Abstract
Yersinia Protein Tyrosine Phosphatase (YopH) is the most efficient enzyme amongst all PTPases and YopH is hyperactive compared to human PTPases, interferes with mammalian cellular pathways to achieve the pathogenicity of Yersinia. Two properties related to the catalytic loop structure differences have been proposed to affect its dynamics and enzyme efficiency. One is the ability of the loop to form stabilizing interactions to bound ligand after loop closure, which has long been recognized. In addition, the loop flexibility/mobility was suggested in a previous study to be a factor as well, based on the observation that incremental changes in PTPase loop structure by single point mutations to alanine often induce incremental changes in enzyme catalytic efficiency. In this study, the temperature jump relaxation spectroscopy (T-jump) has been used to discern the subtle changes of the loop dynamics due to point loop mutations. As expected, our results suggest a correlation between loop dynamics and the size of the residue on the catalytic loop. The stabilization of the enzyme-ligand complex is often enthalpy driven, achieved by formation of additional favorable hydrogen bonding/ionic interactions after loop closure. Interestingly, our T-jump and X-ray crystallography studies on YopH suggest that the elimination of some ligand-protein interactions by mutation does not necessarily destabilize the ligand-enzyme complex after loop closure since the increased entropy in the forms of more mobile protein residues may be sufficient to compensate the free energy loss due to lost interactions and may even lead to enhanced efficiency of the enzyme catalysis. How these competing loop properties may affect loop dynamics and enzyme function are discussed.
Keywords: Loop flexibility, ligand binding, active site interactions, enthalpy, entropy
Introduction
Protein tyrosine phosphatases (PTPase) comprise a large and structurally diverse family of signaling enzymes with a unique signature (H/V)C(X)5R(S/T) motif. It is estimated that more than 100 PTPases are encoded in the human genome. In humans, the cellular protein tyrosine phosphorylation level is regulated by protein-tyrosine kinases and PTPases. Malfunctions of the PTPases have been associated with a number of cancers and also metabolic diseases, including non-insulin dependent diabetes (cf. 1–4). Yersinia pestis is the causative agent of human diseases from gastrointestinal syndromes to the plague.5, 6 It directly injects cytotoxic effector proteins, including YopH, a PTPase, into the cytosol of mammalian cells. YopH is hyperactive compared to human PTPases, and interferes with mammalian cellular pathways to achieve the pathogenicity of Yersinia.
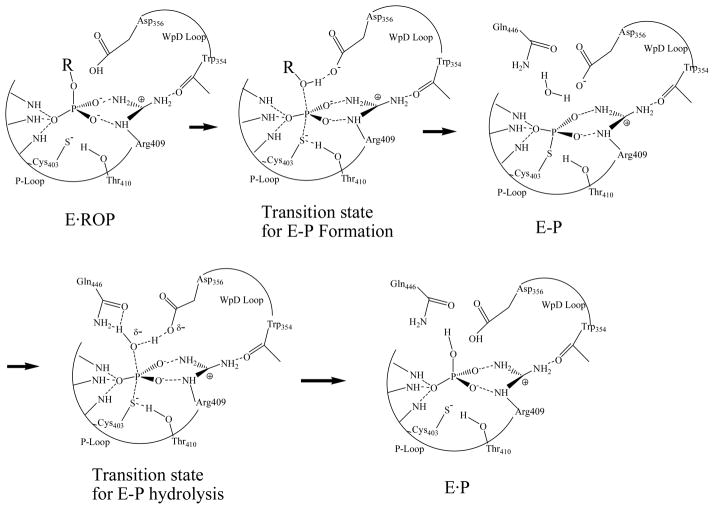
The high activity of YopH has made it a model system for detailed mechanistic studies of the PTPases.7 X-ray structures of PTPase complexed with various ligands to simulate ground state, reaction intermediate, and transition state complexes have been determined.8–13 A catalytic mechanism of PTPases has been proposed based on the analysis of these structures, as well as on the results from kinetic, mutagenesis and computational studies (reviewed in 6, 7, 14). PTPase catalysis involves a minimum of two main chemical steps (see Figure 1). After substrate binding, a protein conformational change involving the so-called WPD loop motion brings the essential Asp356 residing on the loop from almost 8 Å away to the scissile oxygen of the phosphorylated tyrosine (see Figure 2). In the first chemical step, Asp356 is proposed to act as a general acid to transfer a proton to the scissile oxygen of the substrate to initiate the tyrosine leaving group dissociation. The active site Cys403 becomes (or already is) deprotonated to serve as nucleophile to accept the phosphoryl group dissociated from phosphotyrosine substrate. The completion of the first reaction step results in a phosphocysteine intermediate. The Arg409 residue forms a salt bridge with two nonbridging oxygens of the phosphate group throughout the reaction, thus plays a role in both substrate recognition and transition state stabilization. It is believed that the phosphoryl transfer proceeds via a dissociative transition state.15 After fast dissociation of the substrate leaving group, the side chain of Gln446 flips into the active site to stabilize a water molecule that is positioned along the S-P bond direction. In the second chemical step, the OH group of Thr410 interacts with the scissile sulfur of the intermediate to initiate the dissociation of the phosphoryl group from Cys403. The rate limiting step of the overall reaction is believed to be the hydrolysis of the phosphoenzyme at pH 4, whereas at pH 7.5, substrate effects also contribute to the rate limiting step.16
Figure 1.
PTPase catalyzed reaction. E: YopH; P: phosphate; ROP: peptide tyrosine phosphate substrate; E-P: phosphorylated YopH.
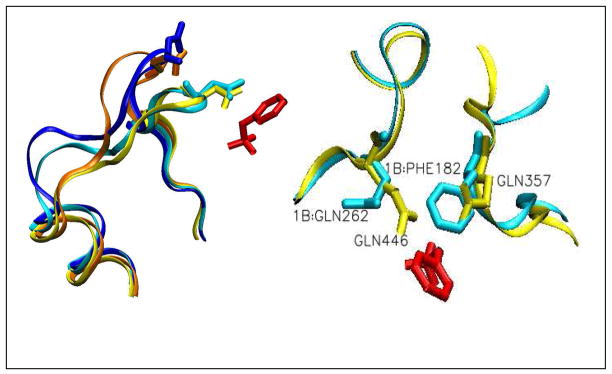
Figure 2.
(Left) The X-ray determined catalytic WPD loop closure movement upon substrate binding in YopH (orange to yellow) and in PTP1B (dark blue to light blue). The catalytic Asp356 (Asp181 for PTP1B) on the loop is also shown. Substrate pTyr is shown in red. (Right) Encounter between Gln357 and Gln446 upon loop closure in YopH. The corresponding residues in PTP1B are Phe182 and Gln262, respectively. The color codes are the same as in the left figure. The figures are constructed from pdb files 1YPT, 1PA9, 2I42, 1OEO and 2HB1.
The importance of the WPD loop structure and dynamics to PTPase catalysis is implied in this proposed reaction mechanism and verified by various experimental observations. For example, in a humans PTPase SHP-1, out of the 7 single Ala mutations in the WPD loop region (underlined residues in LSWPDHGVPSEP sequence) aimed to increase loop flexibility, 5 mutants showed higher kcat up to three fold, one mutant showed no change and only one mutant showed decreased kcat (L417A, by about one fold) 17. The authors of the paper proposed that the rate enhancements these loop Ala mutants were resulted from the increased loop dynamics, although no direct evidence of the increased dynamics was presented.17, 18 The WPD loop sequence in another human PTPase, PTP1B, (WPDFGVPESP, Figure 2, left) is similar to that of the SHP-1 in that it also contains two proline residues. In YopH, the WPD loop sequence is WPDQTAVSSE, (shown in Figure 2, left) which is quite different from its human counterpart. Apparently, the two proline residues in the PTP1B WPD loop can hinder its flexibility, affect its dynamics during catalysis, thus can be partially responsible for the more than 100-fold lower activity of PTP1B compared to YopH when pNPP is used as the substrate. Our previous studies have shown that the enzyme loop dynamics can be quantitatively determined by time-resolved fluorescence techniques such as temperature jump relaxation spectroscopy.19–21 In this study, this technique is applied to YopH mutants to find out if it has sufficient sensitivity to detect the subtle changes of the loop dynamics in the ligand binding process due to single point loop mutation, and how the loop dynamics may affect the enzyme function.
