SUMMARY
Purpose
We sought to determine the association of C/EBPα expression status with clinical, pathologic and molecular characteristics, as well as outcomes, in non-small-cell lung cancer (NSCLC). This is the first comprehensive study of this transcription factor in patients with NSCLC.
Patients and Methods
Our cohort originated from ECOG 3590 (randomized trial of postoperative adjuvant therapy with thoracic radiation or cisplatin and etoposide plus thoracic radiation in patients with completely resected stages II and IIIA NSCLC; and its laboratory correlate, ECOG 4592). 164 tumor samples contained sufficient material for immunohistochemical (IHC) analysis. C/EBPα tumor staining was compared to that of basal bronchial cells (3+). 0 or 1+ (weak) suggested lack of, while 2 or 3+ (strong) suggested C/EBPα expression.
Results
90 tumors (55%) had 0 or 1+ C/EBPα staining, and the remaining 74 (45%) 2 or 3+. Patients with squamous cell carcinomas had a higher percentage of weak C/EBPα IHC staining compared to other histologies (p=0.048) and there was a trend for loss of C/EBPα in poorly differentiated compared to well differentiated tumors (p=0.07). There was no association between C/EBPα IHC and mutations in p53 or K-ras. The median disease-free survival for patients with weak and strong C/EBPα IHC expression was 29.6 and 30.6 months, respectively (p=0.94). The median overall survival between the weak and strong groups was 43.5 and 38.5 months, respectively (p=0.83).
Conclusions
Loss of expression of C/EBPα is seen in over half of stage II and IIIA NSCLC, specifically in squamous cell carcinomas and poorly differentiated tumors. Since down-regulation of C/EBPα is a common event in NSCLC, further elucidation of the involvement of C/EBPα in the pathogenesis and progression of lung cancer may identify novel therapeutic targets.
Keywords: lung cancer; C/EBPα; transcription factor; immunohistochemistry; non-small cell lung cancer; squamous cell carcinoma, and survival
INTRODUCTION
Lung cancer continues to be the leading cause of death from cancer in the United States (1). Complete surgical resection, whenever feasible, is generally recognized as the most effective initial treatment for non–small-cell lung cancer (NSCLC). However less than one third of patients present with early-stage disease amenable to effective surgical intervention (1,2). Following surgery alone, the 5-year survival for patients with pathological stage IB, II and IIIA NSCLC is still disappointing at 60%, 40–50% and 25%, respectively (2). Adjuvant chemotherapy with platinum-based doublets has been shown to improve survival in well-conducted randomized trials (3–6). The overall 5-year survival improvement in these studies ranged from 5 to 15% (3,4,6). Despite the recognized association of clinical and pathologic characteristics (age, weight loss, performance status, tumor-node-metastasis staging system) with prognosis, these parameters alone do not predict which patients are prone to develop recurrent disease and do not identify patients who will benefit from adjuvant therapy (2,4,7). The search for novel prognostic and predictive markers is essential to improve patient selection in early-stage NSCLC for adjuvant treatment strategies, as well as for the discovery of new therapeutic targets.
During the last decade numerous oncogenes and tumor suppressor genes implicated in the pathogenesis of lung cancer have been identified (8). The prognostic significance of these newly described genes has been studied in small retrospective series with conflicting results (9–13). The few studies that employed well-established prospective databases of patients with resected NSCLC have frequently shown a lack of significant prognostic information. As an example, mutations in the K-ras oncogene (implicated in signal transduction pathways) and the p53 tumor suppressor gene (involved in cell-cycle regulation) were not associated with significant prognostic significance in the context of a clinical trial of adjuvant therapy for completely resected NSCLC (7). Recent studies of factors involved in metastatic potential, such as vascular endothelial growth factor (VEGF) (14), as well as recently identified transcription factors, such as early growth response gene 1 (EGR1) (15), and cytokines (16) have suggested new prognostic markers in early-stage NSCLC. However validation in well established prospective patient cohorts is still lacking.
The transcription factor CCAAT/enhancer binding protein alpha (C/EBPα) is a tissue-specific differentiation factor that plays a major role in the terminal differentiation of myeloid cells, hepatocytes, and adipocytes among others (17,18). The association of C/EBPα and cancer has been elucidated in acute myeloid leukemia (AML) (19), where C/EBPα expression is reduced in leukemias carrying the t(8;21) translocation (20) and C/EBPα mutations confer improved prognosis in a small subgroup of AML patients with normal cytogenetics (21,22). In epithelial tissues, C/EBPα is expressed in the breast, colon, prostate, and the respiratory epithelium (23).
