expect that further understanding of the intricacies and varieties of Wnt signaling will help to achieve these important goals.
Acknowledgments
We thank Thomas Schwarz-Romond and Renee van Amerongen for inviting us to write this essay. We thank Elizabeth Matthews for comments on various drafts and for suggesting the name Wnt during the nomenclature discussions over 20 years ago.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aberle H, Bauer A, Stappert J, Kispert A, Kemler R (1997) Beta-catenin is a target for the ubiquitin-proteasome pathway. EMBO J 16: 3797–3804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn VE, Chu ML, Choi HJ, Tran D, Abo A, Weis WI (2011) Structural basis of Wnt signaling inhibition by Dickkopf binding to LRP5/6. Dev Cell 21: 862–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bafico A, Liu G, Yaniv A, Gazit A, Aaronson SA (2001) Novel mechanism of Wnt signalling inhibition mediated by Dickkopf-1 interaction with LRP6/Arrow. Nat Cell Biol 3: 683–686 [DOI] [PubMed] [Google Scholar]
- Baker NE (1987) Molecular cloning of sequences from wingless, a segment polarity gene in Drosophila: the spatial distribution of a transcript in embryos. EMBO J 6: 1765–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banziger C, Soldini D, Schutt C, Zipperlen P, Hausmann G, Basler K (2006) Wntless, a conserved membrane protein dedicated to the secretion of Wnt proteins from signaling cells. Cell 125: 509–522 [DOI] [PubMed] [Google Scholar]
- Barker N, Huch M, Kujala P, M van de Wetering, Snippert HJ, van Es JH, Sato T, Stange DE, Begthel H, M van de Born, Danenberg E, S van de Brink, Korving J, Abo A, Peters PJ, Wright N, Poulsom R, Clevers H (2010) Lgr5+ve stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Stem Cell 6: 25–36 [DOI] [PubMed] [Google Scholar]
- Barker N, van Es JH, Kuipers J, Kujala P, van de Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, Clevers H (2007) Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449: 1003–1007 [DOI] [PubMed] [Google Scholar]
- Bartscherer K, Pelte N, Ingelfinger D, Boutros M (2006) Secretion of Wnt ligands requires Evi, a conserved transmembrane protein. Cell 125: 523–533 [DOI] [PubMed] [Google Scholar]
- Behrens J, Jerchow BA, Wurtele M, Grimm J, Asbrand C, Wirtz R, Kuhl M, Wedlich D, Birchmeier W (1998) Functional interaction of an axin homolog, conductin, with beta-catenin, APC, and GSK3beta. Science 280: 596–599 [DOI] [PubMed] [Google Scholar]
- Behrens J, von Kries JP, Kuhl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W (1996) Functional interaction of beta-catenin with the transcription factor LEF-1. Nature 382: 638–642 [DOI] [PubMed] [Google Scholar]
- Bentvelzen P, Daams JH, Hageman P, Calafat J (1970) Genetic transmission of viruses that incite mammary tumor in mice. Proc Natl Acad Sci USA 67: 377–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhanot P, Brink M, Samos CH, Hsieh JC, Wang Y, Macke JP, Andrew D, Nathans J, Nusse R (1996) A new member of the frizzled family from Drosophila functions as a Wingless receptor. Nature 382: 225–230 [DOI] [PubMed] [Google Scholar]
- Bhanot P, Fish M, Jemison JA, Nusse R, Nathans J, Cadigan KM (1999) Frizzled and Dfrizzled-2 function as redundant receptors for Wingless during Drosophila embryonic development. Development 126: 4175–4186 [DOI] [PubMed] [Google Scholar]
- Bhat KM (1998) Frizzled and frizzled 2 play a partially redundant role in wingless signaling and have similar requirements to wingless in neurogenesis. Cell 95: 1027–1036 [DOI] [PubMed] [Google Scholar]
- Bishop JM (1983) Cellular oncogenes and retroviruses. Annu Rev Biochem 52: 301–354 [DOI] [PubMed] [Google Scholar]
- Bishop JM, Varmus H (1985) Functions and origins of retroviral transforming genes. InRNA Tumor Viruses Weis R, Teich N, Varmus H, Coffin JM (eds)pp249–356Cold Spring Harbor: Cold Spring Harbor Laboratory, [Google Scholar]
- Bittner J (1936) Some possible effects of nursing on the mammary gland tumor incidence in mice. Science 84: 162. [DOI] [PubMed] [Google Scholar]
- Boveri T (2008) Concerning the Origin of Malignant Tumours. The Company of Biologists Limited and Cold Spring Harbor Laboratory Press
- Bridges C, Brehme K (1944) The Mutants of Drosophila Melanogaster. Washington, DC: Carnegie Institute,
- Brown AM, Wildin RS, Prendergast TJ, Varmus HE (1986) A retrovirus vector expressing the putative mammary oncogene int-1 causes partial transformation of a mammary epithelial cell line. Cell 46: 1001–1009 [DOI] [PubMed] [Google Scholar]
- Brown P, Varmus H (1989) Retroviruses. InMobile DNA Berg D, Howe M (eds)Washington, DC: American Society for Microbiology Press, [Google Scholar]
- Brunner E, Peter O, Schweizer L, Basler K (1997) Pangolin encodes a Lef-1 homologue that acts downstream of Armadillo to transduce the Wingless signal in Drosophila. Nature 385: 829–833 [DOI] [PubMed] [Google Scholar]