It has been suggested that one of the most important properties of the mobile loop lies in the stabilization of the system when the active site contains ligand and the loop is closed.22 The stabilizations are frequently found in the form of an enthalpy decrease, e. g. additional favorable interactions between loop residues and the bound-ligand/active-site residues. For example, in triosephosphate isomerase, the Y208F mutation, which eliminates a hydrogen bond present only in the loop closed conformation between the active site Y208 and a loop residue, alters the catalytic loop dynamics significantly, resulting a >1000-fold reduction of the kcat.23, 24 In this study, we investigate a similar feature of the WPD loop that may affect its dynamics and function.
Careful examinations of the X-ray structures of the WPD loop in YopH and PTP1B revealed that Gln357 in YopH, which is located at the center of the WPD loop, forms a hydrogen bond to the invariant Gln446 (Figure 2, right). The corresponding residue in PTP1B, Phe182, seems to push the invariant Gln262 away from the bound substrate (Figure 2, right). Since the invariant G446 (Gln262 in PTP1B) is proposed to stabilize a water molecule/hydroxyl for its nucleophilic attack on the thiophosphate intermediate in the second chemical step (see Figure 1), we have postulated that Gln357 may contribute to the stabilization of the enzyme-substrate complex by forming additional active site contacts, including hydrogen bonds to Gln446, at appropriate times during catalysis to enhance the functional role of Gln446. These interactions could be partially responsible for the higher hydrolysis rate of the phospho-tyrosine, consequently the >100-fold higher kcat of YopH compared to PTP1B.
To determine the effects of the Gln446-Gln357 hydrogen bond on the WPD loop dynamics and enzyme catalysis and to check if alanine mutation of loop residue can actually increase the loop mobility and affects the catalytic efficiency, we applied mutagenesis, temperature-jump relaxation spectroscopy (T-Jump), and X-ray crystallography to study several YopH Q357X mutants, including Q357F, Q357Y and Q357A. The WPD loop dynamics of these mutants in the enzyme-ligand complex formation process have been determined by fluorescence laser induced temperature jump relaxation experiments25–29 which yield, in our hands, a time resolution of nanoseconds. We previously have demonstrated the versatility of applying laser-induced T-jump spectroscopic methods for measuring dynamic processes in enzymes (reviewed in 30, 31).
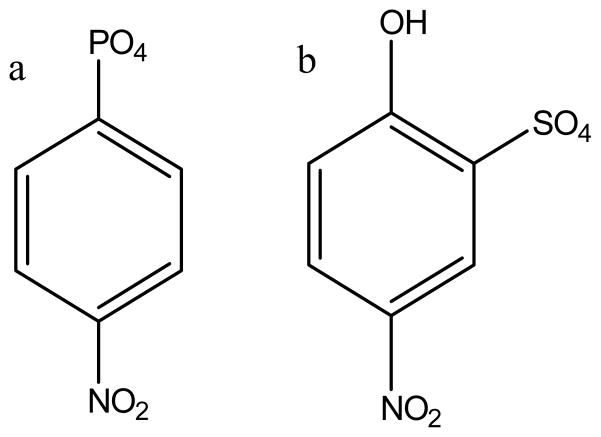
The enzyme system under our current study is the catalytic domain of YopH (residues 162-468)), which contains a single tryptophan (Try354) at the hinge of the WPD loop. The spectral properties of this lone tryptophan can be used to monitor the dynamics of the WPD loop. The WPD loop dynamics in the ligand binding process was determined by using p-nitrocatechol sulfate (pNCS, Figure 3b), a substrate mimic. This molecule has a structure similar to the commonly used substrate p-nitrophenyl phosphate (pNPP, Figure 3a) and is a competitive inhibitor of YopH.13 In a previous study, the kinetic parameters for the pNCS binding to wild type YopH have been determined using fluorescence T-jump relaxation measurements.32 We have extended the T-jump measurements on Q357X mutants in this study to determine how the WDP loop dynamics are affected by the disruption of the Gln446-Gln357 hydrogen bond in the loop closed conformation and how the WDP loop dynamics may correlate with the enzyme activities in these mutants.
Figure 3.
Chemical structure of a) pNPP (p-nitrophenyl phosphate) and b) pNCS (p-nitrocatechol sulfate).
In addition, we have used X-ray crystallography to determine how the hydrogen bonding network near the active site, especially how the catalytically important, Gln446 stabilized water molecule may be affected by Gln357 mutation. This water molecule is observed in the X-ray structures of wild type YopH complexed with vanadate (2I42,11), tungstate/nitrate (1YTW/1YTN,33) and pNCS (1PA9,13) and has well defined B factor similar to the B factors of the surrounding enzyme/ligand atoms. Whereas in the structures of some YopH mutants with diminished enzyme activities, this water is either not observed (1YTS,12) or is shifted significantly from the line of the nucleophilic attack (3F9A,34).
By performing these combined experimental studies, we are aimed to find out the proper techniques to relate loop dynamics with its structure and function, and to understand, both dynamically and structurally, the effects of the residue size on the catalytic loop, as well as the hydrogen bonding interactions originated from Gln357 to the function of the YopH enzyme, including enzyme-ligand complex formation and enzyme catalysis. To our surprise, the elimination of the active site contacts originated from Gln357 does not necessarily result in decreased stability of the enzyme system with the bound ligand. In the case of Q357F mutant, increased catalytic efficiency of the enzyme was actually observed.
Materials and Methods
Chemicals
p-Nitrophenyl phosphate (pNPP) was obtained from Fisher Scientific (Pittsburgh, PA). p-nitrophenolate (pNP) and 4-Nitrocatechol sulfate dipotassium salt monohydrate (pNCS) were purchased from Aldrich (Milwaukee, WI). Commercially available analytical grade or higher materials were used for the buffer preparation and other experiments.
Site-directed Mutagenesis
The plasmid encoding the catalytic domain (residues 162–468) of Yersinia PTP (referred as YopH or Yop51*K162) was a gift from Dr. Z.-Y. Zhang (Department of Biochemistry and Molecular Biology, Indiana University School of Medicine) and was used as the template. Q357X mutants were generated using the QuikChange Site-Directed Mutagenesis Kit according to the standard procedure, using the following primers synthesized by Integrated DNA Technologies (Coralville, IA):
Q357A, 5′-GGCCCGATGCGACCGCAGTCAGC-3′;
Q357F, 5′-GGCCCGATTTTACCGCAGTCAGC-3′;
Q357Y, 5′-GGCCCGATTATACCGCAGTCAGC-3′.
The mutations were confirmed by sequencing at the DNA Facility, Albert Einstein College of Medicine.
Enzyme Preparation
The YopH wild type and Q357A, Q357F, and Q357Y mutants were expressed under the control of the T7 promoter in E coli BL21(DE3) cells. Briefly, 10 mL overnight culture of E. coli BL21(DE3) cells grown from single colony were diluted to 1 L of 2xYT containing 100 μg/mL ampicillin. The cells were grown at 37 °C until the optical density of the culture cells reached 1.6 OD at 600 nm. 0.4 mM isopropyl-D-thiogalactoside (IPTG) was added and the cells were grown for an additional 22 h at room temperature. The cells were harvested by centrifugation at 3200 rpm for 30 min. The resulting pellet was resuspended in 50 mL of 10mM sodium acetate buffer, pH 5.7 with 1 mM EDTA and 1 mM DTT, and was lysed by three French press passages at 4 °C. After removing the residual debris through centrifugation at 20000 rpm for 60 min, the supernatant was directly loaded onto a CM Sepharose FF 50 mL home made column. The protein was eluted with a linear salt gradient using 100 mM acetate pH 5.7 as Buffer A and 100 mM acetate +1.0 M NaCl pH 5.7 as Buffer B. The fractions with the enzyme were pooled and concentrated to about 10 mL and then loaded on a gel filtration column (Superdex 200) using 90% Buffer A and 10% Buffer B as the elution buffer. Homogeneities of the enzymes after two separation steps were estimated to be >95% by SDS-PAGE. The yields of the purified enzymes are: YopH wild type, 65 mg/L; Q357A 60 mg/L; Q357F, 75 mg/L and Q357Y, 55 mg/L. The concentration of the enzyme was determined by the UV-Vis absorbance at 280 nm using Amg/mL (280nm) = 0.49 OD. The purified enzyme was stored in the presence of 1 mM DTT and 1 mM EDTA at 4 °C until use.