In the lung, C/EBPα is highly expressed in type II pneumocytes as well as cells of the bronchial epithelium (24,25) and regulates the expression of several key genes during lung differentiation (26). Abnormal proliferation of type II pneumocytes is seen in the context of the lethal phenotype of C/EBPα −/− knockout mice (27). In the lung-specific conditional C/EBPα knockout mouse developed by our group, a block in type II cell differentiation and increase in respiratory epithelial proliferation is seen (28). We also demonstrated that C/EBPα is down-regulated in a large proportion of lung cancers, and induction of C/EBPα expression in lung cancer cell lines led to growth arrest, apoptosis and cellular differentiation (29). We identified this gene as a novel tumor suppressor gene in lung cancer. In the primary lung cancer samples analyzed by immunohistochemistry (IHC) specific to C/EBPα, almost half of the tumors had absent or low expression of this protein (29). Tada and colleagues also showed the frequent down-regulation of C/EBPα in NSCLC and identified methylation of the upstream promoter of this gene as the mechanism of silencing. Loss of C/EBPα is one of the most common genetic events in NSCLC and therefore supports C/EBPα as a critical tumor suppressor gene in lung cancer (30).
However, it is still unknown if the observed down-regulation of C/EBPα has clinical significance. In order to investigate the association of C/EBPα expression and clinical, pathologic and molecular characteristics, as well as the possible prognostic and predictive role of C/EBPα in lung cancer, we studied the immunohistochemical expression of this protein in the prospective cohort of patients from E3590 (a trial of postoperative adjuvant therapy in patients with resected stages II or IIIA NSCLC) (31).
MATERIAL AND METHODS
Patient Study
The samples examined originated from patients who participated in E3590, a prospective randomized intergroup trial of postoperative adjuvant therapy in patients with completely resected stages II and IIIA NSCLC (31). Patients were required to have complete resection of the primary tumor and mediastinal lymph node sampling or dissection. After stratification for nodal status, weight loss, histology, and lymph node dissection patients were randomized to receive either radiotherapy alone – arm A, or four cycles of cisplatin (60 mg/m2 intravenously on day 1) and etoposide (120 mg/m2 intravenously days 1 to 3) administered concurrently with thoracic radiotherapy (50.4 Gy in 28 daily fractions) – arm B (31). Patients participating in E3590 were also offered enrollment into the ancillary laboratory study, E4592, provided that at least one paraffin block of primary tumor or a portion of tumor tissue removed from the block was available from the patient (7). All patients gave informed consent for both the clinical and laboratory correlative study.
488 patients were entered onto E3590 between April 1991 and February 1997. 217 patients were registered onto the laboratory correlative study. However, 20 samples were never submitted (7). Of the 197 samples available, 164 contained sufficient material for immunohistochemical analysis of C/EBPα expression.
C/EBPα Immunohistochemistry (IHC)
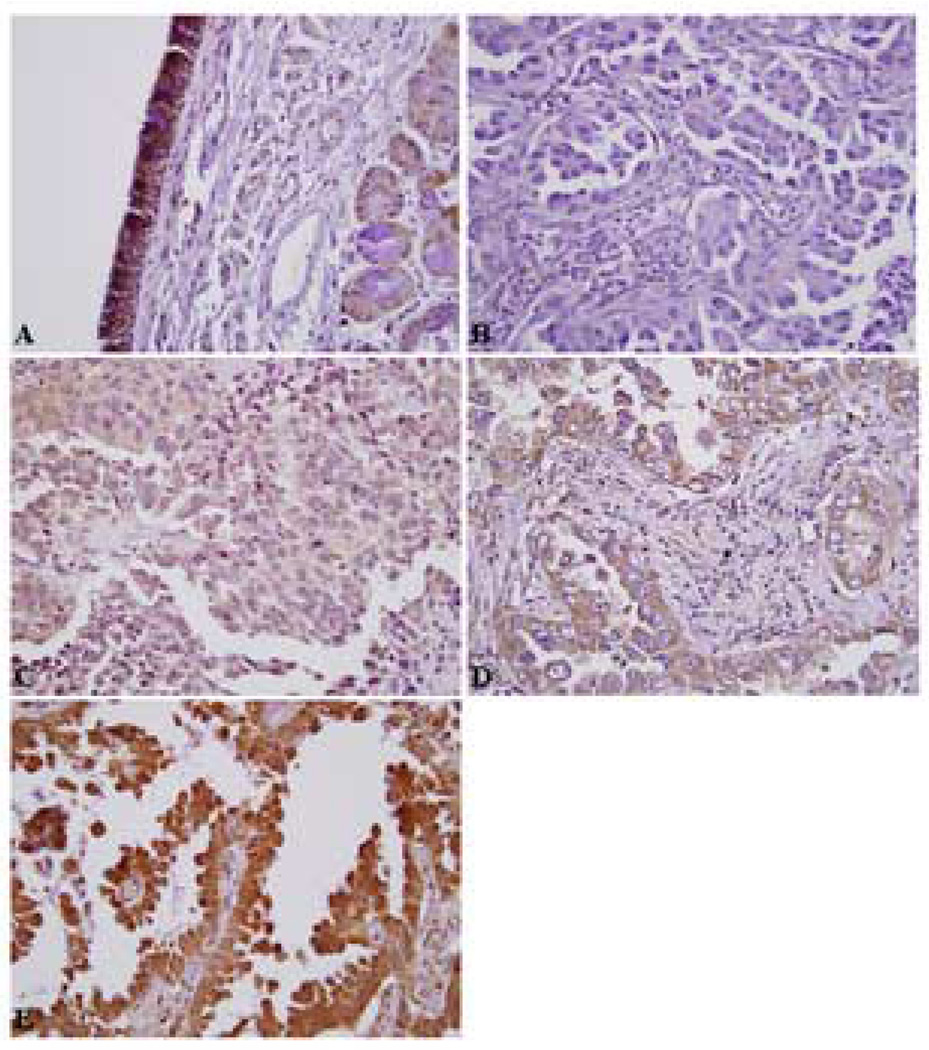