- Cavallo RA, Cox RT, Moline MM, Roose J, Polevoy GA, Clevers H, Peifer M, Bejsovec A (1998) Drosophila Tcf and Groucho interact to repress Wingless signalling activity. Nature 395: 604–608 [DOI] [PubMed] [Google Scholar]
- Chen CM, Struhl G (1999) Wingless transduction by the Frizzled and Frizzled2 proteins of Drosophila. Development 126: 5441–5452 [DOI] [PubMed] [Google Scholar]
- Chen S, Bubeck D, Macdonald BT, Liang WX, Mao JH, Malinauskas T, Llorca O, Aricescu AR, Siebold C, He X, Jones EY (2011) Structural and functional studies of LRP6 ectodomain reveal a platform for Wnt signaling. Dev Cell 21: 848–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Biechele T, Wei Z, Morrone S, Moon RT, Wang L, Xu W (2011) Crystal structures of the extracellular domain of LRP6 and its complex with DKK1. Nat Struct Mol Biol 18: 1204–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JC, Shank PR, Morris VL, Cardiff R, Varmus HE (1979) Integration of the DNA of mouse mammary tumor virus in virus-infected normal and neoplastic tissue of the mouse. Cell 16: 333–345 [DOI] [PubMed] [Google Scholar]
- Cohen JC, Varmus HE (1979) Endogenous mammary tumour virus DNA varies among wild mice and segregates during inbreeding. Nature 278: 418–423 [DOI] [PubMed] [Google Scholar]
- Copeland NG, Jenkins NA (2010) Harnessing transposons for cancer gene discovery. Nat Rev Cancer 10: 696–706 [DOI] [PubMed] [Google Scholar]
- Daniels DL, Weis WI (2005) Beta-catenin directly displaces Groucho/TLE repressors from Tcf/Lef in Wnt-mediated transcription activation. Nat Struct Mol Biol 12: 364–371 [DOI] [PubMed] [Google Scholar]
- Dann CE, Hsieh JC, Rattner A, Sharma D, Nathans J, Leahy DJ (2001) Insights into Wnt binding and signalling from the structures of two Frizzled cysteine-rich domains. Nature 412: 86–90 [DOI] [PubMed] [Google Scholar]
- DasGupta R, Fuchs E (1999) Multiple roles for activated LEF/TCF transcription complexes during hair follicle development and differentiation. Development 126: 4557–4568 [DOI] [PubMed] [Google Scholar]
- Davidson G, Wu W, Shen J, Bilic J, Fenger U, Stannek P, Glinka A, Niehrs C (2005) Casein kinase 1 gamma couples Wnt receptor activation to cytoplasmic signal transduction. Nature 438: 867–872 [DOI] [PubMed] [Google Scholar]
- Dent P, Campbell DG, Hubbard MJ, Cohen P (1989) Multisite phosphorylation of the glycogen-binding subunit of protein phosphatase-1G by cyclic AMP-dependent protein kinase and glycogen synthase kinase-3. FEBS Lett 248: 67–72 [DOI] [PubMed] [Google Scholar]
- Dickson C, Peters G (1987) Potential oncogene product related to growth factors [letter]. Nature 326: 833. [DOI] [PubMed] [Google Scholar]
- Dickson C, Smith R, Brookes S, Peters G (1984) Tumorigenesis by mouse mammary tumor virus: proviral activation of a cellular gene in the common integration region int-2. Cell 37: 529–536 [DOI] [PubMed] [Google Scholar]
- Dominguez I, Itoh K, Sokol SY (1995) Role of glycogen synthase kinase 3 beta as a negative regulator of dorsoventral axis formation in Xenopus embryos. Proc Natl Acad Sci USA 92: 8498–8502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errede B, Cardillo TS, Sherman F, Dubois E, Deschamps J, Wiame JM (1980) Mating signals control expression of mutations resulting from insertion of a transposable repetitive element adjacent to diverse yeast genes. Cell 22: 427–436 [DOI] [PubMed] [Google Scholar]
- Finch PW, He X, Kelley MJ, Uren A, Schaudies RP, Popescu NC, Rudikoff S, Aaronson SA, Varmus HE, Rubin JS (1997) Purification and molecular cloning of a secreted, Frizzled-related antagonist of Wnt action. Proc Natl Acad Sci USA 94: 6770–6775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811 [DOI] [PubMed] [Google Scholar]
- Funayama N, Fagotto F, McCrea P, Gumbiner BM (1995) Embryonic axis induction by the armadillo repeat domain of beta-catenin: evidence for intracellular signaling. J Cell Biol 128: 959–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung YK, Shackleford GM, Brown AM, Sanders GS, Varmus HE (1985) Nucleotide sequence and expression in vitro of cDNA derived from mRNA of int-1, a provirally activated mouse mammary oncogene. Mol Cell Biol 5: 3337–3344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallahan D, Callahan R (1987) Mammary tumorigenesis in feral mice-identification of a new int locus in mouse mammary tumor virus (Czech II)-induced mammary tumors. J Virol 61: 66–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallahan D, Callahan R (1997) The mouse mammary tumor associated gene INT3 is a unique member of the NOTCH gene family (NOTCH4). Oncogene 14: 1883–1890 [DOI] [PubMed] [Google Scholar]
- Gavin BJ, McMahon JA, McMahon AP (1990) Expression of multiple novel Wnt-1/int-1-related genes during fetal and adult mouse development. Genes Dev 4: 2319–2332 [DOI] [PubMed] [Google Scholar]