Steady state kinetic methods
All assays were performed at 23 °C in 50 mM Citrate buffers with 1 mM EDTA, 1 mM DTT in the pH range of 5 – 6.5. The ionic strength of buffers was adjusted to 0.15 M using NaCl. A continuous spectrophotometric assay was employed to determine kcat and Km values by monitoring the absorption increase at 400 nm versus time. The absorption at 400 nm is due to the absorbance of p-nitrophenolate (pNP), the pNPP dephosphorylation product. All measurements were performed on a Rapid Kinetics Beckman-Coulter-DU7400 spectrophotometer with a 1 cm path-length cell. The reaction rate was determined from the first 30 seconds of the reaction. Because the absorbance of the pNP at 400 nm is pH-dependent, its absorption extinction coefficients in different pH buffers were calibrated by pNP standard solutions.35 The Michaelis-Menten kinetic parameters were calculated from a nonlinear fitting of the υ vs. [s] data to the Michaelis-Menten equation using OriginPro 6.1.
Fluorescence Temperature-Jump Measurements
The temperature-jump relaxation studies using fluorescence detection has been described in detail previously. 19, 21, 32, 36 Briefly, a 300 μL sample in water solution is exposed to a pulse of infrared light (1.56 μm wavelength, 90–120 mJ energy, about 1.5 mm diameter spot on the sample, 0.5 mm pathlength). Water absorbs the laser energy, and the temperature jump (T-jump) is induced in approximately 6 ns. In this study, the T-jump values ranged from 6.5 to 8.5 °C. The value of the T-jump is monitored using temperature dependent IR absorption of water. The final temperature after T-jump remains nearly constant within approximately 5 ms. The measurement of the fluorescence intensity of the tryptophan fluorophore is performed by subjecting the sample to excitation at 300 nm from an Innova 200-25/5 argon ion laser (Coherent, Palo Alto, CA) and monitoring the emission at 340 nm. To avoid photo damage to the sample, the excitation light is modulated using a shutter that opens 4 ms before the T-jump pulse, and closes 1 ms after the end of data acquisition. In this study, the relaxation was monitored in the first 14 ms after the T-jump. Each relaxation kinetics was obtained as an average of 3600 acquisitions. The kinetic data were normalized to the fluorescence intensity before the T-jump.
Crystallization, Data Collection, and Structure Determination of YopH/pNCS and Q357F/pNCS complexes
YopH (or Q357F) were concentrated to 40 mg/mL in 10 mM imidazole buffer pH 7.2, 0.2 mM EDTA and 1 mM DTT. Both YopH and Q357F in presence with 5 mM pNCS crystallized in 20% polyethylene glycol 10000, and 0.1 M Hepes pH 7.5 at 18 °C by sitting-drop vapor diffusion. Crystals were transferred to the mother liquor supplemented with 20% glycerol and flash frozen in liquid N2 before data collection. X-ray diffraction data were collected at the X29 beamline of Brookhaven National Laboratory on an ADSC Q315 detector at 100 K. The data were processed using the HKL2000 program suite.
Results
Determination of Kinetic Constants using pNPP as a substrate
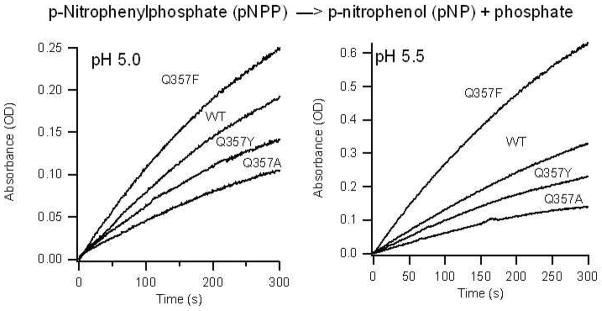
The Michaelis-Menten parameters for YopH wild type and Q357A, Q357F and Q357Y were determined at 23 °C in citrate buffer in the pH range of 5 – 6.5. p-nitrophenyl phosphate (pNPP) was used as the substrate. The enzymatic activity was monitored by the UV absorbance at 400 nm versus time. Since the absorbance of the product p-nitrophenyl is highly dependent on the pH, its extinction coefficients at various pH values at 400 nm are carefully calibrated. Typical reaction curves for YopH/Q357X mutants catalyzed reaction at pH 5 and 5.5 are shown in Figure 4. It is interesting to note that the activity of Q357F is clearly higher than that of the wild type under the same conditions. In Table 1, the kinetic parameters kcat and kcat/Km for YopH and its Q357 mutants in the pH range of 5.0 to 6.5 are summarized.
Figure 4.
YopH catalyzed reactions monitored at 400 nm. The samples were in 0.050 M Citrate buffer with 0.10 M NaCl. All enzyme concentrations were 8.0 nM and the concentration of pNPP was 2.5 mM.
Table 1.
pH dependent Michaelis-Menten parameters of YopH and its Q357X mutants.
| pH 5.0 | pH 5.5 | pH 6.0 | pH 6.5 | |||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| pNP ε400nm (M−1/cm−1) | 253 | 735 | 2673 | 5244 | ||||
|
| ||||||||
| Enzymes | kcat (s−1) | kcat/Km (mM−1S−1) | kcat (s−1) | kcat/Km (mM−1S−1) | kcat (s−1) | kcat/Km (mM−1S−1) | kcat (s−1) | kcat/Km (mM−1S−1) |
| YopH Q357F | 1075 | 446 | 1792 | 231 | 608 | 82 | 456 | 73 |
| YopH wild type | 914 | 331 | 731 | 183 | 250 | 69 | 183 | 34 |
| YopH Q357Y | 740 | 172 | 727 | 105 | 270 | 47 | 122 | 25 |
| YopH Q357A | 856 | 133 | 382 | 62 | 151 | 21 | 61 | 12 |
ε400nm: - extinction coefficient at 400nm. The measurements were conducted in the pH range of 5.0 to 6.5 using pNPP as the substrate. The sample temperature was 23 °C.
The kcat and kcat/Km values for Q357F are up to 2.5-fold higher than those of the wild type. For Q357Y and Q357A mutants, the kcat and kcat/Km values are similar to or up to 3-fold lower than the wild type, respectively. The kcat and kcat/Km values of all four YopH variants are pH dependent. For Q357F, the kcat maximizes at about pH 5.5, while for other YopH variants, the kcat maxima are close to pH 5 or lower (Table 1).
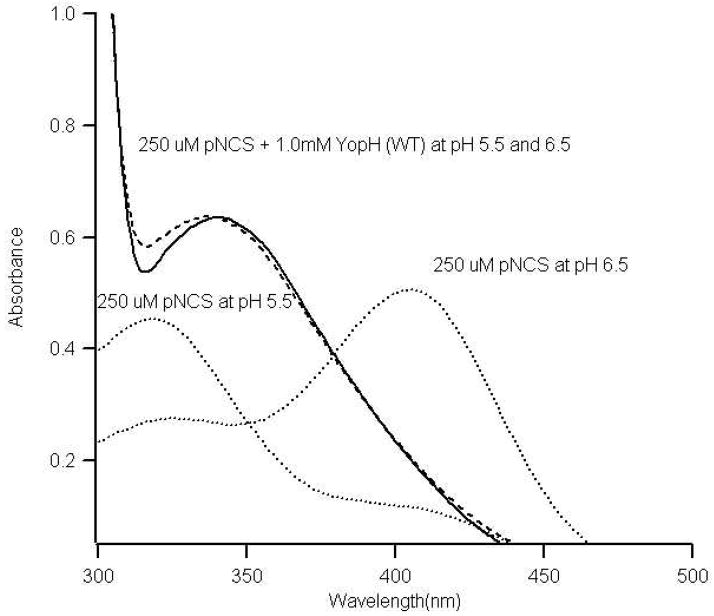
Ionic state of bound pNCS in YopH
The absorbance of pNCS is pH dependent as shown in Figure 5. Titration studies result in a pKa value of 6.3 for its OH group in solution. Upon binding to YopHs, the absorbance of pNCS remains to be the same at pH values between 5.5 and 6.5. The absorbance maximum of the bound pNCS is similar to that of pNCS in solution at pH 5.5, suggesting that YopH bound pNCS contains an OH group and its pKa is increased significantly upon binding to YopH. In the X-ray structure of YopH/pNCS complex, the pNCS OH group is within the hydrogen bonding distance to the carboxyl group of Asp356,13 providing a rational for its increased pKa value. Similar results were obtained for YopH Q357A/pNCS and YopH Q357F/pNCS complexes (data not shown).