Immunohistochemical studies were performed on formalin-fixed, 5-micron thick paraffin-embedded tissue specimens using citrate-microwave antigen retrieval (29). In brief, a dilution (1:500) of a polyclonal rabbit anti-C/EBPα antibody (200 µg/0.1 ml; Santa Cruz Biotechnology, Santa Cruz, CA) was utilized, and the specificity of the stain confirmed by the concomitant use of a specific blocking peptide (1:100 dilution, 100 µg/0.5 ml; Santa Cruz Biotechnology). IHC was performed using Vectastain ABC kits (Vector Laboratories, Burlingame, CA). The positive staining was visualized by incubating slides with diaminobenzadine. Blinded scoring of specimens was performed by an experienced lung pathologist (OK) and an oncologist (BH), comparing the intensity of tumor staining to the staining of non-affected basal bronchial cells, which was deemed 3+. Essentially all slides contained some normal lung tissue and bronchial elements, thereby serving as an internal control. This strategy was used in our initial lung tissue studies (29). Individual samples had homogenous C/EBPα nuclear staining in tumor cells. Two separate groups were categorized for statistical analysis (29). The presence of 2+ (moderate intensity) or 3+ (comparable to the intensity of the bronchial epithelium) staining was suggestive of C/EBPα expression and designated as “strong”; the complete absence of staining (0) or faint levels of staining (1+) were indicative of lack of C/EBPα expression, and labeled “weak”. Examples of each IHC staining category are depicted in Figure 1.
Figure 1.
Immunohistochemical patterns for C/EBPα expression in E3590. Representative images: A) Normal bronchial mucosa with strong C/EBPa staining of bronchial epithelium (3+). B) Lack of C/EBPα staining (0) within tumor cells, C) Faint C/EBPα staining (1+), D) Moderate staining (2+), E) Strong C/EBPα staining within tumor cells (3+, which is comparable to that of the normal bronchial epithelium). (A, 100X; B, C, D and E, 400X).
Statistical Analysis
Fisher’s exact test was utilized to access the association between C/EBPα expression status with clinical, pathologic and molecular characteristics in E3590. A two-sample t-test was used to assess the difference between age and IHC score. All tests were two-sided. P-values ≤ 0.05 were considered statistically significant.
We also analyzed differences in disease-free survival and overall survival between the groups with weak (0 or 1+) or strong (2 or 3+) C/EBPα IHC staining. Overall survival was calculated as the time from randomization to death from any cause. Patients alive were censored at the date of last contact. Disease-free survival was defined as time from randomization to the earliest sign of disease recurrence or death from any cause. Patients alive without a recurrence reported were censored at the date of last contact. Distributions for overall survival time and disease-free survival were estimated with the Kaplan-Meier method (32) and compared with the log-rank test (33).
RESULTS
Patient cohort
Survival data for patients enrolled in E3590 and E4592 have been reported previously (7,31). Of the original 488 patients of E3590, median follow up is close to 10 years. Median survival was 38.3 months. For the subgroup of 217 patients enrolled in E4592, median survival was 40.3 months. There was no statistical difference in survival distributions between E3590 and E4592 (7).
Among the 164 patients analyzed in the current study, the median overall survival of 40.3 months and median disease-free survival of 30.6 months were not different from those of the complete study group and its ancillary laboratory study (7,31).
Clinical, pathologic and molecular characteristics in each C/EBPα IHC group
Of the assessable tumor samples, 90 (55%) had either 0 or 1+ (weak) staining for C/EBPα. The other 74 (45%) were scored as 2+ or 3+ (strong). The clinical, pathologic and molecular characteristics of the weak and strong C/EBPα IHC groups, as well as treatment arm of randomization are listed in Table 1. There was no statistical difference in age, sex (a trend of male sex and the weak C/EBPα IHC group did not reach statistical significance; p=0.16), TNM stage, and treatment arm between the two IHC groups. A history of tobacco use was not systematically collected in E3590 and, therefore, no data are available regarding smoking and C/EBPα IHC in this study.
Table 1.
