Abstract
β-Catenin (Armadillo in Drosophila) is a multitasking and evolutionary conserved molecule that in metazoans exerts a crucial role in a multitude of developmental and homeostatic processes. More specifically, β-catenin is an integral structural component of cadherin-based adherens junctions, and the key nuclear effector of canonical Wnt signalling in the nucleus. Imbalance in the structural and signalling properties of β-catenin often results in disease and deregulated growth connected to cancer and metastasis. Intense research into the life of β-catenin has revealed a complex picture. Here, we try to capture the state of the art: we try to summarize and make some sense of the processes that regulate β-catenin, as well as the plethora of β-catenin binding partners. One focus will be the interaction of β-catenin with different transcription factors and the potential implications of these interactions for direct cross-talk between β-catenin and non-Wnt signalling pathways.
Keywords: β-catenin, cell adhesion, cell signalling, transcription, Wnts
Introduction
Wnt signalling represents one of a few key molecular cascades that regulate cell fate in animals throughout their lifespan. Already during embryonic development Wnt-regulated β-catenin critically contributes to the establishment of the body axis and the orchestration of tissue and organ development. In adult organs, Wnt signalling continues to play indispensable roles in tissue homeostasis, cell renewal, and regeneration. Interaction of Wnt ligands with their receptor complexes triggers several intracellular signalling cascades; these are, traditionally, separated into two types according to the role played by β-catenin. In the canonical Wnt cascade, β-catenin is the key effector responsible for transduction of the signal to the nucleus and it triggers transcription of Wnt-specific genes responsible for the control of cell fate decisions in many cells and tissues. The second type of Wnt signalling is independent of β-catenin signalling function and comprises, among others, the Wnt/PCP (Planar Cell Polarity) and Wnt-dependent/protein kinase C (PKC)-dependent pathways (van Amerongen et al, 2008; Angers and Moon, 2009). We will focus here on β-catenin-dependent signalling and refer the interested reader to the following excellent reviews on the other types of Wnt signalling (van Amerongen et al, 2008; Angers and Moon, 2009).
But β-catenin is not just a component of the Wnt signal cascade (Figure 1). In the late 1980s, β-catenin was independently discovered twice, on the basis of its different functions: structural and signalling. The group of Rolf Kemler isolated β-catenin, together with two other molecules (α-catenin and γ-catenin/plakoglobin), as proteins associated with E-cadherin, the key molecule of Ca2+-dependent cell adhesion. These proteins were named catenins (in Latin catena means chain) to reflect their linking of E-cadherin to cytoskeletal structures (Ozawa et al, 1989). The signalling potential of β-catenin was exposed through its Drosophila orthologue Armadillo: the armadillo gene was discovered in the seminal screens for mutations affecting segmentation of the Drosophila embryo, performed by Eric Wieschaus, Christiane Nüsslein-Volhard, and Gerd Jürgens (Wieschaus et al, 1984). Whereas the wild-type embryonic cuticle contains segments with alternating rows of denticles and naked belts, segments in armadillo mutants form only a lawn of denticles, the naked belts are missing. Such phenotypes resemble wingless null mutants (Wieschaus and Riggleman, 1987). Further analysis of Armadillo performed by the laboratories of Eric Wieschaus, Mark Peifer, and others revealed the conservation of its structural function in adherens junctions (McCrea et al, 1990; Peifer and Wieschaus, 1990; Orsulic and Peifer, 1996). Epistatic analysis determined that the armadillo segmentation function is regulated by Wingless (Riggleman et al, 1990). This finding was a key step in the subsequent characterization of the Wnt/β-catenin (or Wingless/Armadillo, respectively) signalling cascade, and of the functions and mutual interactions of its individual components. Another important part of this mosaic was revealed by the description of the basic pathway leading from the Wingless ligand through Dishevelled to regulation of Armadillo stability by Shaggy/Zeste-white-3 (GSK3 in vertebrates) (Siegfried et al, 1994). Finally in the mid-1990’s several groups independently found that the signalling function of β-catenin/Armadillo in the nucleus is mediated via T-cell factor (TCF)/Lymphoid enhancer-binding factor (Lef) transcription factors, which in association with β-catenin trigger Wnt-mediated transcription (Behrens et al, 1996; Huber et al, 1996; Molenaar et al, 1996; Brunner et al, 1997; van de Wetering et al, 1997). The generation of conditional β-catenin mouse mutants (either knockout or constitutively active) has revealed that a vast array of developmental processes is regulated by the multitasking β-catenin in mammalian embryonic and adult tissues (reviewed in detail by Grigoryan et al, 2008).
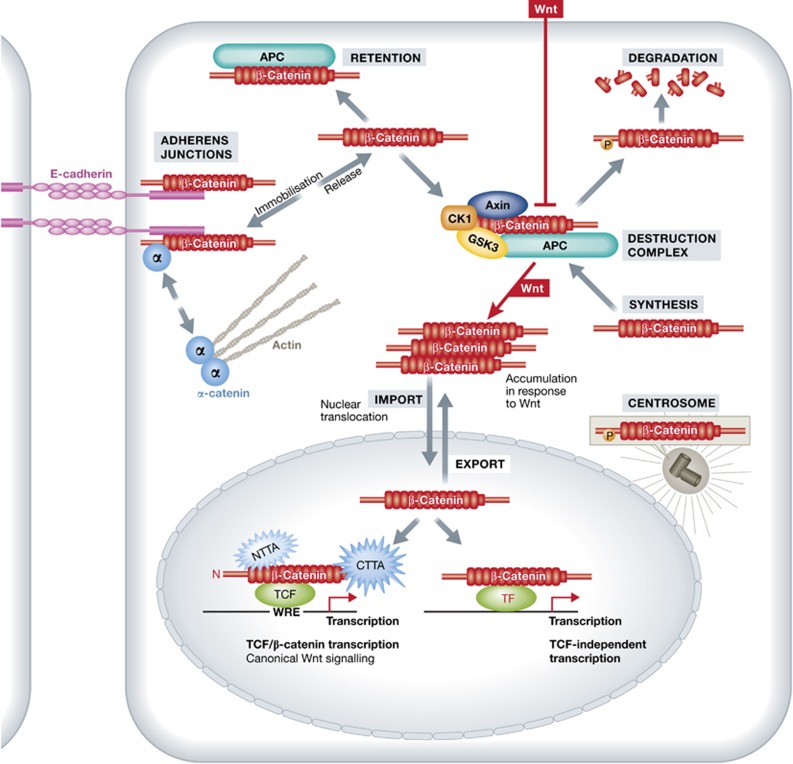
Figure 1.
The life of β-catenin within the cell. Newly synthesized β-catenin is immobilized by E-cadherin at adherens junctions, where it can interact also with α-catenin, thereby indirectly modulating the actin cytoskeleton. β-catenin can be released from the adherens junctions by the activity of protein kinases or by downregulation of E-cadherin. Free excess β-catenin is immediately phosphorylated by the destruction complex and thus marked for subsequent degradation. A portion of β-catenin can be kept in the cytoplasm protected by APC. Wnt signalling blocks the activity of the destruction complex resulting in increased levels of cytolasmic β-catenin, which is translocated to the nucleus. In the nucleus, β-catenin associates with transcription factors from the TCF/Lef family and drives transcription of Wnt/β-catenin target genes. Other factors can also provide β-catenin with a DNA binding platform, often counteracting canonical Wnt signalling. Signalling activity of β-catenin in the nucleus can be regulated by modulating its nuclear import/export. Besides its structural role in the adherens junctions and signalling activity in the nucleus, β-catenin may also play an important function in the centrosome. CTTA, C-Terminal Transcriptional Activators, NTTA, N-Terminal Transcriptional Activators.
The many roles of β-catenin beg the question as to how an evolutionarily conserved pathway can control so many varied processes during animal development and tissue homeostasis via one central molecule? Further, how is the final transcriptional output of β-catenin determined and modulated? Part of the explanation might be found in the plethora of β-catenin binding partners that either affect its transcriptional output or permit its direct cross-talk with other transcription factors and signalling pathways. In this review, we will discuss the following aspects: how is the β-catenin protein regulated, how might different transcriptional co-activators of β-catenin lead to diverse outcomes, how can β-catenin affect the activity of various transcription factors, and finally, what are the potential implications of a direct cross-talk between β-catenin and other signalling pathways?
The structure of β-catenin determines its role as a scaffold molecule
The Wnt/β-catenin pathway is one of a small set of conserved signalling cascades (together with Notch, Hedgehog, TGFβ/BMP, Hippo, and receptor tyrosine kinase-mediated pathways) that regulate animal development from Cnidarians to mammals. β-Catenin’s orchestration of developmental processes is the sum of its dual roles—signalling and structural—as elegantly illustrated by the work of Lyashenko et al (2011): mouse Emryonic Stem Cells (mESC) lacking β-catenin lose their ability to differentiate into a mesodermal germ layer and do not form any neuroepithelium; both defects are connected to defective cell–cell junctions arising during the differentiation processes. Putting back a signalling-defective β-catenin restores the integrity of adherens junction and importantly also the ability of mESC to form neuroepithelial structures and endoderm. However the mesoderm formation is not rescued and thus requires also intact β-catenin transcription (Lyashenko et al, 2011). Similarly in the developing dorsal neural tube and the migrating population of neural crest cells the loss of β-catenin results in more drastic phenotypes than only blocking the β-catenin signalling outputs, further showing that the two roles of β-catenin cause an additive effect (Valenta et al, 2011). Such overlap and possible separation of β-catenin roles was also demonstrated in a seminal analysis in Drosophila more than a decade ago (Orsulic and Peifer, 1996).
How can the β-catenin protein mediate both distinct adhesive and signalling activities? The answer is in the structural composition of β-catenin. The β-catenin protein (781 aa residues in humans) consists of a central region (residues 141–664) made up of 12 imperfect Armadillo repeats (R1–12) that are flanked by distinct N- and C-terminal domains, NTD and CTD, respectively. A specific conserved helix (Helix-C) is located proximally to the CTD, adjacent to the last ARM repeat (residues 667–683) (Xing et al, 2008; Figure 2). The NTD and the CTD may be structurally flexible, whereas the central region forms a relatively rigid scaffold. This scaffold serves as an interaction platform for many β-catenin binding partners, at the membrane, in cytosol, and in the nucleus (Huber et al, 1997).
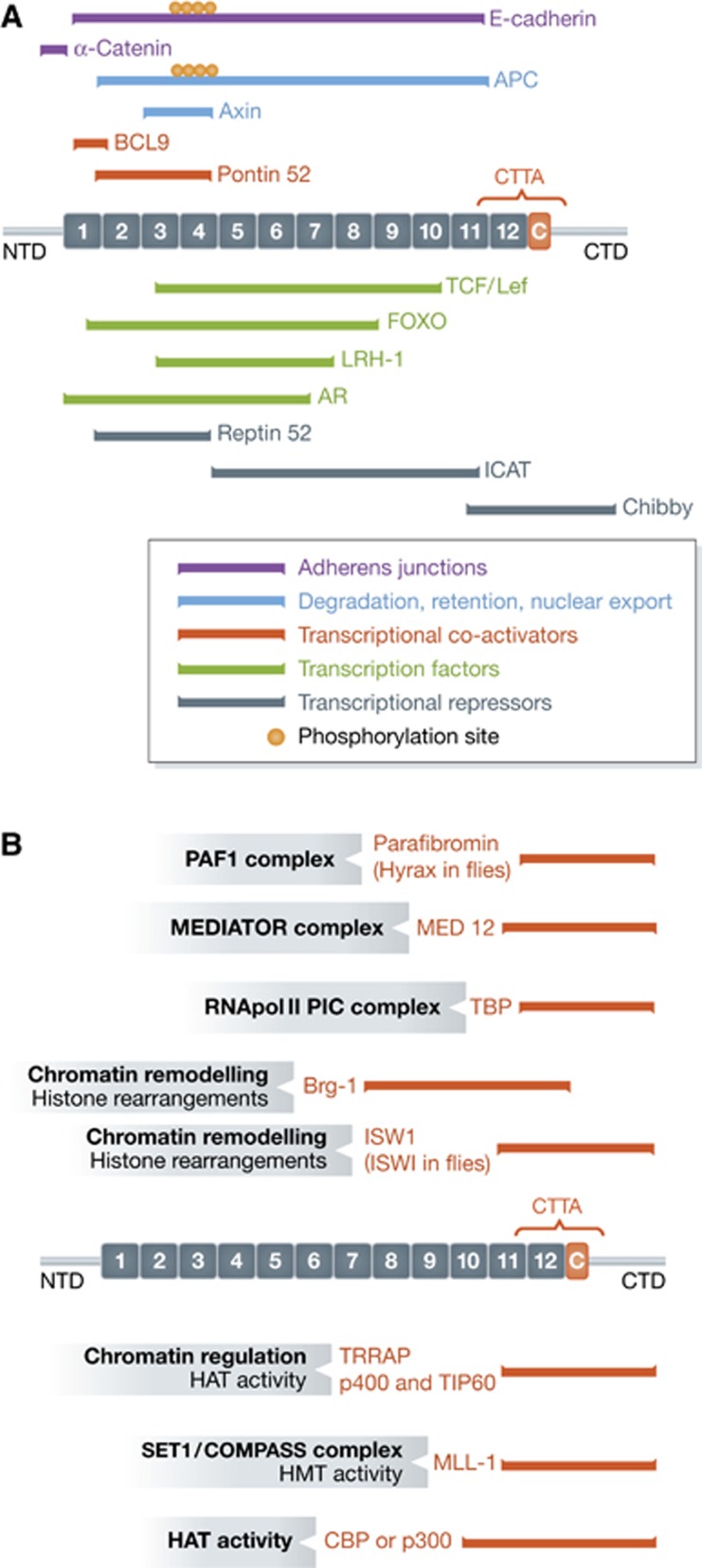
Figure 2.
β-Catenin serves as a binding platform for a multitude of interaction partners in adherens junctions, in the cytoplasm and in the nucleus. (A) The β-catenin protein consists of a central region composed of 12 Armadillo repeats (numbered boxes), flanked by an amino-terminal domain (NTD) and a carboxy-terminal domain (CTD). Between the last Armadillo repeat and the flexible part of the CTD is the conserved helix-C (C). Coloured bars show experimentally validated binding sites for β-catenin interaction partners. Colour code: purple, components of adherens junctions; blue, members of the β-catenin destruction complex; red, transcriptional co-activators; green, transcription factors providing DNA binding; gray, transcriptional inhibitors. C-Terminal Transcriptional Activators (CTTA), the critical domain for their binding is marked by brackets. Little circles indicate phosphorylation sites on either E-cadherin or APC that enhance the interactions. APC, Adenoma Polyposis Coli; TCF/Lef, T-cell factor/Lymphoid enhancer factor; AR, Androgen Receptor; LRH-1, Liver Receptor Homologue-1; ICAT, Inhibitor of β-catenin and TCF; BCL9, B-cell lymphoma-9. (B) The C-terminus of β-catenin serves as a binding factor for a multitude of complexes promoting β-catenin-mediated transcription. Experimentally validated binding motifs for particular proteins are indicated. In the grey boxes, the function is indicated of the particular β-catenin interactor or of a complex, where this binding partner is a member. Brg-1 is also known as SMARCA4, CBP as CREBBP. HAT, histone acetyl-transferase; HMT, histone methyl-transferase; MLL, mixed lineage leukaemia; PAF-1, Polymerase-associated factor-1; PIC, Pre-Initiation Complex; TBP, TATA-box Binding Protein; TRRAP, Transformation/transcription domain-associated protein.
β-Catenin is a founding member of the Armadillo (ARM) repeat protein superfamily. Each ARM repeat of its central region comprises ∼42 residues, forming three helices arranged in triangular shape. Together, all ARM repeats form a superhelix that features a long, positively charged groove. Biochemical and crystal structure analyses revealed that many of β-catenin’s binding partners share overlapping binding sites in the groove of the central β-catenin region: consequently, these partners cannot bind to β-catenin simultaneously. This mutual exclusivity is certainly valid for the key β-catenin interacting molecules: E-cadherin (the main partner in adherens junctions), APC (the main partner in the destruction complex), and TCF/Lef (the main partner in the nucleus). All these β-catenin interactors bind to the core binding site comprising ARM repeats R3–R9, where they form salt bridges with two key amino-acid residues, Lys312 and Lys435. Other ARM repeats are also involved, at least in strengthening the interaction (Graham et al, 2000; Eklof Spink et al, 2001; Huber and Weis, 2001; Poy et al, 2001).
The spatial segregation of the different β-catenin binding partners within the cell may be important for enabling the function of these proteins. However, the competition among them for β-catenin is also important for regulating canonical Wnt signalling. Conformational changes of β-catenin may also help regulate its binding properties: the terminal regions of β-catenin (NTD and especially CTD) have been proposed to fold back on the central region affecting, for example, the binding of TCF/Lef (Castaño et al, 2002; Solanas et al, 2004). Biochemical evidence supports the model that β-catenin-mediated transcription is performed by a monomeric, back-folded form of β-catenin, whereas a cadherin-binding dimeric form is associated with α-catenin in adherens junctions (Gottardi and Gumbiner, 2004). Interestingly, Helix-C within the most N-terminal part of the CTD was shown to be essential for the signalling activity of β-catenin, while being completely dispensable for its role in cell–cell adhesion (Xing et al, 2008). This is not surprising, as a plethora of β-catenin transcriptional co-activators require an intact Helix-C (or region R12-Helix-C) for their proper binding (Figure 2; Mosimann et al, 2009). The crucial role of this part of β-catenin was predicted and experimentally demonstrated more than a decade ago by elegant in-vivo analyses of different mutants of Armadillo (Orsulic and Peifer, 1996). In these studies, mutants of Armadillo (point mutations or deletions) were produced, some interfered with the adhesion function, but not with its role in Wnt signalling, and vice versa: revealing that the two functions of Arm are separable. Some of these mutations specifically hit Helix-C and abrogated the signalling role but perfectly preserved the structural function of Armadillo. Recently, the ability to completely block the signalling output of Armadillo, without affecting its adhesive role, was confirmed and experimentally extended to β-catenin in mice (Valenta et al, 2011). Interestingly, the functional separation of the two roles occurred evolutionarily in Caenorhabdidis elegans. This nematode has at least three specialized β-catenins: the adhesion-specific HMP-2 that binds cadherins but not TCFs, and two signalling ones, BAR-1 and WRM-1, that can bind TCFs and regulate transcription (Liu et al, 2008). Yet another C. elegans protein, SYS-1, has adopted a signalling role, although it is not related to β-catenin on the sequence level. SYS-1 structurally mimics β-catenin (the positively charged groove), interacts with TCFs, and even mediates transcription (Liu et al, 2008). Such examples of functional separation, however, appear to be the exception. In the majority of animal species (including the model organisms Drosophila, Xenopus, and mouse), a single β-catenin carries out the two functions. These functions may be carried out by different protein pools and are orchestrated by mechanisms (e.g., post-translational modifications), which control the spatial separation, retention, or stability of β-catenin. That said, in vertebrates γ-catenin (plakoglobin), a close relative of β-catenin, may in certain cases compensate for the loss of β-catenin’s structural function; in contrast, the signalling role of β-catenin in vivo seems not to be compensated (Huelsken et al, 2000; Grigoryan et al, 2008). The role of plakoglobin in Wnt/β-catenin signalling warrants further clarification.