Figure 5.
UV-Vis spectra of 0.250 mM pNCS in solution and in YopH at pH 6.5 and pH 5.5. The buffer contained 050 M Citrate with 0.10 M NaCl. The concentration of YopH was 1.0 mM to ensure more than 95% pNCS was bound.
WPD loop dynamics in Q357X mutants by T-Jump experiments
To characterize the dynamic properties of the WPD loop in YopH variants, fluorescence T-jump relaxation spectroscopy measurements were performed. In this technique, the equilibrium point of interconverting chemical species is changed by a rapid jump of the solvent temperature, forcing the system to follow the elevated temperature to establish a new equilibrium point:
Fast time resolution can be achieved by using pulsed laser excitation of the solvent. A change in solution temperature then perturbs the equilibrium of the enzyme system, and the relaxation kinetics of the system can then be monitored by fluorescence as the system approaches the new equilibrium. This technique is applied here to study the YopH/pNCS complex formation process. The fluorescence emission signals are from the sole Trp354 of YopH at the hinge of the WPD loop. This emission signal is sensitive to the loop conformation changes and to the pNCS binding, which quenches the tryptophan fluorescence.32
Based on the specific physics known about PTPase, and the data analysis on a series of T-jump studies with different ligand concentrations, a minimum kinetic model for wt YopH/pNCS complex formation has been determined:32
| (1) |
In this model, the apo-enzyme interconverts between loop open and loop closed conformations, and their population ratio can be determined from resonance Raman measurements.37 The ligand binds to the loop open conformation of the apo-enzyme followed by the loop closure to complete the complex formation process.
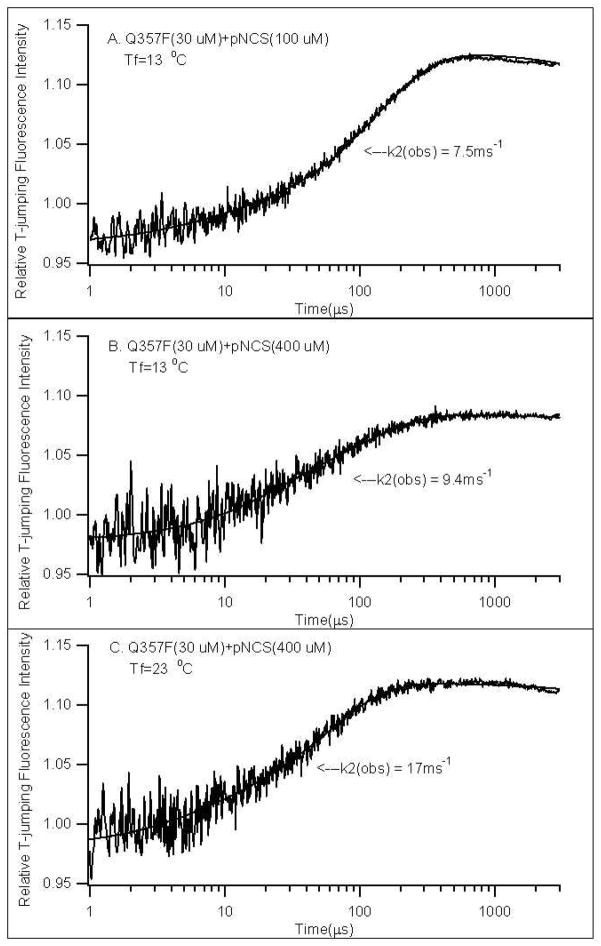
Here we extend the fluorescence T-jump studies to Q357X mutants. Typical relaxation kinetics obtained from our fluorescence T-jump experiments are shown in Figure 6. The relaxation curve faster than a few milliseconds can be fitted by the following equation:
| (2) |
Figure 6.
Typical T-jump fluorescence relaxation profiles of YopH at different concentrations of pNCS and final temperatures. All samples were in 0.050 M Citrate buffer with 0.10 M NaCl, pH 6.5. Curve fitting results with two-exponential functions (Equation 2) are also shown.
The time constant (k1(obs))−1 for the faster phase of the relaxation is in the order of a few microseconds while the slower time constant, (k2(obs))−1, is in the order of a few hundred microseconds. Previous studies of the wild type YopH have shown that the faster phase contains a component from kinetics of the apo-enzyme, and a component from the initial pNCS binding step.32 Since the amplitude of the faster phase of the relaxation curve is low, accurately determining the fast relaxation rate and uncoupling the kinetics of initial ligand binding from the kinetics of the apo-enzyme are difficult due to high noise level. Here we focus our studies on the slower relaxation step (k2(obs))−1, which is associated to the open-close conformational change of the WPD loop after the initial pNCS binding step (Equation 1).
The kinetic model can be solved analytically to determine all three relaxation rates in terms of the microscopic rate constants.29 Under the assumption that the last step in Equation 1 is much slower (kopen, kclose ≪ kon, koff in Eq. 1) than the first two steps, the solution for the slower relaxation rate that corresponds to k2(obs) is derived:
| (3) |
where [E] is the concentration of the free enzyme and [L] is the concentration of the free ligand. This assumption is justified by the good fitting to this solution by the enzyme/ligand concentration dependent kinetic data shown below. Our preliminary Resonance Raman studies on the Q357F/Y/A mutants have shown that in apo-enzyme, the population ratios between loop closed and loop open conformations in these mutants are from 0.6 to 1 under our experimental conditions (Spiro, private communication) similar to that observed for wt YopH.37 Thus, Equation 3 can then be simplified to:
| (4) |
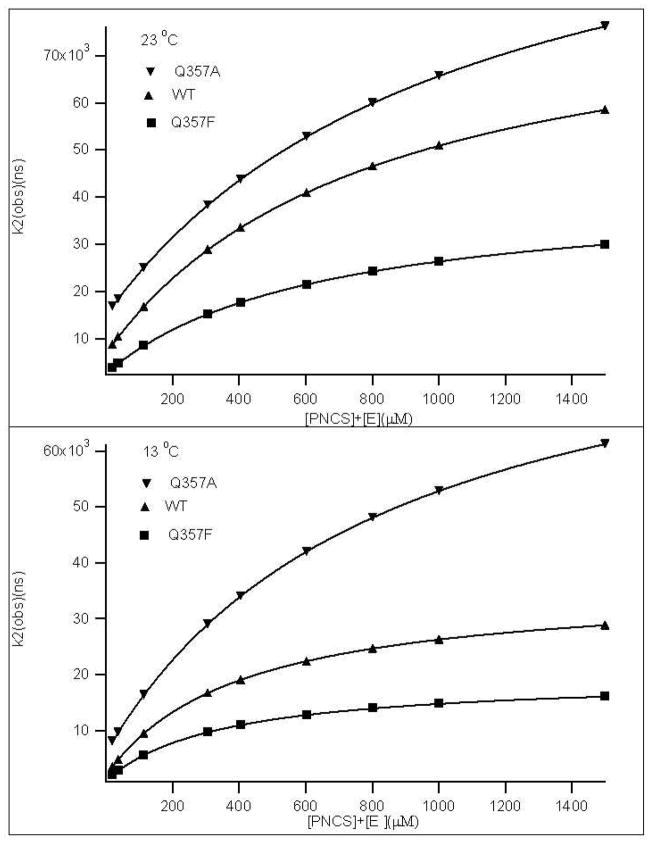
Figure 7 shows the observed relaxation rates, k2(obs), vs the free [pNCS] + [E] concentration for Q357F, Q357A and wt YopH at two final T-jump temperatures. Q357Y was not stable under our T-jump experimental conditions, and thus not further investigated. By fitting the data to Equation 4, the rate constants kopen, kclose, and koff/kon were obtained. The complete fitting parameters kclose, kopen and koff/kon, for wild type, Q357A and Q357F mutant are summarized in Table 2. These results show that the loop motions in Q357F are temperature dependent, about 2–3-fold slower than in wild type. The ratio kopen/kclose is < 0.1 in both Q357A and Q357F, indicating that the loop closed conformer is more than 10 times of the loop open conformer with pNCS bound, similar to that in wild type YopH.32 If 1.6 is used in Equation (4) for curve fitting, the outcomes of the kopen or kclose values will not be affected but the koff/kon value will be increased by 25%. The good fits of our concentration and temperature dependent data to Eq. (4) shown in Figure 7 suggest that our kinetic model (Eq. (1)) and the assumption that kopen, kclose ≪ kon, koff are reasonable.