Comparison of clinical, pathologic and molecular characteristics by C/EBPα immunohistochemistry (IHC) staining
| 0 or 1+ (weak) C/EBPα IHC staining (n=90) |
2 or 3+ (strong) C/EBPα IHC staining (n=74) |
p-value | |
|---|---|---|---|
| Age median (range) in years | 62 (35–81) | 61 (35–77) | 0.50 |
| Sex – no. (%) | |||
| Male | 55 (61) | 37 (50) | 0.16 |
| TNM stage – no. (%) | |||
| II | 37 (41) | 35 (47) | 0.43 |
| IIIA | 53 (59) | 39 (53) | |
| Histology – no. (%) | |||
| Squamous-cell | 37 (41) | 18 (24) | 0.048† |
| Adenocarcinoma | 44 (49) | 42 (57) | |
| Other | 9 (10) | 14 (19) | |
| Treatment arm – no. (%) | |||
| Arm A (radiation) | 47 (52) | 36 (49) | 0.75 |
| Arm B (chemoradiation) | 43 (48) | 38 (51) | |
| p53 mutation – no. (%) | |||
| Wildtype | 52 (58) | 33 (45) | 0.16 |
| Mutant | 38 (42) | 39 (53) | |
| Not done | 0 ( 0) | 2 ( 3) * | |
| K-ras mutation – no. (%) | |||
| Wildtype | 64 (71) | 48 (65) | 0.61 |
| Mutant | 26 (29) | 24 (32) | |
| Not done | 0 ( 0) | 2 ( 3) * |
Not done is excluded in the analysis
Reached statistical significance (≤ 0.05)
Patients with squamous cell histology tended to have a higher percentage of weak C/EBPα IHC staining (37/55, 67%, of the squamous cell carcinoma tumors), when compared to adenocarcinoma (44/86, 51%) and other histologies (p=0.048). Histological differentiation status was available in 88 tumors. Only 1/6 patients with well differentiated tumors had weak C/EBPα IHC, while 22/39 with poorly differentiated cancers were in the weak IHC group. Among the remaining 43 moderately differentiated tumors, 22 had strong C/EBPα IHC. Comparing the well and poorly differentiated cohort, a non-statistical strong trend towards loss of C/EBPα and less differentiated tumor samples was noted (p=0.07).
There was no statistical correlation between C/EBPα expression and mutations in the tumor suppressor gene p53 or in the oncogene K-ras (Table 1). However, in the weak C/EBPα IHC group a non-statistical trend towards wild-type p53 was seen (p=0.16).
Disease-free survival and overall survival in the different C/EBPα IHC groups
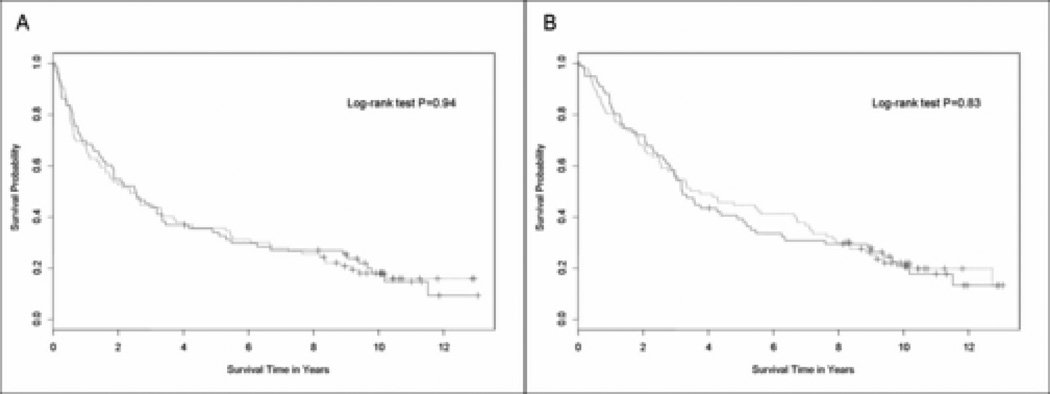
Among the 90 patients with weak C/EBPα IHC staining, median disease-free survival was 29.6 months (95% CI, 17.5 to 45.2 months); and for the 74 patients with strong C/EBPα IHC it was 30.6 months (95% CI, 19.6 to 41.4 months). There was no difference in disease-free survival by C/EBPα IHC (p=0.94 by log-rank) (Figure 2A).
Figure 2.
Survival curves in the weak and strong C/EBPα immunohistochemistry (IHC) groups. A) Disease-free survival and B) Overall survival. The solid line represents the strong IHC (2+ or 3+) group and the dashed line represents the weak IHC group (0 or 1+).
The median overall survival of the weak and strong C/EBPα IHC groups was 43.5 months (95% CI, 30.8 to 80.6 months) and 38.5 months (95% CI, 32.5 to 61.0 months), respectively (p=0.83 by log-rank) (Figure 2B). In April 2006, of the 164 patients, 21% (19/90) were alive in the weak, and 20% (15/74) in the strong C/EBPα IHC group. When each individual IHC staining score was analyzed separately (0, 1+, 2+ or 3+) for disease-free and overall survival there were no statistical differences (data not shown).