- Glinka A, Wu W, Delius H, Monaghan AP, Blumenstock C, Niehrs C (1998) Dickkopf-1 is a member of a new family of secreted proteins and functions in head induction. Nature 391: 357–362 [DOI] [PubMed] [Google Scholar]
- Gong Y, Slee RB, Fukai N, Rawadi G, Roman-Roman S, Reginato AM, Wang H, Cundy T, Glorieux FH, Lev D, Zacharin M, Oexle K, Marcelino J, Suwairi W, Heeger S, Sabatakos G, Apte S, Adkins WN, Allgrove J, Arslan-Kirchner M et al. (2001) LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell 107: 513–523 [DOI] [PubMed] [Google Scholar]
- Graham TA, Weaver C, Mao F, Kimelman D, Xu W (2000) Crystal structure of a beta-catenin/Tcf complex. Cell 103: 885–896 [DOI] [PubMed] [Google Scholar]
- Grant SF, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, Helgason A, Stefansson H, Emilsson V, Helgadottir A, Styrkarsdottir U, Magnusson KP, Walters GB, Palsdottir E, Jonsdottir T, Gudmundsdottir T, Gylfason A, Saemundsdottir J, Wilensky RL, Reilly MP et al. (2006) Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet 38: 320–323 [DOI] [PubMed] [Google Scholar]
- Groden J, Thliveris A, Samowitz W, Carlson M, Gelbert L, Albertsen H, Joslyn G, Stevens J, Spirio L, Robertson M, Sargeant L, Krapcho K, Wolff E, Burt R, Hughes JP, Warrington J, McPherson J, Wasmuth J, Le Paslier D, Abderrahim H et al. (1991) Identification and characterization of the familial adenomatous polyposis coli gene. Cell 66: 589–600 [DOI] [PubMed] [Google Scholar]
- Gubb D, Garcia-Bellido A (1982) A genetic analysis of the determination of cuticular polarity during development in Drosophila melanogaster. J Embryol Exp Morphol 68: 37–57 [PubMed] [Google Scholar]
- Guder C, Pinho S, Nacak TG, Schmidt HA, Hobmayer B, Niehrs C, Holstein TW (2006) An ancient Wnt-Dickkopf antagonism in Hydra. Development 133: 901–911 [DOI] [PubMed] [Google Scholar]
- Gurley KA, Elliott SA, Simakov O, Schmidt HA, Holstein TW, Alvarado AS (2010) Expression of secreted Wnt pathway components reveals unexpected complexity of the planarian amputation response. Dev Biol 347: 24–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward WS, Neel BG, Astrin SM (1981) Activation of a cellular onc gene by promoter insertion in ALV-induced lymphoid leukosis. Nature 290: 475–480 [DOI] [PubMed] [Google Scholar]
- He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, Morin PJ, Vogelstein B, Kinzler KW (1998) Identification of c-MYC as a target of the APC pathway. Science 281: 1509–1512 [DOI] [PubMed] [Google Scholar]
- He X, Saint-Jeannet JP, Woodgett JR, Varmus HE, Dawid IB (1995) Glycogen synthase kinase-3 and dorsoventral patterning in Xenopus embryos. Nature 374: 617–622 [DOI] [PubMed] [Google Scholar]
- Heasman J, Crawford A, Goldstone K, Garner-Hamrick P, Gumbiner B, McCrea P, Kintner C, Noro CY, Wylie C (1994) Overexpression of cadherins and underexpression of beta-catenin inhibit dorsal mesoderm induction in early Xenopus embryos. Cell 79: 791–803 [DOI] [PubMed] [Google Scholar]
- Hofmann K (2000) A superfamily of membrane-bound O-acyltransferases with implications for wnt signaling. Trends Biochem Sci 25: 111–112 [DOI] [PubMed] [Google Scholar]
- Holstein T (2012) The evolution of the Wnt pathway. Cold Spring Harb Perspect Biol (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh JC, Kodjabachian L, Rebbert ML, Rattner A, Smallwood PM, Samos CH, Nusse R, Dawid IB, Nathans J (1999) A new secreted protein that binds to Wnt proteins and inhibits their activities. Nature 398: 431–436 [DOI] [PubMed] [Google Scholar]
- Huber AH, Nelson WJ, Weis WI (1997) Three-dimensional structure of the armadillo repeat region of beta-catenin. Cell 90: 871–882 [DOI] [PubMed] [Google Scholar]
- Huber O, Korn R, McLaughlin J, Ohsugi M, Herrmann BG, Kemler R (1996) Nuclear localization of beta-catenin by interaction with transcription factor LEF-1. Mech Dev 59: 3–10 [DOI] [PubMed] [Google Scholar]
- Ikeda S, Kishida S, Yamamoto H, Murai H, Koyama S, Kikuchi A (1998) Axin, a negative regulator of the Wnt signaling pathway, forms a complex with GSK-3beta and beta-catenin and promotes GSK-3beta-dependent phosphorylation of beta-catenin. EMBO J 17: 1371–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham PW (1988) The molecular genetics of embryonic pattern formation in Drosophila. Nature 335: 25–34 [DOI] [PubMed] [Google Scholar]
- Jiang J, Struhl G (1998) Regulation of the Hedgehog and Wingless signalling pathways by the F-box/WD40-repeat protein Slimb. Nature 391: 493–496 [DOI] [PubMed] [Google Scholar]
- Kadowaki T, Wilder E, Klingensmith J, Zachary K, Perrimon N (1996) The segment polarity gene porcupine encodes a putative multitransmembrane protein involved in Wingless processing. Genes Dev 10: 3116–3128 [DOI] [PubMed] [Google Scholar]