β-Catenin is an evolutionarily conserved protein
The usually single β-catenin gene represents the highly conserved centre piece within the complex expanded Wnt signalling pathway (Figure 3). The Wnt pathway features a multitude of paralogous components, especially at the level of Wnt ligands and their receptors. The complexity at the level of the Wnt ligands and receptors (Frizzled proteins) seems to have arisen early in metazoan evolution. In Cnidarians, 14 Wnt genes have been identified which belong to 12 of the 13 known different Wnt subfamilies, 11 of which are also present in vertebrates, where some additional duplication events within individual subfamilies lead to expression of up to 19 different Wnts in mammals (Kusserow et al, 2005). In sponges (Porifera), which constitute some of the most basal metazoans, the Wnt ligand complexity is not as high, only three (respectively two) Wnt ligands are known (Lapébie et al, 2009; Adamska et al, 2010).
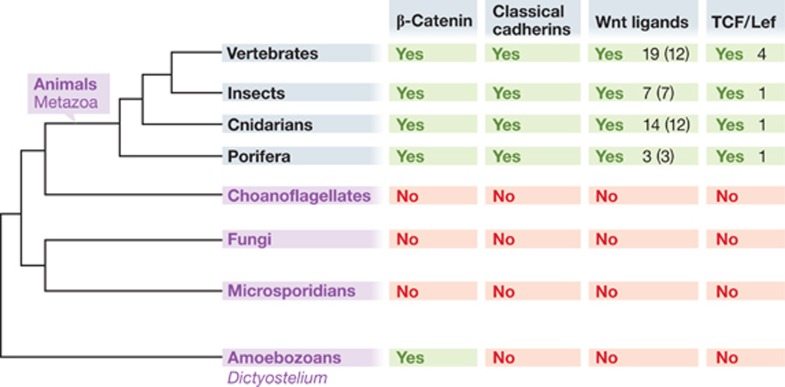
Figure 3.
β-Catenin is an evolutionarily ancient molecule that provided its function before Wnt signalling and classical cadherin-based adhesion appeared. Schematic evolutionary tree showing the relationships among Amoebozoa (represented by Dictyostelium discoideum) and metazoa, as well as the diversity of signalling components. In Dictyostelium discoideum, β-catenin acts as a functional molecule in polarized epithelia. In animals (metazoa), β-catenin plays a dual role as a signalling component of canonical Wnt signalling or as a structural component of cadherin-based cell–cell junctions. The presence of β-catenin and key components of adherens junctions (classical cadherins—containing an intracellular domain binding to β-catenin) and of canonical Wnt signalling (Wnt ligands, TCF/Lef transcription factors) is indicated to the right. In the case of Wnt ligands, the first number indicates how many different Wnt ligands were determined, the second number in brackets indicates how many Wnt subfamilies were determined in a particular group. The number in the case of TCF/Lef refers to how many different TCF/Lef proteins were found. Yes means presence, no absence. The following animal species were compared: Porifera (Sponges): Amphimedon queenslandica, Cnidaria: Nematostella vectans, Insects: Drosophila melanogaster; Vertebrates: Mus musculus.
In sponges, ctenophores, and cnidarians, one gene encodes a β-catenin protein with striking sequence similarity to mammalian β-catenin. Cnidarian β-catenin (e.g., of the polyp Hydra) exhibits >60% of amino-acid identity with mammalian β-catenin, with the main differences being found in the most C-terminal part, which is generally less conserved (Xing et al, 2008; Zhao et al, 2011). The structural identity of the repeat domain of β-catenin is nearly 80%. Moreover, locus conservation among sponges (Amphimedon sp.), cnidarians (sea anemone Nematostella vectans) and later developed metazoans (Deuterostomia; vertebrates belong to this group) includes even the exon/intron features of the β-catenin gene (Holland et al, 2005; Adamska et al, 2010). Highlighting the pivotal role of Wnt/β-catenin signalling also in early metazoans are findings that changes in β-catenin levels dramatically impact their development. In sponges (Porifera), the activation of Wnt/β-catenin signalling by blocking β-catenin degradation leads to the formation of ectopic ostia (canal openings), which can disrupt feeding (Lapébie et al, 2009). In Hydra (Cnidaria), ectopic stabilization of β-catenin results in multiple head and tentacle formation along the body, while the opposite phenotype, a loss of head structures, results from depletion of β-catenin (Broun et al, 2005; Müller et al, 2007; Gee et al, 2010). Such observations suggest that Wnt/β-catenin signalling represents a primeval component, driving metazoan body plan and axis formation since the beginning of animal evolution.
The critical role of Wnt/β-catenin signalling for establishing the primary body axis (Petersen and Reddien, 2009), or more precisely A/P (anterior/posterior) identity, has been long studied in diverse organisms. It is a re-occuring theme and reveals itself either in individual (para)segments, as in the Drosophila embryo, or of whole developing body plans, such as in vertebrates (Sanson, 2001; Niehrs, 2010). A paradigmatic example showing the indispensable role of β-catenin in the process of axial patterning came from experiments in the amphibian Xenopus laevis: ectopic expression of β-catenin in ventral blastomeres of Xenopus embryos induces axis duplication (i.e., the formation of a secondary axis), a phenotype reminiscent of misexpression of several Wnt ligands (Guger and Gumbiner, 1995). Depletion of β-catenin results in the opposite effect, ventralization of the embryo with a shortened A/P axis (Haesman et al, 1994). A/P axis identity and mesoderm formation is controlled by β-catenin-mediated transcription also in mammals, since mouse embryos lacking β-catenin do not develop a proper A/P axis and fail to form mesoderm, resulting in corrupted gastrulation (Haegel et al, 1995; Huelsken et al, 2000).
Interestingly, a homologue of β-catenin was found also in the non-metazoan social amoeba Dictyostelium discoideum (slime mould) (Coates et al, 2002). This contrasts with the lack of Wnt ligands, and indeed of most proteins involved in Wnt signalling, which so far have been identified solely in metazoans but not in any other phyla (Figure 3). In normal circumstances, Dictyostelium discoideum is a unicellular organism; but in response to starvation single cells start to aggregate and form fruiting bodies, which comprise a rigid stalk and on its tip a collection of spores. During the formation of a fruiting body the aggregated cells establish a polarized epithelium that is essential for the subsequent multicellular phase of development. Even though Dictyostelium discoideum, in contrast to metazoans, lacks classical cadherins, the β-catenin orthologue Aardvark is an essential structural molecule in the polarized epithelia. Moreover, to maintain the proper epithelial structure Aardvark cooperates with α-catenin, another key structural molecule known from metazoans. Ddα-catenin is upregulated during the transition to multicellularity and colocalizes with Aardvark and F-actin at the sites of cell–cell contact. Loss or knockdown of either Aardvark/Ddβ-catenin or Ddα-catenin significantly compromises epithelial morphology, polarity, and formation of fruiting bodies (Dickinson et al, 2011). Thus, β-catenin, together with α-catenin and most likely actin, can promote primitive cell–cell contacts even without cadherins. These observations suggest that the principles underlying metazoan multicellularity may be more ancient than previously thought and that the structural role of β-catenin precedes its signalling role in the canonical Wnt pathway—β-catenin and α-catenin were engaged in the formation of polarized epithelia before classical catenins came on the scene in metazoans. However, even in Dictyostelium discoideum, β-catenin is not exclusively a structural molecule. In response to cAMP, a series of events leads to phosphorylation of GSK3, an orthologue of GSKA that phosphorylates Aardvark/Ddβ-catenin. This phosphorylation event activates Aardvark and leads to gene transcription via a so far unknown mechanism, as there are no TCF/Lef transcription factors in Dictyostelium discoideum (Grimson et al, 2000). Even though the mechanism seems different from the core Wnt/β-catenin pathway (phosphorylation leads to activation, not to degradation), at least the interaction between β-catenin and GSK3 is present and may have further evolved into the mode of action typical for canonical Wnt signalling.
Canonical signalling in a nutshell
Without a Wnt signal, the levels of cytoplasmic free β-catenin are kept low (Figure 1). If not bound to E-cadherin, β-catenin is phosphorylated in the cytoplasm by the activity of a multiprotein destruction complex, marking β-catenin for degradation. This complex consists of the scaffold proteins Axin and Adenoma Polyposis Coli (APC), and of the kinases phosphorylating β-catenin, glycogen synthase kinase 3β (GSK3β), casein kinase 1 (CK1) and protein phosphatase 2A (PP2A) (Kimelman and Xu, 2006). The binding of Wnt ligands to the Frizzled transmembrane receptors and LRP5/6 co-receptors (Low density lipoprotein Receptor-related Protein) starts a series of molecular events leading to inhibition of the β-catenin destruction complex. Ligand activated Frizzled receptors recruit the cytoplasmic protein Dishevelled (Dvl) to the receptor complex through direct binding; Dvl subsequently multimerizes and induces formation of so-called LRP-associated Wnt signalosomes (Bilic et al, 2007). Dvl in turn recruits the rate-limiting component Axin (most likely together with associated kinases, such as GSK3) and thus destabilizes the β-catenin destruction complex (Schwarz-Romond et al, 2007). Dvl-mediated phosporylation of LRP5/6 by CK1 is key for proper functioning of the Wnt signalosome as it leads to a block of GSK3 kinase activity (Zeng et al, 2008). Unphosphorylated β-catenin escapes degradation, accumulates in the cytoplasm, and translocates to the nucleus. Nuclear β-catenin associates with DNA-binding transcription factors of the TCF/Lef family. TCF/Lef proteins possess only limited ability to activate transcription. In the absence of Wnt signals, TCF/Lef factors act as transcriptional repressors. The binding of β-catenin converts TCF/Lef proteins into bipartite TCF/β-catenin transcriptional activators, converting the Wnt signal into the transcription of specific target genes (Najdi et al, 2011; Archbold et al, 2012). Albeit elegant, recent work has made clear that such a simplified, linear scheme describing the translation of the Wnt signal via β-catenin into a cell-specific genetic program does not capture the full complexity of β-catenin-mediated functions within Wnt receiving cells.
β-Catenin at the membrane: balancing adhesion and signalling
The majority of β-catenin is located at the cytoplasmic side of the membrane as a component of cadherin-based cell–cell connections in the absence of a Wnt stimulus. Cadherin-based adherens junctions contribute to forming polarized epithelial tissues, a characteristic metazoan feature necessary for maintaining organismal integrity (Meng and Takeichi, 2009). Cadherins are single-pass transmembrane glycoproteins that engage in Ca2+-dependent homotypic interactions via their extracellular regions and that also link to β-catenin through their cytoplasmic tails. Classical cadherins were named for the tissue in which they were most prominently expressed: for example, epithelial cadherin (E-cadherin) in epithelial cells; neural cadherin (N-cadherin) in the nervous system. Many cell types co-express several cadherins (Meng and Takeichi, 2009; Stepniak et al, 2009). β-Catenin can interact with the cytoplasmic domain of all of them, but most studied are the functional consequences of the β-catenin–E-cadherin interaction. The cadherin–β-catenin interaction is constitutive and may occur even in single isolated cells in an adhesion-independent way (Stepniak et al, 2009). Newly synthesized E-cadherin associates with β-catenin while still in the endoplasmic reticulum and the two proteins move together to the cell membrane. Interfering with the binding of β-catenin to E-cadherin results in proteasomal degradation of the cadherin, because β-catenin shields a PEST sequence motif on E-cadherin, which, if free, is recognized by a ubiquitin ligase that marks E-cadherin for degradation (Hinc et al, 1994). Reciprocally, E-cadherin association stabilizes β-catenin by preventing binding of components of the β-catenin destruction complex (namely APC and Axin) (Huber and Weis, 2001).
At cadherin-based cell–cell junctions, β-catenin can bind α-catenin with distal parts of the NTD and the adjacent first ARM repeat of β-catenin close to a hinge region around Arg151 (Figure 2; Pokutta and Weis 2000; Xing et al, 2008). In close proximity, C-terminal to this hinge, is the binding site for BCL9, an important transcriptional co-activator of β-catenin (Hoffmans and Basler, 2004; Sampietro et al, 2006). Hence if the structural conformation of the hinge region could be affected by phosphorylation (see below), α-catenin affinity might be reduced, thus favouring β-catenin to act in a signalling role mediated by BCL9. Conversely, binding of α-catenin may stabilize the hinge, thus promoting β-catenin’s structural function at the cell–cell junctions (Brembeck et al, 2004).
Binding of α-catenin stabilizes β-catenin in the hinged form, and E-cadherin can bind simultaneously (Pokutta and Weis, 2000; Huber and Weis, 2001). Importantly, the β-catenin-binding site on α-catenin overlaps with α-catenin’s homodimerization interface, hence α-catenin can bind to adherens junctions only in its monomeric state (resulting in an α-catenin/β-catenin heterodimer) (Drees et al, 2005; Yamada et al, 2005). As a homodimer, which cannot bind β-catenin, α-catenin interacts with actin and promotes bundling of actin filaments (Benjamin et al, 2010). Consequently, the binding of α-catenin to the cadherin–β-catenin complex negatively regulates its activity on actin polymerization (Yamada et al, 2005). The α-catenin-mediated activity on actin filaments may be, at least partially, spatially independent from adherens junctions (Benjamin et al, 2010). Whatever the case, β-catenin can affect at least indirectly actin properties by balancing the dynamic pools of α-catenin: cadherin/β-catenin bound and free homodimeric α-catenin.
The cadherin–catenin junctions are highly dynamic molecular complexes. Loss of cadherin-mediated cell adhesion can promote β-catenin release and thus its signalling activity. On the other hand, cadherin can act as a membrane trap for a fraction of free β-catenin (Heuberger and Birchmeier, 2010). Several proteases can cleave the intracellular domain of cadherins and thus affect the integrity of adherens junctions and even β-catenin activity. Cleavage of E- and N-cadherin by ADAM10 (A Disintegrin And Metalloprotease 10) reduces cell adhesion in keratinocytes or neuronal cells, leading to nuclear translocation of released β-catenin. In the nucleus β-catenin activates transcription of Wnt/β-catenin-target genes such as c-myc and cyclinD1 (Maretzky et al, 2005; Reiss et al, 2005). Interestingly, the ADAM10 gene was also described as a direct target of β-catenin-mediated transcription and the cleaved cytoplasmic fragment of cadherin (N-cadherin in this case) apparently translocated to the nucleus and promoted β-catenin-mediated transcription (Gavert et al, 2007; Shoval et al, 2007). Both observations suggest a possible positive feedback loop that can amplify β-catenin signalling.
Regulating β-catenin function by phosphorylation
The structural integrity of cadherin-based cell junctions and thus the function of β-catenin can be regulated through phosphorylation by various kinases. Phosphorylating an interaction partner of β-catenin and thereby introducing a negative charge may increase its binding to the positively charged groove formed by the central region of β-catenin. Three serine residues in the cadherin cytoplasmic domain (Ser684, Ser686, and Ser692) are phosphorylated by the protein kinases CK2 (casein kinase 2) and GSK3β. This creates an additional interaction platform with β-catenin that strengthens the cadherin–β-catenin interaction by several hundred-fold (Huber and Weis 2001; Choi et al, 2006, Sampietro et al, 2006). It should be noted that not all phosphorylation events on cadherin might have such enhancing effects, some of them have been reported to reduce β-catenin binding (Qi et al, 2006; Dupre-Crochet et al, 2007).
Conversely, phosphorylation of β-catenin often causes a weakening of the cadherin–β-catenin interaction, directing β-catenin into signalling mode (for details, see Figure 4). Phosphorylation close to the hinge region—Tyr 142 of β-catenin—by Fyn, Fer, or the receptor tyrosine kinase c-Met significantly reduces α-catenin binding and thus impairs the adhesive function of β-catenin (Piedra et al, 2003; Brembeck et al, 2004; Bustos et al, 2006). Even though the Tyr142 site and its surrounding region are highly conserved, the in-vivo significance of the phosphorylation remains debated. In some cell lines, Tyr142 phosphorylation seemed to have an effect on adhesion, shifting β-catenin function towards signalling; in vivo, however, the effects were negligible. In Drosophila, Tyr142 modifications do not have any significant functional consequences (Hoffmans and Basler, 2007). The possibility that Tyr142 phosphorylation regulates specific processes during normal or pathological development in mammals remains open.
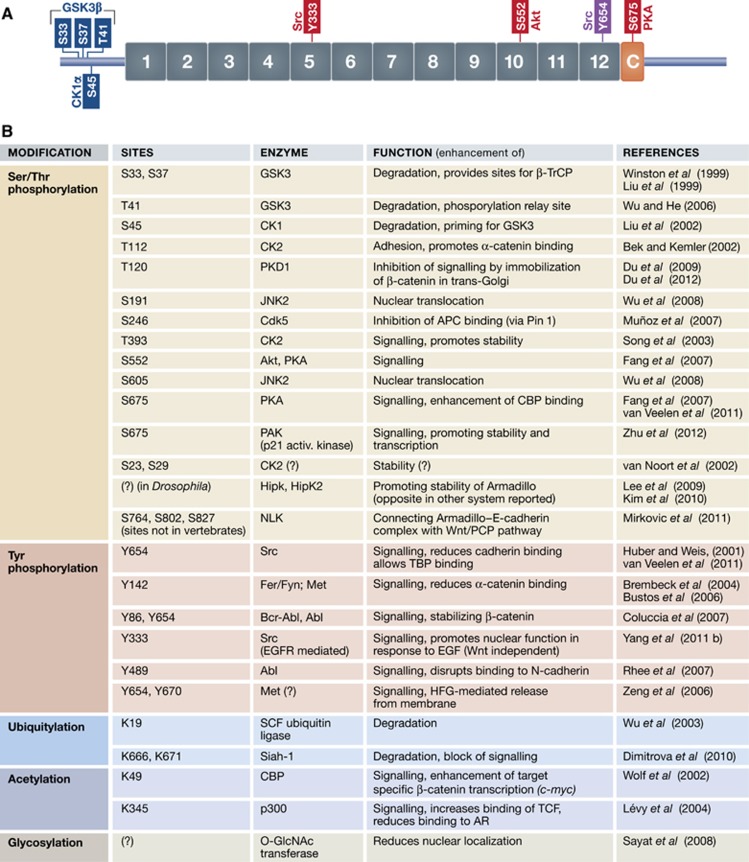
Figure 4.