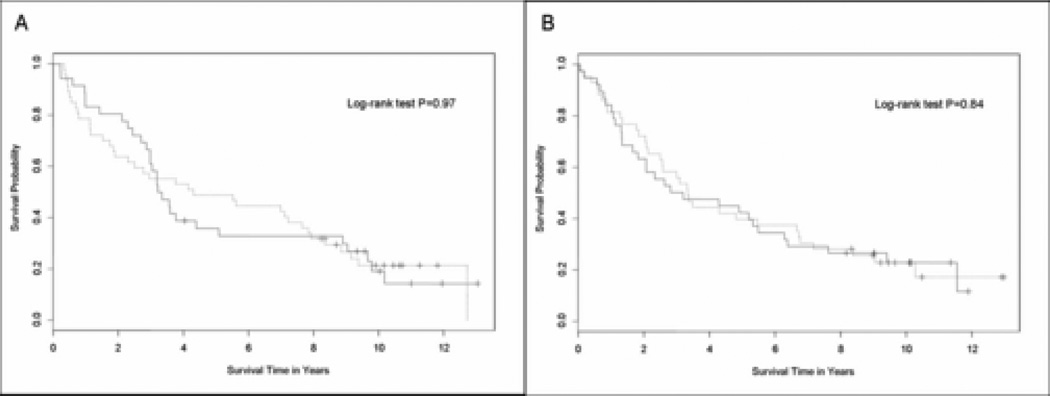
When the C/EBPα IHC weak and strong groups were analyzed by treatment arm (arm A, radiotherapy, and arm B, adjuvant chemoradiotherapy), there was no difference in outcomes. The median overall survival for the weak and strong C/EBPα IHC groups in arm A was 41.6 months (95% CI, 27.6 to 91.6) and 39.4 months (95% CI, 35.8 to 61.0), respectively (p= 0.97) (Figure 3A). In arm B, it was 40 months (95% CI, 30 to 79.6 months) and 36 months (95% CI, 21.8 to 65.5 months), respectively (p= 0.84) (Figure 3B).
Figure 3.
Survival curves in the weak and strong C/EBPα immunohistochemistry (IHC) groups by treatment arm of E3590. A) Overall survival in arm A: radiotherapy; and B) Overall survival in arm B: chemoradiation. The solid line represents the strong IHC (2+ or 3+) group and the dashed line represents the weak IHC group (0 or 1+).
DISCUSSION
The transcription factor C/EBPα is essential for airway epithelial development (26), due to its key role in regulating proliferation and differentiation-dependent gene expression of lung-specific target genes involved in late lung development such as surfactant protein A and D, clara cell secretory protein and the P450 enzyme CYP2B1, among others (34). In addition, hyperproliferation and differentiation arrest of type II pneumocytes were seen in a C/EBPα knockout mice model (27) as well as in the lung-specific conditional mouse model of C/EBPα deletion developed by our group (28). In fact, the prominent lung phenotype of our conditional mouse model resulted in respiratory arrest and death (28). Induction of this gene’s expression in lung cancer cell lines led to growth arrest, apoptosis, and cellular differentiation (29). These results identified C/EBPα as a master regulator of airway epithelial maturation and suggested that the loss of this gene could be an important event in the multistep process of lung carcinogenesis. Furthermore, our group (29) and others (30) have previously identified down-regulation of C/EBPα in a large proportion of lung cancer cell lines and primary tumor samples.
The mechanism of down-regulation of C/EBPα in lung cancer has been recently elucidated. We have shown that C/EBPα mutations are a rare event in lung cancer (only 1 sequence change in 50 primary NSCLC tumor DNA samples analyzed) (35). Tada et al demonstrated the epigenetic modulation of this gene in lung cancer (30). Both in lung cancer cell lines and in patients’ tumors presence of upstream promoter methylation was significantly correlated with lack of C/EBPα RNA levels (30). In the pathologic specimens studied, there was a statistically significant inverse correlation between the density of methylation and loss of C/EBPα using an IHC method similar to ours (30). These data establish IHC as a possible surrogate marker of C/EBPα upstream promoter methylation in NSCLC and validate the use of an IHC method as a marker of C/EBPα expression.
The objective of our project was to assess the frequency of C/EBPα down-regulation in a large number of prospectively collected stage II and IIIA NSCLC specimens and determine the association of C/EBPα expression with baseline clinical, pathologic and molecular characteristics, as well as clinical outcomes. In agreement with previously described IHC studies of C/EBPα in NSCLC patients (29,30), we identified down-regulation (the weak, 0 or 1+ IHC score) of C/EBPα in 55% of the 164 available samples.