- Kennerdell JR, Carthew RW (1998) Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell 95: 1017–1026 [DOI] [PubMed] [Google Scholar]
- Kimelman D, Christian JL, Moon RT (1992) Synergistic principles of development: overlapping patterning systems in Xenopus mesoderm induction. Development 116: 1–9 [DOI] [PubMed] [Google Scholar]
- Kinzler KW, Nilbert MC, Su LK, Vogelstein B, Bryan TM, Levy DB, Smith KJ, Preisinger AC, Hedge P, McKechnie D, Finniear R, Markham A, Groffen J, Boguski M, Altschul S, Horii A, Ano H, Miyoshi Y, Miki Y, Nishsho I et al. (1991) Identification of FAP locus genes from chromosome 5q21. Science 253: 661–665 [DOI] [PubMed] [Google Scholar]
- Kool J, Berns A (2009) High-throughput insertional mutagenesis screens in mice to identify oncogenic networks. Nat Rev Cancer 9: 389–399 [DOI] [PubMed] [Google Scholar]
- Korinek V, Barker N, Moerer P, van Donselaar E, Huls G, Peters PJ, Clevers H (1998) Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat Genet 19: 379–383 [DOI] [PubMed] [Google Scholar]
- Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, Vogelstein B, Clevers H (1997) Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC-/- colon carcinoma. Science 275: 1784–1787 [DOI] [PubMed] [Google Scholar]
- Korteweg R (1936) On the manner in which the disposition to carcinoma of the mammary gland is inherited in mice. Genetics 18: 350–371 [Google Scholar]
- Kusserow A, Pang K, Sturm C, Hrouda M, Lentfer J, Schmidt HA, Technau U, von Haeseler A, Hobmayer B, Martindale MQ, Holstein TW (2005) Unexpected complexity of the Wnt gene family in a sea anemone. Nature 433: 156–160 [DOI] [PubMed] [Google Scholar]
- Lane P (1967) Mouse Newsletter 36: 40 [Google Scholar]
- Lee E, Salic A, Kruger R, Heinrich R, Kirschner MW (2003) The roles of APC and Axin derived from experimental and theoretical analysis of the Wnt pathway. PLoS Biol 1: E10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee FS, Lane TF, Kuo A, Shackleford GM, Leder P (1995) Insertional mutagenesis identifies a member of the Wnt gene family as a candidate oncogene in the mammary epithelium of int-2/Fgf-3 transgenic mice. Proc Natl Acad Sci USA 92: 2268–2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyns L, Bouwmeester T, Kim SH, Piccolo S, De Robertis EM (1997) Frzb-1 is a secreted antagonist of Wnt signaling expressed in the Spemann organizer. Cell 88: 747–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li V, Ng SS, Boersema P, Low T, Karthaus W, Gerlach J, Mohammed S, Hweck A, Maurice M, Mahmoudi T, Clevers H (2012) Wnt signaling inhibits proteasomal β-catenin degradation within a compositionally intact Axin1 complex. Cell (in press) [DOI] [PubMed] [Google Scholar]
- Li Y, Welm B, Podsypanina K, Huang S, Chamorro M, Zhang X, Rowlands T, Egeblad M, Cowin P, Werb Z, Tan LK, Rosen JM, Varmus HE (2003) Evidence that transgenes encoding components of the Wnt signaling pathway preferentially induce mammary cancers from progenitor cells. Proc Natl Acad Sci USA 100: 15853–15858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Thompson S, Priess JR (1995) pop-1 encodes an HMG box protein required for the specification of a mesoderm precursor in early C. elegans embryos. Cell 83: 599–609 [DOI] [PubMed] [Google Scholar]
- Lustig B, Jerchow B, Sachs M, Weiler S, Pietsch T, Karsten U, van de Wetering M, Clevers H, Schlag PM, Birchmeier W, Behrens J (2002) Negative feedback loop of Wnt signaling through upregulation of conductin/axin2 in colorectal and liver tumors. Mol Cell Biol 22: 1184–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons MJ, Moore DH (1962) Purification of the mouse mammary tumour virus. Nature 194: 1141–1142 [DOI] [PubMed] [Google Scholar]
- MacArthur CA, Shankar DB, Shackleford GM (1995) Fgf-8, activated by proviral insertion, cooperates with the Wnt-1 transgene in murine mammary tumorigenesis. J Virol 69: 2501–2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald BT, Tamai K, He X (2009) Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell 17: 9–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T (1999) A ubiquitin ligase complex essential for the NF-kappaB, Wnt/Wingless, and Hedgehog signaling pathways. Genes Dev 13: 505–510 [DOI] [PubMed] [Google Scholar]
- Mao B, Wu W, Li Y, Hoppe D, Stannek P, Glinka A, Niehrs C (2001a) LDL-receptor-related protein 6 is a receptor for Dickkopf proteins. Nature 411: 321–325 [DOI] [PubMed] [Google Scholar]
- Mao J, Wang J, Liu B, Pan W, Farr GH III, Flynn C, Yuan H, Takada S, Kimelman D, Li L, Wu D (2001b) Low-density lipoprotein receptor-related protein-5 binds to Axin and regulates the canonical Wnt signaling pathway. Mol Cell 7: 801–809 [DOI] [PubMed] [Google Scholar]