Figure 7.
The plots of k2(obs) vs the sum of the free ligand and free enzyme concentrations at two final T-jump temperatures for Q357A, Q357F and wt YopH. The T-jump final temperatures were 13 °C, 23 °C. The curve fits of these data to Equation 4 are also shown as smooth curves.
Table 2.
Microscopic rate constants for the loop dynamics in YopH and its Q357A/F mutants at different final T-jump temperatures.
| YopH Q357F | kclose(ms−1) | kopen(ms−1) | kclose/kopen | koff/kon(μM) | ||||
|---|---|---|---|---|---|---|---|---|
| 33 °C | 98±18 | 6±1 | 16 | 821±22 | ||||
| 28 °C | 58±9 | 4±1 | 15 | 484±17 | ||||
| 23 °C | 39±6 | 2.6±0.4 | 15 | 322±13 | ||||
| 13 °C | 19±3 | 1.1± 0.1 | 17 | 175±10 | ||||
|
| ||||||||
| YopH WT | ||||||||
|
| ||||||||
| 33 °C | 230±20 | 16±1 | 14 | 1150±200 | ||||
| 28 °C | 139±9 | 11±1 | 13 | 700±100 | ||||
| 23 °C | 91±4 | 8±1 | 11 | 450±50 | ||||
| 13 °C | 46±3 | 3±1 | 15 | 250±50 | ||||
|
| ||||||||
| YopH Q357A | ||||||||
|
| ||||||||
| 33 °C | 178±20 | 16±3 | 11 | 496±29 | ||||
| 28 °C | 138±18 | 14±2 | 10 | 469±31 | ||||
| 23 °C | 108±15 | 10±2 | 11 | 442±28 | ||||
| 13 °C | 84±12 | 6±1 | 14 | 398±21 | ||||
The values of kclose, kopen and koff/kon are deduced from the curve fit of the data in Figure 7 to Equation 4. Their uncertainties are the RMSDs of the fits.
Comparison of thermodynamic properties of wt and Q357X mutants
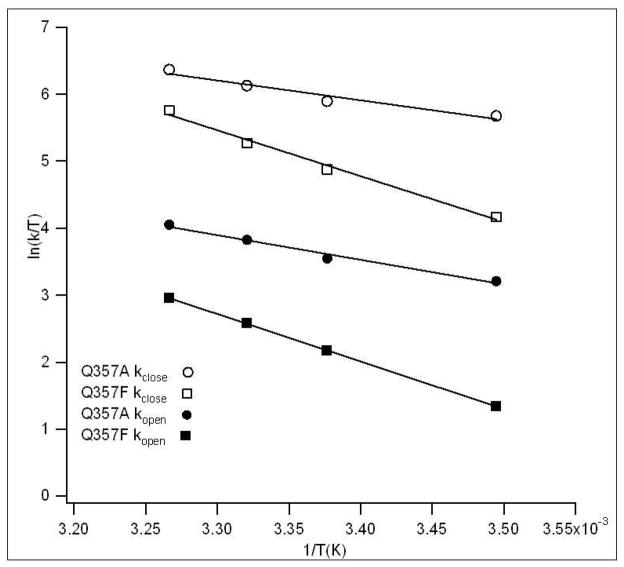
The kclose and kopen values determined from Q357F and Q357A are temperature dependent (Table 2) and they are presented in an Eyring plot and fitted to linear equations as shown in Figure 8. The activation enthalpies associated with the loop open and loop close motions, ΔH‡, can be determined from the slopes of the fittings. These values, along with the values for wt YopH determined previously,32 are summarized in Table 3. In Q357F, the activation enthalpies in both directions differ by ~2kJ/mol compared to wt YopH but in opposite directions. The loop closed form is still favored enthalpically over the loop open conformation but to a less extent ( ~2 kJ/mol vs ~7 kJ/mol) than in wt YopH. On the other hand, the average free energy differences over our experimental temperature range between the loop open and closed conformations are similar in Q357F and wt YopH, as judged by their kclose/kopen ratios under our experimental conditions (see Table 3). Therefore, we can conclude that the change in entropy is somewhat higher in the loop closed conformation of Q357F/pNCS complex than in wt or Q357A YopH/pNCS complex.
Figure 8.
Temperature dependence of the microscopic rate constants for the transitions between loop open and loop closed conformations in Q357F and Q357A, shown in Eyring coordinate. The solid curves are the linear fits for the data.
Table 3.
Activation enthalpies for the loop movements in YopH-variant/pNCS complexes, the enthalpy and free energy differences between loop closed and loop open conformations.
| Enzyme | ΔH‡kclose (kJ/mol) | ΔH‡kopen (kJ/mol) | ΔH (kJ/mol) | ΔG (kJ/mol) |
|---|---|---|---|---|
| Q357F | 57±3 | 59±2 | −2±4 | −6.6±0.5 |
| wild type | 55±3 | 61±3 | −7±5 | −6.2±0.6 |
| Q367A | 24±4 | 30±3 | −6±6 | −5.8±0.5 |
In Q357A, the activation enthalpies in both directions of loop movements after ligand binding are reduced by half compared to wt YopH. The loop closed form is favored enthalpically over the loop open conformation to the same extent as in wt YopH. The free energy differences between the loop open and closed conformations are somewhat reduced in Q357A compared to wt YopH, as judged by the smaller kclose/kopen ratios (see Tables 2 and 3).
X-ray crystallography studies of YopH/pNCS and Q357F/pNCS complexes
The Q357F mutation results in a small kcat increase for an enzyme that is already the most efficient PTPase. Our T-jump dynamics studies suggest that loop movements are slowed by this mutation, resulting a loop closed state with somewhat higher entropy. To provide structural basis for the interpretation of these results, we have determined the X-ray structures of Q357F/pNCS complex.
The structures of YopH/pNCS and Q357F/pNCS complexes were solved by molecular replacement using the crystal structure of ligand-omit YopH (PDB ID: 1PA9) as the search model using the program Molrep. 13, 38 Models without inhibitor were iteratively rebuilt in COOT and refined in Refmac5. 39, 40 Manual inhibitor building was initiated only after the Rfree decreased below 30% and was guided by clear ligand density in Fo−Fc electron density maps contoured at 3σ. Data processing and refinement statistics of Q357F/pNCS are summarized in Table 4.
Table 4.
X-ray structural data collection and refinement statistics
| PDB codes | YopH 3U96 |
|---|---|
| Data collection | |
| Space group | P1 |
| Cell dimension | |
| a, b, c (Å) | 49.86. 53.99, 65.76 |
| α, β, γ (°) | 104.8, 108.8, 97.8 |
| Resolutions (Å) | 20.00 – 1.80 |
| (1.86-1.80) | |
| Rsym (%) | 5.4 (34.6) |
| I/σI | 18.5 (3.2) |
| Completeness (%) | 96.3 (86.9) |
| Redundancy | 3.2 (3.0) |
|
| |
| Refinement | |
| Resolution (Å) | 20.00-1.8 |
| No. unique reflections | 54165 |
| Rwork/Rfree (%) | 19.7/25.9 |
|
| |
| B-factors (Å2) | |
| Protein | |
| (main chain) | 21.2 |
| (side chain) | 23.6 |
| Water | 29.7 |
| Ligand | 33.5 |
|
| |
| No. of Atoms | |
| Protein | 4484 |
| Water | 309 |
| Ligand | 40 |
|
| |
| R.m.s deviations | |
| Bond lengths (Å) | 0.013 |
| Bond angles (°) | 1.38 |
|
| |
| Ramachandran analysis | |
| favored region | 97.7% |
| allowed region | 2.3% |
| disallowed region | 0.0% |
|
| |
| Coordinate Error by Luzzati | |
| plot (Å) | 0.19 |
Numbers in parentheses are for the highest-resolution shell. One crystal was used for each data set.