The functional output of β-catenin is affected by post-translational modifications. (A) A representative scheme showing the sites where β-catenin is phosphorylated: those in blue promote its degradation, those in red and purple enhance the signalling activity. The Y654 site (purple) was experimentally validated in a mouse model in vivo. The protein kinases are denoted that promote each phosphorylation. (B) Table summarizing possible post-translational modifications of β-catenin with their functional consequences.
Another well-studied phosphorylation event affecting the choice of β-catenin’s output occurs on Tyr654 in the last ARM repeat of β-catenin. Tyr654 phosphorylation dramatically reduces the binding to cadherin and may enhance β-catenin-driven transcription (Piedra et al, 2001). The effect of this phosphorylation was not only described in many mammalian cell lines, but was also confirmed in vivo by generating conditional mouse mutants expressing a phospho-mimicking form of β-catenin (β-cateninY654E; van Veelen et al, 2011). Homozygous mutant mice harbouring this mutation die as embryos with phenotypes strongly resembling those caused by aberrantly activated Wnt/β-catenin signalling. Interestingly, heterozygous animals develop intestinal adenomas and show a strong potential to enhance the formation of intestinal tumours caused by a defective β-catenin destruction complex (loss of APC). Tyr654 phosphorylation is effected by receptor tyrosine kinases (e.g., EGFR) or c-src kinase; aberrantly active tyrosine kinases are also implicated, such as Bcr-Abl in chronic myeloid leukaemia cells (Roura et al, 1999; Coluccia et al, 2007). The negative charge of p-Tyr654 clashes with key aspartate residues in cadherin and thus reduces the cadherin–β-catenin interaction (Huber and Weis, 2001). Surprisingly, in contrast to the observations from tissue culture experiments, in the mouse model the reduced affinity of the phospho-mimetic mutant did not dramatically affect the structure of adherens junctions. A fraction of β-catenin was still associated with cadherins at the membrane, suggesting that a compensatory mechanism exists in vivo, or that modifying only Tyr654 is not sufficient to completely destroy the cadherin junctions (van Veelen et al, 2011). Additionally, Tyr654 phosphorylation might affect the conformation of β-catenin. If not phosphorylated, residues of the last ARM repeats, including the area in the vicinity of Tyr654, can form hydrogen bonds with the partially flexible Helix-C. Hence, the C-terminus is folded back onto the ARM repeats blocking the binding of transcriptional co-activators to this area (Xing et al, 2008). Tyr654 phosphorylation evokes a conformational change preventing this folding back of the C-terminus. Helix-C thus stays open and accessible for further phosphorylation by PKA (protein kinase A) on Ser675 within Helix-C. This phosphorylation may stabilize the open structure of Helix-C (Piedra et al, 2001; van Veelen et al, 2011). Together, both phosphorylated Tyr654 and Ser675 increase the recruitment of co-activators of β-catenin-mediated transcription, such as CBP (CREB binding protein) or TBP (TATA binding protein) (Taurin et al, 2006; van Veelen et al, 2011). This two-step phosphorylation thus strongly promotes the signalling activity of β-catenin at the expense of its structural function. Whether the two-step mechanism is conserved, or if in certain circumstances these two phosphorylation events act independently, remains to be resolved.
Tyrosine kinase phosphorylation of β-catenin is balanced by protein tyrosine phosphatases that bind β-catenin and cadherin. The non-receptor protein tyrosine phosphatase PTP1B (protein tyrosine phosphatase 1B) regulates cadherin-based adhesion by binding directly to the cadherin cytoplasmic domain and dephosphorylating β-catenin at Tyr654 (Balsamo et al, 1996; Xu et al, 2004). The activity of PTP1B is regulated by protein kinases (such as Fer), affecting the adhesive function of β-catenin and creating a regulatory circuit, which allows the cell to balance discrepancies at the cell–cell junctions that arise from variable levels of membrane bound β-catenin.
β-Catenin and epithelial–mesenchymal transition
An important developmental process modulated by changes in cell–cell adhesion is epithelial–mesenchymal transition (EMT). During EMT, polarized rather immobile epithelial cells change their morphology to highly motile fibroblastoid mesenchymal cells. EMT plays an important role during embryonic development and drives cancer invasion and formation of metastases (reviewed in Thiery et al, 2009). One of the hallmarks of EMT is the disappearance of E-cadherin from the cellular membrane and its replacement by N-cadherin (the so-called E-to-N-cadherin switch) (Zeisberg and Neilson, 2009). Reduction of E-cadherin is often connected with increased free cytoplasmic levels of β-catenin, which could translate into increased β-catenin transcription if it escapes cytoplasmic degradation (Heuberger and Birchmeier, 2010). In transitions related to cancer progression, activating mutations of β-catenin could enhance transcription, which might have consequences for EMT progression.
The Wnt/β-catenin pathway is one of the signalling pathways controlling EMT: several key transcription factors regulating E-cadherin expression and/or the fate of other epithelial molecules are direct or indirect transcriptional targets of the canonical Wnt pathway. One such target is the transcription factor Twist (Howe et al, 2003), a basic basic-loop-helix transcription factor, that is important to define the mesoderm in invertebrates and vertebrates. In vertebrates, it is active during cell specification and lineage determination and is often upregulated during metastasis formation in cancer (Zeisberg and Neilson, 2009; Ansieau et al, 2010). Twist directly downregulates the expression of E-cadherin while also inducing expression of N-cadherin and fibronectin and thus induces EMT and cellular invasion (Yang et al, 2004, 2007). Like Twist, the transcriptional repressors Snail1 and Snail2 (Slug) are also involved in regulating EMT in a Wnt/β-catenin-dependent manner. Both of them repress E-cadherin transcription (Conacci-Sorrell et al, 2003; Barrallo-Gimeno and Nieto, 2005). Snail2 is upregulated directly by β-catenin-mediated transcription, whereas Snail1 levels increase indirectly upon Wnt stimulation through the block of GSK3β activity. GSK3β normally phosphorylates Snail1 and marks it for degradation (Zhou et al, 2004; Yook et al, 2006). Interestingly, Snail1 can directly interact with β-catenin and enhance Wnt-target gene expression most likely by stimulating canonical Wnt signalling in a positive feedback loop (Stemmer et al, 2008). A final transcription factor regulating EMT that is a direct target of β-catenin-mediated transcription is ZEB1 (also known as δEF1). It has the most consistent inverse expression correlation with E-cadherin across different types of carcinomas. When inducing an EMT, ZEB1 not only transcriptionally represses epithelial markers like E-cadherin but also activates mesenchymal genes (Sánchez-Tilló et al, 2011). It should be mentioned that Twist, Snails, and ZEB1 are regulated by many signalling pathways, of which Wnt/β-catenin is but one. Hence, the cellular context determines which pathways cross-talk on the regulation of these factors and together determine the final output (reviewed in Thiery et al, 2009).
There are many other factors and molecules affecting EMT or cell adhesivity that are regulated by Wnt/β-catenin signalling, and conversely, defects in polarized epithelial adhesivity can influence the transcriptional output of Wnt/β-catenin signalling (reviewed by Heuberger and Birchmeier, 2010).
β-Catenin in the cytoplasm: decision point
In the cytoplasm, free β-catenin (not bound and thus protected by E-cadherin) is normally short-lived: it is recognized by the destruction complex and rapidly targeted for degradation (Kimelman and Xu, 2006). This is true for newly synthesized protein and for β-catenin released from adherens junctions. Free β-catenin is recognized by the key scaffold molecules Axin and APC, both of which can directly interact with β-catenin and also inter se. The scaffold establishes a platform for associated kinases to phosphorylate β-catenin (Kimelman and Xu, 2006; Roberts et al, 2011). CK1α phosphorylates β-catenin at Ser45, priming the sequential phosphorylation of Thr41, Ser37, and Ser33 by GSK3 (preferentially by GSK3β) (Liu et al, 2002; Xing et al, 2003). As a next step, β-catenin bound to APC leaves the destruction complex and interacts with the ubiquitin machinery. Ser33 and Ser37 phosphorylated β-catenin is recognized by β-TrCP and recruited to the Skp1/Cul1/F-box/β-TrCP (SCFβ-TrCP) E3 ubiquitin ligase complex. Ubiquitin-conjugated β-catenin is subsequently degraded by the 26S proteasome (Hart et al, 1999). Alternatively, phospho-β-catenin can be ubiquitinylated by the single unit E-3 ligase Jade 1 (Chitalia et al, 2008). The scaffolding proteins Axin and APC are essential for the GSK3-mediated phosphorylation of β-catenin: although GSK3 can modify a plethora of different proteins within a cell as a free molecule, it modifies β-catenin only if it is associated with Axin and APC (Hur and Zhou, 2010; Wu and Pan, 2010). The rate-limiting protein Axin greatly enhances the activity of GSK3 on β-catenin (Dajani et al, 2003). APC contributes to the establishment of the destruction complex, and stabilizes β-catenin’s phosphorylation status. If N-terminally phosphorylated β-catenin is not associated with APC, after leaving the destruction complex, then it is immediately dephosphorylated by PP2A (Su et al, 2008).
Activation of Wnt signalling leads to the disassembly of the β-catenin destruction complex and GSK3 activity is blocked. Two alternative models to explain how Wnt signalling represses GSK3 activity have been proposed (discussed in detail in Metcalfe and Bienz, 2011). A biochemical explanation is based on a direct competitive block of the GSK3-phosphorylated co-receptors LRP5/LRP6 (Piao et al, 2008; Wu et al, 2009). An alternative explanation is based on the sequestration of GSK3 into multivesicular bodies, thereby preventing an interaction with newly synthesized β-catenin and subsequent phosphorylation (Taleman et al, 2010). Interestingly, in teratocarcinoma cells the activity of GSK3 on β-catenin could be directly inhibited by p38-mitogen activated kinase in a Wnt-dependent manner. Such inhibition results in increased levels of β-catenin and promotes Wnt/β-catenin signalling (Bikkavili et al, 2008). The p38 kinase was also shown to inactivate GSK3β in other Wnt-independent models (Thornton et al, 2008). These findings point to the existence of Wnt-independent mechanisms for dampening the activity of the β-catenin destruction complex. This type of inactivation is different from the well-known inactivation of GSK3 by Akt kinase upon insulin stimulation, which most likely represents a separate regulation of GSK3, without apparent cross-talk to the β-catenin phosphorylation status (Ding et al, 2000; Ng et al, 2009). A Wnt-independent mechanism was demonstrated in some colon cancer cell lines which have (at least partially) functional destruction complexes: treatment of these cells with PDGF results in a dramatic change of morphology connected with EMT. The effects were mediated by the kinase c-Abl, which phosphorylates RNA-helicase p68, which then displaces Axin from the β-catenin destruction complex and thereby blocks phosphorylation of β-catenin by GSK3. As a result, β-catenin is stabilized and promotes transcription triggering EMT (Yang et al, 2006).
Further Wnt-independent mechanisms to regulate the stability of β-catenin have come to light in the recent years. For example, increasing levels of Siah-1 are induced by p53 in response to various stimuli. Siah-1 can bind a multiprotein ubiquitinating complex different from the one involved in the ubiquitination mediated by the destruction complex. Simultaneously, Siah-1 directly associates with the F-box protein Ebi, which is bound to β-catenin and thus mediates β-catenin ubiquitination and subsequent degradation. This mechanism does not require a preceding phosphorylation of β-catenin, but it does involve APC acting in a scaffolding role as it does in the classical β-catenin degradation machinery (Matsuzawa and Reed, 2001; Dimitrova et al, 2010).
In pathological conditions, free β-catenin may also escape from its degradation fate. This can be caused by mutations either inactivating the function of components within the destruction complex (in particular in the APC gene) or preventing β-catenin from being phosphorylated by GSK3. Such mutations result in aberrantly activated β-catenin signalling and contribute to the development of various types of cancer (Polakis, 2007; Lucero et al, 2010).
Surprisingly, not all β-catenin phosphorylated by GSK3 undergoes degradation. A fraction of N-terminally phosphorylated β-catenin was detected in centrosomes, which are microtubule nucleation centres that form the mitotic spindle. Here, β-catenin significantly contributes to centrosomal cohesion and separation at the onset of mitotic spindle formation (Huang et al, 2007). Localization of β-catenin within centrosomes is under the control of the Nek2 kinase during the cell cycle (Bahmanyar et al, 2008). Because the mitotic spindle mediates segregation of chromosomes, β-catenin might be important for the faithful execution of this event. β-Catenin may affect spindle orientation and thus the plane of cell division, a role which was also demonstrated for APC (Quyn et al, 2010; Chilov et al, 2011). Interestingly, besides β-catenin and APC also Axin was found to be an important regulator of centrosomes, suggesting a new role for some components of the β-catenin destruction complex and yet a further structural activity of β-catenin: centrosomal maintenance and separation (Hadjihannas et al, 2010).
β-Catenin in the nucleus: the TCF/β-catenin transcriptional switch
The inhibition or bypassing of β-catenin destruction leads to increased levels of β-catenin, which accumulates in the cytoplasm and then translocates into the nucleus. Although the growing concentration of free cytoplasmic β-catenin seems to be a prerequisite for its subsequent nuclear translocation, it does not completely explain how β-catenin gets into the nucleus to promote transcription. The mechanism of β-catenin nuclear entry is not precisely understood (Henderson and Fagotto, 2002; Städeli et al, 2006). β-Catenin can dynamically shuttle between the cytoplasm and nucleus. Surprisingly, it does not contain any classical nuclear localization signal (NLS) or nuclear export signal (NES) within its polypeptide sequence. Indeed nuclear import of β-catenin was shown to occur importin-karyopterin independently (Fagotto et al, 1998; Yokoya et al, 1999). Recently, β-catenin was shown to directly interact with different nuclear pore complex components (NPCs; Shitashige et al, 2008; Sharma et al, 2012). By transiently and sequentially binding to different NPCs, β-catenin could pass through the nuclear pores. Of the various NPCs interacting with β-catenin, Nup358 seems to be important for docking/undocking of β-catenin to nuclear pores during the process of nuclear translocation (Sharma et al, 2012). ARM repeats R10–12 were revealed as crucial for β-catenin nuclear import (and export). Moreover, Tyr654 in the last ARM repeat was supposed to affect β-catenin import, as a phospho-mimicking mutation strongly enhances nuclear import, indicating yet another way in which p-Tyr654 can promote the signalling activity of β-catenin at the expense of its adhesive function (Sharma et al, 2012).
There are a few other molecular mechanisms facilitating nuclear translocation of β-catenin. The Forkhead-box transcription factor FoxM1 was shown to directly interact with the ARM repeats R11–12 and thereby promotes β-catenin nuclear import in mammalian cells: immortalized neural stem cells or MEF’s lacking FoxM1 display a strong reduction of nuclear β-catenin and thus reduced β-catenin signalling activity (Zhang et al, 2011). FoxM1 might provide its NLS to β-catenin and bipartite β-catenin–FoxM1 complexes could be imported to the nucleus. Alternatively, or additionally, FoxM1 could act as a nuclear anchor preventing export of β-catenin out of the nucleus. The FoxM1–β-catenin interaction is maintained in the nucleus: FoxM1 forms a complex with β-catenin/TCF on the promoters of Wnt target genes. Hence, FoxM1 can serve even as direct modulator of β-catenin-mediated transcription. FoxM1 is upregulated in variety of cancers. In glioblastomas, the activity of FoxM1 most likely contributes to elevated β-catenin signalling activity, since these tumours have a functional β-catenin destruction complex. Reduction of FoxM1 in glioblastomas impeded tumour formation in vivo, in a β-catenin-dependent manner (Zhang et al, 2011). It remains to be shown whether FoxM1 facilitates nuclear import of β-catenin also in other types of mammalian tissues and/or different cancer types.
In murine bone marrow-derived stromal cells, Rac-activated JNK2 phosphorylation of β-catenin was found to be important for nuclear accumulation (Wu et al, 2008). JNK2 phosphorylates β-catenin at serine residues 191 and 605, which promotes β-catenin nuclear localization. Although experiments based on an Rac1 conditional knockout model might suggest that this phosphorylation could affect nuclear import even in other tissues, the evidence is only indirect and must be tested further. It also remains to be addressed if such modification of β-catenin and facilitation of nuclear translocation is conserved.
Once in the nucleus β-catenin can activate transcription of Wnt/β-catenin target genes. Since β-catenin does not possess a DNA binding domain it needs DNA binding partners to bring it to the promoters of its target genes (Huber et al, 1997; Xing et al, 2008). Hence, β-catenin initiates transcription only as a member of bipartite or multimeric complexes wherein one partner provides association with specific response elements on target genes (e.g., Wnt response elements, WREs) and β-catenin acts as the central transcriptional activator.
TCF/Lef (hereafter referred to as TCF) transcription factors serve as the main nuclear partners of β-catenin guiding it to specific DNA loci. Together, they are responsible for the vast majority of Wnt/β-catenin signalling outputs. Invertebrate (and lower deuterostome) genomes encode only one TCF protein (e.g., dTCF/Pangolin in Drosophila) whereas in vertebrates four paralogues, TCF1, TCF3, TCF4, and Lef1, are encoded that are all able to interact with β-catenin. The situation is further complicated by the existence of many splice variants, especially in vertebrates, and some of the isoforms might lose the β-catenin binding potential (Arce et al, 2006; Archbold et al, 2012). In Drosophila, dTCF/Pangolin is absolutely required for Wg/Arm signalling, and similar to armadillo mutants, dTCF/pangolin mutants display defective segmental patterning of the embryonic epidermis (Brunner et al, 1997; van de Wetering et al, 1997; Schweizer et al, 2003). In vertebrates, the redundancy or functional overlap implied by the presence of four TCFs confounds the precise interpretation of mutant phenotypes. Nevertheless, many of the null phenotypes in mice are consistent with a loss of β-catenin signalling. For example, knocking out TCF4 results in the loss of stem cells in intestinal crypts and a combined Lef1/TCF1 double null exhibits defects in paraxial mesoderm (Korinek et al, 1998; Galceran et al, 1999; Fevr et al, 2007). Moreover, the in-vivo expression of TCF-specific transcriptional reporters is strongly affected by changes in β-catenin activity (either knockout or aberrant activation) (DasGupta and Fuchs, 1999; Maretto et al, 2003; Barolo, 2006; Grigoryan et al, 2008).