A significantly greater percentage of squamous cell carcinomas in E3590 had down-regulation of C/EBPα by our immunohistochemical method (67% of the 55 squamous cell carcinoma samples were in the weak C/EBPα IHC group), while C/EBPα was present in approximately half of adenocarcinoma samples. In our initial C/EBPα IHC study of 53 heterogeneous lung cancer samples (there were only 9 squamous carcinomas from total) this pattern was not observed (29), but subsequent analysis of a larger dataset of 122 NSCLCs revealed an association of greater down-regulation of C/EBPα in squamous cell carcinomas (30). It is possible that loss of C/EBPα may be an essential step during squamous cell lung carcinogenesis. Interestingly, loss of C/EBPα seems to be a common finding in squamous cell carcinomas of the skin (36).
Our current study also disclosed that there were no statistically significant associations between mutations in p53 or K-ras and C/EBPα status, however there was a trend towards more p53 mutations in the tumors with presence of C/EBPα. It is possible that in these NSCLCs the tumor suppressive effects of C/EBPα (29) may be overcome by loss of p53 function. These findings merit further investigation.
There was a non-statistical trend towards decreased C/EBPα expression in poorly differentiated tumors. Since only 88 of the 164 tumors analyzed contained data regarding histological differentiation, it is possible that in a larger sample a significant association would emerge. A correlation of C/EBPα expression with differentiation is supported by its role on airway epithelial differentiation as demonstrated by our group and others (27–30), and had been seen in our initial C/EBPα study in lung cancer (29).
We were unable to identify a prognostic role for C/EBPα IHC status in the intergroup E3590 study of resected stage II and III NSCLC. Both disease-free and overall survivals were similar in the groups with weak or strong C/EBPα IHC. E3590 was a randomized trial of adjuvant therapy (31), and we were able to evaluate if C/EBPα IHC could predict response to radiation or platinum-based chemoradiation. C/EBPα IHC was not a predictive marker. One possible hypothesis is that loss of C/EBPα is an early event in the initiation and promotion of lung cancers with down-regulation of this protein and not related to the metastatic potential or chemotherapeutic and radiation-resistant mechanisms of stage II and IIIA NSCLCs (8,37). This hypothesis is currently being tested in mouse models with conditional lung-targeted knockout of C/EBPα (28). It is possible that despite its lack of significance as a prognostic marker in early stage NSCLC tumors, knowledge of a tumor’s C/EBPα expression by IHC may be essential as a predictive marker for response to therapies that aim at this transcription factor’s pathway.
In summary, our current IHC study of stage II and III resected NSCLC from the intergroup study E3590 was able to identify down-regulation of C/EBPα in over half of the tumors, with a higher magnitude of loss of expression in squamous cell carcinomas than adenocarcinomas. A trend was also observed between degree of differentiation and expression status.
Down-regulation of C/EBPα is one of the most common events in lung cancer. In our cohort of patients it was seen more often than p53 and K-ras mutations, which were present in 45 and 24% of samples, respectively (7). Further elucidation of C/EBPα mediated pathways in NSCLC and search for agents that can either re-establish C/EBPα expression or bypass its loss will likely enhance available therapeutic strategies for this malignancy.
Acknowledgments
CONFLICT OF INTEREST STATEMENT:
Dr. Tenen received grant support from the National Institutes of Health (NIH) Specialized Programs of Research Excellence (SPORE) in Lung Cancer PA20-CA090578
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2006. CA Cancer J Clin. 2006;56:106–130. doi: 10.3322/canjclin.56.2.106. [DOI] [PubMed] [Google Scholar]
- 2.Mountain CF. Revisions in the international system for staging lung cancer. Chest. 1997;111:1710–1717. doi: 10.1378/chest.111.6.1710. [DOI] [PubMed] [Google Scholar]
- 3.Arriagada R, Bergman B, Dunant A, Le Chevalier T, Pignon JP, Vansteenkiste J International Adjuvant Lung Cancer Trial Collaborative Group. Cisplatin-based adjuvant chemotherapy in patients with completely resected non-small-cell lung cancer. N Engl J Med. 2004;350(4):351–360. doi: 10.1056/NEJMoa031644. [DOI] [PubMed] [Google Scholar]