Since the structure of YopH/pNCS is very similar to the one in the PDB databank (PDB ID: 1PA9), only the data for Q357F/pNCS are shown and summarized in Table 4.41 For Q357F/pNCS complex, two distinct structures are found: one with loop open conformation and the other with loop closed conformation. Bound pNCS is found at the active site in both structures, and its position is shifted only slightly relative to each other.
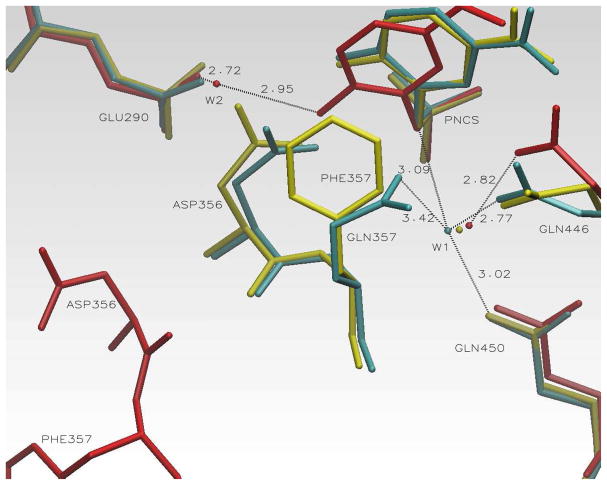
In Figure 9, the structures of YopH/pNCS and both loop open and closed forms of Q357F/pNCS are superimposed to compare the active site contacts in these YopH variants. For clarity, only the WPD loop sides of the pNCS in these structures are shown.
Figure 9.
Some active site contacts observed for YopH/pNCS and Q357F/pNCS complexes. Cyan: YopH/pNCS complex, loop closed form. Hydrogen bonds to a structural water molecule (W1) are shown. Red: Q357F/pNCS complex, loop open form. Hydrogen bonds to the other structural water molecule (W2) are shown. Only one of the hydrogen bonds to W1 is shown. Yellow: Q357F/pNCS complex, loop closed form. W2 in the structure is displaced by the incoming Asp356 carboxyl group.
These structures reveal that, unlike the case in PTP1B, the position of the Gln446 side chain is not pushed away when the Phe357 side chain moves into the active site in the loop closed conformation of Q357F/pNCS complex. In contrast, Phe357 ring is pushed away from Gln446 relative to the Gln357 position in the YopH/pNCS complex. While the hydrogen bond between Gln357 and Gln446 is eliminated by the Q357F mutation, the Phe357 ring and the pNCS ring are partially stacked with each other with significant overlap. Furthermore, the position of the presumed catalytically important water molecule, which is stabilized by hydrogen bond to Gln446 is also little affected by the Q357F mutation. This is due to the fact that this water molecule is stabilized by three hydrogen bonds from Gln446, Gln450 and the bridging oxygen of the sulfate group of pNCS and the disruption of the Gln446-Gln357 hydrogen bond does not affect the interaction network to this water molecule significantly. In fact, this water molecule is also observed in the loop open form of the Q357F/pNCS complex, with the same three hydrogen bonds (see Figure 9).
In the loop open form of the Q357F/pNCS complex, another water molecule stabilized by the hydrogen bonds to the OH group of pNCS and to the carboxyl group of Gln290 is observed. In the loop closed form, this water molecule is replaced by the carboxyl group of the incoming Asp356 residue, similar to that in the YopH/pNCS complex.
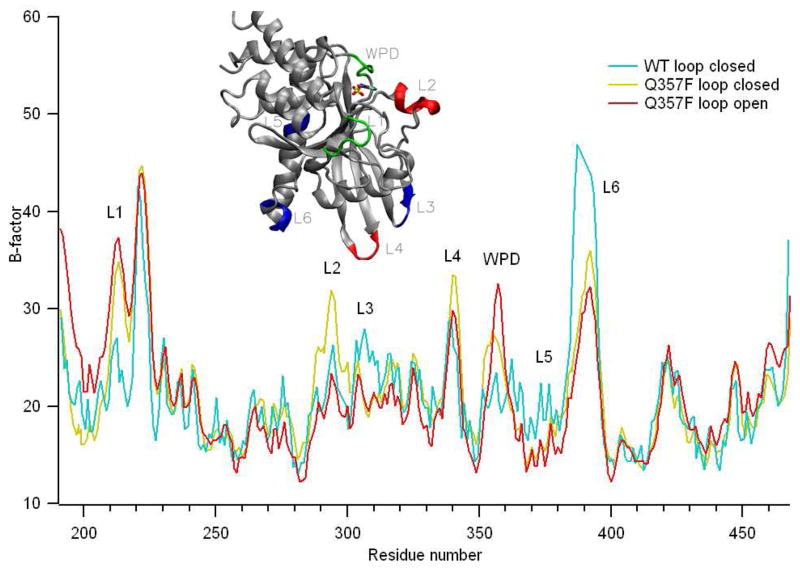
B-factor changes induced by Q357F mutation and loop closure
The fluctuation of individual atoms around their average positions can be derived from the Debye-Waller B-factor used in X-ray diffraction. In macromolecules, B-factors indicate relative mobility of different parts of the structure. Figure 10 shows the plots of B-factors of the Cα atoms vs residue number in YopH/pNCS (cyan), Q357F/pNCS loop open (red) and loop closed forms (yellow). A comparison between B-factors for loop open and loop closed forms of Q357F/pNCS is quite interesting. Loop closure does not result in significantly reduced mobility in any other part of the enzyme except for the WPD loop region. In fact, the atoms in L2 and L4 regions (red segments in the Figure 10 inset) show increased mobility upon loop closure. This provides a structural interpretation for the entropy increase as derived from our T-jump dynamic studies.
Figure 10.
Plots of enzyme backbone Cα B-factor vs residue number for YopH/pNCS and Q357F/pNCS complexes. Cyan: Cα B-factors of YopH/pNCS (from pdb file 1PA9). Red: Q357F/pNCS complex, loop open form. Yellow Q357F/pNCS complex, loop closed form. Inset shows the structure of YopH. The segments with significant differences in B-factors among these three complexes are color coded. See main text for details.
A comparison between the B-factors of loop closed forms of wt YopH and Q357F complexes shows that the Q357F mutation results in decreased mobility in L3, L5 and L6 regions (blue segments in Figure 10 inset) but increased mobility in L1, L2, L4 and WPD regions (red and green segments). The redistribution of the enzyme mobility could play some role in increased kcat for Q357F.
Discussion
pNCS is a YopH specific inhibitor structurally similar to the small molecule substrate pNPP.13 In this study, we have used T-jump relaxation technique to monitor the dynamics of Q357X/pNCS complex formation process and compared these results to previous results obtained for wt YopH/pNCS complex. Upon binding to YopH, the pKa of pNCS hydroxyl group is significantly increased. The pNCS binding affinity to YopH drops drastically at pH values higher than 8,32 presumably due to the ionization of this hydroxyl group. In the X-ray structure of YopH/pNCS complex, three hydrogen bonds are formed to the Gln357 side chain: one to the bridging oxygen of the pNCS SO4 group, and one to the Gln446 side chain and another to a structural water molecule. They are apparently part of the interactions in stabilizing the YopH/pNCS complex.
The Q357F mutation results in a small kcat increase for an enzyme that is already the most efficient PTPase. Although speculative, we believe that this likely results from an increased flexibility of the Q357F protein with bound ligand. Q357X mutations disrupt the hydrogen bonds to Gln357, and the effects of this disruption to the loop movements, to the ligand substate distribution after binding and to the enzyme catalysis have been determined. kopen and kclose become 2–3-fold lower in Q357F compared to wild type YopH but the ratio of kopen and kclose remains ~15, indicating the loop closed conformation is still the dominant form of the Q357F/pNCS complex. The activation enthalpies in Q357F for the loop open or loop close transitions are ~2 kJ/mol higher and lower than the corresponding values in wt YopH, respectively, so that the enthalpy difference between the loop open and loop closed conformations is only 2 kJ/mol in favor of the loop closed conformation, which is about 5 kJ/mol less than that in wild type YopH (Table 3). Apparently, the disruption of the hydrogen bonds to Gln357 in the loop closed form of YopH/pNCS is not fully compensated by the aromatic-aromatic interactions observed in the X-ray structure of Q357F/pNCS. On the other hand, the altered interaction network at the active site by Q357F mutation does not affect the free energy difference between the loop open and loop closed conformations after ligand binding, as shown by the little changed average free energy differences over the temperature range (Table 3). Thus, our results indicate a slightly larger entropy increase upon loop closure in Q357F mutant compared to wt YopH, which is supported by the B-factor analysis of the X-ray structures of the YopH/pNCS and Q357F/pNCS.