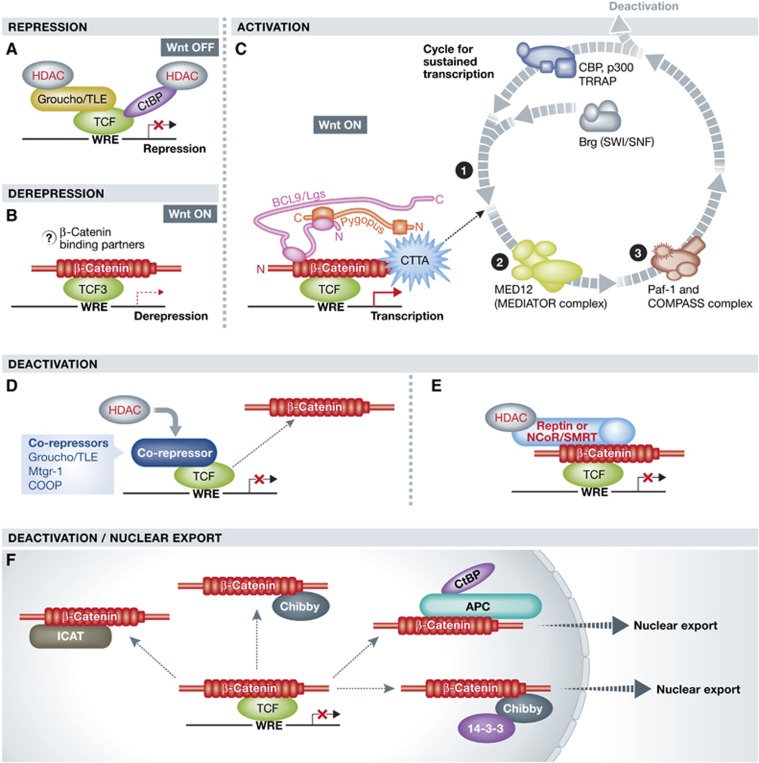
TCFs bind with their high mobility group (HMG) domain to a specific motif in the minor groove of target DNA (CCTTTGAT(G/C)). Such a motif is necessary and sufficient to mediate TCF binding (van de Wetering et al, 1997; van Beest et al, 2000). Additionally, in some TCFs variants there is a cysteine-rich domain known as the C-clamp that can bind to so-called helper sequences, enhancing the association of TCF with target sequences (Atcha et al, 2007; Chang et al, 2008). Without β-catenin, TCFs act as transcriptional repressors by forming a complex with Groucho/TLE repressors. The binding of β-catenin physically displaces Groucho/TLE and converts TCFs into transcriptional activators, translating the Wnt signal into transcription of specific target genes (Cavallo et al, 1998; Daniels and Weis, 2005; Arce et al, 2009). There is a growing list of Wnt/TCF/β-catenin target genes, yet many are subject to complex, context-dependent regulation and expressed in a tissue-specific or temporally restricted manner (Vlad et al, 2008; Bottomly et al, 2010). A comprehensive updated list of targets can be found at http://www.stanford.edu/group/nusselab/cgi-bin/wnt/target_genes.
Whereas in Drosophila, dTCF/Pan can both repress and activate Wnt/β-catenin targets; in vertebrates, it seems that some TCFs are more prone to act as repressors (e.g., TCF3) while others act preferentially together with β-catenin as activators (e.g., Lef1). TCF4 and TCF1 fulfill both modes of action depending on the spatial and temporal context (Wray et al, 2011; Yi et al, 2011; Archbold et al, 2012).
These results provide the basis for a model in which β-catenin provides a platform for the recruitment of a multitude to transcriptional co-activators initiating and propagating Wnt/β-catenin transcription (Figure 2). Nuclear β-catenin serves as a link in a chain that spans from the target enhancer via TCFs and various transcriptional co-activators to RNA polymerase II (Städeli et al, 2006; Mosimann et al, 2009). The majority of the trancriptional co-activators bind to the last ARM repeat of β-catenin and here their interaction with conserved Helix-C seems to be essential (further referred to as C-terminal co-activators). Preventing the interaction between β-catenin and its C-terminal co-activators compromises Wnt/β-catenin transcription (van de Wetering et al, 1997; Cox et al, 1999; Valenta et al, 2011). A small number of β-catenin transcriptional co-activators bind N-terminally to the first ARM repeats, in particular BCL9 (Kramps et al, 2002; Mosimann et al, 2009).
Many of the transcriptional co-activators of β-catenin have the potential to affect chromatin structure either by modifying histones, such as histone acetyltransferases (CBP, p300, and Tip60), or by controlling nucleosome arrangement (SWI/SNF and ISWI). Other binding partners feature properties to promote the association of TCF/β-catenin with the RNA polymerase II complex (members of the Mediator complex and components of the Paf1 complex). It is unlikely that all these multimeric transcriptional co-activator complexes bind to β-catenin simultaneously, especially not to the shared binding region in the C-terminus of β-catenin. The most likely scenario seems to be that the C-terminus serves as a platform for the recruitment and sequential exchange of transcriptional co-activators (Mosimann et al, 2009). Early in such a scenario histone acetyltransferases would acetylate histones and thereby initiate chromatin modifications that relax the chromatin structure. Next or concomitantly, ATP-dependent chromatin remodelling complexes further re-arrange the chromatin structure by shuffling nucleosomes. Then, the Mediator complex initiates the assembly of the RNApolII pre-initiation complex, accompanied by the binding of the Paf1 complex component Parafibromin/Hyrax (Kim et al, 2006; Mosimann et al, 2006; Parker et al, 2008). This sequence would target the Paf1 complex to the proximity of WRE’s where it can facilitate histone H2B monoubiquitylation by the RAD6–BRE1 complex and the recruitment of the COMPASS complex. The latter performs H3K4 methylation, a modification exclusively associated with actively transcribed chromatin. Simultaneously, the Paf1 complex can interact with RNA polymerase II and thus mRNA synthesis can start (Figure 5; Mosimann et al, 2009). This is a simplified model describing the initiation of TCF/β-catenin-mediated transcription and not all of the suggested steps have been experimentally verified. However, ChIP-based time course experiments focussing on β-catenin target promoters support a dynamic cycling mechanism (Sierra et al, 2006; Wang and Jones, 2006). If the entire cohort of identified β-catenin co-factors is needed in every cell type for every gene is debatable. It is likely that there will be cell and gene-specific mechanisms as well. A more detailed discussion is provided by Mosimann et al (2009).
Figure 5.
The TCF/β-catenin transcriptional switch. (A) In an unstimulated (Wnt OFF) situation, TCF/Lef transcription factors associated with Groucho and/or CtBP co-repressors act as transcriptional repressors. (B) As described so far only for TCF3 (Wray et al, 2011; Yi et al, 2011) the binding of β-catenin displaces co-repressors, thereby promoting transcriptional derepression. This mechanism is less well studied than the more classical activation by co-activator recruitment. The question mark indicates that the role of any β-catenin binding partners in this process is not yet clear. (C) β-Catenin converts TCF/Lef into transcriptional activators providing an interacting platform for the multitude of dynamically cycling transcriptional co-activators. (D) TCF/β-catenin-mediated transcription may also be deactivated by transcriptional co-repressors, such as Mtgr-1, COOP, or Groucho/TLE kicking off β-catenin. Some of these co-repressors may act by recruiting HDAC activity. (E) TCF/β-catenin-mediated transcription may also be deactivated by direct interaction with transcriptional co-repressors (Reptin or NCoR/SMRT) recruiting histone deacetylases. (F) Chibby with 14-3-3 protein or APC with CtBP can sequester β-catenin away from target promoters and export it out of the nucleus. Binding of ICAT or Chibby blocks the interaction between TCFs and β-catenin.
The TCF/β-catenin interaction can be modulated to enhance, repress, or switch off β-catenin-mediated transcription. Binding of β-catenin to TCF is prevented by some proteins, such as ICAT (Inhibitor of β-catenin and TCF), which interacts with the central ARM repeat of β-catenin, thereby blocking access to them. Additionally, post-translational modification of either TCF or β-catenin can elicit different effects. The SUMO E3 ligase PIASy has been shown to modify the different TCFs with varied consequences on TCF/β-catenin output: whereas sumoylation of Lef1 leads to its immobilization in nuclear bodies where it cannot interact with β-catenin, sumoylation of TCF4 enhances β-catenin binding and thus transcription (Sachdev et al, 2001; Yamamoto et al, 2003; Ihara et al, 2005). As discussed previously, phosphorylation of β-catenin at Tyr654 and subsequently at Ser675 might render the C-terminal tail of β-catenin more accessible to the multitude of transcriptional co-activators that bind there (van Veelen et al, 2011). The Traf2 and Nck-Interacting Kinase (TNIK) protein kinase was found as direct interaction partner of both TCF4 and β-catenin, positively affecting TCF/β-catenin-mediated transcription in mammalian cells, an association that seems especially important for intestinal progenitor development (Mahmoudi et al, 2009). TCF phosphorylation has both positive and negative effects depending on the cell context (Hikasa and Sokol, 2011; Archbold et al, 2012). β-Catenin can also be acetylated by CBP at Lys49; point mutations that block acetylation result in ectopic transcriptional activation of the target gene c-myc, an effect that seems to be specific to this target, since others were unaffected (Wolf et al, 2002). Interestingly, this lysine residue is often found mutated in various cancers (Wolf et al, 2002). In contrast, modification of Lys345 by p300 has a broader effect and increases the affinity of β-catenin for TCF; β-catenin acetylated at Lys345 was strongly enriched in the colon cancer cells with hyperactivated Wnt/β-catenin transcription (Lévy et al, 2004). Both p300 and CBP are important for β-catenin transcription but the reality is likely to be complicated: they act both on chromatin and on β-catenin, and they probably affect the expression of different subsets of targets. In mouse embryonic stem cells (mESCs), CBP activity keeps the cells in a pluripotent state while p300 drives them into differentiation (Miyabayashi et al, 2007). Whether direct acetylation of β-catenin plays any role in this cell fate switch, and whether the different histone acetylation activities are important, remains to be resolved. Indeed, CBP/p300 may act as bimodal evolutionary conserved buffer of Wnt/β-catenin-mediated transcription (Li et al, 2007b).
The role of BCL9-Pygo co-activators
The transcriptional co-activators binding to β-catenin’s C-terminus are general initiators of transcription and are not β-catenin specific (Mosimann et al, 2009; Jaehning, 2010; Bannister and Kouzarides, 2011; Conaway and Conaway, 2011). The contribution of these general co-factors may, however, be modulated by Wnt-specific transcriptional co-activators. Specific interest has therefore been focussed on the β-catenin-specific co-activator BCL9 (Legless in Drosophila), which binds to the first ARM repeat of β-catenin.
In Drosophila, the Legless (Lgs) protein is essential for Wg signalling and acts by recruiting a further protein, Pygopus, which is believed to provide a transcriptional activation function. Lack of either Lgs or Pygopus phenocopies armadillo null mutants (Kramps et al, 2002; Thompson et al, 2002). The primary function of Lgs is to act as a linker between Armadillo and Pygopus; indeed, a minimal Lgs protein comprising just the domains required to bind Armadillo and Pygopus can rescue a lgs mutant, while overexpression of Lgs alone cannot compensate for Pygopus loss (Kramps et al, 2002; Townsley et al, 2004; Hoffmans et al, 2005).
In mammals, the situation is different. Mammalian genomes encode two paralogues for BCL9 (BCL9 and BCL9L) and also two for Pygopus (Pygo1 and Pygo2). A principle difference between the paralogues is their expression domains; more precise molecular work is pending. Even though BCL9 and BCL9L were shown to promote Wnt/β-catenin signalling in various cell types, their (likely) highly redundant role in the regulation of Wnt/β-catenin-mediated transcription during normal mammalian development remains to be evinced (de la Roche et al, 2008; Mani et al, 2009). The findings to date suggest that mammals have a less strict requirement for the BCL9/Pygo branch compared with Drosophila.
In the mouse, blocking the interaction between β-catenin and BCL9/BCL9L results in embryonic lethality around embryonic day E10.5 (Valenta et al, 2011). This is later than if β-catenin-mediated transcription is completely blocked when death occurs during gastrulation at E7.5. Nevertheless, loss of BCL9/BCL9L binding reduced β-catenin-mediated transcription as determined by a transcriptional reporter. Further contrasting the absolute requirement of Pygopus and Lgs in Drosophila, Pygo1/Pygo2 double mutant animals fully develop in utero but die post-natally. Further results indicate that Pygo can act as a tissue- or context-dependent β-catenin co-activator, for example, during mammary gland development (Gu et al, 2009). More surprisingly, Pygo mutants display some defects unrelated to Wnt/β-catenin signalling (Li et al, 2007; Schwab et al, 2007; Song et al, 2007). An important deduction from these observations is that in the mammalian context BCL9 can act as a transcriptional co-activator, without Pygo, in contrast to the situation in Drosophila. Evidence for this BCL9 function has been reported and suggested to stem from a CTD found in mammalian BCL9 that is missing in Drosophila Lgs (Sustmann et al, 2008). Additionally, Sustmann et al found that BCL9 can, via its C-terminus, directly interact with some of β-catenin's transcriptional co-activators, such as histone acetyltransferases CBP/p300 or TRRAP/GCN5, and strengthen the association of these with the TCF/β-catenin complex.
These results suggest a role for BCL9 in murine Wnt signalling that is more context dependent than it is in Drosophila. A concrete example for this notion comes from intestinal tumourigenesis, where BCL9/BCL9L regulates a subset of Wnt/β-catenin targets important for EMT and invasivity (Deka et al, 2010; Brembeck et al, 2011). This function is most likely provided by BCL9L.
In Drosophila, Pygopus has transcriptional activation activity and was shown to interact with components of the Mediator complex and this interaction may help stabilizing the association of the complex with the C-terminus of Armadillo (Carrera et al, 2008). However, it must be noted that the modus operandi of Pygopus may be more complex and subtle. There is evidence that Pygopus can associate, potentially indirectly, with dTCF/Pangolin in the absence of Wg stimuli and nuclear β-catenin, and may thus serve as pioneering factor and help Armadillo to displace Groucho repressors from dTCF/Pangolin (de la Roche and Bienz, 2007; Mieszczanek et al, 2008). More intriguingly, Pygopus features a PHD domain and retains the associated ability to bind methylated histones. In Drosophila, this interaction is not essential, but in other organisms the binding might contribute to its context-specific role in various tissues (e.g., in the mammary gland) (Fiedler et al, 2008; Kessler et al, 2009; Gu et al, 2009; Gu et al, 2012).
TCF/β-catenin as the platform for broad chromatin remodelling
Wnt stimulation was shown to result in dramatic changes in chromatin structure, not only in close proximity to target promoters (Parker et al, 2008; Mosimann et al, 2009). Experiments in Drosophila analysing histone modifications surrounding the Wnt-target genes naked cuticle and Notum revealed that upon pathway stimulation nucleosomes are rapidly acetylated in a CBP-dependent manner and saturated in a wide region (up to 30 kilobases) circumadjacent to the analysed WREs (Parker et al, 2008). Such widespread chromatin modifications mediated by TCF/β-catenin might help the additional transcription factors to access these regions. These factors may act also independently of TCF/β-catenin as specific master regulators. During the regeneration of haematopoietic lineages after injury, for example, different subclasses of cell-specific genes are transcribed. In many cases, their promoters are co-occupied by SMAD transcription factors together with TCF4, plus cell type-specific master regulators (progenitor specific or erythroid specific); these finally determine the cell type (Trompouki et al, 2011). One possible explanation for such multiple co-operative binding of factors is that TCF, together with β-catenin, modulates the composition of the surrounding chromatin. That such widespread chromatin modifications can take place might be ascribed to the ability of the TCF HMG Box to induce DNA kinks (up to 130°); such bending may play a pivotal architectural role and bring distant parts of DNA closer together (Love et al, 1995; Giese et al, 1997). Another explanation may be that, as is the case for many transcription factors, the consensus DNA binding motif of TCFs is not strict. Indeed, analysis of the binding of TCF to the promoter of the β-catenin target gene c-myc revealed that binding might be polymorphic and some single base substitutions may even increase TCF binding, while others reduce but not completely abolish it (Tuupanen et al, 2009, Wright et al, 2010). Similar variability in the TCF consensus site was seen in a Drosophila study of the Wg target gene naked cuticle (Chang et al, 2008b). Such degeneration in TCF's consensus site would enable TCFs to act as widespread chromatin modulators, either to activate transcription (with β-catenin bringing various transcription promoting complexes, such as HATs or HMTs) or to act as repressors. In the latter case, the TCF partner Groucho/TLE can recruit histone deacetylases (HDACs), rendering the chromatin more compact and transcriptionally inactive (Sekiya and Zaret, 2007). The repressive potential of some TCFs (dTCF and TCF3 or TCF4) may be enhanced by the co-repressor activity of CtBP (C-terminal binding co-repressor) (Valenta et al, 2003; Bhambhani et al, 2011). Hence, β-catenin could bring about derepression just by expelling Groucho/TLE and the associated HDACs, without actively initiating the transcription by recruiting co-activators. This seems to be the case in mESCs where β-catenin derepresses some TCF3-specific-targets (Wray et al, 2011).
Besides promoting acetylation of histones, β-catenin can facilitate via its interacting partners other histone modifications near target promoters, including ubiquitination and subsequently histone H3K4 trimethylation by the MLL1/MLL2 SET1-type HMT complex, which is indispensable for proper β-catenin transcription (Sierra et al, 2006). Recently, SET8 HMT-mediated monomethylation of H3K20 was shown to be an important positive modulator of TCF/β-catenin transcription. SET8 can interact with TCF4, but can perform the methylation only when β-catenin is bound and Groucho/TLE is displaced. Monomethylation of H3K20 was observed upon Wnt stimulation in mammalian cells and it seems to be important for transcription (Li et al, 2011). Interestingly, SET8 was recently found in the complex of transcription factor TWIST promoting its role in downregulating E-cadherin, thereby strongly enhancing EMT (Yang et al, 2011a, 240). Hence, SET8 might not only promote transcription of β-catenin-specific targets (TWIST is a direct β-catenin target) but could also enhance β-catenin transcription by facilitating the release of β-catenin from epithelial junctions via TWIST-mediated EMT.
Not only histone modifications but also various ATP-chromatin remodelling complexes can shuffle histones in the vicinity of Wnt/β-catenin target promoters and thus rearrange the surrounding chromatin. The action of many of them was discussed previously (Mosimann et al, 2009). However, recent evidence suggests that besides the binding of particular chromatin remodelling components to β-catenin, also the exact composition of the relevant complex is crucial. A pertinent example is Imitation Switch (ISWI), a chromatin remodelling ATPse: associated with the Nucleosome Remodelling Factor (NURF) complex ISWI positively stimulates β-catenin mediated transcription, but as a part of ACF (ATP-utilizing Chromatin remodelling and assembly Factor) it acts as a transcriptional repressor (Liu et al, 2008b; Song et al, 2009).
β-Catenin-mediated transcriptional repression
It is becoming increasingly clear that activation of the Wnt/β-catenin pathway not only stimulates transcription but can also repress it. The mechanisms by which the latter is achieved are only just beginning to be understood and seem to be diverse. Targeting either transcriptional repressors such as ISWI-ACF, or activators such as ISWI-NURF, might be one way how β-catenin orchestrates activation or repression. A related way may be to directly recruit repressive factors, such as Reptin or Fhit that associate with the TCF/β-catenin complex and thus repress β-catenin-mediated transcription (Bauer et al, 2000; Weiske et al, 2007).