- 4.Winton T, Livingston R, Johnson D, et al. National Cancer Institute of Canada Clinical Trials Group. National Cancer Institute of the United States Intergroup JBR.10 Trial Investigators. Vinorelbine plus cisplatin vs. observation in resected non-small-cell lung cancer. N Engl J Med. 2005;352(25):2589–2597. doi: 10.1056/NEJMoa043623. [DOI] [PubMed] [Google Scholar]
- 5.Strauss GM, Herndon J, Maddaus MA, et al. Randomized clinical trial of adjuvant chemotherapy with paclitaxel and carboplatin following resection in stage IB non-small cell lung cancer: report of CALGB protocol 9633. J Clin Oncol. 2004;22(14S):621s. [Google Scholar]
- 6.Douillard JY, Rosell R, Delena M, et al. ANITA: phase III adjuvant vinorelbine and cisplatin versus observation in completely resected (stage I–III) non-small cell lung cancer patients: final results after 70-month median follow-up. J Clin Oncol. 2005;23(16S):624s. [Google Scholar]
- 7.Schiller JH, Adak S, Feins RH, et al. Lack of prognostic significance of p53 and K-ras mutations in primary resected non-small-cell lung cancer on E4592: a Laboratory Ancillary Study on an Eastern Cooperative Oncology Group Prospective Randomized Trial of Postoperative Adjuvant Therapy. J Clin Oncol. 2001;19(2):448–457. doi: 10.1200/JCO.2001.19.2.448. [DOI] [PubMed] [Google Scholar]
- 8.Sekido Y, Fong KM, Minna JD. Molecular genetics of lung cancer. Annu Rev Med. 2003;54:73–87. doi: 10.1146/annurev.med.54.101601.152202. [DOI] [PubMed] [Google Scholar]
- 9.Gonzalez-Quevedo R, Iniesta P, Moran A, et al. Cooperative role of telomerase activity and p16 expression in the prognosis of non-small-cell lung cancer. J Clin Oncol. 2002;20(1):254–262. doi: 10.1200/JCO.2002.20.1.254. [DOI] [PubMed] [Google Scholar]
- 10.Mitsudomi T, Hamajima N, Ogawa M, Takahashi T. Prognostic significance of p53 alterations in patients with non-small cell lung cancer: a meta-analysis. Clin Cancer Res. 2000;6(10):4055–4063. [PubMed] [Google Scholar]
- 11.Tammemagi MC, McLaughin JR, Bull SB. Meta-analyses of p53 tumor suppressor gene alterations and clinicopathological features in resected lung cancers. Cancer Epidemiol Biomarkers Prev. 1999;8:625–634. [PubMed] [Google Scholar]
- 12.Slebos RJC, Kibbelaar RE, Dalesio O, et al. K-ras oncogene activation as a prognostic marker in adenocarcinoma of the lung. N Engl J Med. 1990;323:561–565. doi: 10.1056/NEJM199008303230902. [DOI] [PubMed] [Google Scholar]
- 13.Kanematsu T, Yano S, Uehara H, Bando Y, Sone S. Phosphorylation, but not overexpression, of epidermal growth factor receptor is associated with poor prognosis of non-small cell lung cancer patients. Oncol Res. 2003;13(5):289–298. doi: 10.3727/096504003108748348. [DOI] [PubMed] [Google Scholar]
- 14.Huang C, Liu D, Masuya D, Nakashima T, Kameyama K, Ishikawa S, Ueno M, Haba R, Yokomise H. Clinical application of biological markers for treatments of resectable non-small-cell lung cancers. Br J Cancer. 2005;92(7):1231–1239. doi: 10.1038/sj.bjc.6602481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferraro B, Bepler G, Sharma S, Cantor A, Haura EB. EGR1 predicts PTEN and survival in patients with non-small-cell lung cancer. J Clin Oncol. 2005;23(9):1921–1926. doi: 10.1200/JCO.2005.08.127. [DOI] [PubMed] [Google Scholar]
- 16.Lu C, Soria JC, Tang X, Xu XC, Wang L, Mao L, Lotan R, Kemp B, Bekele BN, Feng L, Hong WK, Khuri FR. Prognostic factors in resected stage I non-smallcell lung cancer: a multivariate analysis of six molecular markers. J Clin Oncol. 2004;22(22):4575–4583. doi: 10.1200/JCO.2004.01.091. [DOI] [PubMed] [Google Scholar]
- 17.McKnight SL. McBindall: a better name for CCAAT/enhancer binding proteins? Cell. 2001;107:259–261. doi: 10.1016/s0092-8674(01)00543-8. [DOI] [PubMed] [Google Scholar]
- 18.Lekstrom-Himes J, Xanthopoulos KG. Biological role of the CCAAT/enhancer binding protein family of transcription factors. J Biol Chem. 1998;273:28545–28548. doi: 10.1074/jbc.273.44.28545. [DOI] [PubMed] [Google Scholar]
- 19.Pabst T, Mueller BU, Zhang P, Radomska HS, Narravula S, Schnittger S, Behre G, Hiddemann W, Tenen DG. Dominant-negative mutations of CEBPA, encoding CCAAT/enhancer binding protein-in acute myeloid leukemia. Nat Genet. 2001;17:263–270. doi: 10.1038/85820. [DOI] [PubMed] [Google Scholar]