- McCrea PD, Brieher WM, Gumbiner BM (1993) Induction of a secondary body axis in Xenopus by antibodies to beta-catenin. J Cell Biol 123: 477–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCrea PD, Turck CW, Gumbiner B (1991) A homolog of the armadillo protein in Drosophila (plakoglobin) associated with E-cadherin. Science 254: 1359–1361 [DOI] [PubMed] [Google Scholar]
- McMahon AP, Bradley A (1990) The Wnt-1 (int-1) proto-oncogene is required for development of a large region of the mouse brain. Cell 62: 1073–1085 [DOI] [PubMed] [Google Scholar]
- McMahon AP, Moon RT (1989) Ectopic expression of the proto-oncogene int-1 in Xenopus embryos leads to duplication of the embryonic axis. Cell 58: 1075–1084 [DOI] [PubMed] [Google Scholar]
- Mikels AJ, Nusse R (2006) Purified Wnt5a protein activates or inhibits beta-catenin-TCF signaling depending on receptor context. PLoS Biol 4: e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molenaar M, van de Wetering M, Oosterwegel M, Peterson-Maduro J, Godsave S, Korinek V, Roose J, Destree O, Clevers H (1996) XTcf-3 transcription factor mediates beta-catenin-induced axis formation in Xenopus embryos. Cell 86: 391–399 [DOI] [PubMed] [Google Scholar]
- Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler KW (1997) Activation of b-catenin-Tcf signaling in colon cancer by mutations in b-catenin or APC. Science 275: 1787–1790 [DOI] [PubMed] [Google Scholar]
- Muller HA, Samanta R, Wieschaus E (1999) Wingless signaling in the Drosophila embryo: zygotic requirements and the role of the frizzled genes. Development 126: 577–586 [DOI] [PubMed] [Google Scholar]
- Nelson WJ, Nusse R (2004) Convergence of Wnt, beta-catenin, and cadherin pathways. Science 303: 1483–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehrs C (2010) On growth and form: a Cartesian coordinate system of Wnt and BMP signaling specifies bilaterian body axes. Development (Cambridge, England) 137: 845–857 [DOI] [PubMed] [Google Scholar]
- Noordermeer J, Klingensmith J, Perrimon N, Nusse R (1994) dishevelled and armadillo act in the wingless signalling pathway in Drosophila. Nature 367: 80–83 [DOI] [PubMed] [Google Scholar]
- Nusse R, Brown A, Papkoff J, Scambler P, Shackleford G, McMahon A, Moon R, Varmus H (1991) A new nomenclature for int-1 and related genes: the Wnt gene family. Cell 64: 231. [DOI] [PubMed] [Google Scholar]
- Nusse R, van Ooyen A, Cox D, Fung YK, Varmus HE (1984) Mode of proviral activation of a putative mammary oncogene (int-1) on mouse chromosome 15. Nature 307: 131–136 [DOI] [PubMed] [Google Scholar]
- Nusse R, Varmus HE (1982) Many tumors induced by the mouse mammary tumor virus contain a provirus integrated in the same region of the host genome. Cell 31: 99–109 [DOI] [PubMed] [Google Scholar]
- Nusse R, Varmus HE (1992) Wnt genes. Cell 69: 1073–1087 [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C, Wieschaus E (1980) Mutations affecting segment number and polarity in Drosophila. Nature 287: 795–801 [DOI] [PubMed] [Google Scholar]
- Oishi I, Suzuki H, Onishi N, Takada R, Kani S, Ohkawara B, Koshida I, Suzuki K, Yamada G, Schwabe GC, Mundlos S, Shibuya H, Takada S, Minami Y (2003) The receptor tyrosine kinase Ror2 is involved in non-canonical Wnt5a/JNK signalling pathway. Genes Cells 8: 645–654 [DOI] [PubMed] [Google Scholar]
- Orford K, Crockett C, Jensen JP, Weissman AM, Byers SW (1997) Serine phosphorylation-regulated ubiquitination and degradation of beta-catenin. J Biol Chem 272: 24735–24738 [DOI] [PubMed] [Google Scholar]
- Ozawa M, Baribault H, Kemler R (1989) The cytoplasmic domain of the cell adhesion molecule uvomorulin associates with three independent proteins structurally related in different species. EMBO J 8: 1711–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parr BA, McMahon AP (1995) Dorsalizing signal Wnt-7a required for normal polarity of D-V and A-P axes of mouse limb. Nature 374: 350–353 [DOI] [PubMed] [Google Scholar]
- Parr BA, McMahon AP (1998) Sexually dimorphic development of the mammalian reproductive tract requires Wnt-7a. Nature 395: 707–710 [DOI] [PubMed] [Google Scholar]
- Patthy L (2000) The WIF module. Trends Biochem Sci 25: 12–13 [DOI] [PubMed] [Google Scholar]
- Payne GS, Bishop JM, Varmus HE (1982) Multiple arrangements of viral DNA and an activated host oncogene in bursal lymphomas. Nature 295: 209–214 [DOI] [PubMed] [Google Scholar]
- Payne GS, Courtneidge SA, Crittenden LB, Fadly AM, Bishop JM, Varmus HE (1981) Analysis of avian leukosis virus DNA and RNA in bursal tumours: viral gene expression is not required for maintenance of the tumor state. Cell 23: 311–322 [DOI] [PubMed] [Google Scholar]
- Peifer M, Orsulic S, Pai LM, Loureiro J (1993) A model system for cell adhesion and signal transduction in Drosophila. Development Suppl: 163–176 [PubMed] [Google Scholar]