Binding of β-catenin to some target promoters can directly repress transcription, as reported for the genes E-cadherin in keratinocytes or p16INK4a in melanomas; in both studies, the binding of β-catenin to TCF lead to repression (Jamora et al, 2003; Delmas et al, 2007). In kerationocytes, the direct repressive effect is enhanced indirectly by the repressor Snail in a β-catenin-dependent manner. Another mechanism of direct β-catenin repression was described for the Drosophila stripe gene. Here, the dTCF/Pangolin binding site overlaps with a binding site for Cubitus interruptus (Ci), a transcription factor that is essential for stripe transcription. Binding of TCF/β-catenin most likely displaces Ci and thus represses transcription (Piepenburg et al, 2000). A further example of direct β-catenin-mediated transcriptional repression was also shown in Drosophila cell lines; here, β-catenin represses expression of the extracellular matrix enzyme Ugt36Bc (Blauwkamp et al, 2008). Interestingly, in this case dTCF/β-catenin-mediated repression is targeted to WREs dramatically differing from the TCF consensus, suggesting existence of TCF-repressive motifs. It will be interesting how common this mechanism is and how the different TCF motifs bring about activation or repression.
Deactivation of TCF/β-catenin-mediated transcription
An off switch is essential for fine-tuning and temporal control (Figure 5). Throughout life, the level of β-catenin transcriptional activity needs to be carefully controlled and it should neither be too high nor persist for too long. This aspect of Wnt pathway activity and particularly transcriptional control has not yet been tackled in depth.
One possible way to deactivate a nuclear β-catenin/TCF complex is to change the interaction partners from transcriptional activators to repressors. Such a role was shown for Reptin as noted above (Bauer et al, 2000). The nuclear co-repressor SMRT (Silencing Mediator for Retinoid and Thyroid hormone receptor) and the closely related corepressor NCoR (Nuclear receptor Co-Repressor) act in a similar fashion. In many mammalian cell lines, they were found to bind to β-catenin and change the transcription status via associated HDACs (Song and Gelmann, 2008). Their repressive activity might also be exerted directly at WREs occupied by TCF/β-catenin complexes, where they titrate β-catenin away (Song and Gelmann, 2008). TCF/β-catenin transcription can be deactivated simply by separating TCF and β-catenin, for example, via competition by co-repressors, such as Groucho/TLE, Mtgr-1 (Myeloid translocation gene related-1) or COOP (Moore et al, 2008; Song et al, 2010). Preventing the binding (or re-attachment) between β-catenin and TCFs would have the same effect and could be mediated by specific repressive proteins, such as ICAT (Tago et al, 2000; Daniels and Weis, 2002; Graham et al, 2002; Takemaru et al, 2003). The small protein Chibby deactivates β-catenin-mediated transcription in several ways: First, it binds to Helix-C of β-catenin and thus blocks the binding of the majority of the C-terminal β-catenin transcriptional co-activators; second, it competes with TCFs for β-catenin binding; additionally, Chibby can form a trimolecular complex with β-catenin and the 14-3-3 protein. This complex is exported from the nucleus and thus promotes the nuclear depletion of β-catenin (Takemaru et al, 2003; Li et al, 2008).
Nuclear sequestration and export could also be mediated by nuclear pools of ‘destruction complex’ factors. Nuclear APC does not appear to target β-catenin for destruction (despite the fact that also other components of the destruction complex were found in the nucleus). It was proposed to sequester nuclear β-catenin away from TCF and thus render it inactive (Hamada and Bienz, 2004; Sierra et al, 2006). Nuclear APC and nuclear Axin are reported to mediate nuclear export of β-catenin in various cells (Neufeld et al, 2000; Cong and Varmus, 2004). The repression of the β-catenin target gene c-myc is achieved through the co-operative activity of APC, the transcriptional co-repressors CtBP (C-terminal binding protein) and YY1 (Yin Yang 1). APC together with CtBP is recruited to target promoters, phosphorylation of APC induces its binding to β-catenin, which is subsequently dislodged from TCF. But, it remains open if APC together with either or both CtBP and YY1 can repress or deactivate β-catenin-mediated transcription by a different mechanism (Hamada and Bienz, 2004; Sierra et al, 2006).
TCF/Lef-independent β-catenin-mediated transcription
There is growing evidence that the relationship between β-catenin and TCF is not monogamous and that β-catenin associates with other DNA-binding transcription factors. Most of these transcription factors can start transcription by themselves or in association with other co-factors, in contrast to TCF's ultimate requirement for β-catenin to start transcription. Rather, the potential for interaction with β-catenin provides another layer of control and cross-talk. The interaction with other transcription factors may lead to competition for β-catenin and thus affect TCF/β-catenin-mediated transcription. Some nuclear receptors, for example, the androgen receptor, may act via this mechanism. In colon cancer cell lines, activated androgen receptor may bind to β-catenin, usurping it to enhance its own transcriptional activity at the expense of that of TCF. Interestingly, the androgen receptor can promote nuclear translocation of β-catenin (Pawlowski et al, 2002; Mulholland et al, 2003). Various other nuclear receptors were reported to interact with β-catenin with varied consequences. β-Catenin can synergistically activate nuclear receptor-driven transcription, or the binding of the ligand to the receptor may deactivate or repress β-catenin’s activity (reviewed in detail in Beildeck et al, 2010). A relevant example is the co-operative action between the nuclear receptor LRH1 (Liver Receptor Homologue 1) and the TCF/β-catenin complex. β-Catenin can directly associate with LRH1 and enhance expression of LRH1's target genes (e.g., cyclinE1). On the other hand, LRH1 can serve as transcriptional co-activator of β-catenin; one TCF/β-catenin target whose expression is enhanced in this way is cyclin D1, which together with cyclinE1 promotes cell proliferation in pancreatic and hepatic cells. Moreover, LRH1 was found to be a direct TCF/β-catenin target, suggesting the potential of a positive feedback loop (Botrugno et al, 2004; Wagner et al, 2010). This co-operation between LRH1 and TCFs is evolutionary conserved as it was also found in C. elegans (Asahina et al, 2006).
β-Catenin was shown to play an important role under hypoxic conditions. Hypoxia dramatically enhances the levels of Hypoxia Induced Factor 1α (HIF1α), which sequesters β-catenin away from classical TCF/β-catenin targets and uses it to activate hypoxia-induced (HIF1α) targets, leading to a reduction of TCF/β-catenin-mediated transcription (Kaidi et al, 2007). This role of HIF1α is cell-type specific: whereas in differentiated and cancer cells HIF1α acts as a repressor of TCF/β-catenin activity; in stem cells and progenitor cells HIF1α stimulates TCF/β-catenin-mediated transcription by promoting nuclear translocation β-catenin (via an as-yet unknown mechanism) and by raising expression of TCF (Figure 6; Mazumdar et al, 2010). Reduction in O2 levels can impair mitochondrial functions and contribute to oxidative stress accompanied by generation of reactive oxygen species (ROS). Such conditions activate and upregulate the forkhead transcription factors FOXO, which similar to HIF1α compete with TCF for β-catenin. β-Catenin can directly bind FOXOs and enhance transcription driven by them. This mechanism is evolutionarily conserved (Essers et al, 2005). Such competition between FOXO and TCF may contribute to insulin resistance and diabetes, and it also seems to be important for bone development (Figure 6; Manolagas and Almeida, 2007; Jin et al, 2008; Almeida, 2011). A FOXO β-catenin complex can play an important role in regulating hepatic metabolism and gluconeogenesis (Liu et al, 2011).
Figure 6.
β-catenin in hypoxia and under oxidative stress. (A) Under hypoxic conditions in stem cells activated HIF1α stimulates transcription mediated by β-catenin and Lef1 (and/or TCF1) and by promoting nuclear translocation of β-catenin. (B) In differentiated or cancer cells, HIF1α usurps β-catenin at the expense of TCF/Lef transcription factors, thereby inhibiting Tcf/β-catenin-mediated transcription. β-Catenin potentiates transcription of HIF1α target genes. Among the others targets are the forkhead transcription factors FOXO, which are also upregulated under stress conditions caused by ROS. FOXO transcription factors even sequester β-catenin from TCF/Lef, counteracting TCF/β-catenin-mediated transcription. Growth stimuli (such as Insulin) can activate the protein kinase Akt/PKB, which blocks FOXO activity and may promote phosphorylation of β-catenin, restoring active TCF/β-catenin transcription.
β-Catenin has been reported to bind various other transcription factors (including Sox17 during vertebrate gastrulation) and augment transcription of their target genes (Sinner et al, 2004; Archbold et al, 2012). In mESCs, β-catenin can increase transcription mediated by the key pluripotency transcription factor Oct4. Interestingly, for this augmentation the C-terminally recruited transcriptional co-activators of β-catenin are not essential: a C-terminally truncated β-catenin can also augment Oct4-driven transcription and promote pluripotency (Kelly et al, 2011). Another mechanism by which β-catenin can affect cell fate independent of TCFs was shown in prostatic cancer, where β-catenin affects the transcription of the metastatic suppressor gene KAI1. Expression of this gene is activated by the p50 subunit of the transcription factor NF-κB with the help of the Pontin and Tip60 transcriptional co-activator complexes. If KAI1 is expressed, then prostatic cancer cells have low metastatic potential. But the situation dramatically changes when β-catenin directly binds to p50, as it can recruit the transcriptional co-repressor Reptin, blocking KAI1 transcription and promoting the metastatic potential of prostatic cancer cells (Kim et al, 2005). These observations probably represent only the tip of the iceberg and future studies may find that the signalling role of β-catenin is broader and more complex than we thought when the primary route of Wnt-to-β-catenin-to-TCFs was first described.
Illustrating this point, recently co-operation between TCF/β-catenin signalling and Smad transcription factors was demonstrated in Drosophila. When the TGFβ/BMP pathway is not active, the Drosophila Smads Mad and Medea seem to enhance TCF/β-catenin-mediated transcription through direct interaction with the TCF/β-catenin complexes, creating a competitive circuit between these two pathways (Eivers et al, 2011). Ground-breaking experiments combining analysis of chromatin occupancy, transcriptional activity measurements, and biochemical approaches are beginning to unveil the intricacy of the co-operation. In the regulation of many genes, a close proximity of Smad and TCF binding sites were demonstrated to activate transcription in a coordinated way (Nakano et al, 2010, Archbold et al, 2012). In the haematopoietic system, TCF and β-catenin co-operate with Smads, the transcriptional effectors of TGFβ/BMP signalling, and with other specific cell-type master regulators to determine cell fates (Trompouki et al, 2011). Analogously in intestinal cancer cell lines, ChIP analyses revealed many TCF (β-catenin) binding sites in close proximity to AP-1 binding sites (Bottomly et al, 2010). This observation confirms previous results indicating direct co-operation between TCF/β-catenin and the AP-1 complex (or its component c-Jun) (Toualbi et al, 2007; Sancho et al, 2009). Such cooperation has also been known for many years, albeit in less detail, in the regulation of body axis determination or cellular patterning in the intestine (Archbold et al, 2012).
Future directions—a question of context
β-Catenin is a multifaceted molecule and despite decades of study we are only just beginning to grasp the intricacies of its different functions and how they are co-ordinated. How can β-catenin as a single molecule affect so many phenomena in animals and permit so many different outputs? Of course, Wnt/β-catenin signalling cannot be considered as an isolated entity, the end result is certainly the sum of its co-operation and synergy with other signalling pathways, such as the TGFβ/BMP pathway. The advent of genome-wide systems biology-based approaches signposts one path that future studies should take to investigate this: the language of the cross-talk will be deciphered by examining the chromatin occupancy of different transcription factors and performing expression studies to monitor the effect of perturbing the pathways controlling those factors.
As outlined in this review, part of the solution of the issue of β-catenin’s pleiotropy will also be found in dissecting the roles of the proteins that interact with it. What will likely emerge is that many factors act in a context-dependent manner and be crucial only in specific cell types and maybe even only for a specific subset of genes. Recently, the protein Jerky/Earthbound was reported to modulate β-catenin-TCF activity in a cell type-specific manner (Benchabane et al, 2011; Xin et al, 2011). While this particular example remains to be further validated it is likely that more such ‘specific’ factors will be unearthed. To appreciate the functional significance of an interaction, we must consider the evolutionary conservation of this function. An illustrative example is the role of Lgs/BCL9. In Drosophila, it is essential; in mice it is important, but only in a context-dependent way. What is the situation in other species? The best way to address this will be to carry out expression studies, to determine the genomic finger-prints of Wnt/Wg signalling and then assess the impact of loss of BCL9/Lgs. Such analyses will test how valid the assumption of a universal requirement actually is. In the context of probing the disease relevance of an interaction, analogous finger-printing studies in the relevant tissues in mouse models will be important.
The binding of BCL9 to β-catenin can be specifically abrogated by the point mutation D164A. Future analysis of mice carrying this mutation will give us precise information about the relevance of the BCL9–β-catenin interaction. Structure function studies, or chemical-based screens, will hopefully reveal similar means to surgically block other interactions. The investment of generating model organisms (knockin mice, transgenic Drosophila, etc.) that carry these mutations will pay dividends in terms of the insights revealed.
Thirty years ago, Wnts were discovered and subsequent work has placed a spot-light on β-catenin as an essential downstream effector. To untangle the role of β-catenin, we must consider the pathways that converge on it and the ligands that trigger those pathways: Wnts, R-spondins, and others. Are there subtle differences in the elicited responses and what do they mean? Only by answering such questions we will gain a true understanding of the Wnt/β-catenin pathway. This will serve as a basis for targeted medical approaches in situations where aberrant Wnt/β-catenin signaling plays a role, such as in cancer, metabolic diseases, and impaired tissue regeneration.