- 20.Pabst T, Mueller BU, Harakawa N, Schoch C, Haferlach T, Behre G, Hiddemann W, Zhang D-E, Tenen DG. AML1-ETO downregulates the granulocytic differentiation factor C/EBPα in t(8;21) myeloid leukemia. Nat Med. 2001;7:444–451. doi: 10.1038/86515. [DOI] [PubMed] [Google Scholar]
- 21.Preudhomme C, Sagot C, Boissel N, et al. Favorable prognostic significance of CEBPA mutations in patients with de novo acute myeloid leukemia: a study from the Acute Leukemia French Association (ALFA) Blood. 2002;100:2717–2723. doi: 10.1182/blood-2002-03-0990. [DOI] [PubMed] [Google Scholar]
- 22.Frohling S, Schlenk RF, Stolze I, et al. CEBPA mutations in younger adults with acute myeloid leukemia and normal cytogenetics: prognostic relevance and analysis of cooperating mutations. J Clin Oncol. 2004;22(4):624–633. doi: 10.1200/JCO.2004.06.060. [DOI] [PubMed] [Google Scholar]
- 23.Antonson P, Xanthopoulos KG. Molecular cloning, sequence, and expression patterns of the human gene encoding CCAAT/enhancer binding protein α (C/EBPα) Biochem. Biophys. Res. Commun. 1995;215:106–113. doi: 10.1006/bbrc.1995.2439. [DOI] [PubMed] [Google Scholar]
- 24.Sugahara K, Iyama KI, Kimura T, et al. Mice lacking CCCAAT/enhancer-binding protein-alpha show hyperproliferation of alveolar type II cells and increased surfactant protein mRNAs. Cell Tissue Res. 2001;306:57–63. doi: 10.1007/s004410100420. [DOI] [PubMed] [Google Scholar]
- 25.Nord M, Cassel TN, Braun H, Suske G. Regulation of the Clara cell secretory protein/uteroglobin promoter in lung. Ann N Y Acad Sci. 2000;923:154–165. doi: 10.1111/j.1749-6632.2000.tb05527.x. [DOI] [PubMed] [Google Scholar]
- 26.Cassel TN, Nord M. C/EBP transcription factors in the lung epithelium. Am J Physiol Lung Cell Mol Physiol. 2003;285:L773–L781. doi: 10.1152/ajplung.00023.2003. [DOI] [PubMed] [Google Scholar]
- 27.Flodby P, Barlow C, Kylefjord H, Ahrlund-Richter L, Xanthopoulos KG. Increased hepatic cell proliferation and lung abnormalities in mice deficient in CCAAT/enhancer binding protein alpha. J Biol Chem. 1996;271:24753–24760. doi: 10.1074/jbc.271.40.24753. [DOI] [PubMed] [Google Scholar]
- 28.Basseres DS, Levantini E, Ji H, et al. Respiratory failure due to differentiation arrest and expansion of alveolar cells following lung-specific loss of the transcription factor C/EBPalpha in mice. Mol Cell Biol. 2006;26(3):1109–1123. doi: 10.1128/MCB.26.3.1109-1123.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Halmos B, Huettner CS, Kocher O, et al. Down-regulation and antiproliferative role of C/EBPalpha in lung cancer. Cancer Res. 2002;62:528–534. [PubMed] [Google Scholar]
- 30.Tada Y, Brena RM, Hackanson B, Morrison C, Otterson GA, Plass C. Epigenetic modulation of tumor suppressor CCAAT/enhancer binding protein α activity in lung cancer. J Natl Cancer Inst. 2006;98(6):396–406. doi: 10.1093/jnci/djj093. [DOI] [PubMed] [Google Scholar]
- 31.Keller SM, Adak S, Wagner H, et al. A randomized trial of postoperative adjuvant therapy in patients with completely resected stage II or IIIA non-small-cell lung cancer. Eastern Cooperative Oncology Group. N Engl J Med. 2000;343(17):1217–1222. doi: 10.1056/NEJM200010263431703. [DOI] [PubMed] [Google Scholar]
- 32.Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc. 1958;53:457–481. [Google Scholar]
- 33.Mantel N. Evaluation of survival data and two new rank order statistics arising in its consideration. Cancer Chemother Rep. 1966;50:163–170. [PubMed] [Google Scholar]
- 34.Cassel TN, Nord M. C/EBP transcription factors in the lung epithelium. Am J Physiol Lung Cell Mol Physiol. 2003;285:L773–L781. doi: 10.1152/ajplung.00023.2003. [DOI] [PubMed] [Google Scholar]
- 35.Costa DB, Dayaram T, D’Alo F, et al. C/EBPalpha mutations in lung cancer. Lung Cancer. 2006;53:253–254. doi: 10.1016/j.lungcan.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 36.Shim M, Powers KL, Ewing SJ, Zhu S, Smart RC. Diminished expression of C/EBPalpha in skin carcinomas is linked to oncogenic Ras and reexpression of C/EBPalpha in carcinoma cells inhibits proliferation. Cancer Res. 2005;65:861–867. [PubMed] [Google Scholar]
- 37.Olauseen KA, Dunant A, Fouret P, et al. DNA repair by ERCC1 in non–small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N Engl J Med. 2006;355(10):983–991. doi: 10.1056/NEJMoa060570. [DOI] [PubMed] [Google Scholar]