- Peifer M, Pai LM, Casey M (1994) Phosphorylation of the Drosophila adherens junction protein Armadillo: roles for wingless signal and zeste-white 3 kinase. Dev Biol 166: 543–556 [DOI] [PubMed] [Google Scholar]
- Peifer M, Rauskolb C, Williams M, Riggleman B, Wieschaus E (1991) The segment polarity gene armadillo interacts with the wingless signaling pathway in both embryonic and adult pattern formation. Development 111: 1029–1043 [DOI] [PubMed] [Google Scholar]
- Peifer M, Wieschaus E (1990) The segment polarity gene armadillo encodes a functionally modular protein that is the Drosophila homolog of human plakoglobin. Cell 63: 1167–1176 [DOI] [PubMed] [Google Scholar]
- Perrimon N, Mahowald AP (1987) Multiple functions of segment polarity genes in Drosophila. Dev Biol 119: 587–600 [DOI] [PubMed] [Google Scholar]
- Peters G, Lee AE, Dickson C (1986) Concerted activation of two potential proto-oncogenes in carcinomas induced by mouse mammary tumour virus. Nature 320: 628–631 [DOI] [PubMed] [Google Scholar]
- Petersen CP, Reddien PW (2009) Wnt signaling and the polarity of the primary body axis. Cell 139: 1056–1068 [DOI] [PubMed] [Google Scholar]
- Pierce SB, Kimelman D (1996) Overexpression of Xgsk-3 disrupts anterior ectodermal patterning in Xenopus. Dev Biol 175: 256–264 [DOI] [PubMed] [Google Scholar]
- Pinson KI, Brennan J, Monkley S, Avery BJ, Skarnes WC (2000) An LDL-receptor-related protein mediates Wnt signalling in mice. Nature 407: 535–538 [DOI] [PubMed] [Google Scholar]
- Port F, Basler K (2010) Wnt Trafficking: New Insights into Wnt Maturation, Secretion and Spreading Copenhagen, Denmark: Traffic, [DOI] [PubMed] [Google Scholar]
- Rattner A, Hsieh JC, Smallwood PM, Gilbert DJ, Copeland NG, Jenkins NA, Nathans J (1997) A family of secreted proteins contains homology to the cysteine-rich ligand-binding domain of frizzled receptors. Proc Natl Acad Sci USA 94: 2859–2863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riese J, Yu X, Munnerlyn A, Eresh S, Hsu SC, Grosschedl R, Bienz M (1997) LEF-1, a nuclear factor coordinating signaling inputs from wingless and decapentaplegic. Cell 88: 777–787 [DOI] [PubMed] [Google Scholar]
- Riggleman B, Schedl P, Wieschaus E (1990) Spatial expression of the Drosophila segment polarity gene armadillo is posttranscriptionally regulated by wingless. Cell 63: 549–560 [DOI] [PubMed] [Google Scholar]
- Rijsewijk F, Schuermann M, Wagenaar E, Parren P, Weigel D, Nusse R (1987a) The Drosophila homolog of the mouse mammary oncogene int-1 is identical to the segment polarity gene wingless. Cell 50: 649–657 [DOI] [PubMed] [Google Scholar]
- Rijsewijk F, van Deemter L, Wagenaar E, Sonnenberg A, Nusse R (1987b) Transfection of the int-1 mammary oncogene in cuboidal RAC mammary cell line results in morphological transformation and tumorigenicity. EMBO J 6: 127–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robitaille J, MacDonald ML, Kaykas A, Sheldahl LC, Zeisler J, Dube MP, Zhang LH, Singaraja RR, Guernsey DL, Zheng B, Siebert LF, Hoskin-Mott A, MT Trese, Pimstone SN, Shastry BS, Moon RT, Hayden MR, Goldberg YP, Samuels ME (2002) Mutant frizzled-4 disrupts retinal angiogenesis in familial exudative vitreoretinopathy. Nat Genet 32: 326–330 [DOI] [PubMed] [Google Scholar]
- Rocheleau CE, Downs WD, Lin R, Wittmann C, Bei Y, Cha YH, Ali M, Priess JR, Mello CC (1997) Wnt signaling and an APC-related gene specify endoderm in early C. elegans embryos. Cell 90: 707–716 [DOI] [PubMed] [Google Scholar]
- Rocheleau CE, Yasuda J, Shin TH, Lin R, Sawa H, Okano H, Priess JR, Davis RJ, Mello CC (1999) WRM-1 activates the LIT-1 protein kinase to transduce anterior/posterior polarity signals in C. elegans. Cell 97: 717–726 [DOI] [PubMed] [Google Scholar]
- Roelink H, Wagenaar E, Lopes da Silva S, Nusse R (1990) Wnt-3, a gene activated by proviral insertion in mouse mammary tumors, is homologous to int-1/Wnt-1 and is normally expressed in mouse embryos and adult brain. Proc Natl Acad Sci USA 87: 4519–4523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinfeld B, Albert I, Porfiri E, Fiol C, Munemitsu S, Polakis P (1996) Binding of GSK3beta to the APC-beta-catenin complex and regulation of complex assembly. Science (New York, NY) 272: 1023–1026 [DOI] [PubMed] [Google Scholar]
- Rubinfeld B, Robbins P, El-Gamil M, Albert I, Porfiri E, Polakis P (1997) Stabilization of beta-catenin by genetic defects in melanoma cell lines. Science 275: 1790–1792 [DOI] [PubMed] [Google Scholar]
- Rubinfeld B, Souza B, Albert I, Müller O, Chamberlain SH, Masiarz FR, Munemitsu S, Polakis P (1993) Association of the APC gene product with beta-catenin. Science (New York, NY) 262: 1731–1734 [DOI] [PubMed] [Google Scholar]
- Rulifson EJ, Wu CH, Nusse R (2000) Pathway specificity by the bifunctional receptor frizzled is determined by affinity for wingless. Mol Cell 6: 117–126 [PubMed] [Google Scholar]