The YopH catalyzed reaction can be described by the following simplified scheme: 42
After substrate binding and the catalytic loop is closed, the subsequent steps involve the formation of the phospho-enzyme and the release of the tyrosine leaving group, which are believed to contribute to the rate limiting step near neutral pH and thus directly related to kcat.16 Although free energy is described by enthalpy and entropy components, typically the reduction of the free energy barriers in enzyme catalyzed reactions is attributed to enthalpic components such as hydrogen bonding, ionic interactions and van der Waals contacts, interactions that can stabilize the transition state of the chemical reaction. In this description of enzyme catalysis, the entropic component is generally considered as an energy penalty that is difficult to avoid. Recently, however, it has been found that the entropy component may be similar in overall contribution to the enthalpy component in the protein-ligand interactions and the entropy increase in the form of increased mobility of protein backbone/side-chain atoms can also make positive contributions to increase the ligand binding affinity.43–45 Moreover, much of the literature concerning thermal adaptation of enzyme to differing environments suggests that structural changes bringing about an increase of flexibility in residues outside the binding pocket can decrease or increase kcat of a specific enzyme, depending on the needs of the adaptive process (typically enzymes that operate at low ambient temperatures need to increase the relative kcat to overcome the lowered activity due to the lower operating temperature of the organism). 46
Our dynamic, thermodynamic, and structural studies of YopH and its Q357X mutants clearly show that the free energy barrier from [ES]open state to [ES]close is higher in Q357F than the barrier in wt YopH but the free energy differences between [ES]close and [ES]open conformations are very similar. Furthermore, the entropic component makes a favorable contribution to [ES]close in the [ES]close and [ES]open equilibrium in Q357F but no or slightly unfavorable contribution in wt YopH (see Table 3). Although the loop motions in Q357F become slower than in wild type, kopen is still about 5-fold faster than kcat (compared to 10 times in the wild type). Therefore, loop movements in Q357F do not contribute to the rate limiting step in YopH. The Q357F mutation does, however, result in a [ES]close structure with increased atomic flexibility/dynamic fluctuations near the active site and a few other regions. By building increased flexibility within [ES]close, the dynamic motions of the protein can be matched by the dynamics of the substrate, allowing, in principle, to a more efficient search of more catalytically effective enzyme-substrate conformations. In other words, enhanced motions in the [ES]close state in Q357F could lead to increased number of feasible reactive trajectories traversing the transition states of the reaction and result in increased kcat.
In summary, our current studies have shown that the loop mobility/flexibility can be affected by the mutation of a single loop residue as suggested in previous studies. By increasing or decreasing the size of a single loop residue, the loop dynamics show decreased or increased mobility, respectively, as detected by the T-jump studies. Our results also suggest an interesting possibility that for the loop closure step, a fine tuning of the enthalpy component in the unfavorable direction may cause a larger entropy component change in the favorable direction, resulting in an overall free energy gain to stabilize the enzyme/ligand complex.
Acknowledgments
We thank Dr. Z.-Y. Zhang for providing the plasmid encoding YopH catalytic domain. This research was supported by grants EB001958 and GM068036 from the National Institutes of Health. Data for this study were measured at beamline X29A of the National Synchrotron Light Source. Financial support comes principally from the Offices of Biological and Environmental Research and of Basic Energy Sciences of the US Department of Energy, and from the National Center for Research Resources of the National Institutes of Health grant number P41RR012408.
References
- 1.Zhang Z-Y. Protein-Tyrosine Phosphatases: Biological Function, Structural Characteristics, and Mechanism of Catalysis. Crit Rev in Biochem and Mol Biol. 1998;33(1):1–52. doi: 10.1080/10409239891204161. [DOI] [PubMed] [Google Scholar]
- 2.Kappert K, Peters KG, Bohmer FD, Ostman A. Tyrosine phosphatases in vessel wall signaling. Cardiovascular Research. 2005;65(3):587–598. doi: 10.1016/j.cardiores.2004.08.016. [DOI] [PubMed] [Google Scholar]
- 3.Neel BG, Tonks NK. Protein tyrosine phosphatases in signal transduction. Current Opinion in Cell Biology. 1997;9(2):193–204. doi: 10.1016/s0955-0674(97)80063-4. [DOI] [PubMed] [Google Scholar]
- 4.Barford D, Das AK, Egloff M-P. THE STRUCTURE AND MECHANISM OF PROTEIN PHOSPHATASES: Insights into Catalysis and Regulation. Annual Review of Biophysics and Biomolecular Structure. 1998;27(1):133–164. doi: 10.1146/annurev.biophys.27.1.133. [DOI] [PubMed] [Google Scholar]
- 5.Brubaker RR. Factors promoting acute and chronic diseases caused by yersiniae. Clin Microbiol Rev. 1991;4(3):309–324. doi: 10.1128/cmr.4.3.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Z-Y. Protein tyrosine phosphatases: prospects for therapeutics. Current Opinion in Chemical Biology. 2001;5(4):416–423. doi: 10.1016/s1367-5931(00)00223-4. [DOI] [PubMed] [Google Scholar]
- 7.Zhang ZY. Chemical and mechanistic approaches to the study of protein tyrosine phosphatases. Acc Chem Res. 2003;36(6):385–92. doi: 10.1021/ar020122r. [DOI] [PubMed] [Google Scholar]
- 8.Pannifer ADB, Flint AJ, Tonks NK, Barford D. Visualization of the Cysteinyl-phosphate Intermediate of a Protein-tyrosine Phosphatase by X-ray Crystallography. Journal of Biological Chemistry. 1998;273(17):10454–10462. doi: 10.1074/jbc.273.17.10454. [DOI] [PubMed] [Google Scholar]
- 9.Wan Z-K, Lee J, Xu W, Erbe DV, Joseph-McCarthy D, Follows BC, Zhang Y-L. Monocyclic thiophenes as protein tyrosine phosphatase 1B inhibitors: Capturing interactions with Asp48. Bioorganic & Medicinal Chemistry Letters. 2006;16(18):4941–4945. doi: 10.1016/j.bmcl.2006.06.051. [DOI] [PubMed] [Google Scholar]
- 10.Stuckey JA, Schubert HL, Fauman E, Zhang ZY, Dixon JE, Saper MA. Crystal Structure of Yersinia Protein Tyrosine Phosphatase at 2.5 A and the complex with Tungstate. Nature. 1994;370:571–575. doi: 10.1038/370571a0. [DOI] [PubMed] [Google Scholar]
- 11.Denu JM, Lohse DL, Vijayalakshmi J, Saper MA, Dixon JE. Visualization of intermediate and transition-state structures in protein-tyrosine phosphatase catalysis. Proceedings of the National Academy of Sciences. 1996;93(6):2493–2498. doi: 10.1073/pnas.93.6.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schubert HL, Fauman EB, Stuckey JA, Dixon JE, Saper MA. A ligand-induced conformational change in the yersinia protein tyrosine phosphatase. Protein Science. 1995;4(9):1904–1913. doi: 10.1002/pro.5560040924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun J-P, Wu L, Fedorov AA, Almo SC, Zhang Z-Y. Crystal Structure of the Yersinia Protein-tyrosine Phosphatase YopH Complexed with a Specific Small Molecule Inhibitor. Journal of Biological Chemistry. 2003;278(35):33392–33399. doi: 10.1074/jbc.M304693200. [DOI] [PubMed] [Google Scholar]