Acknowledgments
We thank C Mosimann for critical reading of the manuscript. Work in our laboratory is supported by the European Research Council, the Swiss National Science Foundation, and the Kanton of Zürich.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adamska M, Larroux C, Adamski M, Green K, Lovas E, Koop D, Richards GS, Zwafink C, Degnan BM (2010) Structure and expression of conserved Wnt pathway components in the demosponge Amphimedon queenslandica. Evol Dev 12: 494–518 [DOI] [PubMed] [Google Scholar]
- Almeida M (2011) Unraveling the role of FoxOs in bone--insights from mouse models. Bone 49: 319–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angers S, Moon RT (2009) Proximal events in Wnt signal transduction. Nat Rev Mol Cell Biol 10: 468–477 [DOI] [PubMed] [Google Scholar]
- Ansieau S, Morel AP, Hinkal G, Bastid J, Puisieux A (2010) TWISTing an embryonic transcription factor into an oncoprotein. Oncogene 29: 3173–3184 [DOI] [PubMed] [Google Scholar]
- Arce L, Pate KT, Waterman ML (2009) Groucho binds two conserved regions of LEF-1 for HDAC-dependent repression. BMC Cancer 9: 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arce L, Yokoyama NN, Waterman ML (2006) Diversity of LEF/TCF action in development and disease. Oncogene 25: 7492–7504 [DOI] [PubMed] [Google Scholar]
- Archbold HC, Yang YX, Chen L, Cadigan KM (2012) How do they do Wnt they do?: regulation of transcription by the Wnt/β-catenin pathway. Acta Physiol (Oxf) 204: 74–109 [DOI] [PubMed] [Google Scholar]
- Asahina M, Valenta T, Silhankova M, Korinek V, Jindra M (2006) Crosstalk between a nuclear receptor and beta-catenin signaling decides cell fates in the C. elegans somatic gonad. Dev Cell 11: 203–211 [DOI] [PubMed] [Google Scholar]
- Atcha FA, Syed A, Wu B, Hoverter NP, Yokoyama NN, Ting JH, Munguia JE, Mangalam HJ, Marsh JL, Waterman ML (2007) A unique DNA binding domain converts T-cell factors into strong Wnt effectors. Mol Cell Biol 27: 8352–8363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahmanyar S, Kaplan DD, Deluca JG, Giddings TH Jr, O'Toole ET, Winey M, Salmon ED, Casey PJ, Nelson WJ, Barth AI (2008) Beta-Catenin is a Nek2 substrate involved in centrosome separation. Genes Dev 22: 91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsamo J, Leung T, Ernst H, Zanin MK, Hoffman S, Lilien J (1996) Regulated binding of PTP1B-like phosphatase to N-cadherin: control of cadherin-mediated adhesion by dephosphorylation of beta-catenin. J Cell Biol 134: 801–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T (2011) Regulation of chromatin by histone modifications. Cell Res 21: 381–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barolo S (2006) Transgenic Wnt/TCF pathway reporters: all you need is Lef? Oncogene 25: 7505–7511 [DOI] [PubMed] [Google Scholar]
- Barrallo-Gimeno A, Nieto MA (2005) The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development 132: 3151–3161 [DOI] [PubMed] [Google Scholar]
- Bauer A, Chauvet S, Huber O, Usseglio F, Rothbächer U, Aragnol D, Kemler R, Pradel J (2000) Pontin52 and reptin52 function as antagonistic regulators of beta-catenin signalling activity. EMBO J 19: 6121–6130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens J, von Kries JP, Kühl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W (1996) Functional interaction of beta-catenin with the transcription factor LEF-1. Nature 382: 638–642 [DOI] [PubMed] [Google Scholar]
- Beildeck ME, Gelmann EP, Byers SW (2010) Cross-regulation of signaling pathways: an example of nuclear hormone receptors and the canonical Wnt pathway. Exp Cell Res 316: 1763–1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bek S, Kemler R (2002) Protein kinase CKII regulates the interaction of beta-catenin with alpha-catenin and its protein stability. J Cell Sci 115(Pt 24): 4743–4753 [DOI] [PubMed] [Google Scholar]
- Benchabane H, Xin N, Tian A, Hafler BP, Nguyen K, Ahmed A, Ahmed Y (2011) Jerky/Earthbound facilitates cell-specific Wnt/Wingless signalling by modulating β-catenin-TCF activity. EMBO J 30: 1444–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin JM, Kwiatkowski AV, Yang C, Korobova F, Pokutta S, Svitkina T, Weis WI, Nelson WJ (2010) AlphaE-catenin regulates actin dynamics independently of cadherin-mediated cell-cell adhesion. J Cell Biol 189: 339–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhambhani C, Chang JL, Akey DL, Cadigan KM (2011) The oligomeric state of CtBP determines its role as a transcriptional co-activator and co-repressor of Wingless targets. EMBO J 30: 2031–2043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bikkavilli RK, Feigin ME, Malbon CC (2008) p38 mitogen-activated protein kinase regulates canonical Wnt-beta-catenin signaling by inactivation of GSK3beta. J Cell Sci 121(Part 21): 3598–3607 [DOI] [PubMed] [Google Scholar]
- Bilic J, Huang YL, Davidson G, Zimmermann T, Cruciat CM, Bienz M, Niehrs C (2007) Wnt induces LRP6 signalosomes and promotes dishevelled-dependent LRP6 phosphorylation. Science 316: 1619–1622 [DOI] [PubMed] [Google Scholar]
- Blauwkamp TA, Chang MV, Cadigan KM (2008) Novel TCF-binding sites specify transcriptional repression by Wnt signalling. EMBO J 27: 1436–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botrugno OA, Fayard E, Annicotte JS, Haby C, Brennan T, Wendling O, Tanaka T, Kodama T, Thomas W, Auwerx J, Schoonjans K (2004) Synergy between LRH-1 and beta-catenin induces G1 cyclin-mediated cell proliferation. Mol Cell 15: 499–509 [DOI] [PubMed] [Google Scholar]
- Bottomly D, Kyler SL, McWeeney SK, Yochum GS (2010) Identification of {beta}-catenin binding regions in colon cancer cells using ChIP-Seq. Nucleic Acids Res 38: 5735–5745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brembeck FH, Schwarz-Romond T, Bakkers J, Wilhelm S, Hammerschmidt M, Birchmeier W (2004) Essential role of BCL9-2 in the switch between beta-catenin's adhesive and transcriptional functions. Genes Dev 18: 2225–2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brembeck FH, Wiese M, Zatula N, Grigoryan T, Dai Y, Fritzmann J, Birchmeier W (2011) BCL9-2 promotes early stages of intestinal tumor progression. Gastroenterology 141: 1359–13701370. e1–3 [DOI] [PubMed] [Google Scholar]
- Broun M, Gee L, Reinhardt B, Bode HR (2005) Formation of the head organizer in hydra involves the canonical Wnt pathway. Development 132: 2907–2916 [DOI] [PubMed] [Google Scholar]
- Brunner E, Peter O, Schweizer L, Basler K (1997) Pangolin encodes a Lef-1 homologue that acts downstream of Armadillo to transduce the Wingless signal in Drosophila. Nature 385: 829–833 [DOI] [PubMed] [Google Scholar]
- Bustos VH, Ferrarese A, Venerando A, Marin O, Allende JE, Pinna LA (2006) The first armadillo repeat is involved in the recognition and regulation of beta-catenin phosphorylation by protein kinase CK1. Proc Natl Acad Sci USA 103: 19725–19730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrera I, Janody F, Leeds N, Duveau F, Treisman JE (2008) Pygopus activates Wingless target gene transcription through the mediatorcomplex subunits Med12 and Med13. Proc Natl Acad Sci USA 105: 6644–6649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaño J, Raurell I, Piedra JA, Miravet S, Duñach M, García de Herreros A (2002) Beta-catenin N- and C-terminal tails modulate the coordinated binding of adherens junction proteins to beta-catenin. J Biol Chem 277: 31541–31550 [DOI] [PubMed] [Google Scholar]
- Cavallo RA, Cox RT, Moline MM, Roose J, Polevoy GA, Clevers H, Peifer M, Bejsovec A (1998) Drosophila TCF and Groucho interact to repress Wingless signalling activity. Nature 395: 604–608 [DOI] [PubMed] [Google Scholar]
- Chang JL, Chang MV, Barolo S, Cadigan KM (2008a) Regulation of the feedback antagonist naked cuticle by Wingless signaling. Dev Biol 321: 446–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang MV, Chang JL, Gangopadhyay A, Shearer A, Cadigan KM (2008b) Activation of wingless targets requires bipartite recognition of DNA by TCF. Curr Biol 18: 1877–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilov D, Sinjushina N, Rita H, Taketo MM, Mäkelä TP, Partanen J (2011) Phosphorylated β-catenin localizes to centrosomes of neuronal progenitors and is required for cell polarity and neurogenesis in developing midbrain. Dev Biol 357: 259–268 [DOI] [PubMed] [Google Scholar]
- Chitalia VC, Foy RL, Bachschmid MM, Zeng L, Panchenko MV, Zhou MI, Bharti A, Seldin DC, Lecker SH, Dominguez I, Cohen HT (2008) Jade-1 inhibits Wnt signalling by ubiquitylating beta-catenin and mediates Wnt pathway inhibition by pVHL. Nat Cell Biol 10: 1208–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HJ, Huber AH, Weis WI (2006) Thermodynamics of beta-catenin-ligand interactions: the roles of the N- and C-terminal tails in modulating binding affinity. J Biol Chem 281: 1027–1038 [DOI] [PubMed] [Google Scholar]
- Coates JC, Grimson MJ, Williams RS, Bergman W, Blanton RL, Harwood AJ (2002) Loss of the beta-catenin homologue aardvark causes ectopic stalk formation in Dictyostelium. Mech Dev 116: 117–127 [DOI] [PubMed] [Google Scholar]
- Coluccia AM, Vacca A, Duñach M, Mologni L, Redaelli S, Bustos VH, Benati D, Pinna LA, Gambacorti-Passerini C (2007) Bcr-Abl stabilizes beta-catenin in chronic myeloid leukemia through its tyrosine phosphorylation. EMBO J 26: 1456–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conacci-Sorrell M, Simcha I, Ben-Yedidia T, Blechman J, Savagner P, Ben-Ze'ev A (2003) Autoregulation of E-cadherin expression by cadherin-cadherin interactions: the roles of beta-catenin signaling, Slug, and MAPK. J Cell Biol 163: 847–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaway RC, Conaway JW (2011) Function and regulation of the Mediator complex. Curr Opin Genet Dev 21: 225–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong F, Varmus H (2004) Nuclear-cytoplasmic shuttling of Axin regulates subcellular localization of beta-catenin. Proc Natl Acad Sci USA 101: 2882–2887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox RT, Pai LM, Kirkpatrick C, Stein J, Peifer M (1999) Roles of the C terminus of Armadillo in Wingless signaling in Drosophila. Genetics 153: 319–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dajani R, Fraser E, Roe SM, Yeo M, Good VM, Thompson V, Dale TC, Pearl LH (2003) Structural basis for recruitment of glycogen synthase kinase 3beta to the axin-APC scaffold complex. EMBO J 22: 494–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels DL, Weis WI (2002) ICAT inhibits beta-catenin binding to TCF/Lef-family transcription factors and the general coactivator p300 using independent structural modules. Mol Cell 10: 573–584 [DOI] [PubMed] [Google Scholar]
- Daniels DL, Weis WI (2005) Beta-catenin directly displaces Groucho/TLE repressors from TCF/Lef in Wnt-mediated transcription activation. Nat Struct Mol Biol 12: 364–371 [DOI] [PubMed] [Google Scholar]
- DasGupta R, Fuchs E (1999) Multiple roles for activated LEF/TCF transcription complexes during hair follicle development and differentiation. Development 126: 4557–4568 [DOI] [PubMed] [Google Scholar]
- de la Roche M, Bienz M (2007) Wingless-independent association of Pygopus with dTCF target genes. Curr Biol 17: 556–561 [DOI] [PubMed] [Google Scholar]
- de la Roche M, Worm J, Bienz M (2008) The function of BCL9 in Wnt/beta-catenin signaling and colorectal cancer cells. BMC Cancer 8: 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deka J, Wiedemann N, Anderle P, Murphy-Seiler F, Bultinck J, Eyckerman S, Stehle JC, André S, Vilain N, Zilian O, Robine S, Delorenzi M, Basler K, Aguet M (2010) Bcl9/Bcl9l are critical for Wnt-mediated regulation of stem cell traits in colon epithelium and adenocarcinomas. Cancer Res 70: 6619–6628 [DOI] [PubMed] [Google Scholar]
- Delmas V, Beermann F, Martinozzi S, Carreira S, Ackermann J, Kumasaka M, Denat L, Goodall J, Luciani F, Viros A, Demirkan N, Bastian BC, Goding CR, Larue L (2007) Beta-catenin induces immortalization of melanocytes by suppressing p16INK4a expression and cooperates with N-Ras in melanoma development. Genes Dev 21: 2923–2935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson DJ, Nelson WJ, Weis WI (2011) A polarized epithelium organized by beta- and alpha-catenin predates cadherin and metazoan origins. Science 331: 1336–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova YN, Li J, Lee YT, Rios-Esteves J, Friedman DB, Choi HJ, Weis WI, Wang CY, Chazin WJ (2010) Direct ubiquitination of beta-catenin by Siah-1 and regulation by the exchange factor TBL1. J Biol Chem 285: 13507–13516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding VW, Chen RH, McCormick F (2000) Differential regulation of glycogen synthase kinase 3beta by insulin and Wnt signaling. J Biol Chem 275: 32475–32481 [DOI] [PubMed] [Google Scholar]
- Drees F, Pokutta S, Yamada S, Nelson WJ, Weis WI (2005) Alpha-catenin is a molecular switch that binds E-cadherin-beta-catenin and regulates actin-filament assembly. Cell 123: 903–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du C, Jaggi M, Zhang C, Balaji KC (2009) Protein kinase D1-mediated phosphorylation and subcellular localization of beta-catenin. Cancer Res 69: 1117–1124 [DOI] [PubMed] [Google Scholar]
- Du C, Zhang C, Li Z, Biswas MH, Balaji KC (2012) Beta-catenin phosphorylated at threonine 120 antagonizes generation of active beta-catenin by spatial localization in trans-golgi network. PLoS One 7: e33830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupre-Crochet S, Figueroa A, Hogan C, Ferber EC, Bialucha CU, Adams J, Richardson EC, Fujita Y (2007) Casein kinase 1 is a novel negative regulator of E-cadherin-based cell-cell contacts. Mol Cell Biol 27: 3804–3816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eivers E, Demagny H, Choi RH, De Robertis EM (2011) Phosphorylation of Mad controls competition between wingless and BMP signaling. Sci Signal 4: ra68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eklof Spink K, Fridman SG, Weis WI (2001) Molecular mechanisms of beta-catenin recognition by adenomatous polyposis coli revealed by the structure of an APC-beta-catenin complex. EMBO J 20: 6203–6212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers MA, de Vries-Smits LM, Barker N, Polderman PE, Burgering BM, Korswagen HC (2005) Functional interaction between beta-catenin and FOXO in oxidative stress signaling. Science 308: 1181–1184 [DOI] [PubMed] [Google Scholar]
- Fagotto F, Glück U, Gumbiner BM (1998) Nuclear localization signal-independent and importin/karyopherin-independentnuclear import of beta-catenin. Curr Biol 8: 181–190 [DOI] [PubMed] [Google Scholar]
- Fang D, Hawke D, Zheng Y, Xia Y, Meisenhelder J, Nika H, Mills GB, Kobayashi R, Hunter T, Lu Z (2007) Phosphorylation of beta-catenin by AKT promotes beta-catenin transcriptional activity. J Biol Chem 282: 11221–11229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fevr T, Robine S, Louvard D, Huelsken J (2007) Wnt/beta-catenin is essential for intestinal homeostasis and maintenance of intestinal stem cells. Mol Cell Biol 27: 7551–7559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedler M, Sánchez-Barrena MJ, Nekrasov M, Mieszczanek J, Rybin V, Müller J, Evans P, Bienz M (2008) Decoding of methylated histone H3 tail by the Pygo-BCL9 Wnt signaling complex. Mol Cell 30: 507–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galceran J, Fariñas I, Depew MJ, Clevers H, Grosschedl R (1999) Wnt3a−/−like phenotype and limb deficiency in Lef1(−/−)TCF1(−/−) mice. Genes Dev 13: 709–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavert N, Sheffer M, Raveh S, Spaderna S, Shtutman M, Brabletz T, Barany F, Paty P, Notterman D, Domany E, Ben-Ze'ev A (2007) Expression of L1-CAM and ADAM10 in human colon cancer cells induces metastasis. Cancer Res 67: 7703–7712 [DOI] [PubMed] [Google Scholar]
- Gee L, Hartig J, Law L, Wittlieb J, Khalturin K, Bosch TC, Bode HR. (2010) Beta-catenin plays a central role in setting up the head organizer in hydra. Dev Biol 340: 116–124 [DOI] [PubMed] [Google Scholar]
- Giese K, Pagel J, Grosschedl R (1997) Functional analysis of DNA bending and unwinding by the high mobility group domain of LEF-1. Proc Natl Acad Sci USA 94: 12845–12850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottardi CJ, Gumbiner BM (2004) Distinct molecular forms of beta-catenin are targeted to adhesive or transcriptional complexes. J Cell Biol 167: 339–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham TA, Clements WK, Kimelman D, Xu W (2002) The crystal structure of the beta-catenin/ICAT complex reveals the inhibitory mechanism of ICAT. Mol Cell 10: 563–571 [DOI] [PubMed] [Google Scholar]
- Graham TA, Weaver C, Mao F, Kimelman D, Xu W (2000) Crystal structure of a beta-catenin/TCF complex. Cell 103: 885–896 [DOI] [PubMed] [Google Scholar]
- Grigoryan T, Wend P, Klaus A, Birchmeier W (2008) Deciphering the function of canonical Wnt signals in development and disease: conditional loss- and gain-of-function mutations of beta-catenin in mice. Genes Dev 22: 2308–2341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson MJ, Coates JC, Reynolds JP, Shipman M, Blanton RL, Harwood AJ (2000) Adherens junctions and beta-catenin-mediated cell signalling in a non-metazoan organism. Nature 408: 727–731 [DOI] [PubMed] [Google Scholar]
- Gu B, Sun P, Yuan Y, Moraes RC, Li A, Teng A, Agrawal A, Rhéaume C, Bilanchone V, Veltmaat JM, Takemaru K, Millar S, Lee EY, Lewis MT, Li B, Dai X (2009) Pygo2 expands mammary progenitor cells by facilitating histone H3 K4methylation. J Cell Biol 185: 811–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu B, Watanabe K, Dai X (2012) Pygo2 regulates histone gene expression and H3 K56 acetylation in humanmammary epithelial cells. Cell Cycle 11: 79–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guger KA, Gumbiner BM (1995) Beta-Catenin has Wnt-like activity and mimics the Nieuwkoop signaling center in Xenopus dorsal-ventral patterning. Dev Biol 172: 115–125 [DOI] [PubMed] [Google Scholar]
- Hadjihannas MV, Brückner M, Behrens J (2010) Conductin/axin2 and Wnt signalling regulates centrosome cohesion. EMBO Rep 11: 317–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haegel H, Larue L, Ohsugi M, Fedorov L, Herrenknecht K, Kemler R (1995) Lack of beta-catenin affects mouse development at gastrulation. Development 121: 3529–3537 [DOI] [PubMed] [Google Scholar]
- Hamada F, Bienz M (2004) The APC tumor suppressor binds to C-terminal binding protein to divert nuclear beta-catenin from TCF. Dev Cell 7: 677–685 [DOI] [PubMed] [Google Scholar]
- Hart M, Concordet JP, Lassot I, Albert I, del los Santos R, Durand H, Perret C, Rubinfeld B, Margottin F, Benarous R, Polakis P (1999) The F-box protein beta-TrCP associates with phosphorylated beta-catenin and regulates its activityin the cell. Curr Biol 9: 207–210 [DOI] [PubMed] [Google Scholar]