- Sato T, van Es JH, Snippert HJ, Stange DE, Vries RG, van den Born M, Barker N, Shroyer NF, van de Wetering M, Clevers H (2010) Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 469: 415–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh S, Daigo Y, Furukawa Y, Kato T, Miwa N, Nishiwaki T, Kawasoe T, Ishiguro H, Fujita M, Tokino T, Sasaki Y, Imaoka S, Murata M, Shimano T, Yamaoka Y, Nakamura Y (2000) AXIN1 mutations in hepatocellular carcinomas, and growth suppression in cancer cells by virus-mediated transfer of AXIN1. Nat Genet 24: 245–250 [DOI] [PubMed] [Google Scholar]
- Sawa H, Lobel L, Horvitz HR (1996) The Caenorhabditis elegans gene lin-17, which is required for certain asymmetric cell divisions, encodes a putative seven-transmembrane protein similar to the Drosophila frizzled protein. Genes Dev 10: 2189–2197 [DOI] [PubMed] [Google Scholar]
- Schneider SQ, Bowerman B (2007) beta-Catenin asymmetries after all animal/vegetal-oriented cell divisions in Platynereis dumerilii embryos mediate binary cell-fate specification. Dev Cell 13: 73–86 [DOI] [PubMed] [Google Scholar]
- Schwarz-Romond T, Fiedler M, Shibata N, Butler PJ, Kikuchi A, Higuchi Y, Bienz M (2007) The DIX domain of Dishevelled confers Wnt signaling by dynamic polymerization. Nat Struct Mol Biol 14: 484–492 [DOI] [PubMed] [Google Scholar]
- Semenov MV, Tamai K, Brott BK, Kuhl M, Sokol S, He X (2001) Head inducer Dickkopf-1 is a ligand for Wnt coreceptor LRP6. Curr Biol 11: 951–961 [DOI] [PubMed] [Google Scholar]
- Shackleford GM, Varmus HE (1987) Expression of the proto-oncogene int-1 is restricted to postmeiotic male germ cells and the neural tube of mid-gestational embryos. Cell 50: 89–95 [DOI] [PubMed] [Google Scholar]
- Shackleton M, Vaillant F, Simpson KJ, Stingl J, Smyth GK, Asselin-Labat ML, Wu L, Lindeman GJ, Visvader JE (2006) Generation of a functional mammary gland from a single stem cell. Nature 439: 84–88 [DOI] [PubMed] [Google Scholar]
- Sharma RP, Chopra VL (1976) Effect of the Wingless (wg1) mutation on wing and haltere development in Drosophila melanogaster. Dev Biol 48: 461–465 [DOI] [PubMed] [Google Scholar]
- Sidow A (1992) Diversification of the Wnt gene family on the ancestral lineage of vertebrates. Proc Natl Acad Sci USA 89: 5098–5102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegfried E, Chou TB, Perrimon N (1992) Wingless signaling acts through zeste-white 3, the Drosophila homolog of glycogen synthase kinase-3, to regulate engrailed and establish cell fate. Cell 71: 1167–1179 [DOI] [PubMed] [Google Scholar]
- Siegfried E, Wilder EL, Perrimon N (1994) Components of wingless signalling in Drosophila. Nature 367: 76–80 [DOI] [PubMed] [Google Scholar]
- Su LK, Kinzler KW, Vogelstein B, Preisinger AC, Moser AR, Luongo C, Gould KA, Dove WF (1992) Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science 256: 668–670 [DOI] [PubMed] [Google Scholar]
- Su LK, Vogelstein B, Kinzler KW (1993) Association of the APC tumor suppressor protein with catenins. Science 262: 1734–1737 [DOI] [PubMed] [Google Scholar]
- Sugioka K, Mizumoto K, Sawa H (2011) Wnt regulates spindle asymmetry to generate asymmetric nuclear beta-catenin in C. elegans. Cell 146: 942–954 [DOI] [PubMed] [Google Scholar]
- Takeichi M (1990) Cadherins: a molecular family important in selective cell-cell adhesion. Annu Rev Biochem 59: 237–252 [DOI] [PubMed] [Google Scholar]
- Tamai K, Semenov M, Kato Y, Spokony R, Liu C, Katsuyama Y, Hess F, Saint-Jeannet JP, He X (2000) LDL-receptor-related proteins in Wnt signal transduction. Nature 407: 530–535 [DOI] [PubMed] [Google Scholar]
- Tamai K, Zeng X, Liu C, Zhang X, Harada Y, Chang Z, He X (2004) A mechanism for Wnt coreceptor activation. Mol Cell 13: 149–156 [DOI] [PubMed] [Google Scholar]
- Teich N, Wyke J, Mak T, Bernstein A, Hardy W (1982) Pathogenesis of Retrovirus-indiced disease. InRNA Tumor Viruses Weis R, Teich N, Varmus H, Coffin JM (eds)pp785–998Cold Spring Harbor: Cold Spring Harbor, [Google Scholar]
- ten Berge D, Kurek D, Blauwkamp T, Koole W, Maas A, Eroglu E, Siu RK, Nusse R (2011) Embryonic stem cells require Wnt proteins to prevent differentiation to epiblast stem cells. Nat Cell Biol 13: 1070–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas KR, Capecchi MR (1990) Targeted disruption of the murine int-1 proto-oncogene resulting in severe abnormalities in midbrain and cerebellar development. Nature 346: 847–850 [DOI] [PubMed] [Google Scholar]
- Thomas KR, Musci TS, Neumann PE, Capecchi MR (1991) Swaying is a mutant allele of the proto-oncogene Wnt-1. Cell 67: 969–976 [DOI] [PubMed] [Google Scholar]
- Thorpe CJ, Schlesinger A, Carter JC, Bowerman B (1997) Wnt signaling polarizes an early C. elegans blastomere to distinguish endoderm from mesoderm. Cell 90: 695–705 [DOI] [PubMed] [Google Scholar]