- 14.Zhang ZY. Mechanistic studies on protein tyrosine phosphatases. Prog Nucleic Acid Res Mol Biol. 2003;73:171–220. doi: 10.1016/s0079-6603(03)01006-7. [DOI] [PubMed] [Google Scholar]
- 15.Hengge AC, Sowa GA, Wu L, Zhang Z-Y. Nature of the Transition State of the Protein-Tyrosine Phosphatase-Catalyzed Reaction. Biochemistry. 1995;34:13982–13987. doi: 10.1021/bi00043a003. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Z-Y, Malachowski WP, Van Etten RL, Dixon JE. The Nature of the Rate-Determining Steps of the Yersinia Protein Tyrosine Phosphatase-catalyzed Reactions. J Biol Chem. 1994;269:8140–8145. [PubMed] [Google Scholar]
- 17.Yang J, Niu T, Zhang A, Mishra AK, Zhao ZJ, Zhou GW. Relation between the flexibility of the WPD loop and the activity of the catalytic domain of protein tyrosine phosphatase SHP-1. Journal of Cellular Biochemistry. 2002;84(1):47–55. doi: 10.1002/jcb.1265. [DOI] [PubMed] [Google Scholar]
- 18.Yang J, Liang X, Niu T, Meng W, Zhao Z, Zhou GW. Crystal Structure of the Catalytic Domain of Protein-tyrosine Phosphatase SHP-1. Journal of Biological Chemistry. 1998;273(43):28199–28207. doi: 10.1074/jbc.273.43.28199. [DOI] [PubMed] [Google Scholar]
- 19.Desamero R, Rozovsky S, Zhadin N, McDermott A, Callender R. Active Site loop Motion in Triosephosphate Isormerase: T-Jump relaxation Spectroscopy of Thermal Activation. Biochemistry. 2003;42:2941–2951. doi: 10.1021/bi026994i. [DOI] [PubMed] [Google Scholar]
- 20.McClendon S, Zhadin N, Callender R. The Approach to the Michaelis Complex in Lactate Dehydrogenase: the substrate binding pathway. Biophysical J. 2005;89:2024–2032. doi: 10.1529/biophysj.105.062604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhadin N, Gulotta M, Callender R. Probing the role of dynamics in hydride transfer catalyzed by lactate dehydrogenase. Biophys J. 2008;95(4):1974–84. doi: 10.1529/biophysj.108.132464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sampson NS, Knowles JR. Segmental movement: definition of the structural requirements for loop closure in catalysis by triosephosphate isomerase. Biochemistry. 1992;31(36):8482–8487. doi: 10.1021/bi00151a014. [DOI] [PubMed] [Google Scholar]
- 23.Sampson NS, Knowles JR. Segmental motion in catalysis: investigation of a hydrogen bond critical for loop closure in the reaction of triosephosphate isomerase. Biochemistry. 1992;31(36):8488–8494. doi: 10.1021/bi00151a015. [DOI] [PubMed] [Google Scholar]
- 24.Berlow RB, Igumenova TI, Loria JP. Value of a Hydrogen Bond in Triosephosphate Isomerase Loop Motion. Biochemistry. 2007;46(20):6001–6010. doi: 10.1021/bi700344v. [DOI] [PubMed] [Google Scholar]
- 25.Eigen M, De Maeyer LD. Relaxation Methods. In: Friess SL, Lewis ES, Weissberger A, editors. Technique of Organic Chemistry. Vol. 8. Interscience; New York: 1963. pp. 895–1054. [Google Scholar]
- 26.Eigen M, Hammes GG. Elementary Steps in Enzyme Reactions (as Studied by Relaxation Spectrometry) Advan Enzymol. 1963;25:1. doi: 10.1002/9780470122709.ch1. [DOI] [PubMed] [Google Scholar]
- 27.Fasella P, Giartosio A, Hammes GG. The Interaction of Aspartate Aminotransferase with α-Methylaspartic Acid. Biochemistry. 1966;5(1):197–202. doi: 10.1021/bi00865a026. [DOI] [PubMed] [Google Scholar]
- 28.Hammes GG, Schimmel PR. Relaxation spectra of enzymatic reactions. J Phys Chem. 1967;71(4):917–23. doi: 10.1021/j100863a023. [DOI] [PubMed] [Google Scholar]
- 29.Halford SE. Escherichia coli alkaline phosphatase. Relaxation spectra of ligand binding. Biochem J. 1972;126(3):727–38. doi: 10.1042/bj1260727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Callender R, Dyer RB. Probing protein dynamics using temperature jump relaxation spectroscopy. Cur Opin Struct Biol. 2002;12:628–633. doi: 10.1016/s0959-440x(02)00370-6. [DOI] [PubMed] [Google Scholar]
- 31.Callender RH, Dyer RB. Advances in Time-Resolved Approaches to Characterize the Dynamical Nature of Enzymatic Catalysis. Chem Revs. 2006;106:3031–3042. doi: 10.1021/cr050284b. [DOI] [PubMed] [Google Scholar]
- 32.Khajehpour M, Wu L, Liu S, Zhadin N, Zhang Z-Y, Callender R. Loop Dynamics and Ligand Binding Kinetics in the Reaction Catalyzed by Yersinia Protein Tyrosine Phosphatase. Biochemistry. 2007;46:4370–4378. doi: 10.1021/bi602335x. [DOI] [PubMed] [Google Scholar]
- 33.Fauman EB, Yuvaniyama C, Schubert HL, Stuckey JA, Saper MA. The X-ray Crystal Structures of Yersinia Tyrosine Phosphatase with Bound Tungstate and Nitrate. Journal of Biological Chemistry. 1996;271(31):18780–18788. doi: 10.1074/jbc.271.31.18780. [DOI] [PubMed] [Google Scholar]
- 34.Brandao TAS, Robinson H, Johnson SJ, Hengge AC. Impaired Acid Catalysis by Mutation of a Protein Loop Hinge Residue in a YopH Mutant Revealed by Crystal Structures. Journal of the American Chemical Society. 2009;131(2):778–786. doi: 10.1021/ja807418b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang ZY, Palfey BA, Wu L, Zhao Y. Catalytic function of the conserved hydroxyl group in the protein tyrosine phosphatase signature motif. Biochemistry. 1995;34(50):16389–96. doi: 10.1021/bi00050a020. [DOI] [PubMed] [Google Scholar]
- 36.Ghanem M, Zhadin N, Callender R, Schramm VL. Loop-Tryptophan Human Purine Nucleoside Phosphorylase Reveals Submillisecond Protein Dynamics. Biochemistry. 2009;48(16):3658–3668. doi: 10.1021/bi802339c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Juszczak LJ, Zhang Z-Y, Wu L, Gottfried D, Eads D. Rapid loop Dynamics of the Yersinia Protein Tyrosine Phosphatases. Biochemistry. 1997;36:2227–2236. doi: 10.1021/bi9622130. [DOI] [PubMed] [Google Scholar]
- 38.The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 39.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 40.Potterton E, Briggs P, Turkenburg M, Dodson E. A graphical user interface to the CCP4 program suite. Acta Crystallogr D Biol Crystallogr. 2003;59:1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- 41.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 42.Hengge AC, Edens WA, Elsing H. Transition-State Structures for Phosphoryl-Transfer Reactions of p-Nitrophenyl Phosphate. Journal of the American Chemical Society. 1994;116(12):5045–5049. [Google Scholar]
- 43.Tzeng S-R, Kalodimos CG. Dynamic activation of an allosteric regulatory protein. Nature. 2009;462(7271):368–372. doi: 10.1038/nature08560. [DOI] [PubMed] [Google Scholar]
- 44.Diehl C, Engstrom O, Delaine T, Hakansson M, Genheden S, Modig K, Leffler H, Ryde U, Nilsson UJ, Akke M. Protein Flexibility and Conformational Entropy in Ligand Design Targeting the Carbohydrate Recognition Domain of Galectin-3. Journal of the American Chemical Society. 2010;132(41):14577–14589. doi: 10.1021/ja105852y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hirschi JS, Arora K, Brooks CL, Schramm VL. Conformational Dynamics in Human Purine Nucleoside Phosphorylase with Reactants and Transition-State Analogues. The Journal of Physical Chemistry B. 2010;114(49):16263–16272. doi: 10.1021/jp108056s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hochachka PW, Somero GN. Biochemical adaptation: mechanism and process in physiological evolution. Chapter 7. Oxford University Press; Oxford: 2002. Temperature; pp. 290–378. [Google Scholar]