- Heasman J, Crawford A, Goldstone K, Garner-Hamrick P, Gumbiner B, McCrea P, Kintner C, Noro CY, Wylie C (1994) Overexpression of cadherins and underexpression of beta-catenin inhibit dorsal mesoderm induction in early Xenopus embryos. Cell 79: 791–803 [DOI] [PubMed] [Google Scholar]
- Henderson BR, Fagotto F (2002) The ins and outs of APC and beta-catenin nuclear transport. EMBO Rep 3: 834–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuberger J, Birchmeier W (2010) Interplay of cadherin-mediated cell adhesion and canonical Wnt signaling. Cold Spring Harb Perspect Biol 2: a002915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikasa H, Sokol SY (2011) Phosphorylation of TCF proteins by homeodomain-interacting protein kinase 2. J Biol Chem 286: 12093–12100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinck L, Näthke IS, Papkoff J, Nelson WJ (1994) Dynamics of cadherin/catenin complex formation: novel protein interactions and pathways of complex assembly. J Cell Biol 125: 1327–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmans R, Basler K (2004) Identification and in vivo role of the Armadillo-Legless interaction. Development 131: 4393–4400 [DOI] [PubMed] [Google Scholar]
- Hoffmans R, Basler K (2007) BCL9-2 binds Arm/beta-catenin in a Tyr142-independent manner and requires Pygopus for its function in Wg/Wnt signaling. Mech Dev 124: 59–67 [DOI] [PubMed] [Google Scholar]
- Hoffmans R, Städeli R, Basler K (2005) Pygopus and legless provide essential transcriptional coactivator functions to armadillo/beta-catenin. Curr Biol 15: 1207–1211 [DOI] [PubMed] [Google Scholar]
- Holland LZ, Panfilio KA, Chastain R, Schubert M, Holland ND (2005) Nuclear beta-catenin promotes non-neural ectoderm and posterior cell fates in amphioxus embryos. Dev Dyn 233: 1430–1443 [DOI] [PubMed] [Google Scholar]
- Howe LR, Watanabe O, Leonard J, Brown AM (2003) Twist is up-regulated in response to Wnt1 and inhibits mouse mammary cell differentiation. Cancer Res 63: 1906–1913 [PubMed] [Google Scholar]
- Huang P, Senga T, Hamaguchi M (2007) A novel role of phospho-beta-catenin in microtubule regrowth at centrosome. Oncogene 26: 4357–4371 [DOI] [PubMed] [Google Scholar]
- Huber AH, Nelson WJ, Weis WI (1997) Three-dimensional structure of the armadillo repeat region of beta-catenin. Cell 90: 871–882 [DOI] [PubMed] [Google Scholar]
- Huber AH, Weis WI (2001) The structure of the beta-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by beta-catenin. Cell 105: 391–402 [DOI] [PubMed] [Google Scholar]
- Huber O, Korn R, McLaughlin J, Ohsugi M, Herrmann BG, Kemler R (1996) Nuclear localization of beta-catenin by interaction with transcription factor LEF-1. Mech Dev 59: 3–10 [DOI] [PubMed] [Google Scholar]
- Huelsken J, Vogel R, Brinkmann V, Erdmann B, Birchmeier C, Birchmeier W (2000) Requirement for beta-catenin in anterior-posterior axis formation in mice. J Cell Biol 148: 567–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur EM, Zhou FQ (2010) GSK3 signalling in neural development. Nat Rev Neurosci 11: 539–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihara M, Yamamoto H, Kikuchi A (2005) SUMO-1 modification of PIASy, an E3 ligase, is necessary for PIASy-dependent activation of TCF-4. Mol Cell Biol 25: 3506–3518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaehning JA (2010) The Paf1 complex: platform or player in RNA polymerase II transcription? Biochim Biophys Acta 1799: 379–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamora C, DasGupta R, Kocieniewski P, Fuchs E (2003) Links between signal transduction, transcription and adhesion in epithelial bud development. Nature 422: 317–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin T, George Fantus I, Sun J (2008) Wnt and beyond Wnt: multiple mechanisms control the transcriptional property of beta-catenin. Cell Signal 20: 1697–1704 [DOI] [PubMed] [Google Scholar]
- Kaidi A, Williams AC, Paraskeva C (2007) Interaction between beta-catenin and HIF-1 promotes cellular adaptation to hypoxia. Nat Cell Biol 9: 210–217 [DOI] [PubMed] [Google Scholar]
- Kelly KF, Ng DY, Jayakumaran G, Wood GA, Koide H, Doble BW (2011) β-catenin enhances Oct-4 activity and reinforces pluripotency through a TCF-independent mechanism. Cell Stem Cell 8: 214–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler R, Hausmann G, Basler K (2009) The PHD domain is required to link Drosophila Pygopus to Legless/beta-catenin and not to histone H3. Mech Dev 126: 752–759 [DOI] [PubMed] [Google Scholar]
- Kim EA, Kim JE, Sung KS, Choi DW, Lee BJ, Choi CY (2010) Homeodomain-interacting protein kinase 2 (HIPK2) targets beta-catenin for phosphorylation and proteasomal degradation. Biochem Biophys Res Commun 394: 966–971 [DOI] [PubMed] [Google Scholar]
- Kim JH, Kim B, Cai L, Choi HJ, Ohgi KA, Tran C, Chen C, Chung CH, Huber O, Rose DW, Sawyers CL, Rosenfeld MG, Baek SH (2005) Transcriptional regulation of a metastasis suppressor gene by Tip60 and beta-catenin complexes. Nature 434: 921–926 [DOI] [PubMed] [Google Scholar]
- Kim S, Xu X, Hecht A, Boyer TG (2006) Mediator is a transducer of Wnt/beta-catenin signaling. J Biol Chem 281: 14066–14075 [DOI] [PubMed] [Google Scholar]
- Kimelman D, Xu W (2006) Beta-catenin destruction complex: insights and questions from a structural perspective. Oncogene 25: 7482–7491 [DOI] [PubMed] [Google Scholar]
- Korinek V, Barker N, Moerer P, van Donselaar E, Huls G, Peters PJ, Clevers H (1998) Depletion of epithelial stem-cell compartments in the small intestine of mice lacking TCF-4. Nat Genet 19: 379–383 [DOI] [PubMed] [Google Scholar]
- Kramps T, Peter O, Brunner E, Nellen D, Froesch B, Chatterjee S, Murone M, Züllig S, Basler K (2002) Wnt/wingless signaling requires BCL9/legless-mediated recruitment of pygopus to the nuclear beta-catenin-TCF complex. Cell 109: 47–60 [DOI] [PubMed] [Google Scholar]
- Kusserow A, Pang K, Sturm C, Hrouda M, Lentfer J, Schmidt HA, Technau U, von Haeseler A, Hobmayer B, Martindale MQ, Holstein TW (2005) Unexpected complexity of the Wnt gene family in a sea anemone. Nature 433: 156–160 [DOI] [PubMed] [Google Scholar]
- Lapébie P, Gazave E, Ereskovsky A, Derelle R, Bézac C, Renard E, Houliston E, Borchiellini C (2009) WNT/beta-catenin signalling and epithelial patterning in the homoscleromorph sponge Oscarella. PLoS One 4: e5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, Swarup S, Chen J, Ishitani T, Verheyen EM (2009) Homeodomain-interacting protein kinases (Hipks) promote Wnt/Wg signaling through stabilization of beta-catenin/Arm and stimulation of target gene expression. Development 136: 241–251 [DOI] [PubMed] [Google Scholar]
- Li B, Rhéaume C, Teng A, Bilanchone V, Munguia JE, Hu M, Jessen S, Piccolo S, Waterman ML, Dai X (2007) Developmental phenotypes and reduced Wnt signaling in mice deficient for pygopus 2. Genesis 45: 318–325 [DOI] [PubMed] [Google Scholar]
- Li FQ, Mofunanya A, Harris K, Takemaru K (2008) Chibby cooperates with 14-3-3 to regulate beta-catenin subcellular distribution and signaling activity. J Cell Biol 181: 1141–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Sutter C, Parker DS, Blauwkamp T, Fang M, Cadigan KM (2007b) CBP/p300 are bimodal regulators of Wnt signaling. EMBO J 26: 2284–2294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Nie F, Wang S, Li L (2011) Histone H4 Lys 20 monomethylation by histone methylase SET8 mediates Wnt target gene activation. Proc Natl Acad Sci USA 108: 3116–3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Kato Y, Zhang Z, Do VM, Yankner BA, He X (1999) Beta-Trcp couples beta-catenin phosphorylation-degradation and regulates Xenopus axis formation. Proc Natl Acad Sci USA 96: 6273–6278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Li Y, Semenov M, Han C, Baeg GH, Tan Y, Zhang Z, Lin X, He X (2002) Control of beta-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell 108: 837–847 [DOI] [PubMed] [Google Scholar]
- Liu H, Fergusson MM, Wu JJ, Rovira II, Liu J, Gavrilova O, Lu T, Bao J, Han D, Sack MN, Finkel T (2011) Wnt signaling regulates hepatic metabolism. Sci Signal 4: ra6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Phillips BT, Amaya MF, Kimble J, Xu W (2008) The C. elegans SYS-1 protein is a bona fide beta-catenin. Dev Cell 14: 751–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YI, Chang MV, Li HE, Barolo S, Chang JL, Blauwkamp TA, Cadigan KM (2008b) The chromatin remodelers ISWI and ACF1 directly repress Wingless transcriptional targets. Dev Biol 323: 41–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love JJ, Li X, Case DA, Giese K, Grosschedl R, Wright PE (1995) Structural basis for DNA bending by the architectural transcription factor LEF-1. Nature 376: 791–795 [DOI] [PubMed] [Google Scholar]
- Lucero OM, Dawson DW, Moon RT, Chien AJ (2010) A re-evaluation of the oncogenic nature of Wnt/beta-catenin signaling inmelanoma and other cancers. Curr Oncol Rep 12: 314–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyashenko N, Winter M, Migliorini D, Biechele T, Moon RT, Hartmann C (2011) Differential requirement for the dual functions of β-catenin in embryonic stem cell self-renewal and germ layer formation. Nat Cell Biol 13: 753–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lévy L, Wei Y, Labalette C, Wu Y, Renard CA, Buendia MA, Neuveut C (2004) Acetylation of beta-catenin by p300 regulates beta-catenin-TCF4 interaction. Mol Cell Biol 24: 3404–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoudi T, Li VS, Ng SS, Taouatas N, Vries RG, Mohammed S, Heck AJ, Clevers H (2009) The kinase TNIK is an essential activator of Wnt target genes. EMBO J 28: 3329–3340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani M, Carrasco DE, Zhang Y, Takada K, Gatt ME, Dutta-Simmons J, Ikeda H, Diaz-Griffero F, Pena-Cruz V, Bertagnolli M, Myeroff LL, Markowitz SD, Anderson KC, Carrasco DR (2009) BCL9 promotes tumor progression by conferring enhanced proliferative, metastatic, and angiogenic properties to cancer cells. Cancer Res 69: 7577–7586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolagas SC, Almeida M (2007) Gone with the Wnts: beta-catenin, T-cell factor, forkhead box O, and oxidative stress in age-dependent diseases of bone, lipid, and glucose metabolism. Mol Endocrinol 21: 2605–2614 [DOI] [PubMed] [Google Scholar]
- Maretto S, Cordenonsi M, Dupont S, Braghetta P, Broccoli V, Hassan AB, Volpin D, Bressan GM, Piccolo S (2003) Mapping Wnt/beta-catenin signaling during mouse development and in colorectal tumors. Proc Natl Acad Sci USA 100: 3299–3304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maretzky T, Reiss K, Ludwig A, Buchholz J, Scholz F, Proksch E, de Strooper B, Hartmann D, Saftig P (2005) ADAM10 mediates E-cadherin shedding and regulates epithelial cell-cell adhesion, migration, and beta-catenin translocation. Proc Natl Acad Sci USA 102: 9182–9187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzawa SI, Reed JC (2001) Siah-1, SIP, and Ebi collaborate in a novel pathway for beta-catenin degradation linked to p53 responses. Mol Cell 7: 915–926 [DOI] [PubMed] [Google Scholar]
- Mazumdar J, O'Brien WT, Johnson RS, LaManna JC, Chavez JC, Klein PS, Simon MC (2010) O2 regulates stem cells through Wnt/β-catenin signalling. Nat Cell Biol 12: 1007–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCrea PD, Turck CW, Gumbiner B (1990) A homolog of the armadillo protein in Drosophila (plakoglobin) associated with E-cadherin. Science 254: 1359–1361 [DOI] [PubMed] [Google Scholar]
- Meng W, Takeichi M (2009) Adherens junction: molecular architecture and regulation. Cold Spring Harb Perspect Biol 1: a002899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalfe C, Bienz M (2011) Inhibition of GSK3 by Wnt signalling--two contrasting models. J Cell Sci 124(Part 21): 3537–3544 [DOI] [PubMed] [Google Scholar]
- Mieszczanek J, de la Roche M, Bienz M (2008) A role of Pygopus as an anti-repressor in facilitating Wnt-dependent transcription. Proc Natl Acad Sci USA 105: 19324–19329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkovic I, Gault WJ, Rahnama M, Jenny A, Gaengel K, Bessette D, Gottardi CJ, Verheyen EM, Mlodzik M (2011) Nemo kinase phosphorylates β-catenin to promote ommatidial rotation and connects core PCP factors to E-cadherin-β-catenin. Nat Struct Mol Biol 18: 665–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyabayashi T, Teo JL, Yamamoto M, McMillan M, Nguyen C, Kahn M (2007) Wnt/beta-catenin/CBP signaling maintains long-term murine embryonic stem cell pluripotency. Proc Natl Acad Sci USA 104: 5668–5673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molenaar M, van de Wetering M, Oosterwegel M, Peterson-Maduro J, Godsave S, Korinek V, Roose J, Destrée O, Clevers H (1996) XTCF-3 transcription factor mediates beta-catenin-induced axis formation in Xenopus embryos. Cell 86: 391–399 [DOI] [PubMed] [Google Scholar]
- Moore AC, Amann JM, Williams CS, Tahinci E, Farmer TE, Martinez JA, Yang G, Luce KS, Lee E, Hiebert SW (2008) Myeloid translocation gene family members associate with T-cell factors (TCFs) and influence TCF-dependent transcription. Mol Cell Biol 28: 977–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosimann C, Hausmann G, Basler K (2006) Parafibromin/Hyrax activates Wnt/Wg target gene transcription by direct association with beta-catenin/Armadillo. Cell 125: 327–341 [DOI] [PubMed] [Google Scholar]
- Mosimann C, Hausmann G, Basler K (2009) Beta-catenin hits chromatin: regulation of Wnt target gene activation. Nat Rev Mol Cell Biol 10: 276–286 [DOI] [PubMed] [Google Scholar]
- Mulholland DJ, Read JT, Rennie PS, Cox ME, Nelson CC (2003) Functional localization and competition between the androgen receptor and T-cell factor for nuclear beta-catenin: a means for inhibition of the TCF signaling axis. Oncogene 22: 5602–5613 [DOI] [PubMed] [Google Scholar]
- Muñoz JP, Huichalaf CH, Orellana D, Maccioni RB (2007) Cdk5 modulates beta- and delta-catenin/Pin1 interactions in neuronal cells. J Cell Biochem 100: 738–749 [DOI] [PubMed] [Google Scholar]
- Müller W, Frank U, Teo R, Mokady O, Guette C, Plickert G (2007) Wnt signaling in hydroid development: ectopic heads and giant buds induced by GSK-3beta inhibitors. Int J Dev Biol 51: 211–220 [DOI] [PubMed] [Google Scholar]
- Najdi R, Holcombe RF, Waterman ML (2011) Wnt signaling and colon carcinogenesis: beyond APC. J Carcinog 10: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano N, Itoh S, Watanabe Y, Maeyama K, Itoh F, Kato M (2010) Requirement of TCF7L2 for TGF-beta-dependent transcriptional activation of the TMEPAI gene. J Biol Chem 285: 38023–38033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld KL, Zhang F, Cullen BR, White RL (2000) APC-mediated downregulation of beta-catenin activity involves nuclear sequestration and nuclear export. EMBO Rep 1: 519–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SS, Mahmoudi T, Danenberg E, Bejaoui I, de Lau W, Korswagen HC, Schutte M, Clevers H (2009) Phosphatidylinositol 3-kinase signaling does not activate the wnt cascade. J Biol Chem 284: 35308–35313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehrs C (2010) On growth and form: a Cartesian coordinate system of Wnt and BMP signaling specifies bilaterian body axes. Development 137: 845–857 [DOI] [PubMed] [Google Scholar]
- Orsulic S, Peifer M (1996) An in vivo structure-function study of armadillo, the beta-catenin homologue, reveals both separate and overlapping regions of the protein required for cell adhesion and for wingless signaling. J Cell Biol 134: 1283–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa M, Baribault H, Kemler R (1989) The cytoplasmic domain of the cell adhesion molecule uvomorulin associates with three independent proteins structurally related in different species. EMBO J 8: 1711–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker DS, Ni YY, Chang JL, Li J, Cadigan KM (2008) Wingless signaling induces widespread chromatin remodeling of target loci. Mol Cell Biol 28: 1815–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlowski JE, Ertel JR, Allen MP, Xu M, Butler C, Wilson EM, Wierman ME (2002) Liganded androgen receptor interaction with beta-catenin: nuclear co-localization and modulation of transcriptional activity in neuronal cells. J Biol Chem 277: 20702–20710 [DOI] [PubMed] [Google Scholar]
- Peifer M, Wieschaus E (1990) The segment polarity gene armadillo encodes a functionally modular protein that is the Drosophila homolog of human plakoglobin. Cell 63: 1167–1176 [DOI] [PubMed] [Google Scholar]
- Petersen CP, Reddien PW (2009) Wnt signaling and the polarity of the primary body axis. Cell 139: 1056–1068 [DOI] [PubMed] [Google Scholar]
- Piao S, Lee SH, Kim H, Yum S, Stamos JL, Xu Y, Lee SJ, Lee J, Oh S, Han JK, Park BJ, Weis WI, Ha NC (2008) Direct inhibition of GSK3beta by the phosphorylated cytoplasmic domain of LRP6 in Wnt/beta-catenin signaling. PLoS One 3: e4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piedra J, Martinez D, Castano J, Miravet S, Dunach M, de Herreros AG (2001) Regulation of beta-catenin structure and activity by tyrosine phosphorylation. J Biol Chem 276: 20436–20443 [DOI] [PubMed] [Google Scholar]
- Piedra J, Miravet S, Castaño J, Pálmer HG, Heisterkamp N, García de Herreros A, Duñach M (2003) p120 Catenin-associated Fer and Fyn tyrosine kinases regulate beta-catenin Tyr-142 phosphorylation and beta-catenin-alpha-catenin Interaction. Mol Cell Biol 23: 2287–2297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piepenburg O, Vorbrüggen G, Jäckle H (2000) Drosophila segment borders result from unilateral repression of hedgehog activity by wingless signaling. Mol Cell 6: 203–209 [PubMed] [Google Scholar]
- Pokutta S, Weis WI (2000) Structure of the dimerization and beta-catenin-binding region of alpha-catenin. Mol Cell 5: 533–543 [DOI] [PubMed] [Google Scholar]
- Polakis P (2007) The many ways of Wnt in cancer. Curr Opin Genet Dev 17: 45–51 [DOI] [PubMed] [Google Scholar]
- Poy F, Lepourcelet M, Shivdasani RA, Eck MJ (2001) Structure of a human TCF4-beta-catenin complex. Nat Struct Biol 8: 1053–1057 [DOI] [PubMed] [Google Scholar]
- Qi J, Wang J, Romanyuk O, Siu CH (2006) Involvement of Src family kinases in N-cadherin phosphorylation and beta-catenin dissociation during transendothelial migration of melanoma cells. Mol Biol Cell 17: 1261–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quyn AJ, Appleton PL, Carey FA, Steele RJ, Barker N, Clevers H, Ridgway RA, Sansom OJ, Näthke IS (2010) Spindle orientation bias in gut epithelial stem cell compartments is lost in precancerous tissue. Cell Stem Cell 6: 175–181 [DOI] [PubMed] [Google Scholar]
- Reiss K, Maretzky T, Ludwig A, Tousseyn T, de Strooper B, Hartmann D, Saftig P (2005) ADAM10 cleavage of N-cadherin and regulation of cell-cell adhesion and beta-catenin nuclear signalling. EMBO J 24: 742–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee J, Buchan T, Zukerberg L, Lilien J, Balsamo J (2007) Cables links Robo-bound Abl kinase to N-cadherin-bound beta-catenin to mediate Slit-induced modulation of adhesion and transcription. Nat Cell Biol 9: 883–892 [DOI] [PubMed] [Google Scholar]