- Travis A, Amsterdam A, Belanger C, Grosschedl R (1991) LEF-1, a gene encoding a lymphoid-specific protein with an HMG domain, regulates T-cell receptor alpha enhancer function [corrected]. Genes Dev 5: 880–894 [DOI] [PubMed] [Google Scholar]
- Tsukamoto AS, Grosschedl R, Guzman RC, Parslow T, Varmus HE (1988) Expression of the int-1 gene in transgenic mice is associated with mammary gland hyperplasia and adenocarcinomas in male and female mice. Cell 55: 619–625 [DOI] [PubMed] [Google Scholar]
- van Amerongen R, Bowman A, Nusse R (2012) Developmental stage and time dictate the fate of Wnt/β-catenin responsive stem cells in the mammary gland. Cell Stem Cell (in press) [DOI] [PubMed] [Google Scholar]
- van Amerongen R, Mikels A, Nusse R (2008) Alternative wnt signaling is initiated by distinct receptors. Sci Signal 1: re9. [DOI] [PubMed] [Google Scholar]
- van de Wetering M, Oosterwegel M, Dooijes D, Clevers H (1991) Identification and cloning of TCF-1, a T lymphocyte-specific transcription factor containing a sequence-specific HMG box. EMBO J 10: 123–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leeuwen F, Harryman Samos C, Nusse R (1994) Biological activity of soluble wingless protein in cultured Drosophila imaginal disc cells. Nature 368: 342–344 [DOI] [PubMed] [Google Scholar]
- Van Ooyen A, Kwee V, Nusse R (1985) The nucleotide sequence of the human int-1 mammary oncogene; evolutionary conservation of coding and non-coding sequences. EMBO J 4: 2905–2909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ooyen A, Nusse R (1984) Structure and nucleotide sequence of the putative mammary oncogene int-1; proviral insertions leave the protein-encoding domain intact. Cell 39: 233–240 [DOI] [PubMed] [Google Scholar]
- Veeman MT, Axelrod JD, Moon RT (2003) A second canon. Functions and mechanisms of beta-catenin-independent Wnt signaling. Dev Cell 5: 367–377 [DOI] [PubMed] [Google Scholar]
- Vinson CR, Adler PN (1987) Directional non-cell autonomy and the transmission of polarity information by the frizzled gene of Drosophila. Nature 329: 549–551 [DOI] [PubMed] [Google Scholar]
- Vinson CR, Conover S, Adler PN (1989) A Drosophila tissue polarity locus encodes a protein containing seven potential transmembrane domains. Nature 338: 263–264 [DOI] [PubMed] [Google Scholar]
- Waterman ML, Fischer WH, Jones KA (1991) A thymus-specific member of the HMG protein family regulates the human T cell receptor C alpha enhancer. Genes Dev 5: 656–669 [DOI] [PubMed] [Google Scholar]
- Wehrli M, Dougan ST, Caldwell K, O’Keefe L, Schwartz S, Vaizel-Ohayon D, Schejter E, Tomlinson A, DiNardo S (2000) arrow encodes an LDL-receptor-related protein essential for Wingless signalling. Nature 407: 527–530 [DOI] [PubMed] [Google Scholar]
- Weinberg RA (1983) A molecular basis of cancer. Sci Am 249: 126–142 [DOI] [PubMed] [Google Scholar]
- Wilkinson DG, Bailes JA, McMahon AP (1987) Expression of the proto-oncogene int-1 is restricted to specific neural cells in the developing mouse embryo. Cell 50: 79–88 [DOI] [PubMed] [Google Scholar]
- Willert K, Brown JD, Danenberg E, Duncan AW, Weissman IL, Reya T, Yates JR, Nusse R (2003) Wnt proteins are lipid-modified and can act as stem cell growth factors. Nature 423: 448–452 [DOI] [PubMed] [Google Scholar]
- Yang-Snyder J, Miller JR, Brown JD, Lai CJ, Moon RT (1996) A frizzled homolog functions in a vertebrate Wnt signaling pathway. Curr Biol 6: 1302–1306 [DOI] [PubMed] [Google Scholar]
- Ye X, Wang Y, Rattner A, Nathans J (2011) Genetic mosaic analysis reveals a major role for frizzled 4 and frizzled 8 in controlling ureteric growth in the developing kidney. Development 138: 1161–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa S, McKinnon RD, Kokel M, Thomas JB (2003) Wnt-mediated axon guidance via the Drosophila Derailed receptor. Nature 422: 583–588 [DOI] [PubMed] [Google Scholar]
- Yost C, Torres M, Miller JR, Huang E, Kimelman D, Moon RT (1996) The axis-inducing activity, stability, and subcellular distribution of beta-catenin is regulated in Xenopus embryos by glycogen synthase kinase 3. Genes Dev 10: 1443–1454 [DOI] [PubMed] [Google Scholar]
- Zeng L, Fagotto F, Zhang T, Hsu W, Vasicek TJ, Perry WL III, Lee JJ, Tilghman SM, Gumbiner BM, Costantini F (1997) The mouse Fused locus encodes Axin, an inhibitor of the Wnt signaling pathway that regulates embryonic axis formation. Cell 90: 181–192 [DOI] [PubMed] [Google Scholar]
- Zeng X, Tamai K, Doble B, Li S, Huang H, Habas R, Okamura H, Woodgett J, He X (2005) A dual-kinase mechanism for Wnt co-receptor phosphorylation and activation. Nature 438: 873–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng YA, Nusse R (2010) Wnt proteins are self-renewal factors for mammary stem cells and promote their long-term expansion in culture. Stem Cell 6: 568–577 [DOI] [PMC free article] [PubMed] [Google Scholar]