- Riggleman B, Schedl P, Wieschaus E (1990) Spatial expression of the Drosophila segment polarity gene armadillo is posttranscriptionally regulated by wingless. Cell 63: 549–560 [DOI] [PubMed] [Google Scholar]
- Roberts DM, Pronobis MI, Poulton JS, Waldmann JD, Stephenson EM, Hanna S, Peifer M (2011) Deconstructing the ßcatenin destruction complex: mechanistic roles for the tumor suppressor APC in regulating Wnt signaling. Mol Biol Cell 22: 1845–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roura S, Miravet S, Piedra J, García de Herreros A, Duñach M (1999) Regulation of E-cadherin/Catenin association by tyrosine phosphorylation. J Biol Chem 274: 36734–36740 [DOI] [PubMed] [Google Scholar]
- Sachdev S, Bruhn L, Sieber H, Pichler A, Melchior F, Grosschedl R (2001) PIASy, a nuclear matrix-associated SUMO E3 ligase, represses LEF1 activity by sequestration into nuclear bodies. Genes Dev 15: 3088–3103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampietro J, Dahlberg CL, Cho US, Hinds TR, Kimelman D, Xu W (2006) Crystal structure of a beta-catenin/BCL9/TCF4 complex. Mol Cell 24: 293–300 [DOI] [PubMed] [Google Scholar]
- Sánchez-Tilló E, de Barrios O, Siles L, Cuatrecasas M, Castells A, Postigo A (2011) β-catenin/TCF4 complex induces the epithelial-to-mesenchymal transition (EMT)-activator ZEB1 toregulate tumor invasiveness. Proc Natl Acad Sci USA 108: 19204–19209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancho R, Nateri AS, de Vinuesa AG, Aguilera C, Nye E, Spencer-Dene B, Behrens A (2009) JNK signalling modulates intestinal homeostasis and tumourigenesis in mice. EMBO J 28: 1843–1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanson B (2001) Generating patterns from fields of cells. Examples from Drosophila segmentation. EMBO Rep 2: 1083–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayat R, Leber B, Grubac V, Wiltshire L, Persad S (2008) O-GlcNAc-glycosylation of beta-catenin regulates its nuclear localization and transcriptional activity. Exp Cell Res 314: 2774–2787 [DOI] [PubMed] [Google Scholar]
- Schwab KR, Patterson LT, Hartman HA, Song N, Lang RA, Lin X, Potter SS (2007) Pygo1 and Pygo2 roles in Wnt signaling in mammalian kidney development. BMC Biol 5: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz-Romond T, Metcalfe C, Bienz M (2007) Dynamic recruitment of axin by Dishevelled protein assemblies. J Cell Sci 120(Pt 14): 2402–2412 [DOI] [PubMed] [Google Scholar]
- Schweizer L, Nellen D, Basler K (2003) Requirement for Pangolin/dTCF in Drosophila Wingless signaling. Proc Natl Acad Sci USA 100: 5846–5851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya T, Zaret KS (2007) Repression by Groucho/TLE/Grg proteins: genomic site recruitment generatescompacted chromatin in vitro and impairs activator binding in vivo. Mol Cell 28: 291–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma M, Jamieson C, Johnson M, Molloy MP, Henderson BR (2012) Specific armadillo repeat sequences facilitate β-catenin nuclear transport in live cells via direct binding to nucleoporins Nup62, Nup153, and RanBP2/Nup358. J Biol Chem 287: 819–831 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Shitashige M, Satow R, Honda K, Ono M, Hirohashi S, Yamada T (2008) Regulation of Wnt signaling by the nuclear pore complex. Gastroenterology 134: 1961–1971 [DOI] [PubMed] [Google Scholar]
- Shoval I, Ludwig A, Kalcheim C (2007) Antagonistic roles of full-length N-cadherin and its soluble BMP cleavage product in neural crest delamination. Development 134: 491–501 [DOI] [PubMed] [Google Scholar]
- Siegfried E, Wilder EL, Perrimon N (1994) Components of wingless signalling in Drosophila. Nature 367: 76–80 [DOI] [PubMed] [Google Scholar]
- Sierra J, Yoshida T, Joazeiro CA, Jones KA (2006) The APC tumor suppressor counteracts beta-catenin activation and H3K4 methylation at Wnt target genes. Genes Dev 20: 586–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinner D, Rankin S, Lee M, Zorn AM (2004) Sox17 and beta-catenin cooperate to regulate the transcription of endodermal genes. Development 131: 3069–3080 [DOI] [PubMed] [Google Scholar]
- Solanas G, Miravet S, Casagolda D, Castaño J, Raurell I, Corrionero A, de Herreros AG, Duñach M (2004) Beta-Catenin and plakoglobin N- and C-tails determine ligand specificity. J Biol Chem 279: 49849–49856 [DOI] [PubMed] [Google Scholar]
- Song DH, Dominguez I, Mizuno J, Kaut M, Mohr SC, Seldin DC (2003) CK2 phosphorylation of the armadillo repeat region of beta-catenin potentiates Wnt signaling. J Biol Chem 278: 24018–24025 [DOI] [PubMed] [Google Scholar]
- Song H, Goetze S, Bischof J, Spichiger-Haeusermann C, Kuster M, Brunner E, Basler K (2010) Coop functions as a corepressor of Pangolin and antagonizes Wingless signaling. Genes Dev 24: 881–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Spichiger-Haeusermann C, Basler K (2009) The ISWI-containing NURF complex regulates the output of the canonical Wingless pathway. EMBO Rep 10: 1140–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song LN, Gelmann EP (2008) Silencing mediator for retinoid and thyroid hormone receptor and nuclear receptorcorepressor attenuate transcriptional activation by the beta-catenin-TCF4 complex. J Biol Chem 283: 25988–25999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song N, Schwab KR, Patterson LT, Yamaguchi T, Lin X, Potter SS, Lang RA (2007) Pygopus 2 has a crucial, Wnt pathway-independent function in lens induction. Development 134: 1873–1885 [DOI] [PubMed] [Google Scholar]
- Stemmer V, de Craene B, Berx G, Behrens J (2008) Snail promotes Wnt target gene expression and interacts with beta-catenin. Oncogene 27: 5075–5080 [DOI] [PubMed] [Google Scholar]
- Stepniak E, Radice GL, Vasioukhin V (2009) Adhesive and signaling functions of cadherins and catenins in vertebrate development. Cold Spring Harb Perspect Biol 1: a002949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Städeli R, Hoffmans R, Basler K (2006) Transcription under the control of nuclear Arm/beta-catenin. Curr Biol 16: R378–R385 [DOI] [PubMed] [Google Scholar]
- Su Y, Fu C, Ishikawa S, Stella A, Kojima M, Shitoh K, Schreiber EM, Day BW, Liu B (2008) APC is essential for targeting phosphorylated beta-catenin to the SCFbeta-TrCP ubiquitin ligase. Mol Cell 32: 652–661 [DOI] [PubMed] [Google Scholar]
- Sustmann C, Flach H, Ebert H, Eastman Q, Grosschedl R (2008) Cell-type-specific function of BCL9 involves a transcriptional activation domain thatsynergizes with beta-catenin. Mol Cell Biol 28: 3526–3537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taelman VF, Dobrowolski R, Plouhinec JL, Fuentealba LC, Vorwald PP, Gumper I, Sabatini DD, De Robertis EM (2010) Wnt signaling requires sequestration of glycogen synthase kinase 3 inside multivesicular endosomes. Cell 143: 1136–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tago K, Nakamura T, Nishita M, Hyodo J, Nagai S, Murata Y, Adachi S, Ohwada S, Morishita Y, Shibuya H, Akiyama T (2000) Inhibition of Wnt signaling by ICAT, a novel beta-catenin-interacting protein. Genes Dev 14: 1741–1749 [PMC free article] [PubMed] [Google Scholar]
- Takemaru K, Yamaguchi S, Lee YS, Zhang Y, Carthew RW, Moon RT (2003) Chibby, a nuclear beta-catenin-associated antagonist of the Wnt/Wingless pathway. Nature 422: 905–909 [DOI] [PubMed] [Google Scholar]
- Taurin S, Sandbo N, Qin Y, Browning D, Dulin NO (2006) Phosphorylation of beta-catenin by cyclic AMP-dependent protein kinase. J Biol Chem 281: 9971–9976 [DOI] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RY, Nieto MA (2009) Epithelial-mesenchymal transitions in development and disease. Cell 139: 871–890 [DOI] [PubMed] [Google Scholar]
- Thompson B, Townsley F, Rosin-Arbesfeld R, Musisi H, Bienz M (2002) A new nuclear component of the Wnt signalling pathway. Nat Cell Biol 4: 367–373 [DOI] [PubMed] [Google Scholar]
- Thornton TM, Pedraza-Alva G, Deng B, Wood CD, Aronshtam A, Clements JL, Sabio G, Davis RJ, Matthews DE, Doble B, Rincon M (2008) Phosphorylation by p38 MAPK as an alternative pathway for GSK3beta inactivation. Science 320: 667–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toualbi K, Güller MC, Mauriz JL, Labalette C, Buendia MA, Mauviel A, Bernuau D (2007) Physical and functional cooperation between AP-1 and beta-catenin for the regulation of TCF-dependent genes. Oncogene 26: 3492–3502 [DOI] [PubMed] [Google Scholar]
- Townsley FM, Thompson B, Bienz M (2004) Pygopus residues required for its binding to Legless are critical for transcription and development. J Biol Chem 279: 5177–5183 [DOI] [PubMed] [Google Scholar]
- Trompouki E, Bowman TV, Lawton LN, Fan ZP, Wu DC, DiBiase A, Martin CS, Cech JN, Sessa AK, Leblanc JL, Li P, Durand EM, Mosimann C, Heffner GC, Daley GQ, Paulson RF, Young RA, Zon LI (2011) Lineage regulators direct BMP and Wnt pathways to cell-specific programs duringdifferentiation and regeneration. Cell 147: 577–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuupanen S, Turunen M, Lehtonen R, Hallikas O, Vanharanta S, Kivioja T, Björklund M, Wei G, Yan J, Niittymäki I, Mecklin JP, Järvinen H, Ristimäki A, Di-Bernardo M, East P, Carvajal-Carmona L, Houlston RS, Tomlinson I, Palin K, Ukkonen E et al. (2009) The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nat Genet 41: 885–890 [DOI] [PubMed] [Google Scholar]
- Valenta T, Gay M, Steiner S, Draganova K, Zemke M, Hoffmans R, Cinelli P, Aguet M, Sommer L, Basler K (2011) Probing transcription-specific outputs of β-catenin in vivo. Genes Dev 25: 2631–2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenta T, Lukas J, Korinek V (2003) HMG box transcription factor TCF-4's interaction with CtBP1 controls the expression of the Wnt target Axin2/Conductin in human embryonic kidney cells. Nucleic Acids Res 31: 2369–2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Amerongen R, Mikels A, Nusse R (2008) Alternative wnt signaling is initiated by distinct receptors. Sci Signal 1: re9. [DOI] [PubMed] [Google Scholar]
- van Beest M, Dooijes D, van De Wetering M, Kjaerulff S, Bonvin A, Nielsen O, Clevers H (2000) Sequence-specific high mobility group box factors recognize 10-12-base pair minor groove motifs. J Biol Chem 275: 27266–27273 [DOI] [PubMed] [Google Scholar]
- van de Wetering M, Cavallo R, Dooijes D, van Beest M, van Es J, Loureiro J, Ypma A, Hursh D, Jones T, Bejsovec A, Peifer M, Mortin M, Clevers H (1997) Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell 88: 789–799 [DOI] [PubMed] [Google Scholar]
- van Noort M, van de Wetering M, Clevers H (2002) Identification of two novel regulated serines in the N terminus of beta-catenin. Exp Cell Res 276: 264–272 [DOI] [PubMed] [Google Scholar]
- van Veelen W, Le NH, Helvensteijn W, Blonden L, Theeuwes M, Bakker ER, Franken PF, van Gurp L, Meijlink F, van der Valk MA, Kuipers EJ, Fodde R, Smits R (2011) β-catenin tyrosine 654 phosphorylation increases Wnt signalling and intestinal tumorigenesis. Gut 60: 1204–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlad A, Röhrs S, Klein-Hitpass L, Müller O (2008) The first five years of the Wnt targetome. Cell Signal 20: 795–802 [DOI] [PubMed] [Google Scholar]
- Wagner RT, Xu X, Yi F, Merrill BJ, Cooney AJ (2010) Canonical Wnt/β-catenin regulation of liver receptor homolog-1 mediates pluripotency gene expression. Stem Cells 28: 1794–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Jones KA (2006) CK2 controls the recruitment of Wnt regulators to target genes in vivo. Curr Biol 16: 2239–2244 [DOI] [PubMed] [Google Scholar]
- Weiske J, Albring KF, Huber O (2007) The tumor suppressor Fhit acts as a repressor of beta-catenin transcriptional activity. Proc Natl Acad Sci USA 104: 20344–20349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieschaus E, Nüsslein-Volhard C, Jürgens G (1984) Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster III. Zygotic loci on the X-chromosome and fourth chromosome. Rouxs Arch Dev Biol 193: 296–387 [DOI] [PubMed] [Google Scholar]
- Wieschaus E, Riggleman R (1987) Autonomous requirements for the segment polarity gene armadillo during Drosophila embryogenesis. Cell 49: 177–184 [DOI] [PubMed] [Google Scholar]
- Winston JT, Strack P, Beer-Romero P, Chu CY, Elledge SJ, Harper JW (1999) The SCFbeta-TRCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IkappaBalpha and beta-catenin and stimulates IkappaBalpha ubiquitination in vitro. Genes Dev 13: 270–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf D, Rodova M, Miska EA, Calvet JP, Kouzarides T (2002) Acetylation of beta-catenin by CREB-binding protein (CBP). J Biol Chem 277: 25562–25567 [DOI] [PubMed] [Google Scholar]
- Wray J, Kalkan T, Gomez-Lopez S, Eckardt D, Cook A, Kemler R, Smith A (2011) Inhibition of glycogen synthase kinase-3 alleviates TCF3 repression of the pluripotency network and increases embryonic stem cell resistance to differentiation. Nat Cell Biol 13: 838–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright JB, Brown SJ, Cole MD (2010) Upregulation of c-MYC in cis through a large chromatin loop linked to a cancer risk-associated single-nucleotide polymorphism in colorectal cancer cells. Mol Cell Biol 30: 1411–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Pan W (2010) GSK3: a multifaceted kinase in Wnt signaling. Trends Biochem Sci 35: 161–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, He X (2006) Threonine 41 in beta-catenin serves as a key phosphorylation relay residue in beta-catenin degradation. Biochemistry 45: 5319–5323 [DOI] [PubMed] [Google Scholar]
- Wu G, Huang H, Garcia Abreu J, He X (2009) Inhibition of GSK3 phosphorylation of beta-catenin via phosphorylated PPPSPXS motifs of Wnt coreceptor LRP6. PLoS One 4: e4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Xu G, Schulman BA, Jeffrey PD, Harper JW, Pavletich NP (2003) Structure of a beta-TrCP1-Skp1-beta-catenin complex: destruction motif binding and lysine specificity of the SCF(beta-TrCP1) ubiquitin ligase. Mol Cell 11: 1445–1456 [DOI] [PubMed] [Google Scholar]
- Wu X, Tu X, Joeng KS, Hilton MJ, Williams DA, Long F (2008) Rac1 activation controls nuclear localization of beta-catenin during canonical Wnt signaling. Cell 133: 340–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin N, Benchabane H, Tian A, Nguyen K, Klofas L, Ahmed Y (2011) Erect Wing facilitates context-dependent Wnt/Wingless signaling by recruiting the cell-specific Armadillo-TCF adaptor Earthbound to chromatin. Development 138: 4955–4967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Clements WK, Kimelman D, Xu W (2003) Crystal structure of a beta-catenin/axin complex suggests a mechanism for the beta-catenin destruction complex. Genes Dev 17: 2753–2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Takemaru K, Liu J, Berndt JD, Zheng JJ, Moon RT, Xu W (2008) Crystal structure of a full-length beta-catenin. Structure 16: 478–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Craig AW, Greer P, Miller M, Anastasiadis PZ, Lilien J, Balsamo J (2004) Continuous association of cadherin with beta-catenin requires the non-receptor tyrosine-kinase Fer. J Cell Sci 117(Part 15): 3207–3219 [DOI] [PubMed] [Google Scholar]
- Yamada S, Pokutta S, Drees F, Weis WI, Nelson WJ (2005) Deconstructing the cadherin-catenin-actin complex. Cell 123: 889–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Ihara M, Matsuura Y, Kikuchi A (2003) Sumoylation is involved in beta-catenin-dependent activation of Tcf-4. EMBO J 22: 2047–2059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Sun L, Li Q, Han X, Lei L, Zhang H, Shang Y (2011a) SET8 promotes epithelial-mesenchymal transition and confers TWIST dual transcriptionalactivities. EMBO J 31: 110–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA (2004) Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 117: 927–939 [DOI] [PubMed] [Google Scholar]
- Yang L, Lin C, Liu ZR (2006) P68 RNA helicase mediates PDGF-induced epithelial mesenchymal transition by displacing Axin from beta-catenin. Cell 127: 139–155 [DOI] [PubMed] [Google Scholar]
- Yang W, Xia Y, Ji H, Zheng Y, Liang J, Huang W, Gao X, Aldape K, Lu Z (2011b) Nuclear PKM2 regulates β-catenin transactivation upon EGFR activation. Nature 480: 118–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Zhang X, Gang H, Li X, Li Z, Wang T, Han J, Luo T, Wen F, Wu X (2007) Up-regulation of gastric cancer cell invasion by Twist is accompanied by N-cadherin and fibronectin expression. Biochem Biophys Res Commun 358: 925–930 [DOI] [PubMed] [Google Scholar]
- Yi F, Pereira L, Hoffman JA, Shy BR, Yuen CM, Liu DR, Merrill BJ (2011) Opposing effects of TCF3 and TCF1 control Wnt stimulation of embryonic stem cell self-renewal. Nat Cell Biol 13: 762–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoya F, Imamoto N, Tachibana T, Yoneda Y (1999) Beta-catenin can be transported into the nucleus in a Ran-unassisted manner. Mol Biol Cell 10: 1119–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yook JI, Li XY, Ota I, Hu C, Kim HS, Kim NH, Cha SY, Ryu JK, Choi YJ, Kim J, Fearon ER, Weiss SJ (2006) A Wnt-Axin2-GSK3beta cascade regulates Snail1 activity in breast cancer cells. Nat Cell Biol 8: 1398–1406 [DOI] [PubMed] [Google Scholar]
- Zeisberg M, Neilson EG (2009) Biomarkers for epithelial-mesenchymal transitions. J Clin Invest 119: 1429–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng G, Apte U, Micsenyi A, Bell A, Monga SP (2006) Tyrosine residues 654 and 670 in beta-catenin are crucial in regulation of Met-beta-catenin interactions. Exp Cell Res 312: 3620–3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Huang H, Tamai K, Zhang X, Harada Y, Yokota C, Almeida K, Wang J, Doble B, Woodgett J, Wynshaw-Boris A, Hsieh JC, He X (2008) Initiation of Wnt signaling: control of Wnt coreceptor Lrp6 phosphorylation/activation via frizzled, dishevelled and axin functions. Development 135: 367–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Wei P, Gong A, Chiu WT, Lee HT, Colman H, Huang H, Xue J, Liu M, Wang Y, Sawaya R, Xie K, Yung WK, Medema RH, He X, Huang S (2011) FoxM1 promotes β-catenin nuclear localization and controls Wnt target-gene expression and glioma tumorigenesis. Cancer Cell 20: 427–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao ZM, Reynolds AB, Gaucher EA (2011) The evolutionary history of the catenin gene family during metazoan evolution. BMC Evol Biol 11: 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou BP, Deng J, Xia W, Xu J, Li YM, Gunduz M, Hung MC (2004) Dual regulation of Snail by GSK-3beta-mediated phosphorylation in control of epithelial-mesenchymal transition. Nat Cell Biol 6: 931–940 [DOI] [PubMed] [Google Scholar]
- Zhu G, Wang Y, Huang B, Liang J, Ding Y, Xu A, Wu W (2012) A Rac1/PAK1 cascade controls β-catenin activation in colon cancer cells. Oncogene 31: 1001–1012 [DOI] [PubMed] [Google Scholar]